Abstract
Purpose
Chronic lymphocytic leukemia (CLL) with 17p deletion typically progresses quickly and is refractory to most conventional therapies. However, some del(17p) patients do not progress for years, suggesting that del(17p) is not the only driving event in CLL progression. We hypothesize that other concomitant genetic abnormalities underlie the clinical heterogeneity of del(17p) CLL.
Experimental Design
We profiled the somatic mutations and copy number alterations (CNA) in a large group of del(17p) CLL as well as wild type CLL and analyzed the genetic basis of their clinical heterogeneity.
Results
We found that increased somatic mutation number associates with poor overall survival independent of 17p deletion (p=0.003). TP53 mutation was present in 81% of del(17p) CLL, mostly clonal (82%), and clonal mutations with del17p exhibit shorter overall survival than subclonal mutations with del17p (p= 0.019). Del(17p) CLL has a unique driver mutation profile, including NOTCH1 (15%), RPS15 (12%), DDX3X (8%) and GPS2 (6%). We found that about half of del(17p) CLL cases have recurrent deletions at 3p, 4p, or 9p and that any of these deletions significantly predicts shorter overall survival. In addition, the number of CNAs, but not somatic mutations, predicts shorter time to treatment among patients untreated at sampling. Indolent del(17p) CLLs were characterized by absent or subclonal TP53 mutation and few CNAs, with no difference in somatic mutation number.
Conclusions
We conclude that del(17p) has a unique genomic profile and that clonal TP53 mutations, 3p, 4p or 9p deletions, and genomic complexity are associated with shorter overall survival.
Introduction
Chronic lymphocytic leukemia (CLL) is a common leukemia with a heterogeneous disease course that varies based on the degree of somatic hypermutation in the immunoglobulin heavy-chain variable region (IGHV), and by chromosome abnormalities(1–3). Metaphase analysis of CLL chromosomes revealed that complex karyotype, defined as three or more gross chromosomal abnormalities, is predictive of worse disease outcome(4,5). In 2000, Dohner et al used fluorescence in situ hybridization (FISH) to detect recurrent chromosomal abnormalities and established a hierarchical prognostic model for OS(3) in which 17p deletion is associated with the worst disease outcome. Many studies have since confirmed that del(17p) CLL responds poorly to conventional chemoimmunotherapies and has a median OS of less than 3 years, at least prior to the newly approved targeted therapies(6,7).
Del(17p) is found in 5-10% of patients at diagnosis but in up to 40% of patients relapsing after fludarabine based therapies(8,9). Del(17p) causes loss of one allele of the tumor suppressor TP53 which plays an important role in DNA repair, cell cycle arrest, and apoptosis in response to genotoxic insults(10). Somatic mutations in TP53 occur in the other allele of TP53 in about 80% of del(17p) CLL, resulting in biallelic inactivation(11–14). About 4-30% of TP53 mutations are monoallelic without coexisting del(17p)(11,13,15,16). Similar to 17p deletion, TP53 mutations alone, even when present as very small subclones(17,18), have been associated with worse treatment response, shorter failure free survival (FFS), and shorter OS(11–14). Notably, however, del(17p) CLL exhibits considerable heterogeneity. A subset of CLL patients with del(17p) have stable disease without need for treatment for over 5 years(19,20). Delgado et al. showed that genomic complexity measured by SNP array and IGHV mutational status were the main predictors of OS in CLL patients with TP53 disruption, but their analysis was limited by an absence of somatic mutation data (21).
Recent work by our group and others has used genomic profiling to characterize the landscape of genomic alterations in CLL, including recurrent copy number alterations (CNAs)(22) and somatic mutations in CLL driver genes (23–26). Surprisingly, none of these studies included significant numbers of CLLs with 17p deletion, for example, only 17 in the recent study of 500 CLLs from Puente et al(24). We undertook this study to systematically characterize the coexisting genetic aberrations in the largest cohort of del(17p) CLL investigated to date using WES and array analysis. Our results show that del(17p) CLL has distinct genetic profiles associated with different disease outcomes.
Materials and Methods
Patient Samples
Matched peripheral blood (tumor) and saliva (germline) samples were collected from patients who consented to an IRB-approved tissue banking protocol. B cell enrichment was performed using Easy Sep Human B cell Enrichment Kit (StemCell Technologies Inc, Vancouver, Canada) for samples with WBC count lower than 25K or absolute lymphocyte count <20K. For samples with lymphocyte count >25K, peripheral blood mononuclear cells (PBMC) were separated using conventional Ficoll-Hypaque separation according to the manufacturer's instructions (StemCell Techonologies Inc, Vancouver, Canada). Tumor and saliva DNA were extracted using QIAamp Blood DNA (Qiagen Inc, Valencia, CA) and Oragene DNA (Oragene, Ontario, Canada) kits respectively, according to the manufacturer's directions.
FISH cytogenetics were assessed by the clinical cytogenetics laboratory at the Brigham & Women's Hospital. The cutoff for 17p deletion was 10% cells positive, 13q deletion: 2.5%, trisomy 12: 2%, and 11q deletion: 5%. IGHV status was assessed by the CLL Research Consortium tissue bank or by LabCorp; unmutated IGHV requires >=98% homology to the closest germline match to an immunoglobin heavy chain variable region. CNA data if available were used to determine the 17p deletion status for patients without FISH cytogenetics (n=14, all were 17p wild type).
Only samples that provided an evaluable karyotype by whichever method they were assessed were considered to have available data. For stimulated karyotyping since 2011, 0.8-1.0.mL of either bone marrow or peripheral blood was added to each of two 5.0mL MarrowMax media (Life Technologies, Carlsbad, CA) replicate cultures and stimulated with a B-cell cocktail consisting of: 100μL CpG-ODN-GNKG685 (10μg/mL, Sigma, St Louis, MO), 50μL Pokeweed mitogen (PWM) from Phytolacca americana (10μg/mL, Sigma, St Louis, MO) and 100μL Phorbol 12-myristate 13-acetate (PMA) (40ng/mL, Sigma, St Louis, MO). Prior to 2011, the cells were stimulated only with PWM. In all cases, the cells were cultured at 37°C with 5% CO2 for 3-days, and harvested/banded according to standard procedures. GTG-banded metaphase chromosomes were captured and processed using Cytovision Ultra Workstations (Leica Microsystems, Version 4.5.2). Twenty metaphases from across both of the two cultures were analyzed.
