Abstract
Reoviruses, enteroviruses, and adenoviruses were quantified by culture for various ambient waters in the Milwaukee area. From August 1994 through July 2003, the influent and effluent of a local wastewater treatment plant (WWTP) were tested monthly by a modified U.S. Environmental Protection Agency Information Collection Rule (ICR) organic flocculation cell culture procedure for the detection of culturable viruses. Modification of the ICR procedure included using Caco-2, RD, and HEp-2 cells in addition to BGM cells. Lake Michigan source water for two local drinking water treatment plants (DWTPs) was also tested monthly for culturable viruses by passing 200 liters of source water through a filter and culturing a concentrate representing 100 liters of source water. Reoviruses, enteroviruses, and adenoviruses were detected frequently (105 of 107 samples) and, at times, in high concentration in WWTP influent but were detected less frequently (32 of 107 samples) in plant effluent and at much lower concentrations. Eighteen of 204 samples (8.8%) of source waters for the two DWTPs were positive for virus and exclusively positive for reoviruses at relatively low titers. Both enteroviruses and reoviruses were detected in WWTP influent, most frequently during the second half of the year.
In the spring of 1993, contamination of drinking water with Cryptosporidium led to a large outbreak of diarrheal illness in the Milwaukee metropolitan community (4). This outbreak highlighted the importance of continued surveillance of local environmental waters for Cryptosporidium and other potential pathogens. Although the exact source of Cryptosporidium for this outbreak is still in question, it was deemed essential by the local government to determine the sources of potential pathogens in the local watershed. To this end, the Milwaukee Health Department virus laboratory developed the capability to become a U.S. Environmental Protection Agency (EPA) Information Collection Rule (ICR) approved laboratory for environmental virus culturing. The ICR, promulgated in 1996 and in effect from July 1997 to December 1998, required drinking water treatment plants (DWTPs) serving ≥100,000 customers to test their source water monthly for total culturable viruses (3). The purpose of the ICR was to determine baseline data for the occurrence of protozoan and viral agents in surface waters used as source water for the production of drinking water. The U.S. EPA ICR virus concentration and virus culture procedures were adopted and modified by the addition of various cell types for the testing of source water for two DWTPs and the influent and effluent of a Milwaukee area wastewater treatment plant (WWTP). By including cell types in addition to Buffalo green monkey kidney (BGM) cells and by employing various virus identification procedures, the total culturable viruses could be specifically identified as reoviruses, enteroviruses, or adenoviruses. Routine testing of this type serves multiple purposes: to assess the viral quality of the source water used for production of drinking water, to determine the types and quantities of viruses entering a local WWTP while assessing the plant's efficiency in eliminating those viruses before discharge of effluent into Lake Michigan, and to determine the types and quantities of viruses that the WWTP discharges into water which may be used, in part, to produce drinking water. This testing also delineates the temporal patterns for the occurrence of culturable reoviruses, enteroviruses, and adenoviruses in local environmental waters.
MATERIALS AND METHODS
Sewage collection.
The Jones Island WWTP is located south of downtown Milwaukee and is one of two plants treating sewage for 1.2 million people in the Milwaukee metropolitan area. Each plant serves about one-half of the area residents, with the Jones Island plant serving most of the City of Milwaukee, as well as northern and western suburbs. There is some overlap in the sections of the metropolitan area served by each plant. The treatment process at the Jones Island plant consists of preliminary, primary, and secondary (activated-sludge) treatment, as well as phosphorus removal, and disinfection using chlorine, followed by dechlorination with sulfur dioxide before discharge into Lake Michigan. The plant treats an average of 100 million gallons of sewage per day. Influent and effluent samples were collected from the Jones Island plant on a routine basis, usually on the third Tuesday of the month; stored at 4°C; and transported to the laboratory within 12 h. Samples were obtained monthly from August 1994 through July 2003 (except for September 1994). Influent samples were 1-liter flow-proportioned samples of the three major flows entering the plant. One-liter WWTP effluent samples (after final chlorination and dechlorination with sulfur dioxide) were routinely collected on the same day as the influent samples and subjected to organic flocculation and virus culture by the same procedures used for the influent.
Collection of source water samples.
Both of Milwaukee's DWTPs are located on the western shore of Lake Michigan. The Linnwood DWTP is located north of the Jones Island WWTP and produces 75 million gallons of drinking water per day. The Howard Avenue DWTP is located south of the Jones Island WWTP and produces 52 million gallons of drinking water per day. The Howard Avenue DWTP was associated with the outbreak of Cryptosporidium-caused diarrheal illness in 1993. Each DWTP collected monthly 200-liter Lake Michigan source water samples by pumping source water through a 1-MDS Zetapor Virosorb filter (CUNO Inc., Meriden, Conn.). The filters and housings were then transported to the virus laboratory soon after collection, and processing for virus detection was initiated the same day. Virus was eluted from each filter with 1 liter of 2% beef extract, pH 9.5 (Difco Laboratories, Detroit, Mich.). The 1 liter of eluate was further concentrated by the organic-flocculation procedure used for the WWTP influent and effluent.
