Abstract
A total of 247 clones of 16S rRNA genes from microorganisms captured by 0.2- and 0.1-μm-pore-size filters from sedimentary and granite rock aquifers were amplified and yielded 37 operational taxonomic units (OTUs). Fifteen OTUs captured by 0.1-μm-pore-size filters were affiliated with the candidate divisions OD1 and OP11, representing novel lineages. On the other hand, OTUs captured by 0.2-μm-pore-size filters were largely affiliated with Betaproteobacteria.
The existence of microorganisms passing through 0.2-μm-pore-size filters has been questioned based on the minimum requisite size for a complete set of genome, ribosomes, proteins, and other intracellular components (21). Geologically, 0.05- to 0.2-μm-diameter spheres on mineral surfaces are presumably involved in biomineralization (6, 7). Similarly, <0.2-μm-diameter spheres mineralized with apatite have been observed in medical samples and affiliated with Alphaproteobacteria (1, 4, 17). Certain aquatic bacteria are filterable by 0.2-μm-pore-size filters (5, 9, 10, 18, 24, 30, 31), and the archaeon Nanoarchaeum equitans is also smaller than 0.2 μm (13).
Molecular techniques phylogenetically characterize the microbial communities filterable by 0.2-μm-pore-size filters, and a Mediterranean community was thus affiliated with Alphaproteobacteria, Gammaproteobacteria, and Cytophaga-Flavobacterium-Bacterioides (11). On the other hand, few or no studies have focused on the microorganisms in deep groundwater that are filterable by 0.2-μm-pore-size filters. We have characterized the microorganisms captured by 0.2- and 0.1-μm-pore-size filters in deep groundwaters of the Tono uranium mine, Japan (35.4°N, 137.2°E), where total counts of cells captured by 0.2-μm-pore-size filters are in the range of 105 to 106 ml−1 (22, 23).
Groundwaters were collected from aquifers in sedimentary rock (161 to 163 m below ground level) and granite rock (177 to 227 m below ground level) through separate Teflon tubes after >1,000 passages of the aquifer waters. The temperature and pH of the anaerobic groundwaters are 19.0 to 20.1°C and 7.8 to 9.6, respectively (23). Sterivex filters (0.2-μm pore size; Millipore Corp., Bedford, Mass.) were connected to Teflon tubes to capture >0.2-μm-diameter cells in 10 liters (each) of groundwater. In parallel, 0.1-μm-pore-size Sterivex filters captured microorganisms filterable by 0.2-μm-pore-size filters in 100 liters (each) of the 0.2-μm filtrate.
Bulk DNA was extracted from the 0.2- and 0.1-μm-pore-size Sterivex filters washed with 1 ml of SET buffer (20% sucrose, 50 mM EDTA, and 50 mM Tris-HCl, pH 7.6) after cell lysis according to the method of Somerville et al. (28). The lysates were used for DNA preparation by phenol-chloroform extraction and isopropanol precipitation. The 0.2-μm-pore-size filter-captured cells in 10 liters of sedimentary and granite rock groundwater yielded 0.93 and 1.17 μg of DNA, respectively, while 0.1-μm-pore-size filter-captured cells in 100 liters of the 0.2-μm filtrate yielded 0.70 and 0.62 μg of DNA, respectively.
DNA preparations were used for PCR amplification of 16S rRNA genes using the Archaea-specific primers Arch21F (5′-TTCCGGTTGATCCYGCCGGA-3′) and Arch958R (5′-YCCGGCGTTGAMTCCAATT-3′) and the Bacteria universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (3). Bacterial (archaeal in parentheses) 16S rRNA gene sequences were amplified with ExTaq DNA polymerase (TaKaRa Bio Inc., Otsu, Japan) as follows: 1 cycle of 96°C for 3 min, 56°C for 25 s (53 to 56°C for 40 s), and 72°C for 15 s (40 s) and 27 cycles of 96°C for 30 s (40 s), 56°C for 25 s (53°C for 30 s), and 72°C for 15 s (40 s), followed by 72°C for 10 min. The PCR products were purified by agarose gel electrophoresis and cloned into pCR2.1-TOPO in the Escherichia coli TOP10 transformants (Invitrogen Corp., San Diego, Calif.).