SNP arrays
Matched tumor and normal DNA of 40 CLL patients with 17p deletion were run on the Affymetrix CytoScan HD array according to the manufacturer's protocol. CNAs were inferred using the Affymetrix Chromosome Analysis Suite [version −2.0.1.2 (r5919), genome build hg19]. Tumor and germline DNA of 160 CLL patients, including 15 del(17p) and 145 WT, were run on Affymetrix SNP6.0 arrays, and CNAs were inferred using Nexus Copy Number 7.0 (Biodiscovery, significance threshold set at 1.0E-10). All CNAs and LOHs were manually reviewed to ensure call quality and to remove small germline copy number variants. Neighboring segments with similar copy number ratios were joined as single segments. The final set of copy number segments were checked for probe densities on both the SNP6.0 platform and the Cytoscan HD platform and all segments had at least 60 probes on both platforms. The 160 SNP6.0 arrays have been previously described(22). Sex chromosomes and immunoglobulin switch regions were excluded in the copy number analysis. A cutoff of 25 probes and 200kb for deletion CNAs or 400kb for gain CNAs was used. A size cut off of 10 MB genome-wide and 3 MB within genic regions was used to detect LOH events. Chromothripsis events were identified using the criteria defined by Korbel and Campbell(27). A region was considered to have undergone chromothripsis if it satisfied at least two criteria; the first being the presence of oscillating patterns of retention of heterozygosity alternating with areas of LOH, and second, alternating patterns of copy number states with clustering of at least 5 – 10 breakpoints within a 50 Kb region.
Whole exome sequencing (WES)
Purified genomic DNA of 176 CLL tumor-germline pairs were submitted to the Genomics Platform at the Broad Institute (Cambridge, MA) for WES. Mean target coverage of all samples is 80X. 134 of the 176 whole exomes were included in a previous study(25). Picogreen based dsDNA quantitation (Life Technologies, Carlsbad, CA) was performed to ensure sample purity and DNA fingerprinting was used to confirm the match between the tumor and its cognate normal, prior to WES library preparation. Libraries were sequenced on either an Illumina HiSeq2000 or GA-IIX and aligned reads were generated by the Picard pipeline (28). Somatic mutations and indels were identified using MuTect(29) and indelocator(23), respectively, integrated in the Call Somatic Mutations for Capture workflow and the Call Indels for Capture workflows in the Broad Firehose pipelines. Somatic mutations with less than 8 sequencing reads of the alternative alleles were filtered out in the analysis as previously described(30). ABSOLUTE(31) was used to assess cancer cell fraction (CCF) of somatic mutations. Somatic mutations were considered clonal when the CCF was 85% or greater as previously described (26).
Sanger Sequencing of the TP53 Gene
Exons 4-9 of TP53 were sequenced according to the protocol described in the IARC TP53 database (p53.iarc.fr). Primers 326/327 were used to amplify and sequence exon 4, primers 236/240 for exons 5 and 6, primers 237/238 for exon 7, and primers 314/315 for exons 8 and 9. Sequencing results were aligned to the TP53 reference sequence and manually reviewed.
Statistical Analysis
Overall survival (OS) and failure free survival (FFS) were estimated using the Kaplan-Meier method. FFS was measured from the time of sampling to first treatment after sampling or death, whichever occurred first. OS was measured from the time of sampling to death in order to control for differences in the time of sampling in relation to diagnosis. Patients were censored at the date last known alive. Because genetic changes may evolve over time, all time to event endpoints were measured from time of sampling. For OS and FFS, univariable and multivariable Cox regression analysis was also performed. All Cox models were stratified by treatment status at sampling since previously treated and untreated patients represent two different patient populations with different prognoses. Prior to performing the regression analysis, the proportional hazards assumption for each variable was tested and interaction terms were examined. The linearity assumption for continuous variables was examined using restricted cubic spline estimates of the relationship between the continuous variable and log relative hazard(32). Natural log or log10 transformation was attempted in order to normalize mutation and CNA event parameters. However, due to the influence of some large values in the SNP parameters, all continuous variables from WES and SNP analyses were dichotomized for consistency of presentation and optimal cutoff values were identified using the method of recursive partitioning for survival trees and the restricted cubic spline estimation method (33). These cutoff values are presented in tables and figures throughout the paper. Furthermore, the C-statistic(34) was used to guide and compare the predictive ability of selected parameters from the Cox model. Fisher's Exact test was used for comparison of binary variables and the Wilcoxon-Rank-Sum test was used for comparison of continuous variables. The log-rank test was used to assess the difference in FFS or OS between groups. All statistical tests were two-sided at a significance level of 0.05 and multiplicity was not considered. All statistical analyses were performed using SAS 9.3 (SAS Institute Inc, Cary, NC), and R version 3.2.0 (The Comprehensive R Archive Network, www.cran.r-project.org). The heatmap was generated using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E).
Results
Patient Characteristics
277 CLL patients were included in this study: 69 with del(17p) and 208 without (Table 1, Table S2). Of these 277 patients, 176 were profiled by WES for somatic mutations and 200 were profiled by SNP arrays for CNAs (Table 1). Ninety-nine patients were characterized by both WES and SNP arrays. As is well documented in the literature (3,6,11), del(17p) CLL exhibits different clinical characteristics with markedly fewer mutated IGHV (16% del(17p) vs 64% WT, p<0.0001). 73% of all patients were untreated at sampling, with 34% subsequently undergoing therapy, with a median follow-up time from sampling of 6.0 years among survivors (range 3 days – 8.8 years). As expected, a higher percentage of del(17p) patients had been treated prior to sampling (51% vs 19%, p<0.0001). Among the 35 del(17p) patients who were untreated at sampling, 24 (70%) patients subsequently underwent therapy at a median follow-up of 22 months.
Table 1.
Patient characteristics and summary of CNAs and somatic mutations.
| ALL | wt 17p | del(17p) | p-value | |
|---|---|---|---|---|
| n | 277 (100%) | 208 (75%) | 69 (25%) | |
| Male | 163 (59%) | 118 (57%) | 45 (65%) | 0.26 |
| Age of onset | 55 (32 - 86) | 54 (32 - 78) | 61 (38 - 86) | 2.2e-05 |
| Treated before sampling | 74(27%) | 39(19%) | 35(51%) | 6.4e-07 |
| IGHV mutated | 135(52%) | 125(64%) | 10(16%) | 1.2e-11 |
| Complex karyotype | 42(32%) | 20(21%) | 22(61%) | 4.03e-05 |
| FISH cytogenetics | ||||
| 13q14 loss | 170(61%) | 139(67%) | 31(45%) | 1.9E-4 |
| 11q loss | 37(13%) | 29(14%) | 8(12%) | 0.55 |
| Trisomy 12 | 36(13%) | 25(12%) | 11(16%) | 0.54 |
| Profiled by WESa | 176 (100%) | 123 (70%) | 53 (30%) | |
| Total mutations | 19 (0 - 94) | 18 (0 - 94) | 21 (7 - 68) | 0.0048 |
| Nonsynonymous mutations | 14 (0 - 70) | 13 (0 - 70) | 16 (5 - 54) | 0.0055 |
| Synonymous mutations | 4 (0 - 24) | 4 (0 - 24) | 4 (1 - 14) | 0.14 |
| Subclonal mutations | 9 (0 - 89) | 9 (0 - 89) | 8 (2 - 35) | 0.9 |
| Clonal mutations | 9 (0 - 34) | 7 (0 - 24) | 12 (0 - 34) | 5.8E-4 |
| Profiled by SNPa | 200 (100%) | 145 (72%) | 55 (28%) | |
| # of CNAs | 1 (0 - 36) | 1 (0 - 16) | 7(0 - 36) | 1.5e-16 |
| # of losses | 1 (0 - 35) | 1 (0 - 16) | 6 (0 - 35) | 5e-15 |
| # of gains | 0 (0 - 19) | 0 (0 - 3) | 1 (0 - 19) | 3.3e-08 |
| Lost Mb | 3 (0 - 530) | 1.2(0 - 88) | 97 (0 - 530) | 2.9e-18 |
| Gained Mb | 0 (0 - 340) | 0 (0 - 260) | 5.9 (0 - 340) | 9.9e-6 |
| 8p loss | 21(10%) | 6(4%) | 15(27%) | 1.1e-05 |
| 3p loss | 15(8%) | 0(0%) | 15(27%) | 8.1e-10 |
| 4p loss | 14(7%) | 2(1%) | 12(22%) | 4.1e-06 |
| 9p loss | 15(8%) | 2(1%) | 13(24%) | 1.1e-06 |
| Loss in 3p, 4p, or 9p | 31(16%) | 3(2%) | 28(51%) | 8.6e-16 |
Median values and ranges are presented for the WES and SNP analysis.