Cell cultures.
A number of cell types, in addition to the ICR-mandated BGM cells, were used for culture of wastewaters and source waters. From 1994 through 1998, the BGM cells used were obtained from the U.S. EPA. After 1998, BGM cells were purchased from Biowhittaker, Walkersville, Md. Human rhabdomyosarcoma (RD) cells were also purchased from Biowhittaker. Human epidermoid carcinoma of the larynx (HEp-2) cells and human adenocarcinoma of the colon (Caco-2) cells were obtained from the American Type Culture Collection, Manassas, Va. Culture media, Eagle's minimum essential medium with Earle's salt solution (MEM), and fetal bovine serum (FBS) were purchased from Sigma, St. Louis, Mo. All cell culture media contained HEPES buffer, l-glutamine, penicillin, streptomycin, gentamicin sulfate, and amphotericin B. Cell cultures were grown in CO2 incubators at 35.5°C and 4.5% CO2. Cell stock cultures were grown in 75- or 162-cm2 plastic flasks (Costar, Corning, N.Y.) with 5% FBS-MEM and split weekly. Twenty-four-well plates (Costar) of heteroploid cell types were grown using 5% FBS-MEM and maintained on 2% FBS-MEM. Stock cell cultures and uninoculated 24-well plastic plates of cells were kept in a CO2 incubator separate from inoculated cultures. Cell cultures inoculated with WWTP influent or effluent or DWTP source water concentrates were kept in a different CO2 incubator than were plates inoculated with clinical specimens.
Organic-flocculation procedure.
The concentration of virus from WWTP influent and effluent was accomplished using a modification of the organic-flocculation procedure described in the U.S. EPA Virus Monitoring Protocol for the Information Collection Requirements Rule (12). A brief description of the procedure follows. At room temperature, 1 N HCl was added to a 2% beef extract V solution of influent or effluent to lower the pH to 3.5 ± 0.1. After continued stirring for 30 min, the solution was dispensed into four 250-ml centrifuge bottles and centrifuged at 2,500 × g for 15 min at 4°C. The supernatant was discarded, and all four pellets were resuspended in a total volume of 30 ml of 0.15 M sodium phosphate buffer. After the pH was adjusted to 7.0 to 7.5, the concentrate was refrigerated at 4°C for 15 to 30 min to allow the pellet to dissolve completely. After readjustment of the pH to 9.0 to 9.5 with NaOH and centrifugation at 6,000 × g for 10 min at 4°C, the supernatant was removed and saved. Finally, the supernatant pH was adjusted to 7.0 to 7.5 with HCl, and the supernatant was refrigerated at 4°C until it was used to inoculate cell cultures. Source water 1-MDS filter eluates were concentrated using the same organic-flocculation procedure.
Total culturable virus assay.
Detection of virus in concentrated WWTP samples was accomplished using a modified U.S. EPA ICR cell culture protocol (9, 12). A brief description of our modification of the procedure follows. A 1:8 dilution of the influent concentrate (1:3 dilution for effluent) was prepared with 2% FBS-MEM. Higher dilutions (1:16 and 1:32) were also used for influent during months with high virus titers. The solution was filtered through a 0.2-μm-pore-size sterilizing filter pretreated with 2% beef extract. At times, more than one filter was used due to clogging. The filtered dilution was inoculated into the wells of two 24-well plastic culture plates of BGM cells and one 24-well plate each of Caco-2, RD, and HEp-2 cells. Half a milliliter of dilution was inoculated into each well, or 12 ml per 24-well plate. A total of 60 ml of the filtered dilution was used for five 24-well plates. BGM and RD cells were used for the entire study. Caco-2 cells were used from August 1997 through July 2003, and HEp-2 cells were used from January 1999 through July 2003. For virus adsorption, the plates were incubated for 2 h at 35.5°C. After the incubation period, the inoculum was removed, and each well was washed with 0.5 ml of saline. After aspiration of the saline wash, each well was fed 0.5 ml of 2% FBS-MEM. The plates were incubated at 35.5°C and examined microscopically for cytopathic effect (CPE) daily. Cultures were maintained for 14 days, and no blind passes were done. Enterovirus CPE was typical and was distinct from reovirus CPE, which occurred mainly on BGM cells. For the influent diluted 1:8, the equivalent volume of original influent was 0.05 liter per 24-well plate, with a minimum detection limit of 20 infection-forming units (IFU) per liter, and for the 1:3 dilution of effluent, the equivalent volume of original effluent was 0.135 liter per 24-well plate with a minimum detection limit of 7.5 IFU per liter.
