ABSTRACT
Fungi of the Basidiomycota, representing major pathogen lineages and mushroom-forming species, exhibit diverse means to achieve sexual reproduction, with particularly varied mechanisms to determine compatibilities of haploid mating partners. For species that require mating between distinct genotypes, discrimination is usually based on both the reciprocal exchange of diffusible mating pheromones, rather than sexes, and the interactions of homeodomain protein signals after cell fusion. Both compatibility factors must be heterozygous in the product of mating, and genetic linkage relationships of the mating pheromone/receptor and homeodomain genes largely determine the complex patterns of mating-type variation. Independent segregation of the two compatibility factors can create four haploid mating genotypes from meiosis, referred to as tetrapolarity. This condition is thought to be ancestral to the basidiomycetes. Alternatively, cosegregation by linkage of the two mating factors, or in some cases the absence of the pheromone-based discrimination, yields only two mating types from meiosis, referred to as bipolarity. Several species are now known to have large and highly rearranged chromosomal regions linked to mating-type genes. At the population level, polymorphism of the mating-type genes is an exceptional aspect of some basidiomycete fungi, where selection under outcrossing for rare, intercompatible allelic variants is thought to be responsible for numbers of mating types that may reach several thousand. Advances in genome sequencing and assembly are yielding new insights by comparative approaches among and within basidiomycete species, with the promise to resolve the evolutionary origins and dynamics of mating compatibility genetics in this major eukaryotic lineage.
BREEDING SYSTEMS AND LIFESTYLES IN THE BASIDIOMYCOTA
In the phylum Basidiomycota, a wide variety of lifestyles are represented. These range from well-known and conspicuous wood-decaying mushrooms, plant growth-promoting and mutualistic mycorrhizae, and crop-destroying smut and rust fungi, to yeast-like human pathogens. Lifestyle differences have consequences for the mating and breeding systems of these fungi (see “Glossary,” below, for definitions of specialist terms used in this article), which are reflected in the genetic evolution of mating-type determination. For over a century fungi have been recognized as having diverse breeding systems, from homothallism (i.e., universal compatibility among gametes, including among clonemates) to heterothallism (i.e., mating among haploid gametes carrying different mating-type alleles). The study of breeding systems, for example, led to the discovery of the astounding variability in mating-type alleles among mushrooms, with thousands of different mating types in some species (1), and to the realization that in many fungal pathogens the process of sexual reproduction is closely linked to infection and pathogenicity (2) (Fig. 1). The importance of basidiomycete fungi and their great research tractability, from ecology to genomics, have brought major insights into the diversification of genetic mechanisms used to achieve sexual reproduction.
FIGURE 1.

General life cycles of dimorphic and mushroom-forming basidiomycetes. Three basidiomycetes are pictured where sexual reproduction and a dimorphic switch between a yeast cell and a hyphal form are crucial to infection of plant (A, B) or animal (C) hosts. The haploid yeast forms of the maize smut Ustilago maydis (A) and the anther smut Microbotryum spp. (B) are nonpathogenic and can undergo asexual mitotic vegetative growth. In Microbotryum, the yeast stage is, however, short-lived because mating occurs mostly between cells within the same tetrad. Upon mating with a compatible partner, both fungi switch to an enduring infection hyphal form (dikaryon; n + n) that can invade the host plant. Proliferation and differentiation of U. maydis (A) in the plant culminates with the production of masses of wind-dispersing diploid spores (teliospores; 2n) in large tumor-like tissues, whereas in Microbotryum (B), teliospores are formed in the anthers of infected flowers and transmitted by pollinators onto healthy plants. In the case of Cryptococcus neoformans (C), the single-celled yeast form may be free-living or mycoparasitic. A similar dimorphic switch occurs upon mating of yeast cells of opposite mating type (α or α), ultimately resulting in the infectious propagules (basidiospores) that potentially infect an animal host after dispersal. These infectious structures may also be generated by haploid selfing (depicted with gray background), where fusion occurs between homothallic cells carrying identical MAT alleles (α/α diploid is depicted) and form monokaryotic hyphae with unfused clamp connections (see text for details). In mushroom-forming fungi such as Schizophyllum commune (D), germination of haploid spores yields haploid monokaryons capable of independent growth. When two compatible monokaryons meet, a fertile clamped dikaryon is formed which develops into fruiting bodies (mushrooms) triggered upon suitable environmental cues, where basidia arise. In all these and other basidiomycetes, nuclear fusion (karyogamy) is usually delayed until the formation of basidia (or teliospores). Meiosis ensues, generating four haploid nuclei, which give rise to basidiospores to complete the cycle. Adapted from Morrow and Fraser (2) and Nieuwenhuis et al. (17) with permission of the publishers.
