Abstract
Long noncoding RNAs (lncRNAs) are a relatively well-characterized class of noncoding RNA (ncRNA) molecules, involved in the regulation of various cell processes, including transcription, intracellular trafficking, and chromosome remodeling. Their deregulation has been associated with the development and progression of various cancer types, the fact which makes them suitable as biomarkers for cancer diagnosis and prognosis. In recent years, detection of cancer-associated lncRNAs in body fluids of cancer patients has proven itself as an especially valuable method to effectively diagnose cancer. Cancer diagnosis and prognosis employing circulating lncRNAs are preferential when compared to classical biopsies of tumor tissues, especially due to their noninvasiveness, and have great potential for routine usage in clinical practice. Thus, this review focuses on summarizing the perspectives of lncRNAs as biomarkers in cancer, based on evaluating their expression profiles determined in body fluids of cancer patients.
1. Introduction
Long noncoding RNAs (lncRNAs) belong to a larger group of noncoding RNAs (ncRNAs) and are generally classified as 200 nt–100 kb long transcripts, lacking the open-reading frame [1, 2]. They are usually transcribed by RNA polymerase II and controlled by the transcriptional activators of the SWI/SNF complex. Most of the generated lncRNA transcripts are usually spliced, capped, and polyadenylated in a similar manner as mRNA molecules [3]. lncRNAs represent a large (>80%) and a very heterogeneous group of ncRNAs, with their expression depending largely on the tissue and cellular context [4–7]. Following the discovery of H19 and XIST lncRNAs in 1990s [8, 9], lncRNA per se was initially regarded as a transcriptional noise with practically no or very little function [10]. However, after being identified as a class of RNA molecules in 2002 [11], studies that followed revealed lncRNA importance and indispensability in various cellular processes, including transcription, intracellular trafficking, and chromosome remodeling [3, 12]. In addition, lncRNAs functioning as regulatory factors have been determined for several complex cellular processes, such as cell death, growth, differentiation, identity establishment; controlling apoptosis, epigenetic regulation, genomic imprinting, alternative splicing, regulation of gene expression at posttranscriptional level, chromatin modification, inflammatory pathologies, and, when deregulated, also in various cancer types [13–23].
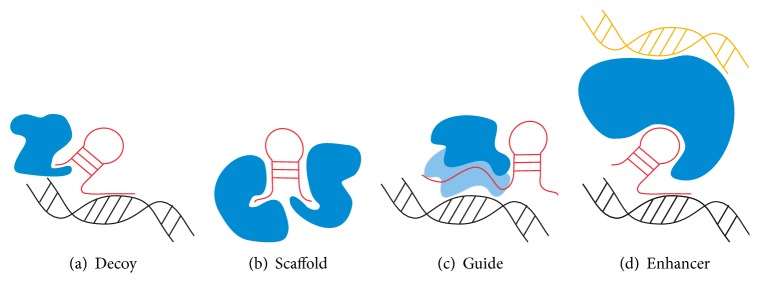
lncRNAs can be present in practically all cell compartments [24]. However, many lncRNAs with high abundance were identified especially in the nucleus and cytoplasm [25, 26]. lncRNA secondary structures, such as stem loops and hairpins, results of posttranscriptional modifications, enable their interaction with proteins and chromatin and are crucial for lncRNA's vast set of functions [12]. Some of the main mechanisms of action that allow lncRNAs to have a crucial role in various cellular processes [27] are presented in Figure 1. In general, lncRNAs may act as scaffolds for grouping protein complexes (Figure 1(b)), guides to recruit proteins (Figure 1(c)), transcriptional enhancers by bending chromatin (Figure 1(d)), decoys to release proteins from chromatin (Figure 1(a)), or antagonists for other regulatory ncRNAs, for example, microRNAs (miRNAs) [12, 28].
Figure 1.
Different mechanisms of long noncoding RNA (lncRNA) action. (a) The lncRNAs can act as decoys, titrating away DNA-binding proteins (e.g., transcription factors); (b) lncRNAs may act as scaffolds to bring two or more proteins to spatial proximity or into a complex; (c) lncRNAs may act as guides to recruit proteins to DNA (e.g., chromatin modification enzymes); (d) lncRNA guidance can also be exerted through chromosome looping in an enhancer-like model in cis. lncRNA (red); DNA (black); section of DNA loop (yellow); DNA-binding proteins (blue and light blue). The figure is adapted from John L. Rinn and Howard Y. Chang [12].
Regardless of the whole human genome analyses that enabled better understanding of lncRNA expression, function, and distribution in the human genome, classification of lncRNAs remains to be unified [25, 29]. lncRNAs can be sorted according to their structure, sequence, function, localization, metabolism, and interaction with protein-coding genes or other DNA elements [29]. Recently, Wang et al. classified different types of lncRNAs according to their genomic location and context, exerted effect on DNA sequences, mechanism of functioning, and targeting mechanism [30]. In addition, lncRNAs can be classified into several categories including sense lncRNAs, antisense lncRNAs, bidirectional lncRNAs, intronic lncRNAs, intergenic lncRNAs, promoter-associated lncRNAs, and untranslated region- (UTR-) associated lncRNAs [25, 26]. Nevertheless, current classification methods remain inadequate and relatively nontransparent. The general long-term goal is to develop a unified, systematic, and comprehensive lncRNA classification and annotation framework, utilizing global system biology and genomics-driven approaches. Also, the development of improved tools is required for the integration of complex data from multiple types of experiments into this framework, revealing associations between coding and noncoding transcripts. Such lncRNA classification would be a prerequisite for an improved overview and more effective access and usage of large-scale lncRNA data in various fields and applications [29].
Association of lncRNAs in carcinogenesis was observed due to their differential expression in tumors when compared to normal tissues [31]. lncRNAs H19, MALAT1, and PCA3 are highly expressed tumor-associated lncRNAs that were characterized before the availability of next generation sequencing technologies [32–34]. It has been demonstrated that tumorigenesis mostly results from ectopic lncRNA expression [35]. lncRNAs regulate several oncogenes and tumor suppressor genes at transcriptional and posttranscriptional levels, affecting proliferation, apoptosis, angiogenesis, invasion, migration, and metastasis of tumor cells [36–39]. Also, lncRNA-mediated regulation of chromatin remodeling is essential for the integrity of nuclear structure [23]. In recent years, next-generation and high-throughput sequencing techniques have enabled a significant breakthrough in lncRNA identification and characterization. This resulted in continuously rising amounts of data elucidating deregulated lncRNAs associated with the development of various cancer types [40–42]. In this review, we primarily focus on describing circulating lncRNAs present in different body fluids which represent a promising category of biomarkers for cancer diagnosis, prognosis, and also treatment.
2. lncRNA-Mediated Epigenetic Modifications
Cancer development and progression can be mediated through multiple mechanisms involving lncRNAs [36, 43–46]. In particular, involvement of lncRNAs has been extensively studied in cancer progression, mainly through epigenetic regulation, activation of oncogenic pathways, and crosstalk with other RNA subtypes [29, 47, 48]. As mentioned before, lncRNAs can interact with chromatin remodeling complexes which usually leads to modifications in the expression of target genes, in either cis or trans [49]. In these processes, lncRNAs usually recruit chromatin modification factors, for example, DNA methyltransferase enzymes [50], resulting in gene expression variations often inherited within cell lineages [51]. One of the first reported and characterized lncRNA involved in cancer progression through genome-wide epigenetic reprogramming was HOTAIR [52–54]. HOTAIR acts through interaction with polycomb repressive complex 2 (PRC2) subunits, a key chromatin remodeling complex involved in gene silencing [55]. When deregulated, HOTAIR recruits PRC2 subunits in promoter regions of tumor suppressor genes which results in their transcriptional repression and chromatin condensation, thus, favoring tumor progression. Studies have shown that beside HOTAIR, ANRIL, and XIST, lncRNAs also recruit PRC2 in a similar fashion [52, 56, 57].