A similar virus culture procedure was used to assess source waters for the two Lake Michigan DWTPs. The standard U.S. EPA ICR total culturable virus procedure (12) was used. Aliquots of concentrate equal to 100 liters of source water were inoculated into 20 25-cm2 flasks of BGM cells and held for 14 days for microscopic observation of CPE. All of the flasks were then frozen and thawed, subcultured to 24-well plates of BGM cells, and observed for another 14 days. For source water concentrate, one 24-well plate each of HEp-2, Caco-2, and RD cells was also inoculated (equivalent to 20 liters of original source water per 24-well plate) and observed for 14 days for the production of CPE. Source water-inoculated 24-well plates were not subjected to blind subculture.
Identification of virus isolates.
Reoviruses were identified by typical CPE on BGM cells. Also, a portion of these reoviruses had their genus identities confirmed by a direct fluorescent antibody (DFA) test using an anti-reovirus polyclonal fluorescent antiserum of caprine origin (VMRD, Inc., Pullman, Wash.). Two hundred forty-five sewage isolates identified as reoviruses by CPE were also tested by the anti-reovirus DFA procedure, and all 245 were confirmed to be reoviruses. Also, of 69 suspected reoviruses from source waters, 35 of 35 tested by DFA were confirmed to be reoviruses. Enteroviruses were identified by typical CPE, and 33% of sewage influent isolates were specifically typed using Lim Benyesh-Melnick neutralizing antibody pools A through H, as previously described (6). Adenoviruses were identified by typical CPE on HEp-2 cells and, at times, adenovirus IFA tests using reagents from Chemicon International, Temecula, Calif.
MPN titer calculation.
A computer program supplied by the U.S. EPA for the ICR was used for most-probable-number (MPN) calculations (9). The program uses the number of replicates inoculated, the number of positive replicates (CPE-positive wells for 24-well cell culture plates), and the inoculation volume to calculate the MPN in IFU. This program also calculates 95% confidence intervals for MPN titers. For WWTP influent and effluent, the titers were calculated as MPN (IFU) per liter; for lake Michigan source water, titers were calculated as MPN (IFU) per 100 liters.
Clinical isolation.
For the comparison of environmental and clinical data, specimens for primary virus isolation were received from Milwaukee area hospitals and clinics, as well as from the Milwaukee County Medical Examiners Office. Local hospitals, which have their own virus laboratories, use this laboratory as a reference laboratory for enterovirus typing and send cell culture enterovirus isolates for typing. All clinical specimens were processed and inoculated onto 24-well plates of seven cell types: Rhesus monkey kidney primary, HEp-2, human foreskin, human embryonic lung, BGM, RD, and Caco-2 cells. These cultures were held for 12 to 14 days and were observed daily for CPE. Over the period of this study, all Milwaukee area clinical enterovirus isolates were typed by this laboratory (9).
QC.
In May 1997, our laboratory was approved by the U.S. EPA to perform ICR virus testing. As part of achieving and maintaining approval status, we passed two on-site inspections and audits and analyzed performance evaluation samples and quality control (QC) samples as required. Also, for our environmental testing, each lot of beef extract used in the organic-flocculation concentration procedure was screened for virus recovery. For the screening, 1 liter of 2% beef extract was inoculated with 200 IFU of poliovirus type 3 (an EPA-approved QC virus) and then processed as described above. The average recovery for eight screenings was 73%.
In addition to the above quality control procedures, the sensitivities of our cell lines were demonstrated by the following: (i) laboratory participation in the College of American Pathologists (Northfield, Ill.) proficiency testing for virus isolation, (ii) the laboratory's ability to isolate a variety of viruses from different types of clinical specimens using the same cell lines used for our environmental testing, and (iii) assay poliovirus type 3-positive control was used with BGM cells for each source water sample tested.
RESULTS
Jones Island WWTP influent virus isolation results.