Diversity and Phylogenetic Relationships: Phylum Basidiomycota
From a phylogenetic perspective, the phylum Basidiomycota is the sister group to the phylum Ascomycota, forming together the subkingdom Dikarya. Three independently evolving lineages are strongly within the Basidiomycota (Fig. 2) (3). The subphylum Agaricomycotina contains most of the described species (ca. 21,000), including many mushrooms as saprophytes or mycorrhizal symbionts of plants, the jelly fungi, and a large diversity of yeasts, some of which are important pathogens of humans (viz., Cryptococcus neoformans) (4, 5). The subphylum Ustilaginomycotina comprises more than 1,700 species, and while many species are pathogens of graminaceous plants (such as the maize smut Ustilago maydis), others are commonly associated with human and animal infections and are known from their asexual (anamorphic) states only (viz., Malassezia spp.) (4, 6, 7). The subphylum Pucciniomycotina is a sister group of the clade containing Ustilaginomycotina and Agaricomycotina and consists of more than 8,400 described species (8). Besides the array of saprobic yeast species usually recovered from soils, aquatic habitats, or the phylloplane (e.g., Rhodotorula spp. and Sporobolomyces spp.) (9), most of the Pucciniomycotina species are plant parasites, such as the obligate pathogenic rust fungi (e.g., Puccinia spp.) or anther-smut fungi (Microbotryum spp.). Through the diversification of mating and dispersal stages, this huge variety of fungal lifestyles is highly integrated with equally diverse sexual cycles and breeding systems, a theme that has long been the subject of study by influential mycologists (1, 10–13).
FIGURE 2.

Phylogeny of the Basidiomycota indicating the breeding system and the number of MAT genes across representative species of the three subphyla. The breeding system and the different taxonomic lineages are color-coded as given in the key and are kept consistent in all figures. Gene numbers shown for each species were obtained either from previous reports (20, 41, 100) or from newly surveyed genome data (marked with a hash sign after the species name). In the Agaricomycetes, values shown in parentheses are putative non-mating-type pheromone receptors. A question mark indicates cases where information on the breeding system is not available or is uncertain (e.g., because the sexual stage of a species is unknown). A schematic representation of the P/R and HD loci is given in Fig. 3 for representative species of each lineage marked with numbers enclosed in white circles. Letters in superscript next to the number of pheromone precursor genes indicate that (a) all genes encode the same mature pheromone peptide or that (b) no CAAX motif was detected in one of the putative pheromone precursors. The species phylogenetic tree was constructed in IQ-TREE (193) using a previously described approach (194). Branch support values are shown in the tree nodes as given in the key and were assessed with the ultrafast bootstrap approximation (UFBoot) and the approximate likelihood ratio test (SH-aLRT), each with 1,000 replicates. The basidiomycete clade is rooted with sequences from Ascomycete fungi.
Sexual Development and Determination of Cell Type Identity
Most fungi are able to undergo both asexual and sexual reproduction and have evolved tightly controlled mechanisms to regulate the process of mating, with respect to timing, and gamete dispersal, recognition, and fusion. In basidiomycetes, the sexual cycle typically involves fusion of genetically distinct homokaryotic hyphae or haploid yeast cells to produce a dikaryon, in which the two haploid parental nuclei are replicated in a coordinated fashion without fusion during hyphal elongation, usually involving the formation of clamp connections (i.e., a hook-like structure formed by hyphal cells to ensure proper distribution of the two genetically distinct nuclei during mitotic cell divisions; see below) (14). Nuclear fusion (karyogamy) then takes place in the basidia or in other specialized structures (e.g., teliospores), after which the diploid nucleus undergoes meiosis to generate haploid basidiospores (meiospores) and complete the life cycle (see Fig. 1 for representative life cycles).