Studies have shown that over 200 lncRNAs participate in imprinting processes where, depending on their parental origins, specific expression of nearby lncRNAs promotes suppression of neighboring genes in cis [58, 59]. Here, instead of acting through, for example, PRC2, lncRNAs recruit DNA methyltransferases directly to modify chromatin conformation and DNA methylation. Among many lncRNAs with such function, several have been characterized, including Kcnq1ot1, TARID, H19, AS1DHRS4, and DACOR1. lncRNAs may also modify nucleosome positioning through SWI/SNF complexes as it was determined for SChLAP1 [60–65]. lncRNA SChLAP1 is overexpressed in a subset of prostate cancers. SChLAP1 can bind directly to hSNF5, one of the core subunits of the SWI/SNF complexes, thus, decreasing their genomic binding. By impairing the proper SWI/SNF regulation of gene expression, SChLAP1 antagonizes tumor suppressive function of the SWI/SNF complexes and promotes tumor cell invasion and metastasis [63, 66]. In addition, NEAT1, UCA1, HIF1A-AS1, and Evf2 also interact with core subunits of SWI/SNF complexes in a similar manner in various cancer types [67]. Other lncRNAs, including Firre, bind chromatin remodelers cohesin and CTCF in order to change the chromatin of whole chromosomes in the process of X chromosome inactivation [68]. lncRNAs may also act as chromatin activators, regulating chromosome looping in their proximity to deposit activating H3K4me3 histone mark on gene promoters [69–71].
3. Circulating lncRNAs as Biomarkers in Cancer
Among the main advantages of lncRNAs that make them suitable as cancer diagnostic and prognostic biomarkers is their high stability while circulating in body fluids, especially when included in exosomes or apoptotic bodies [72]. Studies have shown that despite abundant quantities of ribonucleases in different body fluids, lncRNAs were detected in these samples which could successfully resist ribonuclease degradation activities [35]. In addition, lncRNA deregulation in primary tumor tissues is clearly mirrored in various bodily fluids, including whole blood, plasma, urine, saliva, and gastric juice [73–76]. These lncRNA characteristics present an opportunity to develop effective and convenient lncRNA-based biomarkers that are minimally invasive and may be better tolerated by patients, when compared to conventional biopsies, due to their relative noninvasiveness [77]. Detection of circulating cancer-associated lncRNAs in body fluids could be used in the assessment of cancers at distinguishing tumor patients from healthy people at early stages with both high sensitivity and specificity. In addition, predicting the prognosis of tumor patients and the risk of tumor metastasis and recurrence after surgery could be assessed, along with evaluating operation success [35]. Several individual or combined lncRNAs have demonstrated comparable or, in some cases, even higher diagnostic performance than conventional cancer biomarkers, for different cancer types. lncRNA MALAT1 has been identified, by Kaplan-Meier analysis, as an effective prognostic parameter for patient survival in stage I nonsmall cell lung cancer [78]. Also, the measurement of lncRNA PCA3 in patient urine samples has been shown to allow more sensitive and specific diagnosis of prostate cancer than the widely used prostate-specific antigen (PSA) serum levels [79–81]. CEA, CA125, CA153, and AFP are conventional biomarkers, commonly used for breast cancer diagnosis. lncRNA RP11-445H22.4 is overexpressed in breast cancer tissues and can be detected in serum samples, with a sensitivity of 92% and specificity of 74%, which is significantly better than the performance of above listed conventional biomarkers [82]. In addition, diagnostic performances of lncRNAs TINCR, CCAT2, AOC4P, BANCR, LINC00857, AA174084, and H19 were evaluated in body fluid samples (e.g., plasma and gastric juice) of gastric cancer patients. These lncRNAs had the ability to differentiate gastric cancer patients from healthy individuals and to effectively detect different stages of gastric cancer (from early to metastatic cancer forms). However, despite their overall positive diagnostic performances, similar to those obtained by several conventional cancer biomarkers, false-positive and false-negative detections were observed [19, 76, 83]. Also, similar results were obtained after characterizing lncRNAs MALAT1 and PCA3 as biomarkers in prostate cancer patients [84, 85].
Stability of lncRNAs in body fluids of tumor patients has not been thoroughly explored. Studies revealed that some lncRNAs remained stable in plasma under extreme conditions, including several freeze-thawed cycles and prolonged incubation at elevated temperatures [86]. It has also been demonstrated that lncRNAs remained their stability when using plasma and serum from EDTA vacutainer tubes or from tubes lacking the specific anticoagulant, whereas lncRNA amounts declined when using plasma from heparin vacutainer tubes [84].
Three main mechanisms for lncRNA secretion and transport to the extracellular environment have been proposed. First, extracellular RNAs may package themselves into specific membrane vesicles, such as exosomes and microvesicles, in order to be secreted and resist RNase activity. Studies revealed that exosomes most frequently protect plasma lncRNAs [87–90]. Second, extracellular RNAs can be actively released by tumor tissues and cells [84]. However, elevated values of lncRNAs in plasma may have multiple sources, including cancer-adjacent normal cells, immune cells, and other blood cells [86, 90]. Third, extracellular RNAs may encapsulate themselves into high-density lipoprotein (HDL) or apoptotic bodies or are associated with protein complexes, for example, Argonaute- (Ago-) miRNA complex [91] and nucleophosmin 1- (NPM1-) miRNA complex [92]. However, despite many performed studies, secretion and transport mechanisms of lncRNAs to the circulation system remain poorly understood, mostly because several studies tend to contradict each other. Also, thorough examinations and reports regarding biological functions of lncRNAs in cancers are still lacking [35].
In order to introduce circulating lncRNAs into clinical practice, further studies and improvements should be performed regarding the standardization of sample preparation protocols, endogenous controls of lncRNAs in body fluids and the extraction methods should be uniformed, standards assessing the quality of lncRNAs and the credibility of qPCR results should be more accurate and reliable, and more high-quality research studies should be performed, with selection bias reduced as much as possible [35]. In addition, several technical obstacles remain to be addressed and overcome in the future, to enable a reliable use of circulating lncRNAs as effective cancer biomarkers. Commercial kits employing columns are mostly used for lncRNA extraction from body fluids. Unfortunately, no consistent results have been obtained regarding the differences in the efficiency of column-based methods, indicating that comparison and standardization of lncRNA extraction methods are necessary [93]. Absolute concentration of lncRNAs in body fluids is usually low and frequently requires an RNA amplification step prior analysis, which is time consuming and can be problematic when results are needed promptly [94]. It has also been observed that RNA extracted from plasma and serum samples may be undetectable when using a NanoDrop spectrophotometer for quantifying circulating RNAs [93]. This makes the necessity for the development of highly sensitive methods for quantifying lncRNAs crucial. Also, since the mechanisms of lncRNA secretion are not yet fully understood, the levels of circulating lncRNAs may be affected by other concomitant disease changes, besides tumorigenesis. Thus, overrated amounts of specific lncRNAs associated with a particular disease may be determined [94]. There are also several existing obstacles regarding the techniques, commonly used for quantifying circulating lncRNAs. Quantitative RT-PCR is a well-established method for detecting and quantifying circulating RNAs. However, the cost per sample is relatively high and the throughput of the method low [93]. Recently developed assays, such as the Human Disease-Related lncRNA Profiler (System Biosciences SBI), allow the measurement of a panel of lncRNAs but can detect only annotated lncRNAs. Therefore, only a medium throughput can be attained [93]. Commercial lncRNA microarray platforms can be used to detect only previously described biomarkers already present in the lncRNA databases. Microarrays have a high throughput, but a lower dynamic range and specificity, when compared to qRT-PCR and RNA-seq [93]. RNA-seq can be used for the identification of known and new lncRNA species, with lower cost per sample than microarrays and qRT-PCR. However, a relatively large amount of starting material is required (cca. 1 μg RNA), which is difficult to extract from biological fluids, for example, plasma or serum samples. In addition, current RNA-seq methodology is expensive and complex and requires a special equipment with a trained personnel [93].
Since expression profiles of cancer-associated lncRNAs may be very specific for various cancer types, these specific lncRNAs could be efficiently used as tumor biomarkers in different body fluids in the near future, with vital significance for clinical research [35]. In the following section, we describe some of these lncRNAs.