Figure 1 shows the results of virus culture testing of the Jones Island WWTP influent over the 108-month period August 1994 through July 2003. During this time, reoviruses, enteroviruses, or adenoviruses were detected in 105 of 107 (98.1%) samples. Of these viruses, reoviruses were the most frequently detected (97 of 107 samples [90.6%]), and enteroviruses were detected in 91 of 107 (85.0%) samples. HEp-2 cells were routinely used for the detection of adenoviruses for the 55-month period January 1999 through July 2003, and adenovirus was detected in 35 of these 55 (63.6%) months. Although viruses from these three groups were frequently detected, Fig. 1 indicates a large fluctuation of the titers (MPN per liter) by month. The titers were generally highest for reovirus, with a maximum titer of 12,027 MPN/liter detected in August 1999. The highest enterovirus titer detected was 3,347 MPN/liter in July 1997. Also, Fig. 1 indicates that there are seasonal patterns to the occurrence of reoviruses and enteroviruses in the Jones Island WWTP influent, with both virus groups occurring more frequently during the period July through December. This is a pattern that is expected for enteroviruses, since they have a well-known summer-fall pattern for causing human infections, which has been documented for Milwaukee (8, 9). Only a few articles have been published about the seasonal occurrence of reoviruses in environmental waters. A study of urban river water in Japan also found higher reovirus levels in the cold-weather months of October through January (11).
FIG. 1.
Monthly titers of reoviruses, adenoviruses, and enteroviruses detected by culture for Milwaukee's Jones Island WWTP influent, August 1994 though July 2003.
Table 1 summarizes the total numbers of enteroviruses, reoviruses, and adenoviruses isolated by year during the period of study. Of the 3,903 WWTP influent virus isolates, 2,666 (68.3%) were reoviruses, 1,116 (28.6%) were enteroviruses, and 121 (3.1%) were adenoviruses. Clinical cases of enterovirus and reovirus infection, confirmed by isolation of the virus, during the period August 1994 through July 2003 are also presented in Table 1. Although, a preponderance of environmental virus isolates were reoviruses, the reoviruses were detected far less frequently for clinical cases than enteroviruses (15 and 687 clinical cases, respectively), even though the cell types used for culture of clinical samples could detect viruses of either group. Neither the environmental nor the clinical reoviruses were identified beyond being grouped as reoviruses.
TABLE 1.
Annual number of clinical cases and number of WWTP influent virus isolatesa
| Yr | No. of clinical cases
|
No. of WWTP influent isolates
|
Total WWTP influent isolates plus clinical cases | ||||
|---|---|---|---|---|---|---|---|
| Enterovirus | Reovirus | Enterovirus | Reovirus | Adenovirus: | Total | ||
| 1994b | 65 | 1 | 80 | 42 | 0 | 122 | 188 |
| 1995 | 119 | 3 | 107 | 285 | 0 | 392 | 514 |
| 1996 | 86 | 0 | 62 | 343 | 0 | 405 | 491 |
| 1997 | 99 | 0 | 214 | 200 | 4 | 418 | 517 |
| 1998 | 89 | 3 | 96 | 338 | 0 | 434 | 526 |
| 1999 | 68 | 1 | 140 | 431 | 33 | 604 | 673 |
| 2000 | 42 | 1 | 106 | 333 | 40 | 479 | 522 |
| 2001 | 68 | 0 | 117 | 348 | 25 | 490 | 558 |
| 2002 | 43 | 5 | 180 | 312 | 6 | 498 | 546 |
| 2003c | 8 | 1 | 14 | 34 | 13 | 61 | 70 |
| Total | 687 | 15 | 1,116 | 2,666 | 121 | 3,903 | 4,605 |
Note that the number of virus isolates is the number of CPE-positive wells of 24-well cell culture plates.
August to December. Because no WWTP influent was collected in September 1994, to complete the table, 34 reoviruses were added for September 1994 (average of eight other Septembers) and 22 enteroviruses were added for September 1994 (average of eight other Septembers).
January through July.
Seasonal occurrence of enteroviruses in WWTP influent.
Table 2 presents a summary, by month, of the cumulative number of enterovirus isolates and the percentage of total isolates for the study period of August 1994 through July 2003. Enteroviruses were isolated from WWTP influent during every month of the year, although most (69.6%) of the enteroviruses were isolated during the months of July through October. Table 2 also indicates that clinical enterovirus cases, as expected, were diagnosed most frequently during the July through October period (80.8% of cases). Thus, as was previously documented for Milwaukee (9), enteroviruses were detected in Milwaukee WWTP influent at the same time they were most prevalent clinically. It has also been shown previously that for the most part, the same enterovirus serotypes causing clinical problems are the serotypes detected in WWTP influent each year (9).
TABLE 2.
