Abstract
Bacteria forming light-organ symbiosis with deep-sea chlorophthalmid fishes (Aulopiformes: Chlorophthalmidae) are considered to belong to the species Photobacterium phosphoreum. The identification of these bacteria as P. phosphoreum, however, was based exclusively on phenotypic traits, which may not discriminate between phenetically similar but evolutionarily distinct luminous bacteria. Therefore, to test the species identification of chlorophthalmid symbionts, we carried out a genomotypic (repetitive element palindromic PCR genomic profiling) and phylogenetic analysis on strains isolated from the perirectal light organ of Chlorophthalmus albatrossis. Sequence analysis of the 16S rRNA gene of 10 strains from 5 fish specimens placed these bacteria in a cluster related to but phylogenetically distinct from the type strain of P. phosphoreum, ATCC 11040T, and the type strain of Photobacterium iliopiscarium, ATCC 51760T. Analysis of gyrB resolved the C. albatrossis strains as a strongly supported clade distinct from P. phosphoreum and P. iliopiscarium. Genomic profiling of 109 strains from the 5 C. albatrossis specimens revealed a high level of similarity among strains but allowed identification of genomotypically different types from each fish. Representatives of each type were then analyzed phylogenetically, using sequence of the luxABFE genes. As with gyrB, analysis of luxABFE resolved the C. albatrossis strains as a robustly supported clade distinct from P. phosphoreum. Furthermore, other strains of luminous bacteria reported as P. phosphoreum, i.e., NCIMB 844, from the skin of Merluccius capensis (Merlucciidae), NZ-11D, from the light organ of Nezumia aequalis (Macrouridae), and pjapo.1.1, from the light organ of Physiculus japonicus (Moridae), grouped phylogenetically by gyrB and luxABFE with the C. albatrossis strains, not with ATCC 11040T. These results demonstrate that luminous bacteria symbiotic with C. albatrossis, together with certain other strains of luminous bacteria, form a clade, designated the kishitanii clade, that is related to but evolutionarily distinct from P. phosphoreum. Members of the kishitanii clade may constitute the major or sole bioluminescent symbiont of several families of deep-sea luminous fishes.
The marine luminous bacterium Photobacterium phosphoreum is widely considered to be the bioluminescent light organ symbiont of several deep-sea fishes. Members of six teleost families representing four orders, i.e., Opisthoproctidae (Osmeriformes), Chlorophthalmidae (Aulopiformes), Macrouridae, Steindachneriidae and Moridae (Gadiformes), and Trachichthyidae (Beryciformes), have been reported to harbor luminous bacteria identified as or thought to be P. phosphoreum (2, 14-16, 21-26, 28, 42, 44, 45, 48, 56, 61). As a psychrotrophic species, P. phosphoreum may be adapted thermally to symbiosis with these fishes, which occur in deep, cold marine waters (9, 17, 19).
Detailed taxonomic study has been conducted on bacteria from certain of these fishes (22, 44, 45). In most cases, however, the identification of the bacteria as P. phosphoreum is based on limited or unreported taxonomic data and on the examination of few strains. Furthermore, with the exception of certain strains examined at the DNA sequence level (2, 5, 20, 21), the characterization of these bacteria has been exclusively phenetic. The identity of the bioluminescent symbionts of chlorophthalmid fishes (greeneyes) is a case in point. Chlorophthalmids are reported to harbor P. phosphoreum in a ventral, perirectal light organ (28, 47, 48, 56), but either taxonomic analysis of the bacteria has not been carried out or the taxonomic data on which the identifications were based were not reported; furthermore, no genotypic data have been described for chlorophthalmid bacteria. Therefore, whether or not these bacteria actually are P. phosphoreum is an open question, as is the related question of whether phenetic analysis can provide sufficient information to discriminate among closely related luminous bacteria.
One approach to a better understanding of the phylogenetic status of chlorophthalmid symbionts and other luminous bacteria is through sequence analysis of the luminescence (lux) genes. The lux gene order most common in luminous bacteria is luxCDABEG; luxA and luxB encode the α and β subunits of bacterial luciferase, luxC, luxD, and luxE specify the fatty acid reductase, acyl-transferase, and acyl-protein synthetase components, respectively, of the fatty acid reductase complex of the luminescence system, and luxG specifies a flavin reductase (9, 31). P. phosphoreum and members of the mandapamensis clade of Photobacterium leiognathi, in contrast to other species, bear an additional gene, luxF, which encodes a nonfluorescent flavoprotein, and have a gene order of luxCDABFEG (2, 31, 46). Sequence divergence in luxA has provided insight into the phylogenetic status of uncultured luminous bacteria symbiotic with anomalipod fishes (18) and has formed the basis for a hybridization method for identifying different species of luminous bacteria (35, 57); furthermore, luxA sequence was used in the description of a new species of luminous bacteria (30). Recently, sequence divergence of the luxAB(F)E region was shown to provide inter- and intraspecific phylogenetic resolution among luminous bacteria, allowing phenotypically similar but evolutionarily distinct strains to be distinguished (2). Use of lux sequence for Photobacterium phylogenetic analysis, however, is not feasible for all species; Photobacterium iliopiscarium, for example, a species closely related to P. phosphoreum and indigenous to the intestine of cold-dwelling fish, apparently is nonluminous (38, 54).
In this study, we used genomic profiling with repetitive element palindromic PCR (rep-PCR) (55) and a multilocus phylogenetic analysis based on sequences of the 16S rRNA, gyrB, and luxABFE genes to test the evolutionary relationships of bacteria isolated from the perirectal light organ of Chlorophthalmus albatrossis. The results indicate that the C. albatrossis symbionts, together with certain strains previously identified phenotypically as P. phosphoreum, i.e., NCIMB 844, NZ-11D, and pjapo.1.1, represent a clade of bacteria that is evolutionarily distinct from the type strains of P. phosphoreum and P. iliopiscarium.
MATERIALS AND METHODS
Collection of fish specimens and isolation of bacterial strains.
Specimens of C. albatrossis were collected by trawl net (length, 45 m; mesh size, 20 mm) at a depth of 350 m in the Kumano Sea at 33°57′20"N 136°21′40"W, approximately 40 km east of Owase, Mie Prefecture, Honshu, Japan. The specimens were collected by Ariyasu Makihara, captain of the fishing vessel, Kotobuki-maru, and were kept chilled on ice until sampled microbiologically later on the day of collection. The fish were identified to the species level by reference to Nakabo (32). Ichthyological nomenclature follows the work of Nelson (36). Fish specimen designations follow the work of Dunlap et al. (10). Specimens Calba.1 (standard length [SL], 121 mm) and Calba.2 (SL, 113 mm) were collected on 1 February 2004, and specimens Calba.3 (SL, 137 mm), Calba.4 (SL, 145 mm), and Calba.5 (SL, 144 mm) were collected on 2 February 2004. These specimens have been retained for taxonomic reference.
