Abstract
Lactose is the only soluble and economically feasible carbon source for the production of cellulases or heterologous proteins regulated by cellulase expression signals by Hypocrea jecorina (Trichoderma reesei). We investigated the role of the major β-galactosidase of H. jecorina in lactose metabolism and cellulase induction. A genomic copy of the bga1 gene was cloned, and this copy encodes a 1,023-amino-acid protein with a 20-amino-acid signal sequence. This protein has a molecular mass of 109.3 kDa, belongs to glycosyl hydrolase family 35, and is the major extracellular β-galactosidase during growth on lactose. Its transcript was abundant during growth on l-arabinose and l-arabinitol but was much less common when the organism was grown on lactose, d-galactose, galactitol, d-xylose, and xylitol. Δbga1 strains grow more slowly and accumulate less biomass on lactose, but the cellobiohydrolase I and II gene expression and the final cellulase yields were comparable to those of the parental strain. Overexpression of bga1 under the control of the pyruvate kinase promoter reduced the lag phase, increased growth on lactose, and limited transcription of cellobiohydrolases. We detected an additional extracellular β-galactosidase activity that was not encoded by bga1 but no intracellular β-galactosidase activity. In conclusion, cellulase production on lactose occurs when β-galactosidase activity levels are low but decreases as the β-galactosidase activities increase. The data indicate that bga1-encoded β-galactosidase activity is a critical factor for cellulase production on lactose.
The ascomycete Hypocrea jecorina (anamorph, Trichoderma reesei) is used industrially to produce cellulolytic and hemicellulolytic enzymes, and its strong cellulase promoters are of interest for heterologous protein production by this fungus. Cellulase and hemicellulase formation can be induced by several mono- and disaccharides, including sophorose, xylobiose, lactose, d-xylose, and l-sorbose, most of which are too expensive for industrial fermentations. Thus, the range of technically applicable carbon sources is limited. For cellulase and heterologous protein production with cellulase promoters, lactose (1,4-O-β-d-galactopyranosyl-d-glucose) is virtually the only carbon source that can be used. However, the mechanism by which lactose triggers cellulase formation is not understood. Lactose metabolism is slow, and the cellulase yields on lactose are lower than those on cellulose (27, 28).
A key enzyme in the induction of cellulase is galactokinase, since a galactokinase-negative mutant cannot induce cellulases during growth on lactose (30). Lactose can enter the cell after it is hydrolyzed extracellularly to d-glucose and d-galactose by a β-galactosidase (lactase, β-d-galactoside galactohydrolase; E.C 3.2.1.23), and the monosaccharides are then taken up by the respective permeases. Extracellular β-galactosidases are known from several fungi, including Aspergillus niger, Aspergillus oryzae, and Penicillium canescens (5, 21, 26, 29). Alternatively, the lactose itself can be taken up by a specific lactose permease and hydrolyzed intracellularly. The latter mechanism has been demonstrated in the yeast Kluyveromyces lactis (6, 11), and Aspergillus nidulans is known to contain both an intracellular β-galactosidase (12) and a lactose permease (L. Karaffa, B. Seiboth, and C. P. Kubicek, unpublished data). H. jecorina has been reported to have extracellular β-galactosidase activity (25).
The objectives of the present work were to clone the bga1 gene encoding the extracellular β-galactosidase of H. jecorina and to address the following questions. (i) Does H. jecorina contain other β-galactosidases, in particular intracellular β-galactosidases? (ii) Is the rate of extracellular lactose hydrolysis critical for induction of cellulases by lactose? And (iii) does manipulation of β-galactosidase activity improve cellulase production on lactose? This information is needed to design and control industrial cellulase production with lactose as the carbon source.
MATERIALS AND METHODS
Strains and culture conditions.
H. jecorina strain QM9414 (= ATCC 26921) and strain RUT C30 (= ATCC 56765) and its pyr4 mutant TU-6 (= ATCC MYA-256) (15) were maintained on malt extract agar supplemented with uridine (10 mM) as necessary. Strains were grown in 1-liter flasks on a rotary shaker (250 rpm) at 30°C in the medium described by Mandels and Andreotti (23) with the appropriate carbon source at a final concentration of 10 g/liter.
For transcript analysis, strains were pregrown on glycerol (1%, wt/vol) for 20 h, mycelia were harvested by filtration and washed with tap water, equal amounts of mycelia were transferred to flasks containing the appropriate carbon source (1%, wt/vol), and cultivation was continued for 3 to 6 h. For lactose analysis, expression was analyzed without pregrowth on glycerol.
Escherichia coli strain NM538 (13) was used for library screening, and strains JM109 (Promega, Madison, Wis.) and XL1 Blue (Stratagene, La Jolla, Calif.) were used for plasmid propagation.
Fermentor cultivation.