Monthly cumulative number of WWTP influent virus isolatesa and clinical cases plus percentage of cumulative totals by month, August 1994 through July 2003
| Mo | No. of WWTP influent enterovirus isolates | % Total WWTP influent enterovirus isolates | No. of enterovirus clinical cases | % Total clinical enterovirus cases | No. of WWTP influent reovirus isolates | % Total WWTP influent reovirus isolates |
|---|---|---|---|---|---|---|
| January | 22 | 2.0 | 14 | 2.0 | 134 | 5.0 |
| February | 23 | 2.1 | 4 | 0.6 | 90 | 3.4 |
| March | 15 | 1.3 | 10 | 1.5 | 147 | 5.5 |
| April | 15 | 1.3 | 6 | 0.9 | 151 | 5.7 |
| May | 49 | 4.4 | 8 | 1.2 | 72 | 2.7 |
| June | 96 | 8.6 | 41 | 6.0 | 154 | 5.8 |
| July | 196 | 17.6 | 84 | 12.2 | 448 | 16.8 |
| August | 237 | 21.2 | 191 | 27.8 | 404 | 15.2 |
| September | 201 | 18.0 | 170 | 24.7 | 304 | 11.4 |
| October | 143 | 12.8 | 110 | 16.0 | 310 | 11.6 |
| November | 94 | 8.4 | 31 | 4.5 | 236 | 8.9 |
| December | 25 | 2.2 | 16 | 2.6 | 216 | 8.1 |
| Total | 1,116 | 99.9 | 687 | 100.0 | 2,666 | 100.1 |
Note that number of virus isolates is the number of CPE-positive wells of 24-well cell culture plates.
Seasonal occurrence of reoviruses in WWTP influent.
Table 2 tabulates the cumulative number of WWTP influent reovirus isolates per month for the period August 1994 through July 2003 and also the percentage of total reovirus isolates by month. The table indicates that the reoviruses, like the enteroviruses, were detected in WWTP influent for every month of the year but were detected in the Jones Island WWTP influent more frequently during the latter half of the year. Seventy-two percent of reoviruses were isolated during the period July through December, with 55.0% of total reoviruses being detected for the months of July through October, when enteroviruses were most commonly detected. However, reoviruses continued to be detected in November and December, with 17% of all reoviruses isolated during these 2 months.
WWTP influent versus effluent virus results.
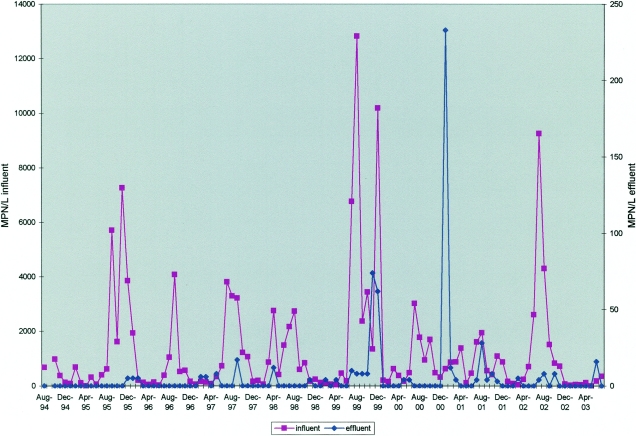
For our study, each month when a 1-liter influent sample was collected, a 1-liter effluent sample was also collected on the same day. Both influent and effluent were processed and inoculated onto cell cultures the same day they were collected. Figure 2 depicts the results of this testing. Culturable viruses were detected in 105 of 107 (98.1%) influent samples. However, culturable viruses were detected in only 32 of 107 (29.9%) effluent samples, and at much lower titers than for influent. The maximum total culturable virus titer for effluent was 233 MPN/liter (221 reoviruses, 8 adenoviruses, and 4 echoviruses) for a sample collected in January 2001. The second-highest total culturable virus titer for effluent was 74 MPN/liter for November 1999. Of the 32 months with culturable virus detected for effluent, reovirus was detected in 28 samples while adenovirus was detected on only two occasions (one month, only adenovirus was detected, and one month, adenovirus and reovirus were detected). For the effluent, enterovirus was detected on four occasions, coxsackievirus B3 was detected twice (both times coxsackievirus B3 was the only virus type detected), and echovirus type 6 and echovirus type 7 were detected once each (1 month, only echovirus type 7 was detected).
FIG. 2.
Monthly titers for total viruses detected by culture for the influent and effluent of Milwaukee's Jones Island WWTP, August 1994 through July 2003.
Table 3 presents data on the monthly average virus load difference between the WWTP influent and effluent for a 9-year period. On average, the effluent virus titer was 2.41 log units lower than the influent virus titer. This represents a 99.6% lower average titer of culturable viruses for the effluent. Since there was no temporal correspondence between the influent and effluent sampling, a virus reduction value would be misleading. Due to variations in the flow and recycle stream and other factors, it is not feasible to obtain paired influent and effluent samples from the Jones Island WWTP that accurately reflect the treatment of a discrete aliquot of sewage as it transits the plant. During most periods, the plant operates in a stable manner. The data used to develop the log difference values for this research on influent and effluent represent >100 sets of paired data obtained over a 108-month period. The average log difference of viruses indicated by these data should not be construed to represent the virus removal at any one time but should be considered representative of the average log removal that occurs during normal plant operation.