To isolate symbiotic bacteria, the perirectal light organ (47) of the freshly caught specimens was dissected aseptically and then was either finely minced directly on plates of LSW-70 agar, containing 10 g tryptone, 5 g yeast extract, 350 ml double-strength artificial seawater (33), 650 ml de-ionized water, and 15 g agar per liter, and streaked for isolated colonies (specimens Calba.1 and Calba.2), or was homogenized in 0.5 ml of sterile artificial 70% seawater containing 25 mM HEPES buffer (pH 7.25) (BSW-70) in a sterile hand-held Ten Broeck tissue homogenizer (8) (specimens Calba.3, Calba.4, and Calba.5); the light-organ homogenates were then serially diluted in BSW-70 and spread on plates of LSW-70 agar. Dilution of the light-organ homogenates, to approximately 10−5, and plating volumes, 25 to 100 μl, yielded several dozen to several hundred colonies per plate after 24 to 48 h of incubation at 18 to 22°C. Ten to thirty individual strains were then picked at random from the plates for each fish specimen, purified on LSW-70 agar, and stored at −75°C in cryoprotective medium (9) under the following bacterial strain designations (10): calba.1.1 to calba.1.10 (from Calba.1); calba.2.1 to calba.2.10 (from Calba.2); calba.3.1 to calba.3.30 (from Calba.3); calba.4.1 to calba.4.30 (from Calba.4); and calba.5.1 to calba.5.30 (from Calba.5).
16S rRNA and gyrB gene amplification and sequencing.
Genomic DNA was purified from 1-ml cultures of strains grown overnight in LSW-70 broth, using the QIAGEN (Valencia, Calif.) DNeasy tissue extraction kit. The 16S rRNA gene was amplified using primers 27f and 1492r (2, 29), Taq polymerase, and reagents of the Eppendorf (Hamburg, Germany) MasterTaq kit with the following protocol: hot start and 2-min denaturing at 95°C; 35 cycles of 20 s at 94°C (denaturing), 15 s at 47°C (primer annealing), and 1.5 min at 72°C (polymerase extension); a single additional extension step of 7 min at 72°C; and snap cooling to 4°C. The 16S rRNA gene product was sequenced with the amplification primers and the internal primers, 772r and 669f (2, 10). For amplification of gyrB, the same protocol as for the 16S rRNA gene was followed except that primers 22f and 1240r (2) were used, and primer annealing was at 48°C. The gyrB product was sequenced using the amplification primers.
Strain genomotyping with rep-PCR.
DNA fingerprint analysis (genomic profiling) was carried out by rep-PCR, as described by Versalovic et al. (55), Rademaker and deBruijn (40), and Di Meo et al. (7). The reaction mixture for rep-PCR (40) contained (per 25-μl reaction volume): 6.2 μl of tissue culture-grade water (Sigma, St. Louis, Mo.), 5 μl of 5× Gitschier buffer, 0.4 μl of bovine serum albumin (10 mg ml−1), 2.5 μl of dimethyl sulfoxide (100%), 3.125 μl of deoxynucleoside triphosphates (10 mM), 1.25 μl (each) of primers REP1R-I (5′-IIIICGICGICATCIGGC-3′) and REP2-I (5′-ICGICTTATCIGGCCTAC-3′) (55) (50 nM), 2.5 μl of MgCl2 (25 mM), and 1 μl of Taq polymerase (5 U μl−1) (Eppendorf). For individual reactions, 23 μl of the mixture was combined with 2 μl of template DNA, prepared as described above. PCR was carried out using the Bio-Rad (Hercules, Calif.) iCycler, with the following conditions for rep-PCR, modified from the method of Di Meo et al. (7): hot start and 2 min of denaturation at 95°C; 30 cycles of 30 s at 92°C (denaturation), 1 min at 40°C (primer annealing), and 8 min at 65°C (polymerase extension); a single additional extension step of 8 min at 65°C; and snap cooling to 4°C. Products of the rep-PCRs were separated on 1.75% agarose gels containing 5 μl of 1% ethidium bromide per 100 ml of agarose in 1× TAE (0.04 M Tris-acetate, 0.002 M EDTA) buffer at 100 V for approximately 4.5 h. Digital images of gels were captured with a Bio-Rad Fluor S Multi-imager and were examined visually for differences in genomic profiles of different strains.
Amplification and sequencing of the luxABFE region.
Primers designed for amplification of the lux region from strains reported to be P. phosphoreum (2) were found to be effective for amplifying the lux region of C. albatrossis symbionts. Specifically, primers Af (CTTTTAGATCCAATGTCAAAAGGCCG) and FEr (TAATATCGTCGATCTCTGTACTTAC), polymerase and reagents as described above for the 16S rRNA gene, and the following protocol were used: hot start and 2 min of denaturing at 95°C; 20 cycles of 20 s at 94°C (denaturing), 15 s at 48°C (primer annealing), and 3 min at 72°C (polymerase extension); 15 cycles in which 5 s were added incrementally to the extension time; a single additional extension step of 7 min at 72°C; and snap cooling to 4°C. The product, approximately 2.6 kb in length, was then used as a template for amplification of two internal regions, using the primer pair ABf (AAACGTCGTGTTGATTATAGCAACG) and ABr (TCCAACGATATGTTAGTGGAAGC) (product AB) and primer pair BFf (TGAAGCTATCACAACTAATTACTG) and BFr (AGTGGTCATCAATATTAATCACATC) (product BF) and the following protocol: hot start and 2 min of denaturing at 95°C; 35 cycles of 20 s at 94°C (denaturing), 15 s at 50°C (primer annealing), and 1 min at 72°C (polymerase extension); a single additional extension step of 7 min at 72°C; and snap cooling to 4°C. The 2.6-kb lux product was sequenced by using the primers Af (above) and Ar (TCAGAGCCATTTGCTTCGAAACCAAG) and FEf (ATGAATAATGCATTAGAAACATTACG) and FEr (above), and the two internal lux products, AB and BF, were sequenced with the amplification primers.
Sequencing and phylogenetic analysis.
For sequences obtained in this study, sequencing of PCR products was carried out by staff of the University of Michigan Sequencing Core using dye terminator cycle sequencing on a Perkin-Elmer (Wellesley, Mass.) ABI 3730 instrument. 16S rRNA gene sequences were aligned by eye, and gyrB and luxABFE sequences were aligned by inferred amino acid sequence. Parsimony analyses were performed with PAUP* (52), using 1,000 heuristic search replicates with tree bisection reconnection branch swapping; branches with a maximum length of 0 were collapsed. Jackknife support was calculated with PAUP*, using 1,000 replicates emulating Jac resampling. Bremer support was calculated with the aid of TreeRot (49).
GenBank accession numbers for previously reported 16S rRNA gene, gyrB, and lux sequences (2, 10) are as follows: ATCC 25587 (AY455870, AY455880, AY456750), lequu.1.1 (AY204492, AY455881, AY341069), lleuc.1.1 (AY204495, AY455882, AY341070), ATCC 27561T (AY341441, AY455883, AY341067), ATCC 33981 (AY341442, AY455884, AY341068), PL-721 (AY341440, AY455885, AY341066), ATCC 11040T (AY341437, AY455875, AY341063), NCIMB 844 (AY341438, AY455876, AY341064), pjapo.1.1 (AY341439, AY455877, AY341065), ATCC 7744T (AY341436, AY455874, AY341062). Previously reported sequences obtained for gyrB of other strains (2) are as follows: ATCC 25915T (AY455890), ATCC 33539T (AY455889), ATCC 51760T (AY455878), ATCC 25521T (AY455879), JCM 10084T (AY455892), and SS9 (AY455891). Other 16S rRNA gene sequences used in this study are as follows: ATCC 25915T (D25307), ATCC 33539T (AB032015), NCIMB 2058 (X78105), ATCC 25521T (X74686), JCM 10084T (D21226), and SS9 (AB003191).
GenBank sequence accession numbers.