4. lncRNAs as Cancer Biomarkers Obtained from Body Fluids
Deregulated expression of lncRNAs is strongly linked to the development of various tumors and can be relatively effectively detected in patient's body fluids for several cancer types [77]. Regarding their involvement in malignant disease development, when comparing to normal tissues of healthy individuals, lncRNAs are generally divided into oncogenic or tumor suppressive, being upregulated or downregulated, respectively [31, 45]. Sets of a number of differentially expressed cancer-associated lncRNAs in a variety of cancers are presented in Tables 1 and 2. Among them, several lncRNAs represent promising noninvasive cancer biomarkers for detection in patient's body fluids, including PCA3, HOTAIR, HULC, MALAT1, H19, LINC00152, RP11-160H22.5, XLOC_014172, LOC149086, AA174084, and UCA1. Moreover, for several of these lncRNAs, it has been already demonstrated that they could be effectively used as diagnostic and prognostic cancer biomarkers in clinical practice.
Table 1.
Upregulated cancer-associated lncRNAs when compared to normal tissues.
| Name | Cancer type | Fold changea | References |
|---|---|---|---|
| Wt1-as | Acute myeloid leukemia | NAb | [111] |
| XIST | Glioma | NA | [112] |
| CRNDE | Glioma | NA | [113] |
| MALAT1 | Glioma | 2.0–5.0 | [114] |
| Colorectal | 2.0–6.0 | [115] | |
| Lung | >40.0 | [116] | |
| Prostate | NA | [84] | |
| Hepatocellular | NA | [117] | |
| Uterus | NA | [118] | |
| LSINCT5 | Breast | 2.0–7.0 | [119] |
| LINC00617 | Breast | >1.5 | [120] |
| RP11-445H22.4 | Breast | 15.0–20.0 | [121] |
| BC200 | Breast | NA | [122] |
| CCHE1 | Cervical | NA | [123] |
| CCAT1-L | Colorectal | NA | [71] |
| POU3F3 | Esophageal | NA | [124] |
| PCAT-1 | Colorectal | NA | [125] |
| Prostate | NA | [126] | |
| HOTAIR | Esophageal | NA | [127] |
| Lung | NA | [128] | |
| Cervical | NA | [129] | |
| Pancreas | NA | [130] | |
| Breast | NA | [52] | |
| Oral | NA | [131] | |
| Hepatocellular | >2.0 | [132] | |
| Glioma | NA | [133, 134] | |
| Colorectal | 5.2 | [101] | |
| CCAT2 | Lung | 7.5 | [135] |
| Colon | NA | [136] | |
| Cervical | NA | [137] | |
| LINC00152 | Gastric | NA | [35, 90] |
| LSINCT-5 | Gastric | NA | [138] |
| HOXA11-AS | Glioma | NA | [139] |
| Linc-POU3F3 | Glioma | >2.6 | [140] |
| ATB | Glioma | 5.0–10.0 | [141] |
| AB073614 | Glioma | NA | [142] |
| RP11-160H22.5 | Hepatocellular | 2.5 | [109] |
| XLOC_014172 | Hepatocellular | 67.7 | [109] |
| LOC149086 | Hepatocellular | 4.6 | [109] |
| BANCR | Hepatocellular | NA | [143] |
| SNHG3 | Hepatocellular | NA | [144] |
| MVIH | Hepatocellular | 3.75 | [145] |
| Lung | NA | [146] | |
| LCAL1 | Lung | NA | [147] |
| LUADT1 | Lung | NA | [148] |
| AFAP1-AS1 | Lung | NA | [149] |
| Colorectal | NA | [150] | |
| Hepatocellular | NA | [151] | |
| Esophageal | >1.0 | [152] | |
| ANRIL | Lung | >1.5 | [153] |
| Hepatocellular | >1.0 | [154] | |
| Bladder | >1.0 | [155] | |
| UCA1 | Lung | NA | [156] |
| Oral | NA | [157] | |
| Bladder | 32.9 | [158] | |
| Colon | NA | [159] | |
| Hepatocellular | NA | [160] | |
| Breast | NA | [161] | |
| Esophageal | >2.0 | [162] | |
| CASC15 | Melanoma | NA | [163] |
| SPRY4-IT1 | Melanoma | >2.0 | [164, 165] |
| Glioma | NA | [166] | |
| H19 | Bladder | NA | [167] |
| Gastric | NA | [83] | |
| Esophageal | NA | [168] | |
| Colorectal | NA | [169] | |
| Glioma | NA | [170] | |
| HULC | Pancreas | NA | [171] |
| Hepatocellular | 32.7 | [102] | |
| Glioma | NA | [172] | |
| PCA3 | Prostate | NA | [34, 99] |
| PCAT5 | Prostate | NA | [173] |
| PCAT18 | Prostate | 8.8–11.1 | [174] |
| PRNCR1 | Prostate | NA | [175] |
| NEAT1 | Glioma | NA | [176, 177] |
| Oral | NA | [74] | |
| Hepatocellular | NA | [178] | |
| Nasopharyngeal | NA | [179] | |
| PVT1 | Thyroid | NA | [180] |
| Gastric | NA | [181] | |
| Colorectal | NA | [182] | |
| SRA | Breast | NA | [183] |
aFold change values, relative to normal controls; bnot available (data is presented in a graphical format in the original report).
Table 2.
Downregulated cancer-associated lncRNAs when compared to normal tissues.
| Name | Cancer type | Fold changea | References |
|---|---|---|---|
| MEG3 | Glioma | NAb | [184, 185] |
| ZFAS1 | Breast | 2.0 | [186] |
| GAS5 | Breast | <1.0 | [187] |
| Glioma | NA | [188] | |
| LOC554202 | Colorectal | NA | [189] |
| CUDR | Gastric | NA | [190] |
| PTENP1 | Gastric | NA | [191] |
| Prostate | NA | [191] | |
| AA174084 | Gastric | 3.2 | [76] |
| LINC00982 | Gastric | 7.7 | [192] |
| TSLC1-AS1 | Glioma | NA | [193] |
| ADAMTS9-AS2 | Glioma | NA | [194] |
| MDC1-AS | Glioma | NA | [195] |
| TUG1 | Glioma | NA | [196] |
| ROR | Glioma | NA | [197] |
| CACS2 | Glioma | NA | [198] |
| PRAL | Hepatocellular | NA | [199] |
| MALAT1 | Lung | 3.3 | [106] |
| AK023948 | Papillary thyroid carcinoma | 5.0 | [200] |
aFold change values, relative to normal controls; bnot available (data is presented in a graphical format in the original report).
PCA3 has been recently approved as a urine biomarker for prostate cancer by the US Food and Drug Administration [73]. This lncRNA allows better sensitivity and specificity when compared to the widely used PSA blood test, mainly because of its significantly higher expression in prostate cancer patients [79–81, 95–97]. A meta-analysis of several studies has determined the validity of PCA3 levels in urine samples for prostate cancer diagnosis, with a summary sensitivity of 62% and specificity of 75%. In the receiver operating characteristic (ROC) curve analysis, this translated to an area under the ROC curve (AUC) of 0.75 [98]. PCA3 has also a prognostic value for prostate cancer, since its expression levels correlate well with tumor aggressiveness [99, 100].
HOTAIR was found to be highly expressed in saliva samples of oral squamous cell carcinoma (OSCC) patients. Since higher expression levels of HOTAIR were determined for metastatic patients, this lncRNA represents a strong candidate for metastatic oral cancer diagnosis [74]. In addition, the association between increased blood levels of HOTAIR and poor prognosis with higher mortality in colorectal cancer patients has been determined. Expression levels of HOTAIR could also predict the survival time of patients. Evaluated diagnostic performance of HOTAIR in peripheral blood cells has shown its sensitivity of 67%, specificity of 92.5%, and AUC of 0.87. Thus, HOTAIR represents an effective negative prognostic biomarker for colorectal cancer in blood samples [101].
HULC can be effectively detected in plasma and peripheral blood cells and is significantly overexpressed in hepatocellular carcinoma patients, thus, representing a prominent novel biomarker for liver cancer. However, no data regarding HULC diagnostic performance are available at this time [102, 103]. HULC detected in blood has also been recently proposed as a diagnostic biomarker for gastric cancer [104].