TABLE 3.
Average monthly total culturable virus (MPN/liter) for Milwaukee's Jones Island WWTP influent and effluent for the period August 1994 through July 2003
| Mo | Influent mean (MPN/liter) | Influent range (MPN/liter) | Influent SD (MPN/liter) | Effluent mean (MPN/liter) | Effluent range (MPN/liter) | Effluent SD (MPN/liter) | Influent-effluent log difference of means |
|---|---|---|---|---|---|---|---|
| January | 380 | 30-1,930 | 608 | 26 | 0-233 | 77 | 1.165 |
| February | 264 | 52-867 | 299 | 3 | 0-12 | 4 | 1.944 |
| March | 322 | 40-876 | 360 | 2 | 0-6 | 3 | 2.206 |
| April | 562 | 0-2758 | 929 | 0.5 | 0-4 | 1 | 3.051 |
| May | 295 | 0-701 | 215 | 3 | 0-12 | 5 | 1.992 |
| June | 692 | 30-2,612 | 849 | 2 | 0-16 | 5 | 2.539 |
| July | 3,086 | 354-9,251 | 3,105 | 2 | 0-10 | 4 | 3.188 |
| August | 3,247 | 621-12,820 | 3,795 | 5 | 0-28 | 9 | 2.813 |
| Septembera | 2,375 | 557-5,713 | 1,732 | 4 | 0-17 | 6 | 2.773 |
| October | 1,284 | 409-3,441 | 920 | 3 | 0-8 | 4 | 2.631 |
| November | 1,457 | 176-7,264 | 2,210 | 9 | 0-74 | 24 | 2.209 |
| December | 1,779 | 73-10,187 | 3,376 | 7 | 0-62 | 21 | 2.405 |
Since neither WWTP influent nor effluent was collected in September 1994, the average MPN/liter for the other eight Septembers was used for September 1994 for table calculations.
The greatest difference in virus titer between influent and effluent was documented for August 1999, when the influent contained 12,820 MPN of culturable viruses/liter and the effluent had an MPN/liter value of 8, for a log difference of 3.204. The smallest titer difference occurred in January 2001, when the titers of influent and effluent were 626 and 233 MPN/liter, respectively, for a log difference of 0.429. Note that during January 2001, the WWTP's secondary treatment system (activated-sludge system) was not operating at full capacity. For safety reasons, it was necessary to shut down part of the plant during emergency demolition of part of the interstate freeway that passes over the plant.
Viruses in source water for two DWTPs.
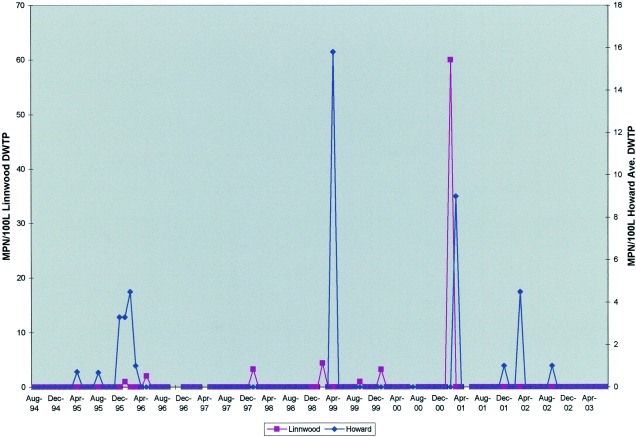
Figure 3 shows that culturable viruses were detected in 18 of 204 (8.8%) samples of Lake Michigan source water for Milwaukee's two DWTPs. For the Linnwood DWTP, viruses were detected in 7 of 103 (6.8%) samples, and for the Howard Avenue DWTP, viruses were detected in 11 of 101 (10.9%) samples. All viruses detected for both DWTPs were reoviruses. The maximum calculated virus titer for the Linnwood DWTP was 59.0 MPN/100 liters in February 2001, and for the Howard Avenue DWTP, it was 15.8 MPN/100 liters in April 1999. Many of the reovirus detections occurred during the colder months. For the Linnwood DWTP, 5 of 7 detections were during the months of December, January, and February, and for the Howard Avenue DWTP, 7 of 11 detections were during the months of December, January, February, and March. The Howard Avenue DWTP was the DWTP associated with the Cryptosporidium outbreak in 1993, and its intake at that time was located within the Milwaukee River plume dispersion pattern, which contained the discharges from the outfall of the Jones Island WWTP. Since 1993, the intake for the Howard Avenue DWTP has been moved 1.25 miles further out into Lake Michigan to escape the influence of the Milwaukee River plume dispersion pattern. There was no obvious correlation between high influent virus titers for the WWTP and detection of virus in source water for either DWTP.