The cloned inserts were grouped by restriction fragment length polymorphism (RFLP) to form operational taxonomic units (OTUs) using the type II restriction enzymes (subtype P) HaeIII and BglII simultaneously. After restriction digestion at 37°C for 4 h, the banding patterns were compared by electrophoresis with a resolution of 1 bp using an Agilent Technologies (Palo Alto, Calif.) 2100 Bioanalyzer, and clones showing the same RFLP pattern were grouped into a single OTU. Cloned inserts were sequenced using a multicapillary DNA sequencer (RISA-384; Shimadzu Corp., Kyoto, Japan) with a Dynamic ET Terminator Cycle Sequencing kit (Amersham Bioscience Corp., Piscataway, N.J.), applying the following primers: 27F, 357F (5′-TACGGGAGGCAGCAG-3′), 926F (5′-ACTCAAAGGAATTGACGG-3′), 1100R (5′-AGGGTTGCGCTCGTTG-3′), and 1492R.
The OTUs with chimeric 16S rRNA gene sequences were removed by the program Chimera Check at the Ribosomal Database Project II and Bellerophon server (14). The remaining sequences were further chimera checked by trisecting (nucleotide positions 28 to 500, 501 to 1000, and 1001 to 1491), with the sequences showing different trisect trees removed (15). The surviving OTUs were searched for homology using the program FASTA at the DNA Database Bank of Japan (DDBJ), aligned with ClustalW (29), and used to construct a phylogenetic tree by the neighbor-joining algorithm (27) with TreeView (25). The 16S rRNA gene sequences of the chimera-checked OTUs were registered in DDBJ as listed in Table 1.
TABLE 1.
Clone number distributions and phylogenetic affiliations of OTUs among the 0.2- and 0.1-μm-pore-size filter-captured microorganisms from sedimentary and granite rock aquifers in the Tono uranium mine
| Filter size (μm) | OTU KNA6- | No. of sediment clones | No. of granite clones | DDBJ accession no. | Putative affiliationa | Closest sequence (>95% similarity) | Similarity (%) | Closest organism (>95% similarity) | Similarity (%) |
|---|---|---|---|---|---|---|---|---|---|
| 0.1 | NB05 | 1 | 1 | AB179661 | Cand. div. OD1 | ||||
| NB07 | 1 | 2 | AB179663 | Cand. div. OD1 | |||||
| NB08 | 3 | AB179664 | Cand. div. OD1 | ||||||
| NB11 | 7 | AB179667 | Cand. div. OD1 | ||||||
| NB12 | 2 | AB179668 | Cand. div. OD1 | ||||||
| NB16 | 2 | AB179672 | Cand. div. OD1 | ||||||
| NB17 | 2 | AB179673 | Cand. div. OD1 | ||||||
| NB23 | 1 | AB179676 | Cand. div. OD1 | ||||||
| NB25 | 1 | AB179678 | Cand. div. OD1 | ||||||
| NB27 | 1 | AB179679 | Cand. div. OD1 | ||||||
| NB29 | 1 | AB179680 | Cand. div. OD1 | ||||||
| NB13 | 3 | AB179669 | Cand. div. OP11 | ||||||
| NB14 | 10 | AB179670 | Cand. div. OP11 | ||||||
| NB18 | 2 | AB179674 | Cand. div. OP11 | ||||||
| NB20 | 2 | AB179675 | Cand. div. OP11 | ||||||
| NB03 | 5 | 24 | AB179659 | β-Proteobacteria | Clone B35 (AY375072) | 98.3 | Aquabacterium parvum B6 (AF035052) | 99.1 | |