MALAT1 represents a promising diagnostic biomarker detectable in blood, to effectively screen lung cancer. One study has shown downregulation of MALAT1 in blood samples of lung cancer patients which was contrary to MALAT1 levels in lung cancer tissues, where it was significantly upregulated. Conversely, MALAT1 showed elevated expression levels in whole blood of metastatic lung cancer patients [105]. Due to its relatively low expression and low detection sensitivity (sensitivity 56%; specificity 96%; AUC 0.79) in diagnosis of non-small-cell lung cancer (NSCLC), MALAT1 is not regarded suitable as an independent biomarker to diagnose lung cancer but should be rather used as a complementary biomarker [106]. In addition to lung cancer, MALAT1 has proven itself as a prominent biomarker with its elevated expression detected in plasma and urine of prostate cancer patients, with a sensitivity and specificity of 58.6% and 84.8%, respectively (AUC 0.836). MALAT1 also helped to predict the outcome of prostate biopsies [84, 107].
Elevated expression profiles of H19 have been determined in plasma samples of gastric cancer patients. H19 has great potential as a promising biomarker due to its high diagnostic value for the detection of gastric cancer (sensitivity 82.9%; specificity 72.9%; AUC 0.838). It has also been more effective in early stage gastric cancer diagnosis than the conventional biomarkers, such as CEA and CA199, with a sensitivity of 85.5%, specificity of 80.1%, and AUC of 0.877 [83].
Expression levels of LINC00152 in plasma were found to be significantly increased in early and advanced gastric cancer patients. This lncRNA had also significantly higher expression profiles in postoperative plasma samples. The diagnostic value of LINC00152 (sensitivity 48.1%; specificity 85.2%; AUC 0.675) was better than those of CEA and CA199 biomarkers, which makes LINC00152 a good candidate as a novel blood-based biomarker for gastric cancer diagnosis [90]. In addition, LINC00152 could also be detected in the gastric juice of patients with gastric cancer [108].
Among the less commonly studied lncRNAs belong RP11-160H22.5, XLOC_014172, and LOC149086 which have been proposed as biomarkers for the diagnosis of hepatocellular carcinoma in patient plasma samples. These three lncRNAs had better scores for hepatocellular carcinoma diagnosis when used in combination, in comparison to each individual lncRNA, with a merged AUC of 0.896, sensitivity of 82%, and specificity of 73% [109]. In addition, XLOC_014172 and LOC149086 lncRNAs had also a good prognostic value for metastasis prediction (sensitivity 91%; specificity 90%; AUC for the combined lncRNAs 0.675) [109].
AA174084 represents a relatively robust but specific biomarker, suitable for the diagnosis of gastric cancer in gastric juice samples (sensitivity 46%; specificity 93%; AUC 0.848). Levels of AA174084 in patient's gastric juices were found to be significantly upregulated when compared to those of healthy individuals. Interestingly, this lncRNA was not suitable for the diagnosis of gastric cancer from plasma samples [76].
UCA1 lncRNA has been identified as a potential biomarker for bladder cancer. Due to its relatively high overall specificity, it has a high potential to discriminate between the bladder/urothelial cancer and other cancer types, or other diseases related to the urinary tract (sensitivity 80.9%; specificity 91.8%; AUC 0.882). UCA1 can be detected in urine samples of bladder cancer patients, mostly in the cellular sediments [110].
Additional, continuously increasing amounts of information regarding cancer-associated lncRNAs, including those detected in body fluids, can be obtained from many existing databases, several of which are presented in Table 3.
Table 3.
Databases containing lncRNA data.
| Name | URL | References |
|---|---|---|
| CHIPbase | http://deepbase.sysu.edu.cn/chipbase/ | [201] |
| C-It-Loci | http://c-it-loci.uni-frankfurt.de/ | [202] |
| Co-LncRNA | http://www.bio-bigdata.com/Co-LncRNA/ | [203] |
| DIANA-LncBase | http://www.microrna.gr/LncBase | [204, 205] |
| Linc2GO | http://www.bioinfo.tsinghua.edu.cn/~liuke/Linc2GO/index.html | [206] |
| Lnc2Cancer | http://www.bio-bigdata.com/lnc2cancer/ | [207] |
| LncACTdb | http://www.bio-bigdata.net/LncACTdb/ | [208] |
| LNCipedia | http://www.lncipedia.org/ | [209, 210] |
| LncRBase | http://bicresources.jcbose.ac.in/zhumur/lncrbase/ | [211] |
| LncRNA2Function | http://mlg.hit.edu.cn/lncrna2function/ | [212] |
| LncRNAdb | http://www.lncrnadb.org/ | [213, 214] |
| LncRNADisease | http://210.73.221.6/lncrnadisease | [215] |
| lncRNASNP | http://bioinfo.life.hust.edu.cn/lncRNASNP/ | [216] |
| LncRNome | http://genome.igib.res.in/lncRNome/ | [217] |
| miRcode | http://www.mircode.org | [218] |
| NONCODE | http://www.noncode.org/ | [219, 220] |
| Starbase 2.0v | http://starbase.sysu.edu.cn/rbpLncRNA.php | [221, 222] |
5. Conclusions and Perspectives
lncRNAs represent a relatively large and heterogeneous group of ncRNAs and are considered as suitable diagnostic and prognostic biomarkers in cancer. In recent years, circulating lncRNAs have proven themselves extremely valuable for the detection of various cancer types. Their usage as biomarkers is convenient not only because samples containing circulating lncRNAs can be easily and noninvasively obtained from cancer patients but also because these lncRNAs remain relatively stable in body fluids. They can be quite easily detected in whole blood, plasma, serum, urine, saliva, and gastric juice samples, by using a variety of common molecular biology techniques, such as qRT-PCR, microarray hybridization, and sequencing (e.g., RNA-seq). Because lncRNAs are usually differentially abundant in different body fluids, mainly depending on the cancer type, effective cancer diagnosis and prognosis currently depend on combining different candidate lncRNAs, together with previously established biomarkers. Some circulating lncRNAs have already been proven as promising and sensitive biomarkers, and there are likely more to come.