Despite the wide variation of sexual cycles in nature, one common underlying feature shared by most fungi is the lack of genetically determined anisogamy: many fungi are isogamous (i.e., where all gametes have the same sizes), and even in anisogamous species, all haploid genotypes produce both types of gamete sizes. This means that there are not individuals of different sexes in fungi (15). Furthermore, many fungi are heterothallic, meaning that syngamy can only occur between gametes of different genetically determined mating types (15–17).
In members of the sister phylum Ascomycota, mating-type identity is governed at a single genetic mating type (MAT) locus (see reference 197). In that case, only two mating types segregate in meiosis, defining what is termed a bipolar system. By contrast, basidiomycetes have evolved a breeding system that relies on two genetic MAT loci. One locus encodes tightly linked pheromones and pheromone receptors (hereafter referred to as the P/R locus), and the other encodes homeodomain-type transcription factors (hereafter, HD locus), determining viability following syngamy. For successful mating and completion of the sexual cycle, haploid cells that conjugate must differ at both MAT loci (18, 19). When the two MAT loci are unlinked, four mating types can be generated by meiosis among the haploid progeny, defining this as a tetrapolar breeding system. Other basidiomycetes have instead a bipolar system controlled by a single MAT locus, either because the P/R and HD loci are linked or because one has lost its function in mating-type determinism. In mushroom-forming species (Agaricomycetes), there has been a generalized diversification of alleles at both MAT loci, in some cases yielding species with hundreds or thousands of possible mating types (1, 20–22). Data compiled from early studies indicate that as many as 65% of the species in the Agaricomycotina are tetrapolar (13, 23), whereas classical mating studies indicate a predominance of bipolar systems in the majority of the Ustilaginomycotina (24) and the Pucciniomycotina (25). In the following sections we summarize current knowledge of the molecular basis of mating-type determination in basidiomycetes and discuss transitions in breeding systems, using examples from throughout the Basidiomycota phylogenetic diversity.
MOLECULAR DETERMINANTS OF MATING TYPE
The Pheromone/Receptor Sensing System
In the Basidiomycota, syngamy is governed by, in its simplest form, small 10- to 15-amino acid-lipopeptide pheromones derived from 35 to 40 amino acid precursors through posttranslation modifications at both the N- and C-termini (26–28). These diffusible pheromones are received by seven transmembrane-domain pheromone receptors, coupled to a G-protein for downstream signal transduction (28, 29). This molecular determination of mating fusion is similar in part to the a- and α-factor P/R system in the ascomycetes and is thought to predate the separation of Asco- and Basidiomycota lineages. There is, however, one remarkable difference between the phyla: only homologues of genes encoding the pair of the a-factor pheromone and Ste3 pheromone receptor of the ascomycete system appear to exist in basidiomycetes. Therefore, instead of the a-factor/Ste3 and α-factor/Ste2 coupled sensing system characteristic of the ascomycetes (described in reference 197), “chemo-sensing” specificity in basidiomycetes is mediated by allelic variants of the same type of genes (18). Mating is often initiated by a reciprocal exchange of pheromones recognized by matching pheromone receptor variants in both mating types, and thus two strains need to carry different alleles of the pheromone and receptor genes at the P/R locus.
All active basidiomycete mating pheromones isolated so far are hydrophobic diffusible lipopeptides that undergo farnesylation at the C-terminal cysteine residue of the CAAX motif (where “C” is cysteine, “A” is often an aliphatic amino acid, and “X” is any residue) and amino-terminal processing. This was first demonstrated in the secreted pheromone (rhodotorucine A) of the red-pigmented yeast Rhodosporidium toruloides (recently renamed Rhodotorula toruloides) (30, 31) and later extended to many other basidiomycetes (26, 32–36), and also for the a-factor of the ascomycete yeast Saccharomyces cerevisiae that now stands as the model paradigm for the fungal kingdom (37, 38).