| NB06 | 2 | 2 | AB179662 | β-Proteobacteria | Clone [Thiobacillus] 44a-B2-21 (AY082471) | 98.7 | Thiobacillus denitrificans NCIMB | 97.5 | |
| NB15 | 1 | 4 | AB179671 | β-Proteobacteria | Clone G14 (AY345397) | 99.0 | Acidovorax sp. “smarlab 133815” (AY093698) | 98.6 | |
| NB24 | 1 | AB179677 | Firmicutes | Clone 2e (AJ223451) | 99.4 | Staphylococcus sp. strain LMG-19417 (AJ276810) | 99.7 | ||
| NB01 | 20 | 2 | AB179658 | Unidentified | |||||
| NB04 | 2 | AB179660 | Unidentified | ||||||
| NB09 | 13 | AB179665 | Unidentified | ||||||
| NB10 | 4 | AB179666 | Unidentified | ||||||
| Total | 32 | 90 | |||||||
| 0.2 | EB01 | 3 | AB179681 | Cand. div. OP3 | |||||
| EB02 | 32 | 47 | AB179682 | β-Proteobacteria | Clone Cart-N1 (AY118150) | 97.3 | |||
| EB03 | 9 | 2 | AB179683 | β-Proteobacteria | Clone [Thiobacillus] 44a-B2-21 (AY082471) | 98.4 | T. denitrificans NCIMB 9548 (AJ243144) | 97.3 | |
| EB04 | 14 | 1 | AB179684 | β-Proteobacteria | Clone GC24 (AF204243) | 95.5 | |||
| EB05 | 3 | 1 | AB179685 | β-Proteobacteria | Clone H20 (AF072920) | 95.5 | |||
| EB07 | 2 | AB179686 | β-Proteobacteria | Clone Cart-N1 (AY118150) | 97.1 | ||||
| EB12 | 1 | AB179689 | β-Proteobacteria | Clone 244ds10 (AY212692) | 96.3 | ||||
| EB08 | 3 | AB179687 | δ-Proteobacteria | Clone GuBH2-AD/TzT-67 (AJ519663) | 97.0 | ||||
| EB15 | 1 | AB179691 | δ-Proteobacteria | ||||||
| EB24 | 1 | AB179694 | δ-Proteobacteria | ||||||
| EB09 | 2 | AB179688 | Acidobacteria | Clone SHA-18 (AJ249099) | 96.0 | ||||
| EB19 | 1 | AB179692 | Acidobacteria | Clone SJA-149 (AJ009495) | 97.2 | ||||
| EB22 | 1 | AB179693 | Green nonsulfur | ||||||
| EB14 | 1 | AB179690 | Nitrospira | ||||||
| Total | 64 | 61 |
Cand. div., candidate division.
This study yielded 247 chimera-checked clones of bacterial 16S rRNA genes. No archaeal clones were obtained despite repeated attempts. Of the 247 clones, 122 were from the microorganisms captured by 0.1-μm-pore-size filters (32 and 90 from sedimentary and granite aquifers, respectively), while 125 were from the microorganisms captured by 0.2-μm-pore-size filters (64 and 61 from sedimentary and granite aquifers, respectively). RFLP of the 122 and 125 clones yielded 23 (0.1-μm-pore-size filter-captured) and 14 (0.2-μm-pore-size filter-captured) OTUs, respectively. No overlaps in the 0.1- and 0.2-μm-pore-size filter-captured OTUs were found, and thus a total of 37 OTUs were obtained (Table 1).
Among the 0.1-μm-pore-size filter-captured OTUs, 15 of 23 OTUs (with 13 of 15 only from the granite aquifer) were related to the candidate divisions OD1 and OP11 of putative bacterial phyla (Fig. 1). The candidate division OP11 phylum consisted of five subdivisions (16), and subdivisions 4 and 5 have recently been considered independent candidate divisions SR1 and OD1, respectively (12). SR1, OD1, and OP11 contain clones from various anaerobic habitats (2, 15, 19, 20, 26, 32, 33), as from the Tono subsurface.