Acknowledgments
The authors would like to thank Nina Hauptman, PhD (Department of Molecular Genetics, Institute of Pathology, Faculty of Medicine, University of Ljubljana, Slovenia), for designing the figure in this review.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Mattick J. S. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Reports. 2001;2(11):986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C., Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell & Bioscience. 2012;2(1):p. 37. doi: 10.1186/2045-3701-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinn J. J., Chang H. Y. Unique features of long non-coding RNA biogenesis and function. Nature Reviews. Genetics. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 4.Harrow J., Frankish A., Gonzalez J. M., et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Research. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alam T., Medvedeva Y. A., Jia H., Brown J. B., Lipovich L., Bajic V. B. Promoter analysis reveals globally differential regulation of human long non-coding RNA and protein-coding genes. PloS One. 2014;9(10, article e109443) doi: 10.1371/journal.pone.0109443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wapinski O., Chang H. Y. Long noncoding RNAs and human disease. Trends in Cell Biology. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Brosnan C. A., Voinnet O. The long and the short of noncoding RNAs. Current Opinion in Cell Biology. 2009;21(3):416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Brannan C. I., Dees E. C., Ingram R. S., Tilghman S. M. The product of the H19 gene may function as an RNA. Molecular and Cellular Biology. 1990;10(1):28–36. doi: 10.1128/MCB.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockdorff N., Ashworth A., Kay G. F., et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–526. doi: 10.1016/0092-8674(92)90519-I. [DOI] [PubMed] [Google Scholar]
- 10.van Bakel H., Hughes T. R. Establishing legitimacy and function in the new transcriptome. Briefings in Functional Genomics & Proteomics. 2009;8(6):424–436. doi: 10.1093/bfgp/elp037. [DOI] [PubMed] [Google Scholar]
- 11.Okazaki Y., Furuno M., Kasukawa T., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 12.Rinn J. L., Chang H. Y. Genome regulation by long noncoding RNAs. Annual Review of Biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W., Alvarez-Dominguez J. R., Lodish H. F. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Reports. 2012;13(11):971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn R. A., Chang H. Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14(6):752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi M. N., Antonangeli F. LncRNAs: new players in apoptosis control. International Journal of Cell Biology. 2014;2014:7. doi: 10.1155/2014/473857.473857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harries L. W. Long non-coding RNAs and human disease. Biochemical Society Transactions. 2012;40(4):902–906. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- 17.Qian K., Liu G., Tang Z., et al. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Archives of Biochemistry and Biophysics. 2017;615:1–9. doi: 10.1016/j.abb.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Liu B., Pan C. F., He Z. C., et al. Long noncoding RNA-LET suppresses tumor growth and EMT in lung adenocarcinoma. BioMed Research International. 2016;2016:9. doi: 10.1155/2016/4693471.4693471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K., Shi H., Xi H., et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer. Theranostics. 2017;7(1):213–227. doi: 10.7150/thno.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z. Y., Yang F., Zhang Y. L., et al. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomarkers : Section A of Disease Markers. 2017;18(1):95–104. doi: 10.3233/CBM-161715. [DOI] [PubMed] [Google Scholar]
- 21.Kapranov P., Cheng J., Dike S., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (New York, N.Y.) 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 22.Spizzo R., Almeida M. I., Colombatti A., Calin G. A. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilusz J. E., Sunwoo H., Spector D. L. Long noncoding RNAs: functional surprises from the RNA world. Genes & Development. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flippot R., Malouf G. G., Su X., Mouawad R., Spano J. P., Khayat D. Cancer subtypes classification using long non-coding RNA. Oncotarget. 2016;7(33):54082–54093. doi: 10.18632/oncotarget.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derrien T., Johnson R., Bussotti G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Research. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponting C. P., Oliver P. L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Guttman M., Rinn J. L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercer T. R., Dinger M. E., Mattick J. S. Long non-coding RNAs: insights into functions. Nature Reviews. Genetics. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 29.St Laurent G., Wahlestedt C., Kapranov P. The landscape of long non-coding RNA classification. Trends in Genetics : TIG. 2015;31(5):239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q., Gao S., Li H., Lv M., Lu C. Long noncoding RNAs (lncRNAs) in triple negative breast cancer. Journal of Cellular Physiology. 2017;9999:1–8. doi: 10.1002/jcp.25830. [DOI] [PubMed] [Google Scholar]
- 31.Bartonicek N., Maag J. L. V., Dinger M. E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Molecular Cancer. 2016;15(1):p. 43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Shields T., Crenshaw T., Hao Y., Moulton T., Tycko B. Imprinting of human H19: allele-specific CpG methylation, loss of the active allele in Wilms tumor, and potential for somatic allele switching. American Journal of Human Genetics. 1993;53(1):113–124. [PMC free article] [PubMed] [Google Scholar]
- 33.Luo J. H., Ren B., Keryanov S., et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology (Baltimore, Md.) 2006;44(4):1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Kok J. B., Verhaegh G. W., Roelofs R. W., et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Research. 2002;62(9):2695–2698. [PubMed] [Google Scholar]
- 35.Shi T., Gao G., Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Disease Markers. 2016;2016:10. doi: 10.1155/2016/9085195.9085195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H., Chen Z., Wang X., Huang Z., He Z., Chen Y. Long non-coding RNA: a new player in cancer. Journal of Hematology & Oncology. 2013;6(1):p. 37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu M. T., Hu J. W., Yin R., Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34(2):613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 38.Xu S., Sui S., Zhang J., et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. International Journal of Clinical and Experimental Pathology. 2015;8(5):4881–4891. [PMC free article] [PubMed] [Google Scholar]
- 39.Ji Q., Liu X., Fu X., et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/beta-catenin signal pathway. PloS One. 2013;8(11, article e78700) doi: 10.1371/journal.pone.0078700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyer M. K., Niknafs Y. S., Malik R., et al. The landscape of long noncoding RNAs in the human transcriptome. Nature Genetics. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan X., Hu Z., Feng Y., et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark M. B., Mercer T. R., Bussotti G., et al. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nature Methods. 2015;12(4):339–342. doi: 10.1038/nmeth.3321. [DOI] [PubMed] [Google Scholar]
- 43.Cheetham S. W., Gruhl F., Mattick J. S., Dinger M. E. Long noncoding RNAs and the genetics of cancer. British Journal of Cancer. 2013;108(12):2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gibb E. A., Brown C. J., Lam W. L. The functional role of long non-coding RNA in human carcinomas. Molecular Cancer. 2011;10(1):p. 38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hauptman N., Glavac D. Long non-coding RNA in cancer. International Journal of Molecular Sciences. 2013;14(3):4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitra S. A., Mitra A. P., Triche T. J. A central role for long non-coding RNA in cancer. Frontiers in Genetics. 2012;3(17):p. 70. doi: 10.3389/fgene.2012.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calin G. A., Liu C. G., Ferracin M., et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 48.Gao Y. F., Wang Z. B., Zhu T., et al. A critical overview of long non-coding RNA in glioma etiology 2016: an update. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(11):14403–14413. doi: 10.1007/s13277-016-5307-4. [DOI] [PubMed] [Google Scholar]
- 49.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature Reviews. Genetics. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 50.Lai F., Shiekhattar R. Where long noncoding RNAs meet DNA methylation. Cell Research. 2014;24(3):263–264. doi: 10.1038/cr.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin C., Zhang Y. Mechanisms of epigenetic inheritance. Current Opinion in Cell Biology. 2007;19(3):266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Gupta R. A., Shah N., Wang K. C., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu L., Zhu G., Zhang C., et al. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Research and Treatment. 2012;136(3):875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 54.Sorensen K. P., Thomassen M., Tan Q., et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Research and Treatment. 2013;142(3):529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 55.Beckedorff F. C., Amaral M. S., Deocesano-Pereira C., Verjovski-Almeida S. Long non-coding RNAs and their implications in cancer epigenetics. Bioscience Reports. 2013;33(4, article e00061) doi: 10.1042/BSR20130054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rinn J. L., Kertesz M., Wang J. K., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanduri C., Whitehead J., Mohammad F. The long and the short of it: RNA-directed chromatin asymmetry in mammalian X-chromosome inactivation. FEBS Letters. 2009;583(5):857–864. doi: 10.1016/j.febslet.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Cao J. The functional role of long non-coding RNAs and epigenetics. Biological Procedures Online. 2014;16(1):p. 11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y., Meng X. M., Huang C., et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Letters. 2014;344(1):20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 60.Mohammad F., Mondal T., Guseva N., Pandey G. K., Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development (Cambridge, England) 2010;137(15):2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 61.Li Q., Su Z., Xu X., et al. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(35):14110–14115. doi: 10.1073/pnas.1116597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merry C. R., Forrest M. E., Sabers J. N., et al. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Human Molecular Genetics. 2015;24(21):6240–6253. doi: 10.1093/hmg/ddv343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee R. S., Roberts C. W. Linking the SWI/SNF complex to prostate cancer. Nature Genetics. 2013;45(11):1268–1269. doi: 10.1038/ng.2805. [DOI] [PubMed] [Google Scholar]