GenBank accession numbers for sequences obtained in this study for the 16S rRNA, gyrB, and luxABFE genes are as follows: NZ-11D (AY642160, AY642171, AY642182), calba.1.1 (AY642161, AY642172, AY642193), calba.1.2 (AY642162, AY642173, AY642183) calba.2.1 (AY642163, AY642174, AY642198), calba.2.2 (AY642164, AY642175, AY642199), calba.3.1 (AY642165, AY642176, AY642200), calba.3.2 (AY642166, AY642177, AY642201), calba.4.1 (AY642167, AY642178, AY642184), calba.4.2 (AY642168, AY642179, AY642215), calba.5.1 (AY642169, AY642180, AY642222), and calba.5.9 (AY642170, AY642181, AY642224). Additional strains were sequenced for luxABFE alone: calba.1.3 (AY642194), calba.1.4 (AY642195), calba.1.6 (AY642196), calba.1.9 (AY642197), calba.3.3 (AY642202), calba.3.4 (AY642187), calba.3.5 (AY642188), calba.3.6 (AY642203), calba.3.7 (AY642204), calba.3.8 (AY642205), calba.3.9 (AY642206), calba.3.10 (AY642207), calba.3.12 (AY642208), calba.3.17 (AY642189), calba.3.19 (AY642209), calba.3.21 (AY642210), calba.3.25 (AY642211), calba.3.26 (AY642212), calba.3.27 (AY642213), calba.3.28 (AY642214), calba.4.3 (AY642216), calba.4.4 (AY642217), calba.4.5 (AY642218), calba.4.6 (AY642219), calba.4.12 (AY642191), calba.4.14 (AY642185), calba.4.15 (AY642192), calba.4.22 (AY642220), calba.4.27 (AY642221), calba.5.3 (AY642186), calba.5.5 (AY642223), calba.5.10 (AY642225), calba.5.14 (AY642226), calba.5.19 (AY642227), and calba.5.21 (AY642190). In addition, the 16S rRNA gene was resequenced for ATCC 51760T (AY643710).
RESULTS
Isolation of bacteria from the C. albatrossis light organ.
Perirectal light organs dissected from fresh specimens of C. albatrossis emitted an intense blue luminescence. Direct streaking for isolated colonies from light organs that were finely minced on agar plates (specimens Calba.1 and Calba.2) yielded substantial numbers of luminous colonies, consistent with previous reports of the presence of luminous bacteria in the C. albatrossis light organ (47, 48). Dilution spread plating of homogenized light organs (specimens Calba.3, Calba.4, and Calba.5) gave rise to several dozen to several hundred luminous colonies per plate, indicating a population size in excess of 107 bacterial cells per light organ. The uniform appearance of the colonies on these primary isolation plates suggested the presence of a single bacterial species, and their small size, off-white color, lack of strong odor, and intense cerulean blue luminescence were similar to characteristics of strains identified previously as P. phosphoreum (e.g., NZ-11D).
16S rRNA and gyrB gene sequence analysis.
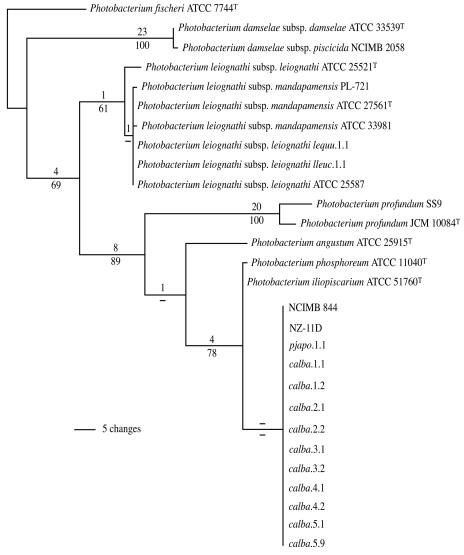
To genotypically identify the C. albatrossis symbionts, we first analyzed the sequence of the 16S rRNA gene. Two strains from each of the five fish specimens were examined, together with type strains and other well-characterized strains of Photobacterium species (Table 1). Consistent in general with reports that chlorophthalmids harbor P. phosphoreum (28, 56), this analysis placed the C. albatrossis strains in a phylogenetically coherent group that is closely affiliated with the P. phosphoreum/P. iliopiscarium species group (2) and distinct from other Photobacterium species (Fig. 1). The C. albatrossis strains were essentially indistinguishable from each other in 16S rRNA gene sequence, and they also were essentially indistinguishable from the NCIMB 844, NZ-11D, and pjapo.1.1 (Table 1), strains previously reported to be P. phosphoreum. Despite a general similarity, however, the C. albatrossis strains differed from the P. phosphoreum type strain, ATCC 11040T, and from the P. iliopiscarium type strain, ATCC 51760T, at several nucleotide positions. The difference in 16S rRNA gene sequence suggested that the C. albatrossis strains might represent a distinct bacterial lineage.
TABLE 1.
Bacterial strains used in this studya
| Species | Strain | Reference(s) |
|---|---|---|
| Photobacterium angustum | ATCC 25915T | 42 |
| Photobacterium damselae | ||
| subsp. damselae | ATCC 33539T | 13 |
| subsp. piscicida | NCIMB 2058 | 13 |
| Photobacterium fischeri | ATCC 7744T | 22 |
| Photobacterium iliopiscarium | ATCC 51760T | 38 |
| Photobacterium leiognathi | ||
| subsp. leiognathi | ATCC 25521T | 4 |
| ATCC 25587 | 4 | |
| lequu.1.1 | 2 | |
| llequc.1.1 | 2 | |
| subsp. mandapamensis | ATCC 27561T | 2, 42 |
| ATCC 33981 | 2, 42 | |
| PL-721 | 2, 34 | |
| Photobacterium “kishitanii clade” | calba.1.1 | This study |
| calba.1.2 | This study | |
| calba.1.3 | This study | |
| calba.1.4 | This study | |
| calba.1.6 | This study | |
| calba.1.9 | This study | |
| calba.2.1 | This study | |
| calba.2.2 | This study | |
| calba.3.1 | This study | |
| calba.3.2 | This study | |
| calba.3.3 | This study | |
| calba.3.4 | This study | |
| calba.3.5 | This study | |
| calba.3.6 | This study | |
| calba.3.7 | This study | |
| calba.3.8 | This study | |
| calba.3.9 | This study | |
| calba.3.10 | This study | |
| calba.3.12 | This study | |
| calba.3.17 | This study | |
| calba.3.19 | This study | |
| calba.3.21 | This study | |
| calba.3.25 | This study | |
| calba.3.26 | This study | |
| calba.3.27 | This study | |
| calba.3.28 | This study | |
| calba.4.1 | This study | |
| calba.4.2 | This study | |
| calba.4.3 | This study | |
| calba.4.4 | This study | |
| calba.4.5 | This study | |
| calba.4.6 | This study | |
| calba.4.12 | This study | |
| calba.4.14 | This study | |
| calba.4.15 | This study | |
| calba.4.22 | This study | |
| calba.4.27 | This study | |
| calba.5.1 | This study | |
| calba.5.2 | This study | |
| calba.5.3 | This study | |
| calba.5.5 | This study | |
| calba.5.9 | This study | |
| calba.5.10 | This study | |
| calba.5.14 | This study | |
| calba.5.19 | This study | |
| calba.5.21 | This study | |
| NCIMB 844b | 2 | |
| NZ-11Db | 44 | |
| pjapo.1.1b | 2 | |
| Photobacterium phosphoreum | ATCC 11040Tb | 22 |
| Photobacterium profundum | JCM 10084T | 37 |
| SS9 | 3 |
For sources of strains other than from this study, see the work of Ast and Dunlap (2) and information below.