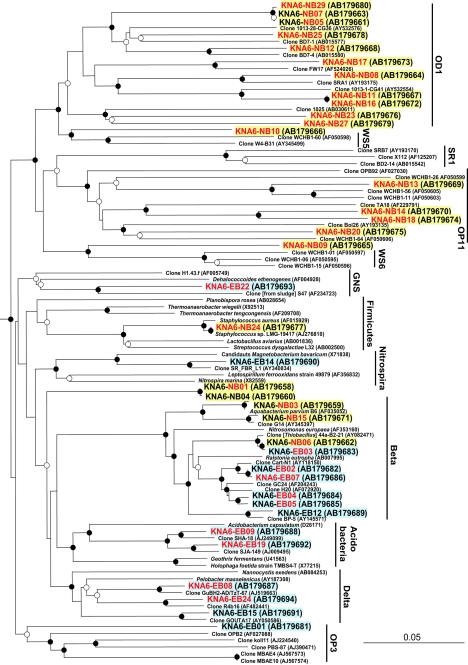
FIG.1.
Neighbor-joining phylogenetic tree based on nearly complete 16S rRNA gene sequences from OTUs in the 0.2- and 0.1-μm-pore-size filter-captured groundwater fractions from sedimentary and granite rock aquifers in the Tono uranium mine. The 16S rRNA gene sequences of the Aquificales Aquifex aeolicus (AJ309733) and Hydrogenobacter hydrogenophilus (Z30242) were used as outgroups but were pruned from the tree. Yellow and light-blue backgrounds indicate the OTUs captured by 0.1- and 0.2-μm-pore-size filters, respectively. Black and red letters indicate the OTUs from sedimentary and granite rock aquifers, respectively. The OTUs written in both black and red are from both sedimentary and granite rock aquifers. Bootstrap values at branching nodes after 1,000 resamplings are represented by filled circles (>75%), open circles (75%>○>50%), and no circles (<50%). GNS, green nonsulfur bacteria.
The OTUs KNA6-NB09 (the common prefix KNA6- is omitted hereafter) and NB10 in particular branched deeply from the candidate divisions WS6 and WS5, respectively, probably forming novel candidate divisions (Fig. 1). Novel lineages among the 0.1-μm-pore-size filter-captured microorganisms were also suggested by four other OTUs. The OTUs NB09 and NB10 were only weakly related to the candidate divisions WS6 and WS5, with similarities of 79.7 and 81.0%, respectively (Table 1). The OTUs NB01 and NB04 were closely related to each other at a similarity of 99.9% and are considered a single group, although NB04 was found only in the sedimentary aquifer while NB01 was recovered from both aquifers (Table 1).
The abundance of 0.1-μm-pore-size filter-captured microorganisms should be only minor compared with that of 0.2-μm-pore-size filter-captured microorganisms in water samples, as shown by the finding of 100 to 101 microorganisms filterable by 0.2-μm-pore-size filters liter−1, as determined by the most-probable-number method, versus 108 to 109 0.2 μm-pore-size filter-captured cells l−1 (acridine orange direct counts) in coastal waters (8). Yet despite possibly being of quantitatively minor importance, the 0.1-μm-pore-size filter-captured microorganisms are a source of novel microbial diversity.
Of 14 OTUs among 0.2-μm-pore-size filter-captured microorganisms, 11 were affiliated with known Betaproteobacteria, Deltaproteobacteria, and Acidobacteria at high similarities of 93.2 to 98.4%. Three OTUs, EB01, EB14, and EB22, branched deeply from the candidate division OP3, Nirospira, and green nonsulfur bacteria, respectively. Although these deep-branching OTUs are likely minor components in the 0.2-μm-pore-size filter-captured populations (Table 1), their presence demonstrates the importance of deep groundwater as a source of unique and novel microorganisms even in the >0.2-μm-diameter regime.
Acknowledgments
We are obliged to Y. Murakami, Japan Nuclear Fuel Development Institute, for technical assistance.
This work was supported by the Special Coordination Fund Archaean Park Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
REFERENCES
- 1.Åkerman, K. K., I. Kuronen, and E. O. Kajander. 1993. Scanning electron microscopy of nanobacteria—novel biofilm producing organisms in blood. Scanning 15(Suppl.):90-91. [Google Scholar]
- 2.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLong, E. F. 1992. Archaea in costal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drancourt, M., V. Jacomo, H. Lépidi, E. Lechevallier, V. Grisoni, C. Coulange, E. Ragni, C. Alasia, B. Dussol, Y. Berland, and D. Raoult. 2003. Attempted isolation of Nanobacterium sp. microorganisms from upper urinary tract stones. J. Clin. Microbiol. 41:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elsaied, H. E., M. Sato, and T. Naganuma. 2001. Viable Cytophaga-like bacterium in the 0.2 μm-filtrate seawater. Syst. Appl. Microbiol. 24:0-5. [DOI] [PubMed] [Google Scholar]
- 6.Folk, R. L. 1992. Bacteria and nanobacteria revealed in hardgrounds, calcite cements, native sulfur, sulfide minerals, and travertines. Proc. Geol. Soc. Amer. Annu. Meet., p. 104.