Overview of Somatic Mutations and CNAs
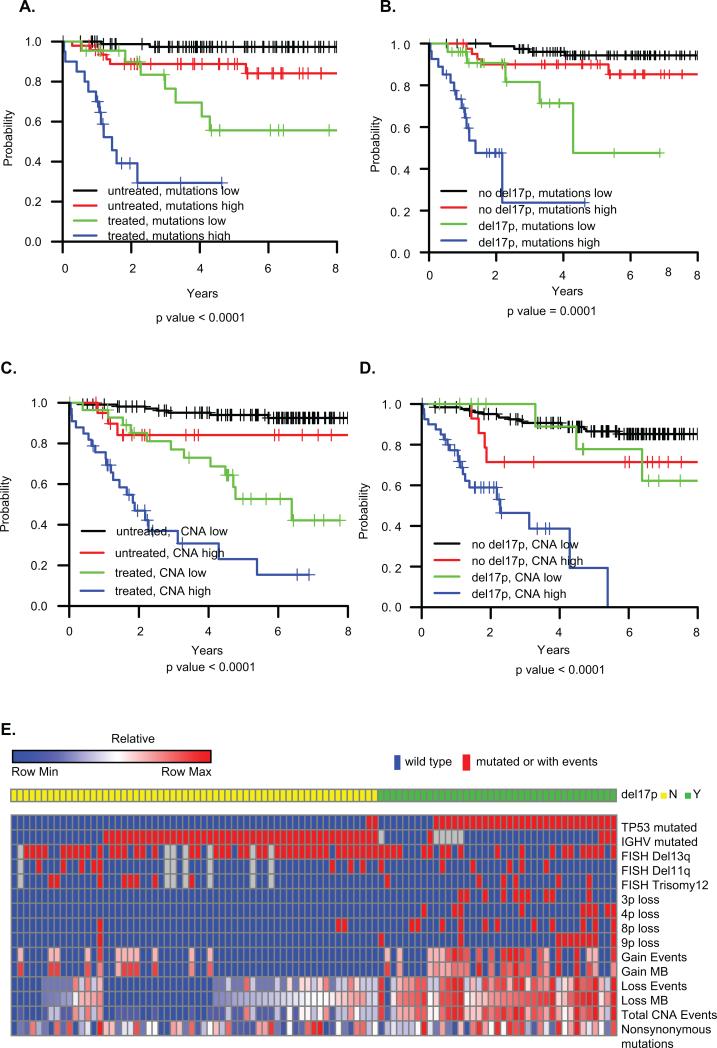
We characterized the somatic mutation spectrum of 17p deleted CLL by WES on paired tumor and germline samples from 176 CLL patients, including 53 with 17p deletions, significantly more than prior studies(23–26,35). Compared to CLL with wild type 17p, del(17p) CLL has more somatic mutations (median 21 vs 18, p = 0.0048), nonsynonymous mutations (median 16 vs 13, p= 0.0055), and clonal mutations (median 12 vs 7, p =5.8E-4) (Table 1). Synonymous mutations and subclonal mutations are not significantly different between del(17p) and wild type. Furthermore, a Cox model stratified by previous treatment status at sampling revealed that increasing total mutations (p = 0.003), clonal mutations (p = 0.003) and nonsynonymous mutations (p = 0.007) are associated with shorter OS in both univariable and multivariable analysis controlling for del(17p) and IGHV mutational status, while synonymous mutations and subclonal mutations are not (p >0.05) (Table 2). The number of mutations stratified OS risk in both the previously treated and untreated groups (Fig 1A), and stratified del(17p) into poor vs. intermediate risk (Fig 1B).
Table 2.
Cox model stratified by previous treatment status at sampling.
| Univariable Analysis | Multivariable Analysis* | |||||||
|---|---|---|---|---|---|---|---|---|
| WES analysis (n=176) | HR | 95% CI | p-value | HR | 95% CI | p-value | ||
| Tot no. of mutations: high vs low | 4.82 | 2.08 | 11.2 | 0.0002 | 3.55 | 1.55 | 8.16 | 0.003 |
| Clonal mutations: high vs low | 6.62 | 1.95 | 22.5 | 0.002 | 7.39 | 2.11 | 25.9 | 0.003 |
| Subclonal mutations: high vs low | 3.35 | 1.34 | 8.38 | 0.01 | 2.25 | 0.87 | 5.82 | 0.1 |
| Nonsynonymous mutations: high vs low | 4.69 | 1.75 | 12.6 | 0.002 | 4 | 1.45 | 11.1 | 0.007 |
| Synonymous mutations: high vs low | 2.79 | 0.957 | 8.14 | 0.06 | 2.45 | 0.828 | 7.24 | 0.1 |
| SNP analysis (n=200) | ||||||||
| Tot no. of events: high vs low | 3.21 | 1.68 | 6.16 | 4e-04 | 2.28 | 1.12 | 4.66 | 0.02 |
| No. of gain events: high vs low | 1.98 | 1.01 | 3.88 | 0.045 | 1.58 | 0.80 | 3.12 | 0.2 |
| No. of loss events: high vs low | 3.63 | 1.9 | 6.95 | 1e-04 | 2.24 | 1.06 | 4.76 | 0.04 |
| Gain MB: high vs low | 2.4 | 1.29 | 4.47 | 0.006 | 1.86 | 0.98 | 3.52 | 0.06 |
| Loss MB: high vs low | 5.26 | 2.61 | 10.6 | 4e-06 | 3.86 | 1.74 | 8.55 | 0.0009 |
Adjusted for IGHV mutational status and del(17p).