Among the basidiomycetes, exceptional pheromone gene structures were revealed in members of the Pucciniomycotina. Where investigated, the pheromone precursor genes each encode tandem copies of the mature peptide moiety (39–43), and an initial step of processing of a spacer region is required to reveal C-terminal CAAX motifs for each repeat (37, 44). In species where pheromones of opposite mating types have been clearly identified (namely, in R. toruloides, Sporidiobolus salmonicolor, Leucosporidium scottii, and Microbotryum spp.), the MAT A1 pheromone precursor typically presents a greater number of repeats of the peptide moiety (up to six copies in some species), whereas the MAT A2 pheromone gene appears to encode only one or two copies of the mature pheromone (39, 42, 43, 45). Interestingly, this increased production of pheromone per mole of precursor in MAT A1 cells, possibly allowing communication with A2 cells at longer distances, is consistent with the observed asymmetric growth of conjugation tubes in both R. toruloides (46) and Microbotryum spp. (43, 47), which occurs earlier and to a greater extent from MAT A2 cells. These findings suggest that the two mating types may have distinct signaling roles during conjugation, despite the absence of morphologically differentiated cells.
In response to the interaction of the pheromone ligand with a suitable pheromone receptor, mating and sexual development are initiated through a heterotrimeric G protein located at the plasma membrane that triggers a variety of downstream signaling processes. In U. maydis, where this has been investigated in greater detail, this signaling is attributed to two interconnected pathways involving a cAMP-dependent protein kinase A and a mitogen-activated protein kinase (28, 48–50). The point of convergence of the two pathways is the pheromone response factor (Prf1), which is an HMG-box transcription factor that recognizes and binds to pheromone response elements located in the regulatory regions of pheromone-induced genes (51, 52). Interestingly, whereas most of the core components of the mitogen-activated protein kinase cascade pathway (e.g., Ste11, Ste7, Fus3) are conserved over wide evolutionary distances, the key downstream transcription factors that ultimately activate or repress their target genes are often not conserved among species and thus are difficult to reveal through candidate gene approaches or sequence comparison.
Homeodomain Transcription Factors: The Second Compatibility Checkpoint
While the P/R system controls syngamy, the production of viable mating products requires compatibility at additional genetic components in the basidiomycetes, which is mediated by genes at the HD locus. In its most basic configuration, the HD locus encodes a pair of proteins of dissimilar homeodomain classes, having protein domains HD1 or HD2, and their coding genes are normally adjacent and divergently transcribed. Development following syngamy depends upon the formation of functional heterodimeric transcription factors where dimerization is restricted to HD1 and HD2 proteins that originate from haploid mates bearing different alleles of the HD locus. This requirement ensures that active heterodimers are only formed in the dikaryotic stage and do not arise in haploid cells. Failure to form heterodimers in the haploid is explained by interfering amino acids at HD1 and HD2 proteins from the same allele that disturb their general cohesiveness (53–55). Studies have shown that the relevant interfaces for discriminating HD1-HD2 interactions and determining allele specificity seem to be located N-terminally to the DNA binding motifs and that this region is usually highly variable between different alleles of both HD1 and HD2 genes (53, 54, 56). Analyses in U. maydis of chimeric and mutant alleles and protein-protein interaction studies using the yeast two-hybrid system revealed that dimerization between HD1 and HD2 proteins involves polar-hydrophobic interactions with variable cohesive contact sites contributing to binding affinity (54, 57, 58). Once formed, the functional heterodimers will bind to gene promoter elements, leading to the induction of the specific sets of genes involved in subsequent differentiation, which can include a morphological switch from yeast-like cells to filamentous growth and pathogenicity, as in the smuts, or fruiting body formation in the mushrooms (28, 41, 58–60).
BREEDING SYSTEMS IN THE BASIDIOMYCOTA
The distinction between heterothallic and homothallic compatibility represents a fundamental breeding system classification in fungi, and both systems are found across the Basidiomycota. In heterothallic basidiomycete species, sexual reproduction is only possible between gametes carrying different alleles at both MAT loci, preventing intrahaploid mating (i.e., selfing at the haploid level) (61). On the other hand, homothallic species produce universally compatible gametes, each being able to undergo intrahaploid mating with their clonemates. We have summarized in Fig. 2 and Fig. 3 data regarding the breeding system and the genomic organization of the mating-type genes in representative species of the three major subphyla of the Basidiomycota, some of which will be highlighted in the following sections.
FIGURE 3a.