- 64.Arab K., Park Y. J., Lindroth A. M., et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Molecular Cell. 2014;55(4):604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 65.Monnier P., Martinet C., Pontis J., Stancheva I., Ait-Si-Ali S., Dandolo L. H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(51):20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prensner J. R., Iyer M. K., Sahu A., et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature Genetics. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang Y., Wang J., Lian Y., et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Molecular Cancer. 2017;16(1):p. 42. doi: 10.1186/s12943-017-0612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang F., Deng X., Ma W., et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biology. 2015;16(1):p. 52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang K. C., Yang Y. W., Liu B., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai F., Orom U. A., Cesaroni M., et al. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature. 2013;494(7438):497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiang J. F., Yin Q. F., Chen T., et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Research. 2014;24(5):513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akers J. C., Gonda D., Kim R., Carter B. S., Chen C. C. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. Journal of Neuro-Oncology. 2013;113(1):1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sartori D. A., Chan D. W. Biomarkers in prostate cancer: what’s new? Current Opinion in Oncology. 2014;26(3):259–264. doi: 10.1097/CCO.0000000000000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang H., Wu Z., Zhang J., Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Molecular Medicine Reports. 2013;7(3):761–766. doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]
- 75.Reis E. M., Verjovski-Almeida S. Perspectives of long non-coding RNAs in cancer diagnostics. Frontiers in Genetics. 2012;3(32):p. 32. doi: 10.3389/fgene.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shao Y., Ye M., Jiang X., et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120(21):3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 77.Silva A., Bullock M., Calin G. The clinical relevance of long non-coding RNAs in cancer. Cancer. 2015;7(4):2169–2182. doi: 10.3390/cancers7040884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ji P., Diederichs S., Wang W., et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 79.Fradet Y., Saad F., Aprikian A., et al. uPM3, a new molecular urine test for the detection of prostate cancer. Urology. 2004;64(2):311–315. doi: 10.1016/j.urology.2004.03.052. discussion 315-316. [DOI] [PubMed] [Google Scholar]
- 80.Tinzl M., Marberger M., Horvath S., Chypre C. DD3PCA3 RNA analysis in urine—a new perspective for detecting prostate cancer. European Urology. 2004;46(2):182–186. doi: 10.1016/j.eururo.2004.06.004. discussion 187. [DOI] [PubMed] [Google Scholar]
- 81.Shappell S. B. Clinical utility of prostate carcinoma molecular diagnostic tests. Reviews in Urology. 2008;10(1):44–69. [PMC free article] [PubMed] [Google Scholar]
- 82.Rasool M., Malik A., Zahid S., et al. Non-coding RNAs in cancer diagnosis and therapy. Non-coding RNA Research. 2016;1(1):69–76. doi: 10.1016/j.ncrna.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou X., Yin C., Dang Y., Ye F., Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Scientific Reports. 2015;5, article 11516 doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ren S., Wang F., Shen J., et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. European Journal of Cancer (Oxford, England : 1990) 2013;49(13):2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 85.Hu B., Yang H., Yang H. Diagnostic value of urine prostate cancer antigen 3 test using a cutoff value of 35 mug/L in patients with prostate cancer. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(9):8573–8580. doi: 10.1007/s13277-014-2109-4. [DOI] [PubMed] [Google Scholar]
- 86.Arita T., Ichikawa D., Konishi H., et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Research. 2013;33(8):3185–3193. [PubMed] [Google Scholar]
- 87.Trajkovic K., Hsu C., Chiantia S., et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science (New York, N.Y.) 2008;319(5867):1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 88.Johnstone R. M. Exosomes biological significance: a concise review. Blood Cells, Molecules & Diseases. 2006;36(2):315–321. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 89.Huang X., Yuan T., Tschannen M., et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14(1):p. 319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Q., Shao Y., Zhang X., et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(3):2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 91.Arroyo J. D., Chevillet J. R., Kroh E. M., et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang K., Zhang S., Weber J., Baxter D., Galas D. J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Research. 2010;38(20):7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qi P., Zhou X., Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Molecular Cancer. 2016;15(1):p. 39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang J. L., Zheng L., Hu Y. W., Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35(3):507–514. doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 95.Lee G. L., Dobi A., Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nature Reviews. Urology. 2011;8(3):123–124. doi: 10.1038/nrurol.2011.10. [DOI] [PubMed] [Google Scholar]
- 96.Shappell S. B., Fulmer J., Arguello D., Wright B. S., Oppenheimer J. R., Putzi M. J. PCA3 urine mRNA testing for prostate carcinoma: patterns of use by community urologists and assay performance in reference laboratory setting. Urology. 2009;73(2):363–368. doi: 10.1016/j.urology.2008.08.459. [DOI] [PubMed] [Google Scholar]
- 97.Hessels D., Klein Gunnewiek J. M., van Oort I., et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. European Urology. 2003;44(1):8–15. doi: 10.1016/S0302-2838(03)00201-X. discussion 15-16. [DOI] [PubMed] [Google Scholar]
- 98.Xue W. J., Ying X. L., Jiang J. H., Xu Y. H. Prostate cancer antigen 3 as a biomarker in the urine for prostate cancer diagnosis: a meta-analysis. Journal of Cancer Research and Therapeutics. 2014;10(Supplement):C218–C221. doi: 10.4103/0973-1482.145881. [DOI] [PubMed] [Google Scholar]
- 99.Merola R., Tomao L., Antenucci A., et al. PCA3 in prostate cancer and tumor aggressiveness detection on 407 high-risk patients: a National Cancer Institute experience. Journal of Experimental & Clinical Cancer Research : CR. 2015;34(1):p. 15. doi: 10.1186/s13046-015-0127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chevli K. K., Duff M., Walter P., et al. Urinary PCA3 as a predictor of prostate cancer in a cohort of 3,073 men undergoing initial prostate biopsy. The Journal of Urology. 2014;191(6):1743–1748. doi: 10.1016/j.juro.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 101.Svoboda M., Slyskova J., Schneiderova M., et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35(7):1510–1515. doi: 10.1093/carcin/bgu055. [DOI] [PubMed] [Google Scholar]
- 102.Panzitt K., Tschernatsch M. M., Guelly C., et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 103.Xie H., Ma H., Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed Research International. 2013;2013:5. doi: 10.1155/2013/136106.136106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao Y., Guo Q., Chen J., Hu J., Wang S., Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncology Reports. 2014;31(1):358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 105.Guo F., Yu F., Wang J., et al. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomedical Reports. 2015;3(3):309–312. doi: 10.3892/br.2015.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weber D. G., Johnen G., Casjens S., et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Research Notes. 2013;6(1):p. 518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang F., Ren S., Chen R., et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5(22):11091–11102. doi: 10.18632/oncotarget.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pang Q., Ge J., Shao Y., et al. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(6):5441–5447. doi: 10.1007/s13277-014-1709-3. [DOI] [PubMed] [Google Scholar]
- 109.Tang J., Jiang R., Deng L., Zhang X., Wang K., Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6(6):4505–4515. doi: 10.18632/oncotarget.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X. S., Zhang Z., Wang H. C., et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clinical Cancer Research : An Official Journal of the American Association for Cancer Research. 2006;12(16):4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 111.Dallosso A. R., Hancock A. L., Malik S., et al. Alternately spliced WT1 antisense transcripts interact with WT1 sense RNA and show epigenetic and splicing defects in cancer. RNA (New York, N.Y.) 2007;13(12):2287–2299. doi: 10.1261/rna.562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yao Y., Ma J., Xue Y., et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Letters. 2015;359(1):75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 113.Wang Y., Wang Y., Li J., Zhang Y., Yin H., Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Letters. 2015;367(2):122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 114.Ma J., Wang P., Yao Y., et al. Knockdown of long non-coding RNA MALAT1 increases the blood-tumor barrier permeability by up-regulating miR-140. Biochimica et Biophysica Acta. 2016;1859(2):324–338. doi: 10.1016/j.bbagrm.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 115.Yang M. H., Hu Z. Y., Xu C., et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochimica et Biophysica Acta. 2015;1852(1):166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang N., Chi Y., Xue J., et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. 2016;7(25):37957–37965. doi: 10.18632/oncotarget.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malakar P., Shilo A., Mogilevsky A., et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Research. 2017;77(5):1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yamada K., Kano J., Tsunoda H., et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Science. 2006;97(2):106–112. doi: 10.1111/j.1349-7006.2006.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Silva J. M., Boczek N. J., Berres M. W., Ma X., Smith D. I. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biology. 2011;8(3):496–505. doi: 10.4161/rna.8.3.14800. [DOI] [PubMed] [Google Scholar]