FIG. 3.
Monthly titers for reoviruses detected by culture for the source water of Milwaukee's Linnwood and Howard Avenue DWTPs, August 1994 through July 2003.
Sensitivity of cell cultures for the isolation of reoviruses and adenoviruses from WWTP influent.
A comparison of the sensitivity of various cell types for the isolation of reoviruses from WWTP influent can be drawn from data accumulated during the period January 1999 through December 2003. Table 4 indicates that during this period, four cell types were routinely inoculated with WWTP influent concentrate and that of these four cell types, reoviruses were isolated on BGM and HEp-2 cells but not Caco-2 or RD cells. Of the 1,598 reovirus isolates, 1,537 (96.2%) produced CPE on BGM cells and 61 (3.8%) grew on HEp-2 cells. For purposes of comparison, when calculating for one BGM cell 24-well plate and one 24-well plate of HEp-2 cells, the comparative number of isolates would be 769 to 61 for BGM and HEp-2 cells, respectively, or 12.6 times more reovirus isolates on BGM cells. A statistical comparison of BGM and HEp-2 cell reovirus isolates (chi-square value, 848; 1 degree of freedom; P ≤ 0.001) indicates that BGM cells are the cells of choice for isolation of reoviruses from WWTP influent. No added value was obtained by using HEp-2 cells for isolation of reoviruses from WWTP influent, since all samples with reovirus detected on HEp-2 cells had more reoviruses detected on BGM cells.
TABLE 4.
Isolation of WWTP influent reoviruses and adenoviruses on various cell types for the period January 1999 through December 2003
| Cell type | No. of reovirus isolatesa | % Total reovirus isolates | No. of adenovirus isolatesa | % Total adenovirus isolates |
|---|---|---|---|---|
| BGM | 1,537 | 96.2 | 0 | 0 |
| RD | 0 | 0 | 0 | 0 |
| Caco-2 | 0 | 0 | 15 | 12.8 |
| HEp-2 | 61 | 3.8 | 102 | 87.2 |
| Total | 1,598 | 100.0 | 117 | 100.0 |
The number of isolates is the number of CPE-positive wells of 24-well cell culture plates.
Table 4 also provides data on the effectiveness of various cell types for the isolation of adenoviruses from WWTP influent for the period January 1999 through December 2003. Adenoviruses were isolated on HEp-2 and Caco-2 cells but not BGM or RD cells. Of the 117 adenovirus isolates, 102 (87.2%) were detected on HEp-2 cells and 15 (12.8%) were detected on Caco-2 cells. Statistically (chi-square = 67.43; P < 0.001), HEp-2 cells were more efficient then Caco-2 cells for the isolation of adenoviruses from WWTP influent.
Usefulness of adding other cell types to ICR-required BGM cells for isolation of viruses from DWTP source water.
In an attempt to improve the recovery of viruses from source water, we added 24-well plates of HEp-2, Caco-2, and RD cells to the U.S. EPA-required 25-cm2 flasks of BGM cells for cultivation of viruses from source water concentrate from both DWTPs. These extra cell types were used for the period January 1999 through July 2003 (55 months), and during this period, an equivalent of 20 liters of original source water was inoculated onto each 24-well plate (equivalent to 0.83 liters of source water per well). No viral CPE was detected for any of the extra cell types for any of the wells inoculated. During this same period, reovirus was detected in 10 of 110 source water samples using 25-cm2 BGM cell culture flasks. Thus, no increased isolation of viruses from source water was demonstrated by adding the extra cell types.
DISCUSSION
This study has increased our understanding of the occurrence and the seasonal fluctuation of viruses in Milwaukee area environmental waters. Viruses were detected very frequently in the WWTP influent, with reoviruses, enteroviruses, and adenoviruses detected in descending order of magnitude. Reoviruses were also the most commonly detected viruses for the WWTP effluent, but at much lower titers then were observed for the influent. For the WWTP influent, reoviruses were detected most frequently during the months of July through December, the same time that enteroviruses were detected in WWTP influent. Other studies in Japan have detected reoviruses during this same time of year in river water contaminated by a sewage treatment plant (11). The study in Japan also detected reoviruses, enteroviruses, and adenoviruses in the same declining order of magnitude that was observed in the present study.