Schematic showing the genomic structure and diversity of MAT loci in representative basidiomycete lineages. The genomic organization of the homeodomain (HD) and pheromone/receptor (P/R) MAT loci is shown for selected species of (A) the Agaricomycotina and (B) the Ustilaginomycotina and Pucciniomycotina. Arrows indicate genes and their direction of transcription. Putative MAT loci are shaded in light brown, and MAT genes are colored as indicated in the key with different color grades representing different alleles (or paralogs). When known, conserved genes flanking MAT loci (colored light yellow or light blue) are shown within each lineage. Genes that encode components of the pheromone response pathway are shown in pink and are in many cases within the MAT locus. Putative homologs of a protein required for posttranslational modification of pheromone precursors (isoprenyl cysteine methyltransferase [ICMT]) are colored purple and appear near the P/R locus in some species. P/R loci no longer determining mating-type specificity in bipolar Agaricomycetes are depicted with a gray background. In M. lychnidis-dioicae, the two mating-type chromosomes are highly rearranged and enriched in transposable elements, so that only a small number of genes is depicted. Of note, whereas in M. nashicola P/R and HD genes are far apart on the same chromosome, in E. vaccinii and M. endogenum the two sets of genes are closer together. Other genes or genomic features are colored or represented as given in the key. For citations and additional details, see text. Gene names or their associated protein accession numbers are shown as they appear in their respective genome databases, except in species of Ustilaginomycotina and Tremellomycetes, where names were given based on sequence identity to the closest homolog in U. maydis and C. neoformans, respectively.
Tetrapolar Systems: U. maydis and Other Smut Relatives
The maize smut U. maydis (order Ustilaginales; Fig. 2) is one of the most intensively studied basidiomycete models with respect to the organization and function of mating-type genes (62). The fungus has a dimorphic life cycle with a yeast-like haploid saprophytic phase switching to filamentous pathogenic growth in the dikaryotic stage following syngamy (Fig. 1A). In this species, the process of mating and completion of the sexual cycle is regulated by a tetrapolar breeding system, i.e., with the P/R (or a) locus unlinked from the HD (or b) locus. The P/R locus exists in two allelic forms (a1 and a2), which are defined by structurally dissimilar DNA regions of 4.5 kb for the a1 and 8 kb for the a2, each comprising one pheromone receptor gene (PRA1 or PRA2) and one pheromone precursor gene (MFA1 or MFA2) (Fig. 3B). The Pra1 receptor only responds to the Mfa2 pheromone, while Pra2 can only recognize Mfa1. The observed size difference between the two allelic forms of the P/R locus is due to the presence in the a2 allele of two additional genes (lga2 and rga2) that direct uniparental mitochondrial DNA inheritance, as well as a third, nonfunctional pheromone-encoding gene (26, 33, 52, 63).
Interestingly, the presence of this pseudogene led to early speculations about the existence of a complex scenario ancestral to the Ustilaginales in which the P/R locus occurred in more than two allelic forms, with this additional cryptic pheromone gene being recognized by a distinct receptor encoded by a third unidentified P/R allele (52). This hypothesis was substantiated upon characterization of the breeding system of the closely related head smut fungus Sporisorium reilianum. Compared to U. maydis, the P/R locus of S. reilianum exists in three alleles, each containing two distinct pheromone genes and one pheromone receptor (64) (Fig. 3B). Importantly, each of the mature pheromones in one allele interacts specifically with only one of the receptors encoded by the other two alleles. For instance, the pheromones encoded by the MFA1.2 and MFA1.3 genes at the a1 locus interact with the Pra2 and the Pra3 receptors encoded by the a2 and a3 alleles, respectively (Fig. 3B). This leads to a scenario in which each mating type can detect (and be detected by) the other two, but not itself. Additional support for this ancestral triallelic P/R system model was more recently attained in a study where the complete P/R loci of additional smut species spanning about 100 million years of evolution in the order Ustilaginales were sequenced and compared (65). Interestingly, the three P/R alleles previously described in S. reilianum were conserved and generally syntenic among members of the same order. This supports a potential scenario in which the biallelic P/R system as observed in U. maydis resulted from the loss of the a3 allele and loss of one of the two pheromone genes, which is now lacking in extant a1 and a2 alleles. Accordingly, a phylogenetic analysis using pheromone receptor sequences from a wider taxonomic range (Fig. 4) shows that all Ustilaginomycotina receptors group together and within one of the three allelic classes previously defined (PRA1, PRA2, PRA3) (65). This result denotes that the triallelic P/R system is probably as old as the stem age of the Ustilaginomycotina, about 450 million years (66).
FIGURE 4.