- 7.Folk, R. L. 1993. SEM imaging of bacteria and nanobacteria in carbonate sediments and rocks. J. Sedim. Petrol. 63:990-999. [Google Scholar]
- 8.Fukuba, T., M. Sato, H. E. Elsaied, and T. Naganuma. 2002. Overlooked microbial agents in aquaculture: nanobacteria, p. 99-107. In C.-S. Lee and P. O'Bryen (ed.), Microbial approaches to aquatic nutrition within environmentally sound aquaculture production systems. World Aquaculture Society, Baton Rouge, La.
- 9.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn, M. W., H. Lünsdorf, Q. L. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haller, C. M., S. Rölleke, D. Vybiral, A. Witte, and B. Velimirov. 1999. Investigation of 0.2 μm filterable bacteria from the Western Mediterranean Sea using a molecular approach: dominance of potential starvation forms. FEMS Microbiol. Ecol. 31:153-161. [DOI] [PubMed] [Google Scholar]
- 12.Harris, J. K., S. T. Kelley, and N. R. Pace. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63-67. [DOI] [PubMed] [Google Scholar]
- 14.Hugenholtz, P., and T. Huber. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 53:289-293. [DOI] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajander, E. O., and N. Ciftçioglu. 1998. Nanobacteria: an alternative mechanism for pathogenic intra- and extracellular calcification and stone formation. Proc. Natl. Acad. Sci. USA 95:8274-8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolter, R., D. A. Siegele, and A. Tormo. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855-874. [DOI] [PubMed] [Google Scholar]
- 19.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 21.Maniloff, J. 1997. Nanobacteria: size limits and evidence. Science 276:1776. [DOI] [PubMed] [Google Scholar]
- 22.Murakami, Y., Y. Fujita, T. Naganuma, and T. Iwatsuki. 2002. Abundance and viability of the groundwater microbial communities from borehole in Tono uranium deposit area, central Japan. Microb. Environ. 17:63-74. [Google Scholar]
- 23.Murakami, Y., Y. Fujita, T. Iwatsuki, and T. Naganuma. 2003. Abundance and viability of subsurface microbial communities in sedimentary and igneous rock aquifers, p. 115-140. In M. Taniguchi, K. Wang, and T. Gamo (ed.), Land and marine hydrogeology. Elsevier B.V., Amsterdam, The Netherlands.
- 24.Nyström, T., N. H. Albertson, K. Flärdh, and S. Kjelleberg. 1990. Physiological and molecular adaptation to starvation and recovery from starvation by the marine Vibrio sp. S14. FEMS Microbiol. Ecol. 74:129-140. [Google Scholar]
- 25.Page, R. D. M. 1996. Tree View: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 26.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Somerville, C. C., I. T. Knight, W. L. Straube, and R. R. Colwell. 1989. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl. Environ. Microbiol. 55:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1989. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrella, F., and R. Y. Morita. 1981. Microcultural study of bacteria size changes and microcolony and ultramicrocolony formation by heterotrophic bacteria in seawater. Appl. Environ. Microbiol. 41:518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velimirov, B. 2001. Nanobacteria, ultramicrobacteria and starvation forms: a search for the smallest metabolizing bacterium. Microbes Environ. 16:67-77. [Google Scholar]
- 32.Watanabe, K., Y. Kodama, and S. Harayama. 2001. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44:253-262. [DOI] [PubMed] [Google Scholar]
- 33.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 1997. Bacterial diversity of a Carolina bay as determined by 16S rDNA gene analysis: confirmation of novel taxa. Appl. Environ. Microbiol. 63:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]