The optimal cutoff values for separating the high and low groups were identified using the method of recursive partitioning for survival trees. Cutoff values for total number of mutations: 21; clonal mutations: 8; subclonal mutations: 22; nonsynonymous mutations: 15; synonymous mutations: 3. For CNAs, total number of events: 4; number of gain events: 2; number of loss events: 5; gain MB: 0.675MB; loss MB: 25MB.
Figure 1. Overall survival and heatmap characteristics of patients.
A) OS by total number of mutations and treatment status at sampling; (B) OS by total number of mutations and del(17p) status. (C) OS by total CNA events and treatment status at sampling. (D) OS by total CNA events and del(17p) status. Mutations low: mutations < 21; mutations high: mutations >=21. CNAs low: CNA < 4; CNA high: CNA >=4. The optimal cutoff values for total number of mutations and CNAs were identified using the method of recursive partitioning for survival trees. (E) Heatmap characteristics of the 99 patients with both SNP and WES profiles. The first 9 rows are indicated by presence (red) or absence (blue) of TP53 mutation, IGHV mutation or abnormal cytogenetics by FISH. CNA events, weights and nonsynonymous mutations represent percentiles of ranks across 99 observations. i.e., the highest value in each row is denoted as 1 (100th percentile) and the lowest value is denoted as 0.01 (1th percentile).
CNA data were obtained for 55 del(17p) CLL and 145 WT CLL, with a median of 1 CNA per patient in the entire cohort similar to our own and others’ previous reports (22,36,37). 17p deletion has a minimally deleted region (MDR) of 34kb which as expected targets the TP53 gene (Table S1 and Figure S1). Figure S2 provides an overview of CNA events, showing many fewer CNAs in patients who did not require therapy in the follow-up period. More CNAs were seen in del(17p) than WT (median 7 vs 1, p<0.0001, see Table 1). In addition, del(17p) CLL has a longer total length of deleted (97Mb vs 1.2Mb, p<0.0001) and gained DNA (5.9Mb vs 0Mb, p<0.0001). High total number of CNAs, as well as number of loss events and total length of loss, were all associated with shorter OS in the entire cohort in both univariable and multivariable analysis stratified by treatment status at sampling and adjusted for 17p deletion and IGHV mutational status (Table 2 and Fig 1C)(22,37). The total number of CNA events further stratified OS risk in both del(17p) and WT CLL (Fig 1D). In del(17p) CLL in our cohort, mutated IGHV was associated with fewer CNAs (p<0.01) but not somatic mutations (p>0.1), and was predictive of longer OS (P<0.01).
Using 99 samples that were profiled by both WES and SNP (Heatmap, Fig 1E), we tested whether the number of somatic mutations was correlated with the number of CNAs. Neither number of CNAs, nor total length of gains or losses, correlated with the number of somatic mutations in individual CLLs (Figure S3A-C), suggesting that mutations and CNAs result from different mechanisms in CLL. Since the number of somatic mutations and CNAs are not correlated, we examined whether one is more predictive of survival than the other. The C-index from univariable Cox regression analysis stratified by treatment status at sampling was 0.765 for the total number of CNA events and 0.750 for the total number of mutations (Table S3), suggesting that both are very predictive of overall survival as single variables. When we included both variables in a Cox model, the C-index was substantially increased to 0.850 (Table S3 and Figure S3D) indicating that both are independently significant in predicting overall survival as noted. It is notable that patients with a high number of both CNAs and mutations all died within 3 years (i.e. 0% 3 year OS) and eleven of twelve of these patients had del(17p).
TP53 Disruption in CLL
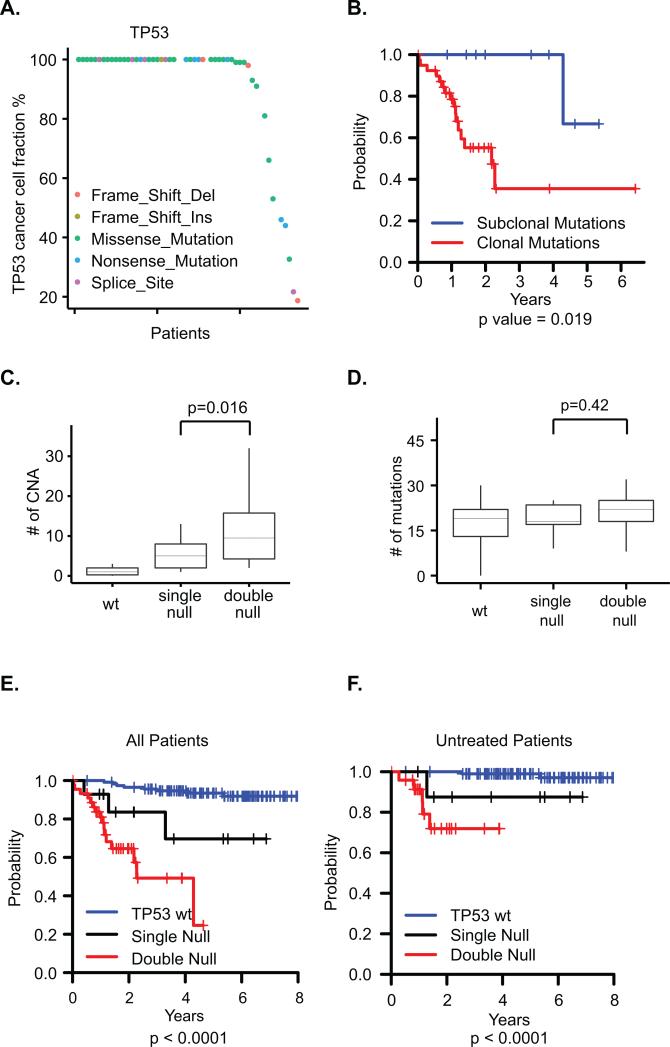
As expected, somatic TP53 mutation is the most abundant mutation in del(17p) CLL, seen in 43 patients (81%). Additionally TP53 mutations were found in 6 (5%) patients without 17p deletion, two of whom have copy neutral LOH at 17p and are therefore considered to have biallelic loss (Figure S4A). We found that most TP53 mutations in this del17p cohort are clonal (40 of 49, 82%, see Fig 2A), similar to a previously reported rate of clonal TP53 mutations in del(17p) patients(17). Del(17p) patients with subclonal TP53 mutations had a longer OS than patients with clonal TP53 mutations (p = 0.019, Fig 2B). In contrast, SF3B1 mutations (n=22), the second most abundant mutation in our cohort(23), are mostly subclonal, and clonality of SF3B1 is not associated with OS (Figure S4BC).
Figure 2. Analysis of TP53 disruptions.
A. Cancer cell fractions of the TP53 mutations in each patient. B. Kaplan-Meier curves of OS by clonality status of TP53 mutations. C. Relationship of CNAs and TP53 disruptions. wt: no TP53 mutation or deletion; single null: monoallelic TP53 mutation or del(17p) deletion; double null: biallelic disruption. D. Relationship of somatic mutations and TP53 disruptions. E-F. Dose-dependent impact of TP53 disruption on overall survival in all patients (E) or previously untreated patients (F).