The defined medium of Vogel (40) was used, except that lactose (10 g/liter) was used instead of sucrose as the carbon source and (NH4)2SO4 (1.65 g/liter) was used instead of NH4NO3 as the nitrogen source. Vogel's salts were prepared as a 50× solution and sterilized by membrane (pore size, 0.22 μm) filtration. The media for shake flask cultures contained Stabileze QM (methyl vinyl ether-maleic anhydride copolymer cross-linked with 1,9-decadien; ISP Technologies, Wayne, N.J.) or Junlon (18) to promote filamentous growth; the concentrations of Stabileze QM and Junlon used were 0.75 g/liter for cultures used for growth rate and enzyme determinations and 1.5 g/liter for precultures used for bioreactors. To prepare precultures, 40-ml batch cultures were grown in 250-ml Erlenmeyer flasks or 250-ml Nephlos flasks (36). The flasks were inoculated with 106 spores/ml (final concentration). The cultures were incubated on a rotary shaker at 200 rpm (throw, 2.5 cm) at 30°C. Fermentations were carried out in either an Applikon bioreactor (full working volume, 2.3 liters; FT Applikon Ltd., Tewkesbury, United Kingdom) or a B. Braun BIOSTAT M (full working volume, 2.1 liters; Sartorius BBI Systems, Melsungen, Germany) as previously described (42). The medium contained 10 g of lactose per liter as the sole carbon source. Cultures were inoculated with five 40-ml shake flask cultures which had been growing in medium containing 10 g of the same carbon source per liter for approximately 3 days. Fermentor cultures were maintained at 28 ± 1°C and pH 5.5 ± 0.1, agitated at 900 to 1,000 rpm with three six-blade (diameter, 48 mm) Rushton turbine impellers, and aerated with 0.7 liter of air liter of culture−1 min−1. Foaming was controlled by adding a mixture of polypropylene glycols (PPG) having different molecular weights that included PPG 1025 (BDH, VWR International, Poole, United Kingdom), PPG 2025 (BDH), and FoamMaster PPG (mixed molecular weight; Henkel Performance Chemicals, Leeds, United Kingdom) at a ratio of 2:2:1 (41), if necessary. For cultures grown in the BIOSTAT M, the PPG mixture was added as an aqueous suspension (25%, vol/vol), while for cultures grown in the Applikon bioreactor it was added as a 100% suspension.
Cloning and sequence analysis of bga1.
β-Galactosidases from the following fungi and bacterium were used for an alignment to identify conserved regions: A. niger (GenBank accession no. P29853), A. oryzae (5), P. canescens (GenBank accession no. Q12660), and Bacillus circulans (GenBank accession no. JC5618). We used the degenerate primers 5′-GGCGGATCCCARAARTAYGTNACNTGGGAYGA-3′ and 5′-GGCGGATCCTAYTCRTTYTCNRKYTG-3′, corresponding to the amino acid sequences QKYVTWDD and Q(P/I)ENEY, with BamHI restriction sites (underlined) at their 5′ ends for PCR amplification; QM9414 chromosomal DNA was used as the template. The following PCR cycling conditions were used: (i) 96°C for 10 min, after which 1 U of enzyme (Dynazyme II; Finnzymes, Espoo, Finland) per 100 μl of reaction mixture was added; (ii) 96°C for 4 min; (iii) three cycles consisting of 96°C for 1 min, 37 or 42°C for 30 s, and 72°C for 1 min; (iv) 24 cycles consisting of 96°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and (v) 4°C indefinitely. PCR products (∼500 bp) were isolated, cut with BamHI, and subcloned into similarly cleaved pBluescript SK(−) (Stratagene). The inserts were sequenced, and two identical clones (543 bp) with 76% amino acid identity to the β-galactosidases of A. niger (accession no. P29853) and P. canescens (accession no. Q12660) were used as probes to screen a genomic EMBL3 λ phage library (37) of QM9414. The bga1 gene was located on a 5-kb HindIII subclone, which was ligated into pUC18, resulting in pNS41. The 3′ end of bga1 was located on a ∼2.8-kb XhoI-ClaI subclone, which was ligated to pBluescript SK(+), resulting in pBGAClaI. A bga1 cDNA fragment was cloned from a λ ZAP II library of H. jecorina RUT-C30 (33) and sequenced after in vivo excision into the phagemid vector pBluescript SK(−).
The genomic subclones and the deduced protein were analyzed with BLAST programs (1) and Expasy (http://www.expasy.org/). Consensus binding sequences in the bga1 5′ region were identified manually.
Construction of plasmids for bga1 expression and deletion.
The upstream and downstream regions of the bga1 coding sequence were amplified by PCR that introduced an SmaI site between the two regions in which the A. nidulans amdS gene encoding acetamidase (19) was cloned. Thus, the 5′ region was amplified from pNS41 by using oligonucleotides bGaldel1 (5′-GTCTCTTTCCCGGGTCCCAATTGGCCGTTTC-3′) and M13forward, and the 3′ region was amplified from pBGAClaI by using bGaldel2 (5′-CAATTGGGACCCGGGAAAGAGACTTGGGATTTTC-3′) and M13reverse. Standard conditions were used for PCR. The resulting fragments were PstI/SmaI digested and ligated in the PstI-restricted vector pUC19-SmaI, resulting in pDBGA. pUC19-SmaI is identical to pUC19 (43) except that the SmaI site was eliminated by Acc65I and BamHI digestion. Finally, the 4-kb SalI-Acc65I amdS fragment of A. nidulans was blunted and ligated into the SmaI site of pDBGA, resulting in pDBGAamdS.
For bga1 overexpression, the coding region and 3′ region were placed under the control of the pyruvate kinase promoter fragment in pRLMex30 (22). A 250-bp region of the N terminus of the bga1 gene coding region was amplified by PCR with bGalXbaI (5′-GTGATCTAGAACATGATGAGACCCGTCTC-3′) and bGalHindIII (5′-TCTCAAGCTTGTGGAACACATCAAGATACAG-3′), and the amplicon was XbaI/HindIII digested and cloned into the respective sites of pRLMex30, resulting in pPKI-NBGA. Subsequently, a ∼4.8-kb ClaI-BglII bga1 fragment comprising the majority of the bga1 coding and terminator region was cloned into ClaI-HindIII-digested vector pPKI-NBGA, resulting in pPKI-BGA. The PCR-amplified portion of the sequence was verified by sequencing.
Construction of strains with altered bga1 expression.
H. jecorina was transformed as previously described (14). For bga1 gene replacement a 6.6-kb EcoRI fragment was excised from pDBGAamdS and purified from an agarose gel. Transformants were selected for growth on acetamide (10 mM) as the sole carbon source. Retransformation of a Δbga1 strain was done by cotransformation of a 6-kb HindIII-BglII bga1 fragment together with pRLMex30. Strains were selected for resistance to hygromycin (50 μg/ml). For bga1 amplification, strain TU-6 was cotransformed with plasmid pPKI-BGA and a 2.7-kb SalI pyr4 fragment of pFG1 (14).