NCIMB 844 was isolated in 1958 from the skin of the hake, Merluccius capensis (Gadiformes: Merlucciidae), from the area of Cape Town, South Africa (D. L. Georgala, personal communication); NZ-11D was isolated in 1976 from the light organ of N. aequalis (Gadiformes: Macrouridae) from the Atlantic Ocean off West Africa (44); pjapo.1.1 was isolated in 1982 from the light organ of P. japonicus (Gadiformes: Moridae) from the area of Manazuru, Kanagawa Prefecture, Japan (2); and ATCC 11040T was isolated by J. C. Hoogerheide in 1934 from the surface of a fish from the North Sea (M. Figge and L. Robertson, personal communication).
FIG. 1.
Phylogram representing 1 of 2,948 equally most parsimonious hypotheses resulting from analysis of 16S rRNA gene sequences of Photobacterium strains (111 informative characters; length = 291; consistency index [CI] = 0.790; retention index [RI] = 0.895). A strict consensus of these hypotheses results in unresolved polytomies in the P. leiognathi clade and the clade comprising P. iliopiscarium, P. phosphoreum, and the C. albatrossis symbionts. Numbers above branches are Bremer support, and numbers below the branches are jackknife resampling values; dashes represent values less than 50%. For strain and host species information, see Table 1.
Most of the nucleotide differences occurred in a region containing sequence ambiguity among several of the strains, from nucleotides 70 to 104 (Escherichia coli reference numbering) (Table 2), which form a stem-loop. Sequence chromatograms exhibited secondary nucleotide peaks in this region. The complementarity of the ambiguous nucleotides along the stem structure suggests that the sequence ambiguity is not a PCR or sequencing artifact, and reamplification and resequencing did not eliminate these peaks. The ambiguity therefore apparently results from the presence of multiple copies of the rrn operon in some strains, as seen in several other bacteria (1, 27), the sequences of which differ in this stem-loop region.
TABLE 2.
Comparison of a portion of the 16S rRNA gene sequence of Photobacterium strains
| Strain | 16S rRNA gene sequencea |
|---|---|
| ATCC 51760T | ...TAACAG ATTGATAGC TT GCTATCAAT GCTGACGAGCG... |
| ATCC 11040T | ...TAACAG AWWGAWAGC TT GCTWTCWWT GCTGACGAGCG... |
| NCIMB 844 | ...TAACAG AAARRAAGC TT GCTTYYTTT GCTGACGAGCG... |
| pjapo.1.1 | ...TAACAG AWWRRWAGC TT GCTWYYWWT GCTGACGAGCG... |
| NZ-11D | ...TAACAG AAAAGAAGC TT GCTTCTTTT GCTGACGAGCG... |
| calba.1.1b | ...TAACAG AAAAGAAGC TT GCTTCTTTT GCTGACGAGCG... |
| calba.1.2 | ...TAACAG AWWRRWAGC TT GCTWYYWWT GCTGACGAGCG... |
| calba.2.1 | ...TAACAG AWWRRWAGC TT GCTWYYWWT GCTGACGAGCG... |
| E. coli K-12 | ...TAACAG -GAAGAAGC TT GCTTCTTT- GCTGACGAGTG... |
Nucleotide positions 70 to 104 of the E. coli reference numbering system. Underlining in the ATCC 51760T sequence indicates the region participating in stem formation, and dashes in the E. coli K-12 sequence indicate gaps introduced to allow alignment with the Photobacterium sequences. W = A or T, R = A or G, and Y = C or T.
Sequences for calba.2.2, calba.3.1, calba.3.2, calba.4.1, calba.4.2, calba.5.1, and calba.5.9 were identical in this region to the sequence of calba.1.1.
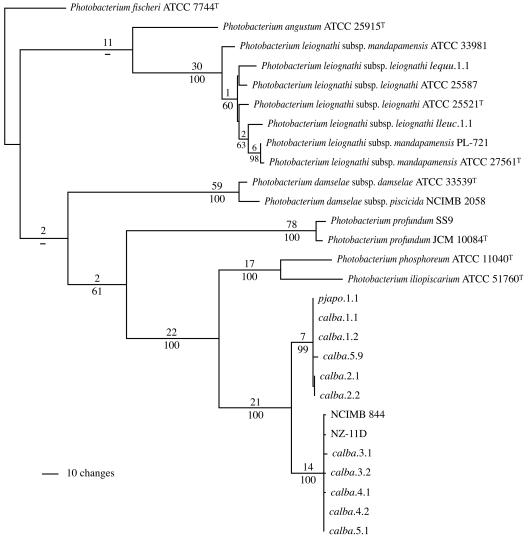
To test the phylogenetic significance of the difference in 16S rRNA gene sequence, we analyzed the gyrB gene of these strains. Sequence analysis of gyrB has proven effective for phylogenetic analysis of other groups of closely related bacteria, providing greater resolution than the 16S rRNA gene (e.g., see references 59 and 60). Analysis of gyrB resolved the C. albatrossis strains as a robustly supported clade distinct from P. phosphoreum ATCC 11040T and P. iliopiscarium ATCC 51760T (Fig. 2). The resolution provided by gyrB was substantially greater than that provided by the 16S rRNA gene. Furthermore, NCIMB 844, NZ-11D, and pjapo.1.1 again grouped with the C. albatrossis strains, not with the P. phosphoreum type strain. The Bremer support and jackknife resampling values indicate strong support for this separation. These results indicate that the C. albatrossis strains, together with NCIMB 844, NZ-11D, and pjapo.1.1, form a clade that is distinct from the type strains of P. phosphoreum and P. iliopiscarium.
FIG. 2.
Phylogram representing one of 24 equally most parsimonious hypotheses resulting from analysis of the gyrB gene sequences of Photobacterium strains shown in Fig. 1 (387 informative characters; length = 896; CI = 0.615; RI = 0.855). A strict consensus of these hypotheses results in an unresolved polytomy in the P. leiognathi clade but retains the distinction among P. iliopiscarium, P. phosphoreum, and the two subgroups of C. albatrossis symbiont strains. Numbers and dashes above and below the branches and abbreviations are as in Fig. 1.
Genomotypic diversity of C. albatrossis symbionts.
To test this conclusion, we next analyzed several additional strains from the C. albatrossis symbiont populations. The possibility existed that the 10 strains chosen for 16S rRNA and gyrB gene sequencing happened to represent one extreme of the phylogenetic variation within the species P. phosphoreum. To consider this possibility, we used genomic profiling by rep-PCR, an effective method for detecting genetic variation among bacteria (55), to identify genomotypically different strains for phylogenetic analysis. Nine strains from Calba.1, 10 strains from Calba.2, and 30 strains each from specimens Calba.3, Calba.4, and Calba.5 were profiled.