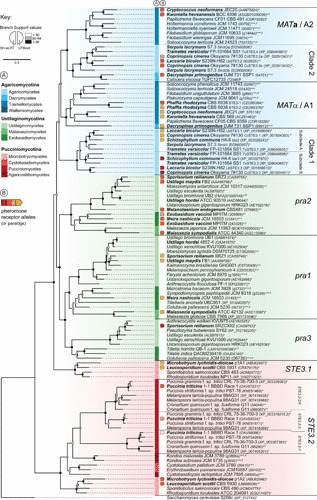
Phylogeny of Basidiomycota pheromone receptor proteins. Amino acid sequences identified by BLAST from publicly available databases or from genome projects were retrieved for representative species of the tree subphyla of the Basidiomycota. A total of 106 sequences were manually inspected, amended where necessary, and aligned with MAFFT (195), and poorly aligned regions were trimmed with trimAl (196). The phylogenetic tree and branch support were obtained as in Fig. 2, and the tree was rooted with S. cerevisiae Ste3p. GenBank accession numbers (*), Joint Genome Institute protein identifiers (**), and RIKEN/NBRP identifiers (***) are given after the strain name, with letters in superscript indicating (a) genomes assembled from available raw sequencing data and inspected locally, (b) genomic contigs/scaffolds lacking gene annotation, and (c) genes whose annotation was corrected. Species highlighted in boldface are shown in Fig. 3, with arrows before their names indicating the allelic version (or paralog) of the pheromone receptor as colored in Fig. 3. Of note, the STE3.1 and STE3.2 alleles in the Microbotryomycetes (Pucciniomycotina) displayed the deepest allelic divergence and trans-specific polymorphism, with the STE3.1 alleles of the different species branching together rather than each of these alleles clustering with the STE3.2 allele from the same species (42, 45, 137).
Following fusion of compatible partners, heterozygosity at the HD locus is required for dikaryon maintenance and filamentous growth in U. maydis (54, 67). The two divergently transcribed HD genes encode homeodomain proteins known as bE (for bEast; HD1-type) and bW (for bWest; HD2-type). However, in contrast to the biallelic and triallelic P/R locus of U. maydis and S. reilianum, respectively, the HD locus is highly multiallelic: in U. maydis more than 30 intercompatible allelic versions were found in nature (68; DeVay, cited in 69), while in S. reilianum five alleles have been identified so far (64). The homeodomains and C-terminal regions of the allelic HD1 and HD2 proteins are highly conserved, whereas the sequences N-terminal to the homeodomain regions are highly variable and required for the interaction of nonallelic HD1 and HD2 proteins (54).
Variation in Tetrapolar Systems beyond Ustilago/Sporisorium
Outside the Ustilaginomycotina, the number of tetrapolar species characterized for molecular polymorphisms at the MAT loci has increased in recent years (42, 45, 70–74). Kwoniella heveanensis (70) and Cryptococcus amylolentus (71) in the Agaricomycotina and L. scottii (42) in the Pucciniomycotina are examples where the P/R locus is biallelic, while the HD locus displays greater polymorphism (Fig. 2 and 3). In all three, the P/R and HD loci localize to different chromosomes. Sequencing of the P/R locus in each species showed their biallelic nature but, in comparison to U. maydis and S. reilianum, the P/R locus in K. heveanensis and L. scottii covers a relatively larger genomic region (about 32 kb and 50 kb, respectively; Fig. 3A and 3B), which is highly rearranged between mating types. Included here are the genes encoding MAT-specific pheromones (MFA or RHA) and receptors (STE3), as well as genes previously shown to be required for the onset of filamentation in dimorphic species (e.g., STE20, STE12) (75–77) and lineage-specific genes not obviously involved in mating (Fig. 3). The HD locus in K. heveanensis and C. amylolentus contains only the divergently transcribed homeodomain genes, HD1 and HD2 (named SXI1 and SXI2) (70, 71). In L. scottii, in contrast to K. heveanensis and C. amylolentus, the boundaries of the HD locus may have extended further outside from the HD1/HD2 gene module, given the evidence of increased sequence divergence in a region spanning about ∼80 kb that encompasses the HD1/HD2 genes (42). Sequence analysis of this variable region from different K. heveanensis isolates provided evidence that the locus is multiallelic with at least six different HD1/HD2 alleles (70). The same holds true for L. scottii, but with 28 HD alleles being revealed upon inspection of 43 isolates (42).