Nucleic acid isolation and hybridization.
Total RNA was isolated as described by Chomczynski and Sacchi (7). Standard methods (3) were used for electrophoresis, blotting, and hybridization of nucleic acids. The probes used for hybridization were a 1.4-kb BglI cbh1 (cellobiohydrolase CBHI/Cel7A-encoding [32]) fragment, a 1.3-kb HaeII cbh2 (cellobiohydrolase CBHII/Cel6A-encoding [accession no. M55080]) fragment, a 1.9-kb Acc65I act1 (actin-encoding [accession no. X75421]) fragment, and the complete bga1 cDNA.
Preparation of cell extracts.
Cell extracts for enzyme activity assays were prepared from H. jecorina strains grown for 26 h on Mandels-Andreotti medium with the lactose or glycerol as the carbon source. The mycelia were harvested by filtration through Miracloth (Calbiochem, VWR International, Vienna, Austria), washed with cold sterile tap water, blotted dry with paper towels, and ground to a fine powder under liquid N2 in a mortar and pestle. One gram (wet weight) of ground mycelium was mixed with 3 ml of extraction buffer (0.1 M Tris-HCl [pH 7.5], 1 mM EDTA, 5 mM β-mercaptoethanol), suspended, and homogenized by sonication 10 times for 30 s at 2°C, with intermittent 2-min cooling periods. The resulting homogenate was centrifuged at 10,000 × g for 20 min at 4°C. The supernatant (average protein concentration, 8 to 15 mg/ml) was used as a cell extract for measuring intracellular β-galactosidase activity. After repeated washing of the remaining pellet with 0.1% Triton X-100, the cell debris was resuspended in 3 ml of citrate buffer (pH 5) per g (dry weight) of the original mycelium and was used as a source to measure cell wall-bound β-galactosidase activity.
Enzyme assay.
β-Galactosidase activity was determined with o-nitrophenyl-β-d-galactopyranoside as the substrate. Cell extracts, supernatant, and cell wall-bound samples were diluted to 700 μl with 0.1 M phosphate buffer (pH 5 or 7) and added to 300 μl of 10 mM o-nitrophenyl-β-d-galactopyranoside. After incubation for 10 to 60 min at 30°C, the reaction was stopped by addition of 3 ml of 1 M Na2CO3. Assay mixtures containing cell wall samples were centrifuged (10,000 × g for 10 min at room temperature) before measurement. The A405 was measured against a blank when the reaction had been stopped at zero time. Enzyme activities were expressed in units, and 1 U corresponded to the conversion of 10 μmol of substrate per min under the conditions used.
β-Glucosidase and cellulase activities were assayed as described previously (39). Culture supernatant was filtered through 0.22-μm-pore-size filters (Millipore GesmbH, Vienna, Austria), and aliquots were stored at −20°C for subsequent analysis. Total cellulase activity was determined by using p-nitrophenyl-β-d-lactopyranoside instead of 4-methylumbelliferyl-β-d-lactoside. Briefly, p-nitrophenyl-β-d-lactopyranoside (400 μl; 1 mg/ml in 50 mM citric acid buffer [pH 5.0]) was incubated with 50 μl of culture supernatant (diluted as necessary) or a standard (500 to 10 mg/liter) in an Eppendorf tube for 1 h at 37°C, and the reaction was terminated by adding 500 μl of 0.1 M borax. The p-nitrophenyl released during the reaction was measured by determining changes in the A405.
β-Glucosidase activity was measured by using the protocol described above, except that p-nitrophenyl-β-d-glucopyranoside was used as the substrate (20).
Determination of fungal growth and biomass.
To determine hyphal growth on agar plates, plates were inoculated with a small piece of agar in the center of each 11-cm plate. The biomass in submerged cultures was measured by filtering two 10-ml portions of culture onto predried and preweighed Whatman no. 1 filter papers. The harvested biomass was dried to constant weight (70°C for at least 3 days or 30 min in a microwave oven at 260 W), after each filter was washed with at least 20 ml of deionized water.
Growth rates in bioreactors were estimated from the CO2 output by using an ADC 7000 infrared gas analyzer (The Analytical Development Co. Ltd., Hoddesdon, United Kingdom).
Biochemical analyses.
Protein concentrations were determined by the Bio-Rad protein assay (Bio-Rad Laboratories, Munich, Germany). The protocols described by Ausubel et al. (3) were used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (with 10% polyacrylamide gels) and Western blotting. Monoclonal antibodies were used to detect cellobiohydrolases CBHI/Cel7A and CBHII/Cel6A (24).
Nucleotide sequence accession number.
The bga1 sequence has been deposited in the GenBank database under accession number AJ549427.
RESULTS
Cloning and characterization of bga1 and its protein product.
A 5-kb genomic subclone, which included the complete structural gene, was isolated following hybridization to a probe based on conserved regions of fungal β-galactosidases. This subclone contains a 3,523-bp open reading frame interrupted by seven introns (55, 57, 75, 69, 66, 64, and 65 bp) encoding a 1,023-amino-acid polypeptide that includes a 20-amino-acid N-terminal signal sequence, as predicted by iPSORT (4). The mature Bga1 protein has a calculated molecular mass of 109.3 kDa and is a member of glycosyl hydrolase family 35 (16) with 10 putative N-glycosylation sites. A BLASTP search revealed high degrees of identity to β-galactosidases from other fungi (e.g., 57% identity to Talaromyces emersonii β-galactosidase [accession no. AAL32052.2]; 54% identity to A. niger β-galactosidase [accession no. P29853]).