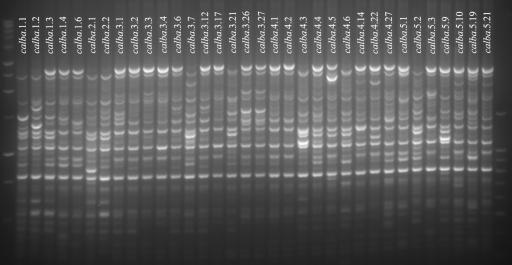
Genomic profiling revealed the presence of two or three numerically dominant strain types for each fish specimen, consistent with an oligoclonal structure for symbiont populations (10). The genomic profiles of the different strains from individual fish specimens and across specimens, however, appeared generally similar (data not shown), which made distinguishing strains from different fish specimens in gel-to-gel comparisons difficult. To circumvent this problem, we chose the strains from each fish that appeared to differ in genomic profile and reexamined them together (Fig. 3). This interspecimen analysis affirmed the general similarity of the genomic profiles of these strains; some bands were seen to be in common to all the strains, and several bands were shared among many of the strains. Through this analysis, several genomotypically different strains were identified.
FIG. 3.
Rep-PCR genomic profiling of C. albatrossis strains. Strains that appeared different on individual rep-PCR gels for each of the five specimens of C. albatrossis were reanalyzed together here. Flanking unlabeled lanes are 1-kb and 100-bp DNA size standard ladders.
We also assessed the genomotypic similarity of the C. albatrossis strains to P. phosphoreum and P. iliopiscarium by profiling with rep-PCR the genomes of ATCC 11040T, ATCC 51760T, and other strains. Several genomotypically different C. albatrossis strains from the interspecimen analysis above were used in this comparison to provide a wide representation of the genomic diversity of these bacteria. The profiles of ATCC 11040T and ATCC 51760T showed no obvious similarity to those of the C. albatrossis strains or to each other (Fig. 4). In contrast, the profiles of NCIMB 844, NZ-11D, and pjapo.1.1 appeared generally similar to those of the C. albatrossis strains. These results provide an additional indication of the similarity among the C. albatrossis strains and NCIMB 844, NZ-11D, and pjapo.1.1 and of the overall dissimilarity of these strains to ATCC 11040T and ATCC 51760T.
FIG. 4.
Rep-PCR genomic profile comparison of Photobacterium strains. Representative genomotypically different strains from Fig. 3 were examined here in comparison with ATCC 11040T, NCIMB 844, NZ-11D, pjapo.1.1, and ATCC 51760T. The two unlabeled lanes at the left are 1-kb and 100-bp DNA size standard ladders.
Phylogenetic analysis based on luxABFE.
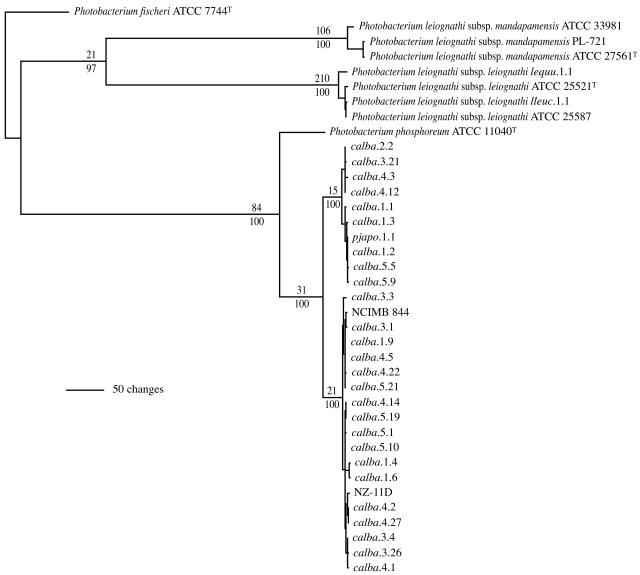
The identification of genomotypically different strains allowed a more comprehensive phylogenetic analysis of the C. albatrossis bacteria. We sought to identify strains that clustered phylogenetically with ATCC 11040T, if present, and to test the phylogenetic validity of the distinction between C. albatrossis strains and ATCC 11040T revealed by gyrB and to a lesser extent by the 16S rRNA gene. For this purpose, the luxABFE locus was examined. Sequence analysis of the four lux genes, together with the luxAB, luxBF, and luxFE intergenic spacer regions, provides robust inter- and intraspecific phylogenetic resolution in luminous bacteria (2). ATCC 51760T, because it apparently is a nonluminous species, was not included in this analysis.
Analysis of luxABFE resolved the C. albatrossis strains in a manner very similar to that of gyrB. None of the strains grouped with ATCC 11040T. All the C. albatrossis strains, together with NCIMB 844, NZ-11D, and pjapo.1.1, clustered together to form a strongly supported clade that was distinct from P. phosphoreum ATCC 11040T (Fig. 5). Furthermore, as seen for gyrB, the C. albatrossis strains, together with NCIMB 844, NZ-11D, and pjapo.1.1, formed two closely related but distinguishable subgroups, and subgroup membership of strains was the same for gyrB and lux. Combined analysis of the 16S rRNA, gyrB, and luxABFE genes resulted in essentially the same phylogenetic hypothesis as either gyrB or luxABFE alone (data not shown). These results demonstrate that the symbionts of C. albatrossis, together with certain strains of luminous bacteria previously thought to be members of P. phosphoreum, form a clade that is closely related to but evolutionarily distinct from the P. phosphoreum type strain.
FIG. 5.
Phylogram representing 1 of 18 equally most parsimonious hypotheses resulting from analysis of luxAB(F)E gene sequences of Photobacterium strains (946 informative characters; length = 1,444; CI = 0.866; RI = 0.951). The 18 hypotheses differ only in the resolution within P. leiognathi subsp. leiognathi, and within (but not between) the two subgroups comprising the C. albatrossis strains. The analysis includes C. albatrossis strains from Fig. 1 and 2 and additional strains identified by genomic profiling as genomotypically distinct. Excluded from the analysis, for computational simplicity and visual clarity, were the several strains found to be identical in luxABFE sequence. The strain(s) with luxABFE sequence identical to those included in the analysis were as follows: calba.1.2 = (same sequence as) calba.2.1 and calba.4.4; calba.1.9 = calba.3.2, calba.3.6, calba.3.10, calba.3.12, calba.3.19 and calba.3.25; calba.2.2 = calba.3.7, calba.3.8, calba.4.6, calba.4.12 and calba.4.15; calba.3.3 = calba.3.9; calba.3.4 = calba.3.5 and calba.3.17; calba.3.26 = calba.3.27 and calba.3.28; calba.4.14 = calba.5.3; and calba.5.5 = calba.5.14. Numbers above and below the branches and abbreviations are as in Fig. 1.
DISCUSSION
The results of this study demonstrate that the bioluminescent bacteria colonizing the light organ of the bentho-pelagic fish C. albatrossis (Aulopiformes: Chlorophthalmidae) represent a phylogenetic lineage distinct from that of P. phosphoreum. Previously, luminous bacteria symbiotic with chlorophthalmids were thought to be members of P. phosphoreum. This species identification, however, was based on general cellular characteristics, inference from other deep-sea symbiotic associations, and unreported taxonomic data (28, 47, 48, 56), leaving incompletely defined the species identity and phylogenetic affinity of these bacteria. Here, we used sequence analysis of the 16S rRNA, gyrB, and luxABFE genes to reveal that despite phenotypic similarities, especially with P. phosphoreum, the C. albatrossis strains, together with NCIMB 844, NZ-11D, and pjapo.1.1, form a clade that is distinct from P. phosphoreum and P. iliopiscarium. We designate this clade the kishitanii clade in recognition of the Japanese scientist Teijiro Kishitani, who first isolated luminous bacteria from the light organ of Physiculus japonicus (Gadiformes: Moridae) (26). The deep phylogenetic division separating this clade from P. phosphoreum and P. iliopiscarium, which are well-characterized species, indicates that separate species status may be warranted for the kishitanii clade. Linked to the issue of species status is the question of whether the luminous bacteria symbiotic with several other deep-sea fishes, also in some cases identified phenotypically as P. phosphoreum (9, 17, 19), all might belong to the kishitanii clade.