- 120.Li H., Zhu L., Xu L., et al. Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Molecular Carcinogenesis. 2017;56(1):3–17. doi: 10.1002/mc.22338. [DOI] [PubMed] [Google Scholar]
- 121.Xu N., Chen F., Wang F., et al. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: a Chinese population-based study. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(10):7659–7665. doi: 10.1007/s13277-015-3469-0. [DOI] [PubMed] [Google Scholar]
- 122.Iacoangeli A., Lin Y., Morley E. J., et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25(11):2125–2133. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 123.Yang M., Zhai X., Xia B., Wang Y., Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(10):7615–7622. doi: 10.1007/s13277-015-3465-4. [DOI] [PubMed] [Google Scholar]
- 124.Tong Y. S., Wang X. W., Zhou X. L., et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Molecular Cancer. 2015;14(1):p. 3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ge X., Chen Y., Liao X., et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Medical Oncology (Northwood, London, England) 2013;30(2):p. 588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 126.Prensner J. R., Chen W., Han S., et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia (New York, N.Y.) 2014;16(11):900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang W., He X., Zheng Z., et al. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Molecular Cancer. 2017;16(1):p. 75. doi: 10.1186/s12943-017-0643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu X. H., Liu Z. L., Sun M., Liu J., Wang Z. X., De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13(1):p. 464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee M., Kim H. J., Kim S. W., et al. The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget. 2016;7(28):44558–44571. doi: 10.18632/oncotarget.10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim K., Jutooru I., Chadalapaka G., et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu Y., Zhang L., Zhang L., et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. International Journal of Oncology. 2015;46(6):2586–2594. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 132.Geng Y. J., Xie S. L., Li Q., Ma J., Wang G. Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. The Journal of International Medical Research. 2011;39(6):2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 133.Zhang J. X., Han L., Bao Z. S., et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro-Oncology. 2013;15(12):1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ke J., Yao Y. L., Zheng J., et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6(26):21934–21949. doi: 10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Qiu M., Xu Y., Yang X., et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(6):5375–5380. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 136.Ling H., Spizzo R., Atlasi Y., et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Research. 2013;23(9):1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu L., Jin L., Zhang W., Zhang L. Roles of long non-coding RNA CCAT2 in cervical cancer cell growth and apoptosis. Medical Science Monitor : International Medical Journal of Experimental and Clinical Research. 2016;22:875–879. doi: 10.12659/MSM.897754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xu M. D., Qi P., Weng W. W., et al. Long non-coding RNA LSINCT5 predicts negative prognosis and exhibits oncogenic activity in gastric cancer. Medicine. 2014;93(28, article e303) doi: 10.1097/MD.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Q., Zhang J., Liu Y., et al. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Letters. 2016;373(2):251–259. doi: 10.1016/j.canlet.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 140.Guo H., Wu L., Yang Q., Ye M., Zhu X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene. 2015;554(1):114–119. doi: 10.1016/j.gene.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 141.Ma C. C., Xiong Z., Zhu G. N., et al. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. Journal of Experimental & Clinical Cancer Research : CR. 2016;35(1):p. 90. doi: 10.1186/s13046-016-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hu L., Lv Q. L., Chen S. H., et al. Up-regulation of long non-coding RNA AB073614 predicts a poor prognosis in patients with glioma. International Journal of Environmental Research and Public Health. 2016;13(4):p. 433. doi: 10.3390/ijerph13040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou T., Gao Y. Increased expression of LncRNA BANCR and its prognostic significance in human hepatocellular carcinoma. World Journal of Surgical Oncology. 2015;14(1):p. 8. doi: 10.1186/s12957-015-0757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang T., Cao C., Wu D., Liu L. SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(2):2379–2385. doi: 10.1007/s13277-015-4052-4. [DOI] [PubMed] [Google Scholar]
- 145.Shi Y., Song Q., Yu S., Hu D., Zhuang X. Microvascular invasion in hepatocellular carcinoma overexpression promotes cell proliferation and inhibits cell apoptosis of hepatocellular carcinoma via inhibiting miR-199a expression. OncoTargets and Therapy. 2015;8:2303–2310. doi: 10.2147/OTT.S86807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nie F. Q., Zhu Q., Xu T. P., et al. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(8):7587–7594. doi: 10.1007/s13277-014-2009-7. [DOI] [PubMed] [Google Scholar]
- 147.White N. M., Cabanski C. R., Silva-Fisher J. M., Dang H. X., Govindan R., Maher C. A. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biology. 2014;15(8):p. 429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Qiu M., Xu Y., Wang J., et al. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death & Disease. 2015;6(8, article e1858) doi: 10.1038/cddis.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zeng Z., Bo H., Gong Z., et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(1):729–737. doi: 10.1007/s13277-015-3860-x. [DOI] [PubMed] [Google Scholar]
- 150.Han X., Wang L., Ning Y., Li S., Wang Z. Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes metastasis in colorectal cancer. Biological Research. 2016;49(1):p. 36. doi: 10.1186/s40659-016-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lu X., Zhou C., Li R., et al. Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. Tumour Biology. 2016;37(7):9699–9707. doi: 10.1007/s13277-016-4858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Luo H. L., Huang M. D., Guo J. N., et al. AFAP1-AS1 is upregulated and promotes esophageal squamous cell carcinoma cell proliferation and inhibits cell apoptosis. Cancer Medicine. 2016;5(10):2879–2885. doi: 10.1002/cam4.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nie F. Q., Sun M., Yang J. S., et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Molecular Cancer Therapeutics. 2015;14(1):268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 154.Huang M., Chen W., Qi F., et al. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. Journal of Hematology & Oncology. 2015;8(50) doi: 10.1186/s13045-015-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhu H., Li X., Song Y., Zhang P., Xiao Y., Xing Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochemical and Biophysical Research Communications. 2015;467(2):223–228. doi: 10.1016/j.bbrc.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 156.Nie W., Ge H. J., Yang X. Q., et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Letters. 2016;371(1):99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 157.Yang Y. T., Wang Y. F., Lai J. Y., et al. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/β-catenin signaling pathway. Cancer Science. 2016;107(11):1581–1589. doi: 10.1111/cas.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Srivastava A. K., Singh P. K., Rath S. K., Dalela D., Goel M. M., Bhatt M. L. Appraisal of diagnostic ability of UCA1 as a biomarker of carcinoma of the urinary bladder. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(11):11435–11442. doi: 10.1007/s13277-014-2474-z. [DOI] [PubMed] [Google Scholar]
- 159.Han Y., Yang Y. N., Yuan H. H., et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46(5):396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 160.Xiao J. N., Yan T. H., Yu R. M., et al. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. Journal of Cancer Research and Clinical Oncology. 2017;143(6):981–990. doi: 10.1007/s00432-017-2370-1. [DOI] [PubMed] [Google Scholar]
- 161.Tuo Y. L., Li X. M., Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. European Review for Medical and Pharmacological Sciences. 2015;19(18):3403–3411. [PubMed] [Google Scholar]
- 162.Li J. Y., Ma X., Zhang C. B. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. International Journal of Clinical and Experimental Pathology. 2014;7(11):7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 163.Lessard L., Liu M., Marzese D. M., et al. The CASC15 long intergenic noncoding RNA locus is involved in melanoma progression and phenotype switching. The Journal of Investigative Dermatology. 2015;135(10):2464–2474. doi: 10.1038/jid.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Khaitan D., Dinger M. E., Mazar J., et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Research. 2011;71(11):3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 165.Mazar J., Zhao W., Khalil A. M., et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5(19):8959–8969. doi: 10.18632/oncotarget.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu H., Lv Z., Guo E. Knockdown of long noncoding RNA SPRY4-IT1 suppresses glioma cell proliferation, metastasis and epithelial-mesenchymal transition. International Journal of Clinical and Experimental Pathology. 2015;8(8):9140–9146. [PMC free article] [PubMed] [Google Scholar]
- 167.Luo M., Li Z., Wang W., Zeng Y., Liu Z., Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Letters. 2013;333(2):213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 168.Tan D., Wu Y., Hu L., et al. Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Diseases of the Esophagus : Official Journal of the International Society for Diseases of the Esophagus. 2017;30(1):1–9. doi: 10.1111/dote.12481. [DOI] [PubMed] [Google Scholar]