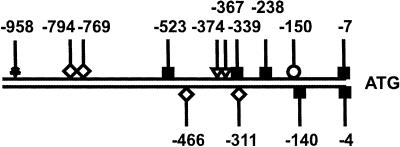
A sequence analysis of ∼1 kb of the bga1 promoter region resulted in identification of a number of consensus binding sites for fungal transcription factors (Fig. 1), including six sites for the CCAAT-binding Hap2/3/5 complex (44), three single sites and one double site for the carbon catabolite repressor Cre1 (SYRGGRG) (17, 34), and one site each for the cellulase and xylanase repressor Ace1 (AGGCA) (2) and the transcriptional activator AmyR (CGGN8CGG) (35). No consensus binding sites for XlnR, the A. niger transcriptional activator of xylanase and cellulase biosynthesis (38) which is also involved in regulation of A. niger lacA (8), or for Saccharomyces cerevisiae Gal4 (CGGN11CCG) (10), the transcriptional activator of the Leloir pathway genes, were found.
FIG. 1.
Presence of consensus binding sites for known fungal transcription factors in the bga1 upstream noncoding region. The positions are relative to the position of the translation start codon (ATG). The positions above the lines are the sites of the consensus sequences on the coding strand, and the positions below the lines are the sites on the noncoding strand. Symbols: ⋄, Cre1; ▪, Hap2/3/5; ▿, Ace1; ○, AmyR; ★, Msn2/4.
Regulation of bga1 gene transcription.
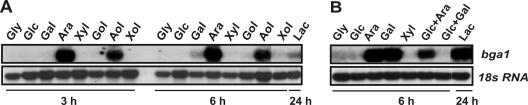
We performed Northern analyses of the bga1 transcript during growth of H. jecorina on a variety of carbon sources (Fig. 2). The bga1 transcript was most abundant during growth of H. jecorina on l-arabinose and l-arabinitol and was present at lower levels during growth on d-galactose and d-xylose and their corresponding polyols. The expression on lactose was low relative to that on d-galactose or d-xylose, and no transcript accumulated during growth on d-glucose and glycerol. Since these findings could be indicative of carbon catabolite repression, we also investigated bga1 transcription in the cre1 mutant RUT-C30 (17). In this Cre1-derepressed strain basal levels of the bga1 transcripts accumulated during growth on d-glucose, glycerol, and d-xylose, and high levels were found during growth on d-galactose and lactose, indicating that Cre1-dependent carbon catabolite repression interferes with bga1 transcription depending on the carbon source at either the basal or the induced level of transcription. Transfer to d-glucose together with l-arabinose or d-galactose showed that the presence of glucose interfered with the transcriptional activation only during growth on d-galactose.
FIG. 2.
Regulation of bga1 gene transcription during growth on various carbon sources of QM9414 (A) and RUT-C30 (B). Samples were obtained after 3 and 6 h of growth after transfer to different carbon sources (1%, wt/vol). In the case of lactose a sample was taken after 24 h of batch growth (conidial inoculum). Abbreviations: Gly, glycerol; Glc, d-glucose; Gal, d-galactose; Ara, l-arabinose; Xyl, d-xylose; Gol, galactitol; Aol, l-arabinitol; Xol, xylitol; Lac, lactose. 18S RNA served as a loading control.
Role of bga1 in growth on lactose.
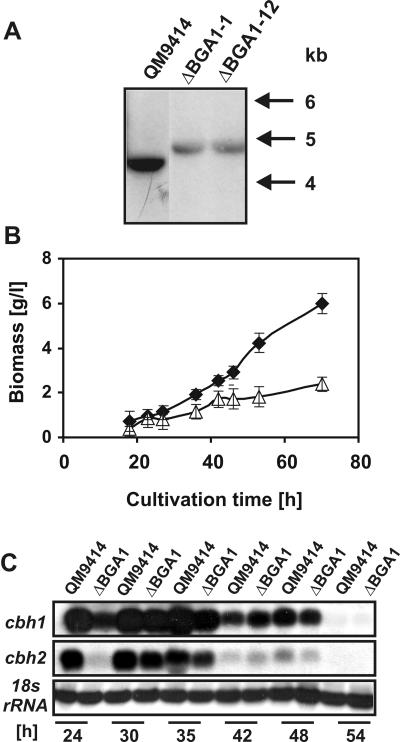
We produced a bga1 deletion mutant by replacing the coding region of bga1 with the A. nidulans acetamidase (amdS) gene in QM9414 and selecting for transformants that could grow on acetamide as a sole nitrogen source. These transformants were analyzed by Southern analysis (Fig. 3A). Hybridization with the fragment of bga1 which was replaced in the deletion vector by amdS gave a signal in QM9414 but not in the Δbga1 strains, confirming its deletion. On agar plates, the Δbga1 deletion strains grew at the wild-type rate with d-galactose as the carbon source but at lower rates with lactose or d-glucose. The effect observed with d-glucose was not due to ectopic insertion of the transforming DNA at other loci because the wild-type phenotype could be completely restored by retransformation of the bga1 deletion strains with the bga1 gene (data not shown).
FIG. 3.
Effect of bga1 gene deletion on growth and enzyme formation. (A) Southern analysis of parental strain QM9414 and two bga1 deletion mutants, ΔBGA1-1 and ΔBGA1-12. Genomic DNA was digested with HindIII and probed with a 5-kb HindIII fragment of the bga1 gene. Homologous insertion of the bga1 deletion cassette into the endogenous bga1 locus resulted in an increase (about 500 kb) in the size of the hybridizing band from 5 kb in the parent strain to 5.5 kb in the bga1 deletion mutants. (B) Growth of QM9414 (♦) and strain ΔBGA1 (▵) in shake flasks with peptone (0.1%, wt/vol) and lactose (1%, wt/vol) as carbon sources. The error bars indicate standard deviations for three different experiments. (C) Northern analysis of cbh1 and cbh2 transcript levels during growth on lactose. 18S RNA served as a loading control.