The few differences in 16S rRNA gene sequence among P. phosphoreum, P. iliopiscarium, and members of the kishitanii clade might be considered a reason for placing these bacteria within a single species. However, the ability of 16S rRNA gene sequence data to resolve closely related bacteria is limited (12, 43, 53), and despite the similarity in 16S rRNA gene sequence, ATCC 11040T and ATCC 51760T represent separate species, differing in a number of attributes (22, 38, 41, 54). We demonstrate here through analysis of gyrB the validity of this species-level separation, and we extend it to a third member of the P. phosphoreum/P. iliopiscarium species group (2), the kishitanii clade. The divergence in gyrB provides good phylogenetic resolution of the members of this group, as it does for certain other bacteria (59, 60). In contrast to these findings, however, we noted recently that gyrB diverged insufficiently to resolve two other closely related luminous bacteria, the leiognathi and mandapamensis clades of Photobacterium leiognathi (2). Thus, the extent of divergence of a given genetic locus, and therefore its value for definitive phylogenetic analysis, varies among species. A more robust test of phylogenetic relationship can be gained through examination of multiple genetic loci (39, 50, 51) compared to any single locus, and for that reason we have also examined here luxABFE, a rapidly diverging locus that provides excellent inter- and intraspecific resolution in luminous bacteria (2). Analysis of luxABFE sequence divergence between ATCC 11040T and members of the kishitanii clade affirms the results obtained with gyrB.
Methods considered standard for bacterial identification and species description, i.e., chemotaxonomic traits, mol% G+C values, DNA reassociation values, and sequence analysis of the 16S rRNA gene, apparently are not entirely adequate for detecting evolutionarily significant differences among luminous bacteria. As shown here and previously (2), strains of Photobacterium that are difficult to distinguish phenotypically nonetheless are revealed as representing evolutionarily distinct lineages when examined genotypically. It follows that additional clades are likely to exist among currently recognized species and species groups in Photobacterium. Attempts to identify additional clades will benefit from examination of large numbers of strains, to define the extent of sequence divergence within and between clades necessary for evolutionarily valid distinctions to be made. Equally important will be analysis of multiple genetic loci, to assess the robustness of phylogenetic inference based on individual loci, and examination of rapidly diverging loci, to trace the evolutionary history of a clade and its close relatives. Previous studies have noted differences between the P. phosphoreum type strain, ATCC 11040T, and other strains identified phenotypically as members of this species (6, 22, 41, 45), possibly presaging the genotypic results presented here. More recently, a study of strains of luminous bacteria from the skin of migrating salmon revealed a possible parallel to results presented here; analysis of the luxA gene sequence showed an apparent phylogenetic distinction between NCIMB 844 and the strains from salmon skin, which were identified phenotypically as P. phosphoreum, whereas analysis of the 16S rRNA gene did not distinguish those strains from ATCC 11040T (5). With respect to the possibility of ecological specificity for different species of luminous bacteria, PCR-based methods for identification and ecological analysis (2, 35, 57) may find increasing application as details of sequence divergence among discrete lineages become better defined, allowing lineage-specific precision in the design of probes and primers.
Genomic profiling of strains with rep-PCR allowed a rapid assessment of the genomotypic diversity among the bacteria colonizing the C. albatrossis light organ and provided a basis for choosing different strains for detailed phylogenetic analysis. The profiling results indicate substantial similarity among strains, both within and between the bacterial populations present in different specimens of the fish. The C. albatrossis specimens examined here were of two different size classes, indicating different ages, caught on two different days. They were, however, from the same location, so it is possible that they acquired their symbiont strains from the same regional oceanic population of genomotypically similar strains. However, the general similarity of genomic profiles of NCIMB 844, NZ-11D, and pjapo.1.1 to the C. albatrossis strains is intriguing in this regard because the fish from which these three strains were isolated were each from a different family (belonging to a different order) and were each caught in different locations at different times (Table 1). What these fishes and C. albatrossis have in common is the deep-sea habitat. Genomic similarity therefore might be typical of luminous bacteria from deep-sea fishes. In support of this notion, 16S rRNA gene restriction fragment length polymorphism analysis and rep-PCR genomic profiling of strains from bioluminescent symbiosis with various deep-sea fish species reveal an apparently limited diversity (58; P. V. Dunlap, M. M. Pearce, and J. C. Ast, unpublished data). This limited diversity contrasts with the extensive genomotypic diversity identified for P. leiognathi from leiognathids, a shallow-dwelling, coastal fish (10). It is possible that environmental differences between the deep-sea and shallow, coastal habitats (e.g., differences in temperature, nutrients, irradiation, environmental stability, etc.) play a role in this apparent contrast in genomic diversity. Consistent with this possibility, we find a high degree of genomotypic diversity in Photobacterium fischeri, a luminous bacterium whose symbiotic host animals, monocentrid fishes and sepiolid squids, dwell in shallow, coastal waters (P. V. Dunlap, J. C. Ast, and M. M. Pearce, unpublished data).
Genomic profiling also provided a general indication of genetic differences between members of the kishitanii clade and ATCC 11040T and ATCC 51760T. These differences appear to be consistent with the results of the sequence-based phylogenetic analysis. Beyond these general indications, however, use of genomic profiling to assess the extent of similarity among these bacteria may be problematic. Numerical analyses of differences between strains based on DNA banding patterns would be difficult to interpret phylogenetically regardless of the analytical method used, due to the unsubstantiated assumption of homology among DNA bands of the same size and the lack of information provided by the limited number of bands typically resolved. We therefore have restricted our use of genomic profiling to the identification of genomotypically distinct strains for nucleotide sequence-based phylogenetic analyses.
The data presented here do not exclude the possibility that some chlorophthalmid fishes, either other specimens of C. albatrossis or other chlorophthalmid species, harbor symbiotic bacteria that are members of P. phosphoreum. However, the large number of strains examined and their isolation from several specimens of C. albatrossis suggest that this possibility is unlikely. Furthermore, the specificity that generally characterizes bioluminescent symbiosis, with the members of a given family of fishes or squids usually harboring a single kind of luminous bacteria as their light organ symbiotic bacteria (11, 17), indicates that all chlorophthalmids might harbor only members of the kishitanii clade. It is important to note in this regard that certain strains of bacteria previously thought to be P. phosphoreum cluster phylogenetically with the C. albatrossis strains. Two of these other strains, NZ-11D and pjapo.1.1, are from symbiosis with members of other families of deep-sea fishes, Nezumia aequalis (Macrouridae) (44) and P. japonicus (Moridae) (2), respectively. A third strain, Og61, from Opisthoproctus grimaldii (Opisthoproctidae), is identical in 16S rRNA gene sequence to NZ-11D (21). These observations suggest that bacteria symbiotic with deep-sea fishes of the families Chlorophthalmidae (Aulopiformes), Opisthoproctidae (Osmeriformes), Macrouridae, Steindachneriidae and Moridae (Gadiformes), and Trachichthyidae (Beryciformes) all might be members of the kishitanii clade. It follows that P. phosphoreum might not occur as the light organ symbiont of deep-sea fishes.