Whether mutation of the second TP53 allele impacts clinical outcome in del(17p) CLL remains an open question. Our cohort contains patients with biallelic disruption of TP53 by somatic TP53 mutation, with either 17p deletion (n= 43) or copy neutral loss of heterozygosity (cnLOH) (n = 2), as well as monoallelic disruptions by either 17p deletion or TP53 mutation (n=14) (Figure S4A). Interestingly, most biallelic disruptions have clonal TP53 mutations (37 clonal vs 8 subclonal). We tested whether there is a dose-dependent effect of TP53 disruption on CNAs or somatic mutations in CLL and found more CNA events in biallelic disruption (median 9.5 in double null vs 5 in single null, p = 0.016, Fig 2C). No significant difference in number of somatic mutations was seen based on mono- or biallelic TP53 disruption (Fig 2D), again suggesting that somatic mutations arise by a different mechanism than CNAs in CLL.
Mono- vs biallelic TP53 disruption was also predictive of OS. With wild type TP53 having the best prognosis and biallelic TP53 disruption having the worst prognosis, patients with monoallelic disruption of TP53 by either del(17p) or somatic mutation showed intermediate OS (p<0.0001, Fig 2E). This dose-dependent pattern remained consistent when the analysis was restricted to previously untreated patients (p<0.0001, Fig 2F).
Driver Genes in Del(17p) CLL
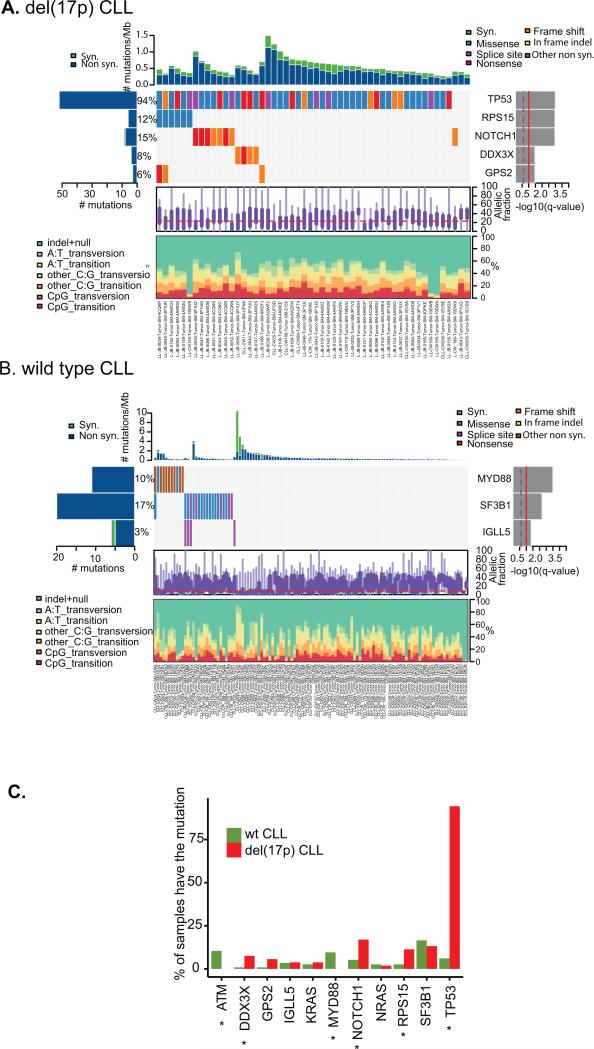
To identify driver genes in del(17p) CLL, we ran the MutSigCV algorithm(30) on the 53 del(17p) CLL in our WES cohort. In addition to TP53, several other genes are significantly mutated, including NOTCH1 (8 mutations, 15% of the cohort), RPS15 (6, 12%), DDX3X (4, 8%), and GPS2 (3, 6%) (Fig 3A). In contrast, MutSigCV identified SF3B1 (18, 17%), MYD88 (11, 10%), and IGLL5 (4, 3%) as significantly mutated genes in 17p wild-type CLL (Fig 3B). We therefore compared the mutation frequencies of known CLL driver genes in 17p vs WT CLL, and found that mutations in ATM and MYD88 occurred exclusively in 17p WT CLL, while mutations in DDX3X, NOTCH1, RPS15, and TP53 occurred with higher frequency in del(17p) CLL (P<0.05) (Fig 3C). Other mutations like SF3B1 and IGLL5 distributed without significant difference.
Figure 3. Driver genes in del(17p) CLL.
A. Comut plot of del(17p) CLL. B. Comut plot of wild type CLL. C. Mutation frequencies of known CLL driver genes in del(17p) and wt 17p CLL. * indicates p < 0.05.
NOTCH1 and DDX3X have been previously implicated in CLL, and NOTCH1 mutation has been correlated with poor outcome and risk of Richter's transformation (24,38,39). The RPS15 gene has recently been identified as a new CLL driver gene with a mutation frequency of 4.3% in 538 CLL cases (26) and 19.5% in relapsing CLL (40). In our cohort enriched for del(17p) CLL, we identify that the RPS15 gene is more frequently mutated in del(17p) CLL than in WT(12% vs 2.5%, p<0.05). We confirmed all RPS15 mutations by Sanger sequencing (Figure S5A). In the two samples with RNAseq data available, we also confirmed that the mutations are expressed in the tumor mRNA (Figure S5B). RPS15 encodes the ribosomal protein S15. Interestingly, all of the RPS15 mutations occur in the far carboxyl end of the protein, ranging from amino acid 131 to 145 (Figure S5C and Table S4), suggesting that mutation of this domain may be important in CLL development. Presence of a RPS15 mutation was not associated with OS however (Figure S5D).
Recurrent Deletions and Chromothripsis in Del(17p) CLL
To identify driver CNA events enriched in del(17p) CLL, we performed Gistic analysis(41) and found recurrent chromosomal deletions at 3p, 4p, 8p, and 9p in del17p CLL (Figure S6). We have previously reported that 8p deletion is associated with del(17p) and worse prognosis(22) (Figure S7). Here we also note that over half of the del(17p) CLLs have at least one of 3p, 4p or 9p deletions, yet WT CLL rarely shows these (Table 1, Fig 1E). Each of these deletions is significantly associated with shorter OS in the entire cohort (p<0.0001) and any one of these deletions stratifies OS in del(17p) CLL (Figure S8). The minimally deleted regions (MDR) of 3p, 4p, 8p, and 9p and the genes affected are included in Table S1 and Figures S9-12.