We grew the Δbga1 strains in shake flasks and monitored the increase in biomass (Fig. 3B). In the presence of peptone, growth of the Δbga1 strain started at the same time (10 to 15 h after inoculation), and the growth rate was approximately constant throughout the incubation experiment. However, the biomass concentration of the Δbga1 strain was somewhat less than one-half of the biomass concentration of the parent strain (2.3 versus 6.0 g/liter). The two strains also were grown in the absence of peptone in controlled fermentor cultures. In this case, the loss of function of bga1 was dramatic and resulted in a significantly delayed onset of growth on lactose (125 versus 40 h). The final decrease in biomass yield (2.5 versus 4.8 g/liter) was comparable to that in shake flasks, however.
Activity of Bga1.
We compared the β-galactosidase activities in the culture supernatant, cell extract, and mycelial debris after homogenization of QM9414 and the Δbga1 strain after growth for 24 h on lactose. More than 95% of the total extracellular β-galactosidase activity of QM9414 was secreted into the culture supernatant, while only a small percentage was associated with the cell walls (0.86 versus 0.006 U/g [dry weight] of mycelium). Most of the activity present in the culture medium was lost in the Δbga1 mutant (0.07 U/g [dry weight] of mycelium), indicating that H. jecorina has a second β-galactosidase which accounts for ∼10% of the total extracellular β-galactosidase activity hydrolyzing aryl-β-galactoside. No intracellular β-galactosidase activity was detected during growth on lactose at either pH 5.0 or pH 7.0.
Lactose-dependent induction of cellulase gene expression in the Δbga1 strain.
We cultivated QM9414 and the Δbga1 strain on lactose and measured the total cellulase and β-glucosidase activities and the accumulation of the cellobiohydrolase CBHI/Cel7A protein in the supernatant. The appearance of cellulase and β-glucosidase activities in the medium was delayed in the bga1 mutant, but the final levels were comparable when they were related to the biomass of the parent strain. Consistent with these data, the level of the CBHI/Cel7A protein that accumulated was only about one-half of the wild-type level in the Δbga1 strain. These findings suggested that loss of the bga1-encoded β-galactosidase function does not have a major effect on cellulase gene expression. We measured the accumulation of the transcripts of two major cellulase genes, cbh1 and cbh2, in both strains (Fig. 3C). The levels of both cellulase transcripts that accumulated were about the same in the two strains, except at 24 h, when the cbh1 and cbh2 transcript levels were lower in Δbga1. This difference could have been due to slower growth by the Δbga1 strain on lactose. Thus, the induction of both genes—apart from some growth-related retardation—was not affected in the bga1 deletion strain.
Overexpression of bga1 and cellulase formation.
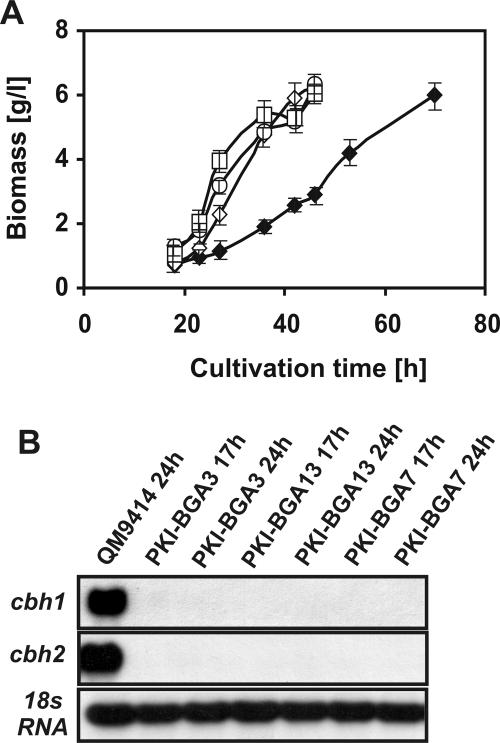
We constructed a series of H. jecorina strains that expressed bga1 under the expression signals of the H. jecorina pki1 (pyruvate kinase-encoding gene) promoter region and the bga1 terminator region and compared their β-galactosidase activities after 24 h of growth on glycerol. The different transformants produced between 3 and 18 U/g (dry weight). Therefore, the best overexpressing transformants had up to >30-fold-greater β-galactosidase activity than lactose-grown cultures of QM9414. Three of these transformants (PKI-BGA3, PKI-BGA7, and PKI-BGA13), which secreted different β-galactosidase activities (3, 5, and 12 U/g [dry weight], respectively), were grown on lactose in shake flasks and evaluated for cellulase formation and cellulase gene transcription. All three transformants had a significantly higher rate of growth on lactose plus peptone (0.27 to 0.29 g [dry weight]/h) and reached the final dry weight (6.4 g/liter) after ∼50 h of cultivation (Fig. 4A). All three transformants also had lost the ability to induce cbh1 and cbh2 gene expression on lactose as a carbon source (Fig. 4B), although we observed induction on cellulose in a manner and at a level comparable to the manner and level in the parental strain (data not shown). We also grew strain PKI-BGA13 in the fermentor in the absence of peptone and observed similar results (i.e., a reduced initial lag phase and 24-h-earlier production of the maximal amount of biomass).
FIG. 4.
bga1 overexpression improves lactose metabolism but impairs cellobiohydrolase expression. (A) Growth of H. jecorina QM9414 (♦) and strains PKI-BGA3 (○), PKI-BGA7 (⋄), and PKI-BGA13 (□) on lactose in shake flasks. The error bars indicate standard deviations for three different experiments. (B) Effect of bga1 overexpression on cbh1 and cbh2 accumulation during growth on lactose.