The kishitanii clade may warrant separate species status. P. phosphoreum and P. iliopiscarium are established species, which the gyrB results presented here support, and ATCC 11040T and ATCC 51760T are more closely related to each other than either is to any member of the kishitanii clade. Furthermore, the kishitanii clade is cleanly resolved as a coherent, well-sampled lineage separated from P. phosphoreum and P. iliopiscarium by a large number of nucleotide changes. To test separate species status for the kishitanii clade, a better sampling of strains of P. phosphoreum and P. iliopiscarium is needed. At present, ATCC 11040T stands alone as the sole known representative of the P. phosphoreum lineage, and very few strains of P. iliopiscarium have been described. Certainly, species-level distinctions based on one or a few strains can be problematic. The possibility exists, for example, that the discovery of additional strains of these species would lead to identification of phylogenetically intermediate strains, a continuum of variation that would blur the apparent distinctions between these three lineages. This scenario would indicate the need for a broader species definition of P. phosphoreum or possibly of P. iliopiscarium and inclusion of the members of the kishitanii clade in that species. Alternatively, the discovery of strains that form coherent clusters with ATCC 11040T and ATCC 51760T would affirm the validity of these two species and establish the species-level separateness of the kishitanii clade.
Acknowledgments
We thank Ariyasu Makihara, captain of the fishing vessel Kotobuki-maru, for providing the specimens of C. albatrossis used in this study. Akihito Iwata, Mie Prefecture Office, Owase, Japan, and Seishi Kimura, Fisheries Research Laboratory, Mie University, Shima, Japan, kindly facilitated the acquisition of these specimens. Information on the ecological origin of NCIMB 844 was provided by Doug Georgala. Information on the ecological origin of ATCC 11040T was provided by Marian Figge, Netherlands Culture Collection of Bacteria, and Lesley Robertson, Kluyver Laboratory for Biotechnology, Delft University of Technology. DNA sequencing was carried out by staff of the University of Michigan Sequencing Core.
Support was provided by NSF grant DEB 0413441.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ast, J. C., and P. V. Dunlap. 2004. Phylogenetic analysis of the lux operon distinguishes two evolutionarily distinct clades of Photobacterium leiognathi. Arch. Microbiol. 181:352-361. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, D. H., M. Wright, A. A. Yayanos, and M. Silverman. 1989. Isolation of a gene regulated by hydrostatic pressure in a deep-sea bacterium. Nature 342:572-574. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert, H., R. Chatelain, and J. M. Bassot. 1967. Étude d'un Photobacterium isolé de l'organe lumineux des poissons Leiognathidae. Ann. Inst. Pasteur (Paris) 112:520-524. [PubMed] [Google Scholar]
- 5.Budsberg, K. J., C. F. Wimpee, and J. F. Braddock. 2003. Isolation and identification of Photobacterium phosphoreum from an unexpected niche: migrating salmon. Appl. Environ. Microbiol. 69:6938-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalgaard, P., O. Mejlholm, and H. H. Huss. 1996. Conductance method for quantitative determination of Photobacterium phosphoreum in fish products. J. Appl. Bacteriol. 81:57-64. [Google Scholar]
- 7.Di Meo, C. A., A. E. Wilbur, W. E. Holben, R. A. Feldman, R. C. Vrijenhoek, and S. C. Cary. 2000. Genetic variation among endosymbionts of widely distributed vestimentiferan tubeworms. Appl. Environ. Microbiol. 66:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlap, P. V. 1985. Physiological and morphological state of the symbiotic bacteria from light organs of ponyfish. Biol. Bull. 167:410-425. [DOI] [PubMed] [Google Scholar]
- 9.Dunlap, P. V., and K. Kita-Tsukamoto. 2001. Luminous bacteria, chapter 328. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The Prokaryotes, an evolving electronic resource for the microbiological community. Academic Press, New York, N.Y.
- 10.Dunlap, P. V., A. Jiemjit, J. C. Ast, M. M. Pearce, R. R. Marques, and C. R. Lavilla-Pitogo. 2004. Genomic polymorphism in symbiotic populations of Photobacterium leiognathi. Environ. Microbiol. 6:145-158. [DOI] [PubMed] [Google Scholar]
- 11.Fidopiastis, P. M., S. von Boletzky, and E. G. Ruby. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Sys. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 13.Gauthier, G., B. Lafay, R. Ruimy, V. Breittmayer, J. L. Nicolas, M. Gauthier, and R. Christen. 1995. Small-subunit rRNA sequences and whole DNA relatedness concur for the reassignment of Pasteurella piscicida (Snieszko et al.) Janssen and Surgalla to the genus Photobacterium as Photobacterium damsela subsp. piscicida comb. nov. Int. J. Syst. Bacteriol. 45:139-144. [DOI] [PubMed] [Google Scholar]
- 14.Haneda, Y. 1951. The luminescence of some deep-sea fishes of the families Gadidae and Macrouridae. Pac. Sci. 5:372-378. [Google Scholar]
- 15.Haneda, Y. 1957. Observations on luminescence in the deep sea fish, Paratrachichthys prosthemius. Sci. Rep. Yokosuka City Mus. 2:15-23. [Google Scholar]
- 16.Haneda, Y., and S. Yoshiba. 1970. On a luminous substance of the anacanthine fish Steindachneria argentea from the Gulf of Mexico. Sci. Rep. Yokosuka City Mus. 16:1-4. [Google Scholar]
- 17.Hastings J. W., and K. H. Nealson. 1981. The symbiotic luminous bacteria, p. 1332-1345. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The Prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Springer-Verlag, Berlin, Germany.
- 18.Haygood, M. G. 1990. Relationship of the luminous bacterial symbiont of the Caribbean flashlight fish, Kryptophanaron alfredi (family Anomalopidae) to other luminous bacteria based on bacterial luciferase (luxA) genes. Arch. Microbiol. 154:496-503. [DOI] [PubMed] [Google Scholar]
- 19.Haygood, M. G. 1993. Light organ symbioses in fishes. Crit. Rev. Microbiol. 19:191-216. [DOI] [PubMed] [Google Scholar]
- 20.Haygood, M. G., and D. L. Distel. 1993. Bioluminescent symbionts of flashlight fishes and deep-sea anglerfishes form unique lineages related to the genus Vibrio. Nature 363:154-156. [DOI] [PubMed] [Google Scholar]
- 21.Haygood, M. G., D. L. Distel, and P. J. Herring. 1992. Polymerase chain reaction and 16S rRNA gene sequences from the luminous bacterial symbionts of two deep-sea anglerfishes. J. Mar. Biol. Assoc. U K 71:149-159. [Google Scholar]
- 22.Hendrie, M. S., W. Hodgkiss, and J. M. Shewan. 1970. The identification, taxonomy and classification of luminous bacteria. J. Gen. Microbiol. 64:151-169. [Google Scholar]
- 23.Herring, P. J. 1975. Bacterial bioluminescence in some argentinoid fishes, p. 563-572. In H. Barnes (ed.), Proceedings of the 9th European Marine Biology Symposium. Aberdeen University Press, Aberdeen, United Kingdom.
- 24.Herring, P. J. 1982. Aspects of the bioluminescence of fishes. Oceanogr. Mar. Biol. Annu. Rev. 20:415-470. [Google Scholar]
- 25.Herring, P. J., and J. G. Morin. 1978. Bioluminescence in fishes, p. 273-329. In P. J. Herring (ed.), Bioluminescence in action. Academic Press, London, United Kingdom.