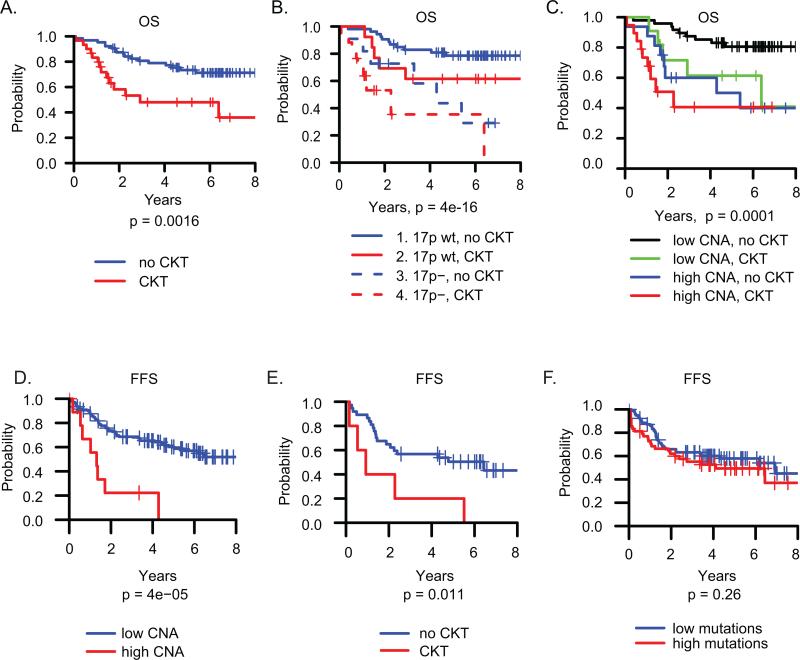
Previous studies have associated complex karyotype (CKT) with poor outcome(4,5,42,43). We had karyotype information on 94 of the 200 patients profiled by SNP array. In this subgroup, and consistent with previous studies(4,5,42,43), CLL patients with CKT had shorter OS than patients without CKT (Fig 4A, p = 0.0016). 63% of del17p CLL had CKT as compared to only 22% of WT (p=0.0002). Patients with both del(17p) and CKT together had worse OS, suggesting possibly additive risks (Fig 4B). As expected, we found that more CNA events measured by SNP array is associated with CKT (Figure S13). OS was similar among patients with either a high CNA count, CKT, or both, indicating that high CNA number and CKT reflect the same phenomenon and are not additive risk factors (Fig 4C).
Figure 4. Overall survival (OS) and failure free survival (FFS).
A. OS by CKT vs no CKT (n=94). B. OS by del(17p) and CKT status. C. OS by CKT vs CNA numbers. Cutoff for high vs low CNAs is 4. D-F: FFS for previously untreated patients (n = 122). D. FFS by number of CNA events. The cutoff for high vs low is 4 CNA events. E. FFS by CKT status. F. FFS by number of total mutations, with cutoff for low vs high mutations 21.
Chromothripsis has been previously reported in CLL, with a suggestion of association with del(17p)(44,45). We found 3 cases of chromothripsis, affecting chromosomes 8, 11q and 15q. 17p deletions were found in 2 cases, together with clonal TP53 mutations, and the third case had cnLOH in 17p (Figure S14). We sequenced the TP53 gene in the latter tumor sample with cnLOH and found a clonal missense mutation p.179H>R in exon 5 of TP53 (Figure S15). Thus all three cases had biallelic inactivation of TP53. The median number of CNA events was markedly higher in these three patients than in del(17p) patients overall (24 vs 11). All three patients had a 3p deletion, and one had deletions in all of 3p, 4p, 8p, and 9p. The latter patient also harbored a potential chromoanasynthesis event adjacent to the chromothripsis region (Figure S14B). Chromoanasynthesis is characterized by stretches of euploidy, alternating with segments of higher ploidy, and is not accompanied by LOH events(46). All three patients had aggressive disease, with two treated before sampling and the third treated shortly after (6 days).
CNAs but not mutations predicts the need for treatment
A central question about the clinical heterogeneity of CLL, and 17p deleted CLL in particular, is what underlies the variable progression to treatment among treatment naïve patients. For example, among 23 del(17p) patients treatment naive at sampling in our SNP cohort, 7 (30%) have not yet required treatment with a median follow up of 57 months, while 16 (70%) underwent therapy soon after sampling. A visual inspection of the CNAs in the different treatment groups reveals that the never treated groups are largely devoid of large chromosomal deletions or gains with the exception of trisomy 12, as we have previously reported (Figure S2)(22). This finding holds true in del(17p) CLL as well (Figure S2). The never treated del(17p) CLLs (n = 7) have a median of 1 CNAs per sample, while the del(17p) CLLs treated after sampling have a median of 9 CNAs per sample, similar to the median in those treated prior to sampling (median =8). IGHV mutational status has also been suggested to impact the outcome of del(17p) patients(20,21), and mutated IGHV is overrepresented in our indolent del(17p) patients (57% mutated (4 of 7) vs 10% in all other del(17p) patients). We further analyzed the association between CNA events and the time from sampling to next treatment or death (failure free survival (FFS)). In the entire cohort of previously untreated patients at sampling, excluding 16 patients who were treated within 30 days of sampling (n=122), increasing CNA events are associated with shorter FFS (Fig 4D). This result remains true in a multivariable model adjusting for del17p and IGHV mutational status (p=0.0002). Complex karyotype, but not total somatic mutations, is also predictive of shorter FFS (Fig 4E-F).
Discussion
Our somatic mutation analysis of 53 del(17p) CLLs is the largest cohort of del(17p) CLL to undergo WES to date(23–25,47) and allowed us to identify increased somatic nonsynonymous mutations and a novel driver gene profile in del(17p) CLL. Clonal mutations are also increased, but this may be related to the older age of del17p patients in this cohort (median 65 vs 60). Additionally, we have found that total number of mutations, particularly nonsynonymous mutations, is associated with shorter OS independent of 17p deletion status, but has a larger impact on OS in del(17p) CLL than WT. No association was observed between the number of somatic mutations and the number of CNAs, suggesting that the mechanisms by which they arise may be distinct. Somatic mutations are point mutations and small insertions or deletions, which reflect failed base or nucleotide excision repair or aberrant translesion synthesis. CNAs involve losses or gains of long stretches of DNA which are likely a result of failed repair of double-strand DNA breaks. Our results suggest a greater impact of TP53 inactivation on CNAs than somatic mutations.
Prior work has demonstrated that somatic point mutations of TP53, even in small subclones, associate with poor prognosis in CLL(17,18). These studies have focused on all CLL with and without del(17p), and have defined clonal mutations as variant allele frequency (VAF) greater than 12%, which translates to cancer cell fractions (CCF) greater than 12% to 24% depending on the 17p deletion status. Therefore these studies suggested that very small fractions of TP53 mutation carry similar risk to TP53 mutations with higher VAF>12% in all CLL cases, but did not address the impact of clonality of somatic TP53 mutations in the context of del(17p). In fact, Rossi et al in his work showed that subclonal TP53 mutations with 17p deletion or another clonal TP53 mutation have shorter overall survival than subclonal or clonal TP53 mutations alone, suggesting that TP53 mutations in the context of 17p deletion represent a worse disease group, consistent with our results (17). As we present here, most TP53 mutations are present in most cells in del17p patients (CCF>0.85) and only 2 patients in our cohort have a VAF less than 0.24. It is clear from our analysis that, in the context of del(17p), TP53 mutations are associated with different clinical outcome when the mutations are in most cells (CCF>0.85) vs fewer cells (CCF<=0.85). Our result showing mostly clonal TP53 mutations with del(17p) likely reflects selection pressure favoring more clonal mutations in the context of del(17p).