DISCUSSION
We identified a gene (bga1) of H. jecorina that encodes the major extracellular β-galactosidase during growth of the fungus on lactose. An alignment of the H. jecorina Bga1 protein sequence with the two β-galactosidases from Neurospora crassa (accession no. NCU00642.1 and NCUO4623.1) and the two β-galactosidases from A. nidulans (accession no. AN0756.2 and AN0980.2) demonstrated that the areas of similarity are distributed throughout the whole protein sequences. Thus, the β-galactosidases in N. crassa and A. nidulans are likely the result of a gene duplication, which appears to have been absent in H. jecorina. The N-terminal half (up to amino acid 370) of these β-galactosidases also is conserved in filamentous fungi and other organisms. The C-terminal half of these proteins is found only in two bacterial β-galactosidases (Clostridium acetobutylicum β-galactosidase [accession no. NP_349128.1] and Streptomyces avermitilis β-galactosidase [accession no. NP_826933.1]), but it is absent from the β-galactosidases of other bacteria, plants, and mammals in the National Center for Biotechnology Information database. The function of the C terminus in the fungal β-galactosidase proteins is therefore obscure, as no domains indicative of a special role could be identified in silico (Seiboth and Kubicek, unpublished data).
No intracellular β-galactosidase activity was found. The intracellular β-galactosidases of fungi are rather stable enzymes (12), and we have been able to extract a number of other more labile enzymes in active forms from H. jecorina without having to use protease inhibitors by the same method (e.g., oxidoreductases involved in d-xylose and l-arabinose metabolism). It is therefore unlikely that the lack of an intracellular β-galactosidase is due to proteolytic or other inactivation during extraction.
This finding is consistent with results of a TBLASTN search (Expect, 1e−5) of the H. jecorina genome database with the intracellular β-galactosidase of K. lactis (accession no. P00723), a family 2 glycosyl hydrolase, as the query, which produced no hits. In contrast, the genomes of A. nidulans and N. crassa (http://www.broad.mit.edu/annotation/fungi/) each contain two putative family 2 β-galactosidases (accession no. AN3201.2, AN2463.2, NCU05956.1, and NCU00810.1).
The lack of an intracellular β-galactosidase in H. jecorina implies that lactose is hydrolyzed extracellularly. This conclusion is consistent with the preliminary finding that germinated spores of H. jecorina grown on lactose can take up d-glucose and d-galactose (the hydrolysis products of lactose) but not lactose (Vehmaanperä, unpublished data). However, genes encoding putative orthologs of the lactose permeases of K. lactis (accession no. P07921) are found in the genome databases of fungi, including T. reesei (E value, 5e−84), A. nidulans (E value, 2e−81), and N. crassa (E value, 2e−79). Thus, it will be interesting to see whether this gene is expressed during growth on lactose in H. jecorina and whether its gene product catalyzes the transport of lactose.
Genetic manipulation of Bga1 activity by deletion or overexpression perceptibly affects growth on lactose during the early stages of cultivation. This effect is even more dramatic in the Δbga1 strain in the absence of peptone; however, there is no obvious correlation between changes in Bga1 activity and the growth rate on lactose. Reduction of the β-galactosidase activity by ∼90% only halved the growth rate, while a >10-fold increase in activity only tripled it.
These results suggest the presence of additional β-galactosidases which either do not hydrolyze aryl-β-galactosides or have only very low activity with them. The residual 5% of β-galactosidase activity found in the Δbga1 strain could result from a second β-galactosidase. A putative candidate was found in the T. reesei genome database (accession no. fg.V.1.C3000108), and this candidate showed a reasonable degree of identity to a β-galactosidase of Bacteroides thetaiotaomicron (accession no. AAO78619.1; E value, 1e−67) and in addition contained a signal peptide at its N terminus. An additional hit for a β-galactosidase was obtained in a TBLASTN search, but the nucleotide sequence of this contig was 100% identical to the DNA sequence of the Haemophilus somnus β-galactosidase/β-glucuronidase (accession no. ZP_00122729) and occurred on a very small contig. Therefore, we considered this match to be an artifact. Presumably, this second β-galactosidase has an unusual amino acid sequence, or its activity is a secondary activity of another glycosidase.
The occurrence of a β-galactosidase in a saprophytic ascomycete like H. jecorina is somewhat paradoxical, as lactose does not occur in its natural environment, and the induction caused by lactose may be artificial. It is more likely that plant polysaccharides containing β-galactosidic linkages are the true substrates for the β-galactosidase. These polysaccharides also may contain l-arabinose, since bga1 expression was triggered by both d-galactose and l-arabinose. In A. niger the extracellular β-galactosidase-encoding gene lacA was shown to be induced by the plant cell wall heteropolysaccharide pectin and several pectin monomers, including d-galacturonic acid, l-arabinose, and d-galactose, indicating that it may be involved in pectin degradation (9).
Overexpression of the bga1 gene product, but not its deletion, eliminated cellulase gene expression on lactose. This result suggests that increased β-galactosidase activity interferes with cellulase induction. One simple explanation for this finding is that the β-galactosidase activity eliminates the inducer. However, we have no evidence that lactose is the inducer, and the lack of intracellular β-galactosidase activity, which is necessary for intracellular lactose catabolism and the termination of induction, argues against this possibility. Alternatively, increased concentrations of lactose hydrolysis products (i.e., d-glucose and d-galactose), which accumulate in the medium, could result in catabolite repression of cellulase gene expression. However, the carbon catabolite repression of cbh1 and other cellulase genes is only partial (45), and carbon catabolite repression alone should not result in a complete loss of cbh1 gene transcription, like that seen in the bga1-overexpressing mutants. Also, growing the H. jecorina cre1 loss-of-function strain RUT-C30 in the simultaneous presence of equimolar concentrations of d-glucose and d-galactose (S. Gyamphi and C. P. Kubicek, unpublished data) or d-galactose alone (30) does not result in cellulase production, which makes Cre1-dependent carbon catabolite repression a less likely explanation.