Routine testing of sewage influent and effluent indicated that the Jones Island WWTP was effective in removing viruses. Over a 9-year period, the effluent virus titers were typically 2 to 3 log units lower than the titers for influent. Although calculation of log removal based on reduction of viral loads from discrete aliquots of wastewater was not possible, the average log removal (based on 108 paired influent-effluent values) was 2.41. A previous analysis of some of these data showed that removal of pathogens (including viruses) and indicators from this waste stream is significantly correlated with removal of solids by the wastewater treatment process (3).
When a total culturable virus assay procedure was used for two DWTPs' source waters, the only viruses isolated from Lake Michigan were reoviruses. A predominance of reoviruses has been reported for many types of source water (1, 2, 5, 7, 10). Reoviruses were isolated from 8.8% of Milwaukee's two DWTP source water samples, and at relatively low titers.
An analysis of cell type sensitivity for isolation of viruses from WWTP influent indicates that there is a benefit to using cell types in addition to BGM cells for influent testing. Using Caco-2 cells increases the detection of enteroviruses (9), and adding HEp-2 cells improves the recovery of adenoviruses. However, adding extra cell types for virus detection from source water did not increase the recovery of viruses. Nevertheless, using cell types in addition to BGM cells has the potential to expand virus culture beyond the total culturable viruses of the ICR and to allow culture detection and identification of viruses on the U.S. EPA Contaminate Candidate List (13) (echovirus, coxsackievirus, and adenovirus).
Routine use of our modified U.S. EPA ICR virus concentration and cultivation procedures for testing environmental waters is feasible and can add to our knowledge of the presence and concentration of reoviruses, enteroviruses, and adenoviruses in local environmental waters.
REFERENCES
- 1.Berg, G., H. Bodily, E. Lennette, J. Melnick, and T. G, Metcalf. 1976. Viruses in water. American Public Health Assoc., Washington, D.C.
- 2.Berg, G. 1987. Methods for recovering viruses from the environment. CRC Press Inc., Boca Raton, Fla.
- 3.MacDonald, J. A., M. S. Gradus, G. V. Sedmak, A. Singh, R. J. Moser, and D. L. Landis. 1998. Pathogen and indicator organism reduction at a large activated sludge wastewater treatment plant, vol. 6, p. 413-423. Proc. 1998 Water Environ. Fed. Tech. Exposition Conf., Orlando, Fla.
- 4.MacKenzie, W. R., N. Hoxie, M. Proctor, S. Gradus, K. Blair, D. Peterson, J. Kazmierzak, K. Fox, D. Addis, J. Rose, and J. Davis. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 331:161-167. [DOI] [PubMed] [Google Scholar]
- 5.Matsuura, K., M. Ishikura, T. Nakayama, S. Hasegawa, O. Morita, and H. Uetake. 1988. Ecological studies on reovirus pollution of rivers in Toyama Prefecture. Microbiol. Immunol. 32:1221-1234. [DOI] [PubMed] [Google Scholar]
- 6.Melnick, J. L., V. Rennick, B. Hampil, N. J. Schmidt, and H. H. Ho. 1973. Lyophilized combination pools of enterovirus equine antisera: preparation and test procedure for identification of field strains of 42 enteroviruses. Bull. W. H. O. 48:263-268. [PMC free article] [PubMed] [Google Scholar]
- 7.Metcalf, T. G., J. E. Melnick, and M. K. Estes. 1995. Environmental virology: from detection of viruses in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu. Rev. Microbiol. 49:461-487. [DOI] [PubMed] [Google Scholar]
- 8.Sedmak, G., C. Abel, B. Voight, and H. Wisniewski. 1981. Seasonal occurrence of viruses in the Milwaukee area. Wisconsin Med. J. 80:31-35. [PubMed] [Google Scholar]
- 9.Sedmak, G., D. Bina, and J. MacDonald. 2003. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 69:7181-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spinner, M., and G. DiGiovanni. 2001. Detection and identification of mammalian reoviruses in surface water by combined cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:3016-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tani, N., Y. Dohi, N. Kurumatani, and K. Yonemasu. 1995. Seasonal distribution of adenoviruses, enteroviruses and reoviruses in urban river water. Microbiol. Immunol. 39:577-580. [DOI] [PubMed] [Google Scholar]
- 12.U.S. Environmental Protection Agency. 1996. ICR microbial laboratory manual, publication no. EPA/600/R-95/178. Government Printing Office, Washington, D.C.
- 13.U.S. Environmental Protection Agency. 1998. Announcement of the drinking water contaminant candidate list. Fed. Regist. 63:0273-10287. [Google Scholar]