- 169.Yang W., Ning N., Jin X. The lncRNA H19 promotes cell proliferation by competitively binding to miR-200a and derepressing beta-catenin expression in colorectal cancer. BioMed Research International. 2017;2017:8. doi: 10.1155/2017/2767484.2767484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shi Y., Wang Y., Luan W., et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PloS One. 2014;9(1, article e86295) doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Peng W., Gao W., Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Medical Oncology (Northwood, London, England) 2014;31(12):p. 346. doi: 10.1007/s12032-014-0346-4. [DOI] [PubMed] [Google Scholar]
- 172.Zhu Y., Zhang X., Qi L., et al. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016;7(12):14429–14440. doi: 10.18632/oncotarget.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ylipaa A., Kivinummi K., Kohvakka A., et al. Transcriptome sequencing reveals PCAT5 as a novel ERG-regulated long noncoding RNA in prostate cancer. Cancer Research. 2015;75(19):4026–4031. doi: 10.1158/0008-5472.CAN-15-0217. [DOI] [PubMed] [Google Scholar]
- 174.Crea F., Watahiki A., Quagliata L., et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chung S., Nakagawa H., Uemura M., et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Science. 2011;102(1):245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 176.He C., Jiang B., Ma J., Li Q. Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. APMIS : Acta Pathologica, Microbiologica, et Immunologica Scandinavica. 2016;124(3):169–174. doi: 10.1111/apm.12480. [DOI] [PubMed] [Google Scholar]
- 177.Zhen L., Yun-Hui L., Hong-Yu D., Jun M., Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(1):673–683. doi: 10.1007/s13277-015-3843-y. [DOI] [PubMed] [Google Scholar]
- 178.Guo S., Chen W., Luo Y., et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. International Journal of Clinical and Experimental Pathology. 2015;8(5):5395–5402. [PMC free article] [PubMed] [Google Scholar]
- 179.Lu Y., Li T., Wei G., et al. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(9):11733–11741. doi: 10.1007/s13277-015-4773-4. [DOI] [PubMed] [Google Scholar]
- 180.Zhou Q., Chen J., Feng J., Wang J. Long noncoding RNA PVT1 modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR) Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(3):3105–3113. doi: 10.1007/s13277-015-4149-9. [DOI] [PubMed] [Google Scholar]
- 181.Kong R., Zhang E. B., Yin D. D., et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Molecular Cancer. 2015;14(1):p. 82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Takahashi Y., Sawada G., Kurashige J., et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. British Journal of Cancer. 2014;110(1):164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yan R., Wang K., Peng R., et al. Genetic variants in lncRNA SRA and risk of breast cancer. Oncotarget. 2016;7(16):22486–22496. doi: 10.18632/oncotarget.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang P., Ren Z., Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. Journal of Cellular Biochemistry. 2012;113(6):1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 185.Li J., Bian E. B., He X. J., et al. Epigenetic repression of long non-coding RNA MEG3 mediated by DNMT1 represses the p53 pathway in gliomas. International Journal of Oncology. 2016;48(2):723–733. doi: 10.3892/ijo.2015.3285. [DOI] [PubMed] [Google Scholar]
- 186.Askarian-Amiri M. E., Crawford J., French J. D., et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA (New York, N.Y.) 2011;17(5):878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mourtada-Maarabouni M., Pickard M. R., Hedge V. L., Farzaneh F., Williams G. T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 188.Zhao X., Wang P., Liu J., et al. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Molecular Therapy : The Journal of the American Society of Gene Therapy. 2015;23(12):1899–1911. doi: 10.1038/mt.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ding J., Lu B., Wang J., et al. Long non-coding RNA Loc554202 induces apoptosis in colorectal cancer cells via the caspase cleavage cascades. Journal of Experimental & Clinical Cancer Research : CR. 2015;34(1, article 100) doi: 10.1186/s13046-015-0217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dong L., Qi P., Xu M. D., et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. International Journal of Cancer. 2015;137(5):1128–1135. doi: 10.1002/ijc.29484. [DOI] [PubMed] [Google Scholar]
- 191.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W. J., Pandolfi P. P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fei Z. H., Yu X. J., Zhou M., Su H. F., Zheng Z., Xie C. Y. Upregulated expression of long non-coding RNA LINC00982 regulates cell proliferation and its clinical relevance in patients with gastric cancer. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(2):1983–1993. doi: 10.1007/s13277-015-3979-9. [DOI] [PubMed] [Google Scholar]
- 193.Qin X., Yao J., Geng P., Fu X., Xue J., Zhang Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. International Journal of Clinical and Experimental Pathology. 2014;7(6):3065–3072. [PMC free article] [PubMed] [Google Scholar]
- 194.Yao J., Zhou B., Zhang J., et al. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(8):7935–7944. doi: 10.1007/s13277-014-1949-2. [DOI] [PubMed] [Google Scholar]
- 195.Yue H., Zhu J., Xie S., Li F., Xu Q. MDC1-AS, an antisense long noncoding RNA, regulates cell proliferation of glioma. Biomedicine & Pharmacotherapy. 2016;81:203–209. doi: 10.1016/j.biopha.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 196.Li J., Zhang M., An G., Ma Q. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Experimental Biology and Medicine (Maywood, N.J.) 2016;241(6):644–649. doi: 10.1177/1535370215622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Feng S., Yao J., Chen Y., et al. Expression and functional role of reprogramming-related long noncoding RNA (lincRNA-ROR) in glioma. Journal of Molecular Neuroscience : MN. 2015;56(3):623–630. doi: 10.1007/s12031-014-0488-z. [DOI] [PubMed] [Google Scholar]
- 198.Wang P., Liu Y. H., Yao Y. L., et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cellular Signalling. 2015;27(2):275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 199.Zhou C. C., Yang F., Yuan S. X., et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology (Baltimore, Md.) 2016;63(3):850–863. doi: 10.1002/hep.28393. [DOI] [PubMed] [Google Scholar]
- 200.He H., Nagy R., Liyanarachchi S., et al. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Research. 2009;69(2):625–631. doi: 10.1158/0008-5472.CAN-08-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang J. H., Li J. H., Jiang S., Zhou H., Qu L. H. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Research. 2013;41(Database issue):D177–D187. doi: 10.1093/nar/gks1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Weirick T., John D., Dimmeler S., Uchida S. C-It-Loci: a knowledge database for tissue-enriched loci. Bioinformatics (Oxford, England) 2015;31(21):3537–3543. doi: 10.1093/bioinformatics/btv410. [DOI] [PubMed] [Google Scholar]
- 203.Zhao Z., Bai J., Wu A., et al. Co-LncRNA: investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-Seq data. Database : The Journal of Biological Databases and Curation. 2015;2015, article bav082 doi: 10.1093/database/bav082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Paraskevopoulou M. D., Georgakilas G., Kostoulas N., et al. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Research. 2013;41(Database issue):D239–D245. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Paraskevopoulou M. D., Vlachos I. S., Karagkouni D., et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Research. 2016;44(D1):D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liu K., Yan Z., Li Y., Sun Z. Linc2GO: a human LincRNA function annotation resource based on ceRNA hypothesis. Bioinformatics (Oxford, England) 2013;29(17):2221–2222. doi: 10.1093/bioinformatics/btt361. [DOI] [PubMed] [Google Scholar]
- 207.Ning S., Zhang J., Wang P., et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Research. 2016;44(D1):D980–D985. doi: 10.1093/nar/gkv1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang P., Ning S., Zhang Y., et al. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Research. 2015;43(7):3478–3489. doi: 10.1093/nar/gkv233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Volders P. J., Helsens K., Wang X., et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Research. 2013;41(Database issue):D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Volders P. J., Verheggen K., Menschaert G., et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Research. 2015;43(8):4363–4364. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chakraborty S., Deb A., Maji R. K., Saha S., Ghosh Z. LncRBase: an enriched resource for lncRNA information. PloS One. 2014;9(9, article e108010) doi: 10.1371/journal.pone.0108010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jiang Q., Ma R., Wang J., et al. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics. 2015;16(Supplement 3):p. S2. doi: 10.1186/1471-2164-16-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Amaral P. P., Clark M. B., Gascoigne D. K., Dinger M. E., Mattick J. S. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Research. 2011;39(Database issue):D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Quek X. C., Thomson D. W., Maag J. L., et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Research. 2015;43(Database issue):D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen G., Wang Z., Wang D., et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Research. 2013;41(Database issue):D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gong J., Liu W., Zhang J., Miao X., Guo A. Y. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Research. 2015;43(Database issue):D181–D186. doi: 10.1093/nar/gku1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bhartiya D., Pal K., Ghosh S., et al. lncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database : The Journal of Biological Databases and Curation. 2013;2013:p. bat034. doi: 10.1093/database/bat034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Jeggari A., Marks D. S., Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics (Oxford, England) 2012;28(15):2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liu C., Bai B., Skogerbo G., et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Research. 2005;33(Database issue):D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhao Y., Li H., Fang S., et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Research. 2016;44(D1):D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yang J. H., Li J. H., Shao P., Zhou H., Chen Y. Q., Qu L. H. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Research. 2011;39(Database issue):D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li J. H., Liu S., Zhou H., Qu L. H., Yang J. H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research. 2014;42(Database issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]