Another possible explanation is that d-galactose uptake could be inhibited by d-glucose. H. jecorina transports d-galactose with a general hexose transporter that can be inhibited by an excess of other hexoses (e.g., d-glucose and d-mannose) (Vehmaanperä, unpublished). However, d-glucose and d-galactose are present in the medium at comparable concentrations, and the affinities of the transporter for them are in the same concentration range, so a complete block of d-galactose uptake by d-glucose uptake is unlikely.
We recently showed that eliminating the galactokinase step in the Leloir pathway also eliminates cellulase induction by lactose, although growth on lactose still occurs (30). We interpreted these results to mean that either d-galactose, bound to galactokinase, or d-galactose-1-phosphate was the inducer of cellulase formation on lactose. We also suggested that this induction occurred only at a narrow range of concentrations of either of these intermediates, because d-galactose itself does not induce cellulase gene expression, even in a Cre1-negative background. This model is also consistent with the present data if the overexpression of bga1 influences the intracellular steady-state levels of either d-galactose or d-galactose-1-phosphate, which leads to a shutdown of cellulose transcription. Although the Leloir pathway genes of H. jecorina are not subject to glucose repression (30, 31), which may take place because of the increased rate of lactose hydrolysis, competition for substrates and coenzymes (e.g., ATP) may occur.
The data presented in this paper show that the H. jecorina bga1-encoded β-galactosidase activity has a major impact on the growth rate during lactose-based fermentations and on the performance of cellulase formation by H. jecorina in this process. Consequently, further attempts to manipulate the expression of genes involved in this process are needed to improve the production rate and yield of mutant strains on an industrial scale.
Acknowledgments
This study was supported by the Fifth (EC) Framework program (Quality of Life and Management of Living Resources; projects EUROFUNG 2 and QLK3-1999-00729) and by grant P 16143 from the Austrian Science Foundation to C.P.K. The H. jecorina/T. reesei genome sequencing project was funded by the United States Department of Energy.
We thank Arja Mäntylä for useful discussions.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aro, N., M. Ilmen, A. Saloheimo, and M. Penttilä. 2003. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 69:56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Stuhl. 2003. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 4.Bannai, H., Y. Tamada, O. Maruyama, K. Nakai, and S. Miyano. 2002. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18:298-305. [DOI] [PubMed] [Google Scholar]
- 5.Berka, R. M., J. A. Hucul, and M. Ward. April 1998. Increased production of β-galactosidase in Aspergillus oryzae. U.S. patent 5736374-A 2.
- 6.Chang, Y. D., and R. C. Dickson. 1988. Primary structure of the lactose permease gene from the yeast Kluyveromyces lactis. Presence of an unusual transcript structure. J. Biol. Chem. 263:16696-16703. [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.de Vries, R. P., H. C. van den Broeck, E. Dekkers, P. Manzanares, L. H. de Graaff, and J. Visser. 1999. Differential expression of three α-galactosidase genes and a single β-galactosidase gene from Aspergillus niger. Appl. Environ. Microbiol. 65:2453-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries, R. P., J. Jansen, G. Aguilar, L. Parenicova, V. Joosten, F. Wulfert, J. A. Benen, and J. Visser. 2002. Expression profiling of pectinolytic genes from Aspergillus niger. FEBS Lett. 530:41-47. [DOI] [PubMed] [Google Scholar]
- 10.Dhawale, S., and A. Lane. 1993. Compilation of sequence-specific DNA-binding proteins implicated in transcriptional control in fungi. Nucleic Acids Res. 21:5537-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson, R. C., and K. Barr. 1983. Characterization of lactose transport in Kluyveromyces lactis. J. Bacteriol. 154:1245-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fekete, E., L. Karaffa, E. Sandor, B. Seiboth, S. Biro, A. Szentirmai, and C. P. Kubicek. 2002. Regulation of formation of the intracellular β-galactosidase activity of Aspergillus nidulans. Arch. Microbiol. 179:7-14. [DOI] [PubMed] [Google Scholar]
- 13.Frischauf, A. M., H. Lehrach, A. Poustka, and N. Murray. 1983. λ replacement vectors carrying polylinker sequences. J. Mol. Biol. 170:827-842. [DOI] [PubMed] [Google Scholar]
- 14.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaf. 1990. Cloning of the Trichoderma reesei pyrG-gene and its use as a homologous marker for a high-frequency transformation system. Curr. Genet. 18:447-451. [DOI] [PubMed] [Google Scholar]
- 15.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaff. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18:71-76. [DOI] [PubMed] [Google Scholar]
- 16.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilmen, M., C. Thrane, and M. Penttilä. 1996. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251:451-460. [DOI] [PubMed] [Google Scholar]
- 18.Jones, P., D. Moore, and A. P. J. Trinci. 1988. Effects of Junlon and Hostacerin on the electrokinetic properties of spores of Aspergillus niger, Phanerochaete chrysosporium and Geotrichum candidum. J. Gen. Microbiol. 134:235-240. [Google Scholar]
- 19.Kelly, J. M., and M. J. Hynes. 1985. Transformation of Aspergillus niger by the amdS gene of Aspergillus nidulans. EMBO J. 4:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubicek, C. P. 1981. Release of carboxymethyl-cellulase and β-glucosidase from the cell walls of Trichoderma reesei. Eur. J. Appl. Microbiol. Biotechnol. 13:226-231. [Google Scholar]
- 21.Kumar, V., S. Ramakrishnan, T. T. Teeri, J. K. Knowles, and B. S. Hartley. 1992. Saccharomyces cerevisiae cells secreting an Aspergillus niger β-galactosidase grow on whey permeate. Bio/Technology 10:82-85. [DOI] [PubMed] [Google Scholar]
- 22.Mach, R. L., M. Schindler, and C. P. Kubicek. 1994. Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr. Genet. 25:567-570. [DOI] [PubMed] [Google Scholar]
- 23.Mandels, M. M., and R. E. Andreotti. 1978. The cellulose to cellulase fermentation. Proc. Biochem. 13:6-13. [Google Scholar]
- 24.Mischak, H., F. Hofer, R. Messner, E. Weissinger, M. Hayn, P. Tomme, H. Esterbauer, E. Kuchler, M. Claeyssens, and C. P. Kubicek. 1989. Monoclonal antibodies against different domains of cellobiohydrolase I and II from Trichoderma reesei. Biochim. Biophys. Acta 990:1-7. [DOI] [PubMed] [Google Scholar]
- 25.Nevalainen, K. M. H., and E. T. Palva. 1978. Production of extracellular enzymes in mutants isolated from Trichoderma viride unable to hydrolyze cellulose. Appl. Environ. Microbiol. 35:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikolaev, I. V., S. M. Epishin, E. S. Zakharova, S. V. Kotenko, and P. Vinetskii Iu. 1992. Molecular cloning of the gene for secreted β-galactosidase of the filamentous fungus Penicillium canescens. Mol. Biol. (Moscow) 26:869-875. [PubMed] [Google Scholar]
- 27.Penttilä, M. E. 1998. Heterologous protein production in Trichoderma, p. 356-383. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium. Enzymes, biological control and commercial applications. Taylor and Francis Ltd., London, United Kingdom.