From an evolutionary standpoint, it is noteworthy that extant tetrapolar dimorphic basidiomycetes have more commonly a biallelic P/R locus (26, 70–72, 74, 78). For these, the proportion of compatible mates in the population cannot exceed 50%, irrespective of the number of alleles at the HD locus. However, given that compatibility at the HD locus can only be assessed after syngamy, increased HD allele diversity should be naturally selected when outcrossing is the rule, to allow more frequent dikaryon viability after syngamy and, in this way, limit the amount of wasted mating opportunities.
Tetrapolar Systems with Multiple Alleles in the Agaricomycetes
A vast repertoire of mating types has been found in most members of the class Agaricomycetes (Fig. 2), such as the wood-rotting mushroom Schizophyllum commune (Fig. 1D) with more than 15,000 intercompatible mating types, and the ink cap mushroom Coprinopsis cinerea with an estimated 12,000 distinct mating types (1). Such diversity is likely due to selection for rare alleles under outcrossing: any new specificity will be compatible with all extant mating types and therefore may increase in frequency under this selective advantage (15, 79). Studies have shown that two hallmarks of the MAT loci of these and other heterothallic Agaricomycetes account for such astonishing numbers of mating types within a single species. First, a single locus may itself be extremely polymorphic, in some cases with hundreds of specificities (1, 13, 16); second, for some species, multiallelism in both P/R and HD loci is generated by rounds of segmental duplication and diversification of MAT genes, resulting in independent but functionally redundant subloci, upon which recombination can act to give rise to novel allele specificities and, by definition, new mating types (80).
Recent advances in genome sequencing are now allowing detailed comparisons of MAT loci among diverse species, and a comparative overview of these genomic regions in several Agaricomycetes is presented in Fig. 3A. A great body of literature exists on the mating-type systems and distribution in Agaricomycetes, preventing a full discussion, and the reader is referred to some exceptional and recent reviews (16, 20). Here, we shall concentrate mainly on the model mushroom C. cinerea, which has long served as an invaluable model to understand the molecular underpinnings of multiple mating types and multiallelic MAT loci in the Agaricomycetes. Where relevant, reference will be made to other mushroom species, but many of the principles that govern the evolution of multiple mating types in C. cinerea seem also to hold in other species.
Initial studies in C. cinerea using classical genetics determined that the HD locus is multiallelic and composed of two subloci (Aα and Aβ; Fig. 3A) that can recombine at detectable frequencies despite their close proximity in the genome (81, 82). Molecular genetic studies and genome sequencing later revealed that, whereas the Aα sublocus encodes one HD1/HD2 gene pair, the Aβ sublocus encodes up to three pairs and is located at a distance of ∼7 kb from the Aα sublocus (Fig. 3A) (83–86). Recombination between the two subloci is possible through the ∼7 kb-long intergenic region that is highly conserved between alleles. Sequence and functional analysis of five HD alleles led to a theoretical archetypal structure for the C. cinerea HD locus with three paralogous HD1/HD2 gene pairs (known as the a, b, and d pairs). This implies that molecular mechanisms akin to gene deletion have been at work during evolution of the HD locus, because the extant alleles have fewer than the six HD1/HD2 genes postulated in the model (exemplified by allele A43 in Fig. 3A) (83–85). At present, four, seven, and three distinct alleles have been identified for the a, b, and d pairs, respectively (56, 85), generating 84 HD alleles considering all possible permutations. Although this is about half the number estimated by John Raper, 160 HD alleles in nature (1), the identification of one additional allele per group would be sufficient to reach Raper’s estimates. However, the actual numbers of alleles in nature are of course unknown.