We demonstrate a novel and distinct pattern of driver genes among the increased somatic nonsynonymous mutations in del(17p) CLL. The association of NOTCH1 mutation with del(17p) has been previously reported(48). While we were preparing this manuscript, Ljungstrom et al reported recurrent RPS15 mutations in relapsing CLL and the association with del(17p)(40). Here we also identify RPS15 mutations enriched in del(17p) CLL, with 12% frequency compared to less than 3% in WT CLL. We found RPS15 mutations do not carry additional survival risk in del(17p) CLL, which is in agreement with Ljungstrom et al's result (40). The RPS15 mutations identified in our del(17p) enriched cohort cluster in a 15 amino acid region in the carboxyl terminus, suggesting potential gain of function activity. Ljungstrom et al showed that RPS15 mutations interact with MDM2 and MDMX to cause reduced stability of TP53. It is paradoxical to find most RPS15 mutations in del(17p) patients if the RPS15 mutants have direct functional interaction with wild type TP53, which is most often absent from del(17p) patients. In fact we identified a case of RPS15 mutation with biallelic disruption of TP53 without any TP53 protein expression by Western blot. We therefore postulate that RPS15 mutations may also have a functional consequence on other targets, possibly ribosomal, that cooperate less directly with the TP53 pathway.
In addition to our mutational analysis, and consistent with our own and others’ prior work (20–22,49), we clearly show that increasing CNAs of any type associate with worse OS, in all CLL and in del(17p) CLL, and that biallelic TP53 inactivation is associated with increased CNAs compared to monoallelic inactivation, which may be mechanistically related to loss of TP53 function. We identify a novel association between del17p CLL and three high-risk CNAs affecting 3p, 4p, and 9p, the targets of which are as yet unknown but worthy of investigation. We further show that increasing CNAs correlate with complex karyotype, and either one has similar negative impact on OS, even in 17p deleted CLL. Prior studies have had limited ability to separate the effect of complex karyotype from 17p deletion, probably because they so commonly occur together. Our data are particularly interesting in light of recent reports that early relapse on ibrutinib can be predicted by 17p deletion, or by complex karyotype, but with numbers too small to easily segregate their separate effects(7,50). Further follow-up of the RESONATE(51) study, which has both FISH and karyotype data on a large cohort of patients treated with ibrutinib, may also help elucidate this question.
Finally, our results shed light on why some del(17p)patients may remain untreated for extended periods. Del(17p) patients not treated during the 5 year follow-up of this study mostly have subclonal (4 subclonal and 1 clonal out of 7 cases) or no TP53 mutation (2 out of 7 cases). Correspondingly, these patients have very few CNAs, consistent with the observation that increasing CNAs occur in the context of greater TP53 inactivationMutated IGHV also appeared to be enriched in this group, while the number of somatic mutations was similar to the rest of the cohort. Thus the primary factor seems to be the very stable genomes of these patients, with few CNAs in the context of and possibly the result of only mono-allelic or subclonal inactivation of TP53. This information can be clinically useful in our assessment of del(17p) CLL patients.
Strengths of our study include assembling the largest cohort of del(17p) patients analyzed by whole exome sequencing and SNP arrays to date, with comparison to a large control cohort. With this large cohort, we were able to perform multivariable analysis for overall survival. Weaknesses include the heterogeneity of the patient population and genomic sampling times, with not all patients undergoing all evaluations. We presented the analysis by optimal cutoff values for CNA events and number of mutations. The primary goal of exploring threshold values in the figures is to graphically illustrate our findings with interpretable overall survival curves - by low vs high CNAs or mutations. These cut-off values, however, would need to be validated in external independent studies, particularly if a different platform or analytic pipeline were used. That being said, the log-transformed continuous variable results are also highly significant and thus robust whether dichotomized or not and will be generalizable to del(17p) patients in various stages of disease.
In summary we present the largest comprehensive genomic analysis of del(17p) CLL to date, showing that clonal and biallelic TP53 mutation as well as increasing CNAs and increasing somatic mutations are associated with worse OS, although CNAs are a more significant predictor in all CLLs regardless of del17p. We identify a unique driver gene profile in del(17p) CLL, including NOTCH1 and RPS15, underscoring its unique pathogenesis which will ultimately need to be understood in order to lead to effective therapy.
Supplementary Material
Translational Relevance.
CLL with 17p deletion represents the most challenging disease group, with poor overall survival, yet significant heterogeneity in survival. We hypothesized that the concurrent genomic landscape of del17p CLL would predict this heterogeneity in outcome, and therefore undertook to characterize the somatic mutation and copy number landscape in this subgroup of CLL, which has been under-represented in all prior sequencing studies. Our results demonstrate that subclonal rather than clonal mutation of the second TP53 allele and lack of genomic complexity of any type are characteristic of patients with longer overall survival, who may not require therapy for an extended period. These results have direct implications for clinical prognostication.
Acknowledgements
This work was supported by American Cancer Society Grant RSG-13-002-01-CCE to JRB, as well as the National Comprehensive Cancer Network. JRB also acknowledges support from the Leukemia Lymphoma Society, and the Melton and Rosenbach Funds. We acknowledge the support provided by Affymetrix Inc. to SRS. We thank Mr. Abiram Srivatsa for his assistance in the initial stages of analysis. We also thank all members of the Broad Institute's Biological Samples, Genetic Analysis, and Genome Sequencing Platforms, who made this work possible (NHGRI-U54HG003067).
Footnotes
Authorship Contributions
LY, CL, GG, SRS, and JRB designed the research; LY, SK, PB, SF and SR performed the experiments; LY, SK, PB, WD, KH, BT,AA and SRS collected the data; LY, HTK, DW, RI, JK, GG, BT, AA, SRS, JRB analyzed the data; LY, HTK, GG, SRS, and JRB wrote the manuscript; all authors approved the manuscript.