- 28.Persson, I., F. Tjerneld, and B. Hahn-Hägerdal. 1991. Fungal cellulolytic enzyme production: a review. Proc. Biochem. 26:65-74. [Google Scholar]
- 29.Ramakrishnan, S., and B. S. Hartley. 1993. Fermentation of lactose by yeast cells secreting recombinant fungal lactase. Appl. Environ. Microbiol. 59:4230-4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiboth, B., L. Hartl, M. Pail, E. Fekete, L. Karaffa, and C. P. Kubicek. 2004. The galactokinase of Hypocrea jecorina is essential for cellulase induction by lactose but dispensable for growth on d-galactose. Mol. Microbiol. 51:1015-1025. [DOI] [PubMed] [Google Scholar]
- 31.Seiboth, B., G. Hofmann, and C. P. Kubicek. 2002. Lactose metabolism and cellulase production in Hypocrea jecorina: the gal7 gene, encoding galactose-1-phosphate uridylyltransferase, is essential for growth on galactose but not for cellulase induction. Mol. Genet. Genomics 267:124-132. [DOI] [PubMed] [Google Scholar]
- 32.Shoemaker, S., V. Schweickart, M. Ladner, D. Gelfand, S. Kwok, K. Myambo, and M. Innis. 1983. Molecular cloning of exo-cellbiohydrolase from Trichoderma reesei strain L27. Bio/Technology 1:691-696. [Google Scholar]
- 33.Stalbrand, H., A. Saloheimo, J. Vehmaanpera, B. Henrissat, and M. Penttila. 1995. Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei β-mannanase gene containing a cellulose binding domain. Appl. Environ. Microbiol. 61:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strauss, J., R. L. Mach, S. Zeilinger, G. Hartler, G. Stoffler, M. Wolschek, and C. P. Kubicek. 1995. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 376:103-107. [DOI] [PubMed] [Google Scholar]
- 35.Tani, S., T. Itoh, M. Kato, T. Kobayashi, and N. Tsukagoshi. 2001. In vivo and in vitro analyses of the AmyR binding site of the Aspergillus nidulans agdA promoter; requirement of the CGG direct repeat for induction and high affinity binding of AmyR. Biosci. Biotechnol. Biochem. 65:1568-1574. [DOI] [PubMed] [Google Scholar]
- 36.Trinci, A. P. 1972. Culture turbidity as a parameter of mould growth. Trans. Br. Mycol. Soc. 58:467-473. [Google Scholar]
- 37.Vanhanen, S., M. Penttila, P. Lehtovaara, and J. Knowles. 1989. Isolation and characterization of the 3-phosphoglycerate kinase gene (pgk) from the filamentous fungus Trichoderma reesei. Curr. Genet. 15:181-186. [DOI] [PubMed] [Google Scholar]
- 38.van Peij, N. N., M. M. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Tilbeurgh, H., M. Claeyssens, and C. K. de Bruyne. 1982. The use of 4-methylumbelliferyl and other chromophoric glycosides in the study of cellulolytic enzymes. FEBS Lett. 149:152-156. [Google Scholar]
- 40.Vogel, H. J. 1956. A convenient growth medium for Neurospora (medium N). Microb. Genet. Bull. 13:42-44. [Google Scholar]
- 41.Wiebe, M. G., G. D. Robson, J. Shuster, and A. P. Trinci. 2001. Evolution of a recombinant (glucoamylase-producing) strain of Fusarium venenatum A3/5 in chemostat culture. Biotechnol. Bioeng. 73:146-156. [DOI] [PubMed] [Google Scholar]
- 42.Wiebe, M. G., and A. P. J. Trinci. 1991. Dilution rate as a determinant of mycelial morphology in continuous culture. Biotechnol. Bioeng. 38:75-81. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 44.Zeilinger, S., A. Ebner, T. Marosits, R. Mach, and C. P. Kubicek. 2001. The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase II-gene) activating element. Mol. Genet. Genomics 266:56-63. [DOI] [PubMed] [Google Scholar]
- 45.Zeilinger, S., M. Schmoll, M. Pail, R. L. Mach, and C. P. Kubicek. 2003. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol. Genet. Genomics 270:46-55. [DOI] [PubMed] [Google Scholar]