- 26.Kishitani, T. 1930. Studien über die Leuchtsymbiose in Physiculus japonicus Hilgendorf, mit der Beilage der zwei neuen arten der Leuchtbacterien. Sci. Rep. Tohoku Univ. 5:801-823. [Google Scholar]
- 27.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwae, T., S. Fukasawa, T. Sasaki, and M. Kurata. 1982. Immunochemical comparisons among lipopolysaccharides from symbiotic luminous bacteria isolated from several luminous marine animals. Microbiol. Immunol. 26:1181-1186. [DOI] [PubMed] [Google Scholar]
- 29.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley & Sons, New York, N.Y.
- 30.Makemson, J. C., N. R. Fulayfil, W. Landry, L. M. Van Ert, C. F. Wimpee, E. A. Widder, and J. F. Case. 1997. Shewanella woodyi sp. nov., an exclusively respiratory luminous bacterium isolated from the Alboran Sea. Int. J. Syst. Bacteriol. 47:1034-1039. [DOI] [PubMed] [Google Scholar]
- 31.Meighen, E. A., and P. V. Dunlap. 1993. Physiological, biochemical and genetic control of bacterial bioluminescence. Adv. Microb. Physiol. 34:1-67. [DOI] [PubMed] [Google Scholar]
- 32.Nakabo, T. 2002. Family 111. Chlorophthalmidae, p. 362-363. In T. Nakabo (ed.), Fishes of Japan with pictoral keys to the species, English ed., vol. 1 and 2. Tokai University Press, Tokyo, Japan.
- 33.Nealson, K. H. 1978. Isolation, identification and manipulation of luminous bacteria. Methods Enzymol. 57:153-166. [Google Scholar]
- 34.Nealson, K. H., and J. W. Hastings. 1977. Low oxygen is optimal for luciferase synthesis in some bacteria. Arch. Microbiol. 112:9-16. [DOI] [PubMed] [Google Scholar]
- 35.Nealson, K. H., B. Wimpee, and C. F. Wimpee. 1993. Identification of Vibrio splendidus as a member of the planktonic luminous bacteria from the Persian Gulf and Kuwait region with luxA probes. Appl. Environ. Microbiol. 59:2684-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson, J. S. 1994. Fishes of the world, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 37.Nogi, Y., N. Masui, and C. Kato. 1998. Photobacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 2:1-7. [DOI] [PubMed] [Google Scholar]
- 38.Onarheim, A. M., R. Wiik, J. Burghardt, and E. Stackebrandt. 1994. Characterization and identification of two Vibrio species indigenous to the intestine of fish in cold sea water; description of Vibrio iliopiscarius sp. nov. Syst. Appl. Microbiol. 17:370-379. [Google Scholar]
- 39.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 40.Rademaker, J. L. W., and F. J. de Bruijn. 1997. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer assisted pattern analysis, p. 151-171. In G. Caetano-Anolles and P. M. Gresshoff (ed.), DNA markers: protocols, applications, and overviews. Wiley, New York, N.Y.
- 41.Reichelt, J. L., and P. Baumann. 1973. Taxonomy of the marine, luminous bacteria. Arch. Mikrobiol. 94:283-330. [DOI] [PubMed] [Google Scholar]
- 42.Reichelt, J. L., P. Baumann, and L. Baumann. 1976. Study of genetic relationships among marine species of the genera Beneckea and Photobacterium by means of in vitro DNA/DNA hybridization. Arch. Microbiol. 110:101-120. [DOI] [PubMed] [Google Scholar]
- 43.Roselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 44.Ruby, E. G., and J. G. Morin. 1978. Specificity of symbiosis between deep-sea fish and psychrotrophic luminous bacteria. Deep-Sea Res. 25:161-171. [Google Scholar]
- 45.Singleton, R. J., and T. M. Skerman. 1973. A taxonomic study by computer analysis of marine bacteria from New Zealand waters. J. R. Soc. New Zealand 3:129-140. [Google Scholar]
- 46.Soly, R. R., J. A. Mancini, S. R. Ferri, M. Boylan, and E. A. Meighen. 1988. A new lux gene in bioluminescent bacteria codes for a protein homologous to the bacterial luciferase subunits. Biochem. Biophys. Res. Commun. 155:351-358. [DOI] [PubMed] [Google Scholar]
- 47.Somiya, H. 1977. Bacterial bioluminescence in chlorophthalmid deep-sea fish: a possible interrelationship between the light organ and the eyes. Experientia 33:906-908. [Google Scholar]
- 48.Somiya, H. 1981. On the bacterial-associated light organ in Chlorophthalmus, p. 561-567. In M. A. DeLuca and W. D. McElroy (ed.), Bioluminescence and chemiluminescence basic chemistry and analytical applications, Academic Press, New York, N.Y.
- 49.Sorenson, M. D. 1999. TreeRot, version 2. Boston University, Boston, Mass.
- 50.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kämpfer, M. C. J. Maiden, X. Nesme, R. Rosselló-Mora, J. Swings, H. G. Trüper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 51.Staley, J. T., and J. J. Gosink. 1999. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53:189-215. [DOI] [PubMed] [Google Scholar]
- 52.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0B10. Sinauer Associates, Sunderland, Mass.
- 53.Thompson, C. C., F. L. Thompson, K. Vandemeulebroecke, B. Hoste, P. Dawyndt, and J. Swings. 2004. Use of recA as an alternative phylogenetic marker in the family Vibrionaceae. Int. J. Syst. Evol. Microbiol. 54:919-924. [DOI] [PubMed] [Google Scholar]
- 54.Urakawa, H., K. Kita-Tsukamoto, and K. Ohwada. 1999. Reassessment of the taxonomic position of Vibrio iliopiscarius (Onarheim et al. 1994) and proposal for Photobacterium iliopiscarium comb. nov. Int. J. Syst. Bacteriol. 49:257-260. [DOI] [PubMed] [Google Scholar]
- 55.Versalovic, J., J. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 56.Vydryakova, G. A., A. M. Kuznetsov, G. A. Primakova, Y. V. Chugaeva, and A. M. Fish. 1995. Luminescent bacterial symbionts and commensals of luminescent and nonluminescent animals of the Indian Ocean. Microbiology 64:589-592. [Google Scholar]
- 57.Wimpee, C. F., T.-L. Nadeau, and K. H. Nealson. 1991. Development of species-specific hybridization probes for marine luminous bacteria using in vitro DNA amplification. Appl. Environ. Microbiol. 57:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfe, C. J., and M. G. Haygood. 1991. Restriction fragment length polymorphism analysis reveals high levels of genetic divergence among the light organ symbionts of flashlight fish. Biol. Bull. 181:135-143. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto, S., H. Kasai, D. L. Arnold, R. W. Jackson, A. Vivian, and S. Harayama. 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotides sequences of gyrB and rpoD genes. Microbiology 146:2385-2395. [DOI] [PubMed] [Google Scholar]
- 60.Yáñez, M. A., V. Catalán, D. Apráiz, M. J. Figueras, and A. J. Martínez-Murcia. 2003. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 53:875-883. [DOI] [PubMed] [Google Scholar]
- 61.Yasaki, Y., and Y. Haneda. 1935. On the luminescence of the deep-sea fishes, family Macrouridae. Oyo-Dobustugaku Zasshi (Jpn. J. Appl. Zool.). 7:165-177. [Google Scholar]