References
- 1.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 2.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 3.Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 4.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21:2442–51. doi: 10.1038/sj.leu.2404935. [DOI] [PubMed] [Google Scholar]
- 5.Baliakas P, Iskas M, Gardiner A, Davis Z, Plevova K, Nguyen-Khac F, et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: a systematic reappraisal of classic cytogenetic data. Am J Hematol. 2014;89:249–55. doi: 10.1002/ajh.23618. [DOI] [PubMed] [Google Scholar]
- 6.Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Döhner K, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123:3247–54. doi: 10.1182/blood-2014-01-546150. [DOI] [PubMed] [Google Scholar]
- 7.Byrd JC, Furman RR, Coutre SE, Burger JA, Blum KA, Coleman M, et al. Three-year follow-up of treatment-naïve and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125:2497–506. doi: 10.1182/blood-2014-10-606038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zenz T, Häbe S, Denzel T, Mohr J, Winkler D, Bühler A, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–97. doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 9.Tam CS, O'Brien S, Lerner S, Khouri I, Ferrajoli A, Faderl S, et al. The natural history of fludarabine-refractory chronic lymphocytic leukemia patients who fail alemtuzumab or have bulky lymphadenopathy. Leuk Lymphoma. 2007;48:1931–9. doi: 10.1080/10428190701573257. [DOI] [PubMed] [Google Scholar]
- 10.Trbusek M, Malcikova J. TP53 aberrations in chronic lymphocytic leukemia. Adv Exp Med Biol. 2013;792:109–31. doi: 10.1007/978-1-4614-8051-8_5. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29:2223–9. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- 12.Zenz T, Kröber A, Scherer K, Häbe S, Bühler A, Benner A, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112:3322–9. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- 13.Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S, et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15:995–1004. doi: 10.1158/1078-0432.CCR-08-1630. [DOI] [PubMed] [Google Scholar]
- 14.Trbusek M, Smardova J, Malcikova J, Sebejova L, Dobes P, Svitakova M, et al. Missense mutations located in structural p53 DNA-binding motifs are associated with extremely poor survival in chronic lymphocytic leukemia. J Clin Oncol. 2011;29:2703–8. doi: 10.1200/JCO.2011.34.7872. [DOI] [PubMed] [Google Scholar]
- 15.Zenz T, Eichhorst B, Busch R, Denzel T, Häbe S, Winkler D, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473–9. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]
- 16.Malcikova J, Smardova J, Rocnova L, Tichy B, Kuglik P, Vranova V, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood. 2009;114:5307–14. doi: 10.1182/blood-2009-07-234708. [DOI] [PubMed] [Google Scholar]
- 17.Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Famà R, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–47. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1 and ATM mutations in chronic lymphocytic leukemia. Blood. 2016 doi: 10.1182/blood-2015-07-659144. blood – 2015–07 – 659144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best OG, Gardiner AC, Davis ZA, Tracy I, Ibbotson RE, Majid A, et al. A subset of Binet stage A CLL patients with TP53 abnormalities and mutated IGHV genes have stable disease. Leukemia. 2009;23:212–4. doi: 10.1038/leu.2008.260. [DOI] [PubMed] [Google Scholar]
- 20.Tam CS, Shanafelt TD, Wierda WG, Abruzzo LV, Van Dyke DL, O'Brien S, et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the M. D. Anderson and Mayo Clinic experience. Blood. 2009;114:957–64. doi: 10.1182/blood-2009-03-210591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado J, Salaverria I, Baumann T, Martínez-Trillos A, Lee E, Jiménez L, et al. Genomic complexity and IGHV mutational status are key predictors of outcome of chronic lymphocytic leukemia patients with TP53 disruption. Haematologica. 2014;99:e231–4. doi: 10.3324/haematol.2014.108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown JR, Hanna M, Tesar B, Werner L, Pochet N, Asara JM, et al. Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia. Clin Cancer Res. 2012;18:3791–802. doi: 10.1158/1078-0432.CCR-11-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 25.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–30. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–36. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 28.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank E, Harrell . Regression Modeling Strategies: with Applications to Linear Models, Logistic Regression, and Survival Analysis. 1st ed. Springer-Verlag; New York: 2001. [Google Scholar]
- 33.Therneau Terry M., Grambsch Patricia M., Fleming Thomas R. Martingale-Based Residuals for Survival Models. Biometrika. 1990;77:147–60. [Google Scholar]
- 34.Harrell FE, Lee KL, Califf RM, Pryor DB, Rosati RA. Regression modelling strategies for improved prognostic prediction. Stat Med. 1984;3:143–52. doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- 35.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edelmann J, Holzmann K, Miller F, Winkler D, Bühler A, Zenz T, et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012;120:4783–94. doi: 10.1182/blood-2012-04-423517. [DOI] [PubMed] [Google Scholar]
- 37.Ouillette P, Malek S. Acquired genomic copy number aberrations in CLL. Adv Exp Med Biol. 2013;792:47–86. doi: 10.1007/978-1-4614-8051-8_3. [DOI] [PubMed] [Google Scholar]
- 38.Villamor N, Conde L, Martínez-Trillos A, Cazorla M, Navarro A, Beà S, et al. NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia. 2013;27:1100–6. doi: 10.1038/leu.2012.357. [DOI] [PubMed] [Google Scholar]
- 39.Rossi D, Rasi S, Fabbri G, Spina V, Fangazio M, Forconi F, et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood. 2012;119:521–9. doi: 10.1182/blood-2011-09-379966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ljungström V, Cortese D, Young E, Pandzic T, Mansouri L, Plevova K, et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood. 2016;127:1007–16. doi: 10.1182/blood-2015-10-674572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juliusson G, Merup M. Cytogenetics in chronic lymphocytic leukemia. Semin Oncol. 1998;25:19–26. [PubMed] [Google Scholar]
- 43.Strefford JC. The genomic landscape of chronic lymphocytic leukaemia: biological and clinical implications. Br J Haematol. 2015;169:14–31. doi: 10.1111/bjh.13254. [DOI] [PubMed] [Google Scholar]
- 44.Pei J, Jhanwar SC, Testa JR. Chromothripsis in a Case of TP53-Deficient Chronic Lymphocytic Leukemia. Leuk Res Rep. 2012;1:4–6. doi: 10.1016/j.lrr.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P, Erez A, Nagamani SCS, Dhar SU, Kołodziejska KE, Dharmadhikari AV, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 48.López C, Delgado J, Costa D, Conde L, Ghita G, Villamor N, et al. Different distribution of NOTCH1 mutations in chronic lymphocytic leukemia with isolated trisomy 12 or associated with other chromosomal alterations. Genes Chromosomes Cancer. 2012;51:881–9. doi: 10.1002/gcc.21972. [DOI] [PubMed] [Google Scholar]
- 49.Delgado J, Espinet B, Oliveira AC, Abrisqueta P, de la Serna J, Collado R, et al. Chronic lymphocytic leukaemia with 17p deletion: a retrospective analysis of prognostic factors and therapy results. Br J Haematol. 2012;157:67–74. doi: 10.1111/j.1365-2141.2011.09000.x. [DOI] [PubMed] [Google Scholar]
- 50.Thompson PA, O'Brien SM, Wierda WG, Ferrajoli A, Stingo F, Smith SC, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121:3612–21. doi: 10.1002/cncr.29566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown JR, Hillmen P, O'Brien S, Barrientos JC, Reddy N, Coutre S, et al. Updated Efficacy Including Genetic and Clinical Subgroup Analysis and Overall Safety in the Phase 3 RESONATETM Trial of Ibrutinib Versus Ofatumumab in Previously Treated Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma. Blood. 2014;124:3331–3331. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.