Similarly, the P/R locus in C. cinerea is multiallelic and consists of three tandemly arranged but independent groups of paralogous genes (groups or subloci 1 to 3). The sequencing of several alleles revealed that each group contains multiallelic and functionally redundant genes encoding at least one pheromone receptor and up to four pheromone genes (Fig. 3A) (20). In any given group of paralogous genes, the relative order, direction of transcription, and number of genes may vary and, importantly, each group has freely interchangeable alleles (80). In C. cinerea, from 13 sequenced P/R alleles, two, five, and seven alleles were identified for groups 1, 2, and 3, respectively. Together, this gives rise to 70 possible combinations, approaching the number of 79 P/R alleles estimated to exist in nature (1). Phylogenetic analyses of C. cinerea receptors of the three paralogous gene groups and from different alleles added yet another layer of complexity in the evolutionary history. Strikingly, the different allelic versions of the pheromone receptors as defined by their position within the P/R locus do not always fall into the same clade (20, 80), an observation that led some authors to conclude that gene shuffling occurred within the locus during evolution (80). This pattern has also been observed in S. commune (87) and in Ganoderma lucidum (88). Such relocations may have restricted intralocus recombination at some point during evolution, thereby contributing to the fixation of some of the alleles in the populations.
With additional pheromone receptors from other Agaricomycetes added to the phylogenetic analysis, the origin of the paralogous groups of genes within the P/R locus can be traced back to an early stage of the evolution of this taxonomic lineage, with a duplication event initially separating these genes into two distinct evolutionary groups (clades 1 and 2 in Fig. 4). More recent duplications (e.g., CcSTE3.1 and CcSTE3.3 in C. cinerea or LbSTE3.1 and LbSTE3.3 in Laccaria bicolor) (Fig. 4) followed by sequence diversification and recombination (80) are suggested to have led to the high numbers of alleles presently estimated from natural populations of several mushroom species (16, 20, 41, and references therein).
In C. cinerea and other Agaricomycetes, each encounter of two monokaryotic hyphae results in promiscuous cell-cell fusion irrespective of whether or not they are compatible at MAT (18, 89). However, for the development and maintenance of the dikaryon, nuclei of interacting partners must harbor different allelic versions of genes in at least one sublocus of both P/R and HD loci (18, 89–91). Classical genetic analyses of these semicompatible interactions in C. cinerea and S. commune have shown that the P/R system is involved in events that follow hyphal fusion, namely in the reciprocal exchange and migration of nuclei and clamp cell fusion (Fig. 5). On the other hand, genes encoded at the HD locus repress asexual sporulation (92), regulate pairing of nuclei within the dikaryotic tip cell, and coordinate nuclear division, clamp cell formation, and septation from the subapical cell (1, 18, 93) (Fig. 5). If the two mates share identical alleles at P/R and HD loci, no nuclear exchange occurs and the fungal individuals resume monokaryotic growth.
FIGURE 5.

Roles of P/R and HD genes on the formation and maintenance of the dikaryon in C. cinerea. Pheromone signaling is not required to attract mates, and hyphal fusion is mating-type independent (diagram 1). Upon fusion, nuclei enter the mycelium of the other mate and migrate until they reach a hyphal tip cell (diagram 2). During hyphal tip elongation, the two types of haploid nuclei (depicted in white and black, representing different MAT genotypes) pair at the tip cell (diagram 3), and at the place where mitosis will take place, a hook-like structure (called a clamp connection) is formed (diagram 4). The two nuclei divide synchronously: one of the nuclei divides in the direction of the clamp cell that is growing backward toward the main hyphae, while the other divides along the main hyphal axis (diagram 5). Septa are generated between the dividing nuclei. This way one nucleus stays temporarily entrapped in the clamp cell, one nucleus of the other type is enclosed in the newly formed subapical cell, and a nucleus of each type is maintained in the emerging hyphal tip cell (diagram 6). The clamp cell fuses with the subapical cell and releases the entrapped nucleus from the clamp cell, restoring the dikaryotic state of the subapical cell (diagram 7). In C. cinerea, steps controlled by P/R (diagrams 2 and 7) and HD (diagrams 3 to 6) genes are colored red and green, respectively. See Casselton et al. (18) and Kües (90) for details. The micrograph at the bottom was obtained from Stajich et al. (86), with permission.
Additional pheromone receptor-like genes (i.e., STE3-like) have been identified from numerous Agaricomycetes using sequence similarity-based approaches (Fig. 3A), and many fall within one of the three clades of Agaricomycete mating-type receptors (subclades A and B of clade 1 as well as clade 2) (Fig. 4). However, their function remains largely elusive, because reports suggest that these are non-mating-type-specific receptors (94, 95), and experimental evidence from S. commune suggests that they are not sufficient per se to induce mating-specific development (96). Many of these non-mating-type-specific receptors are located in the close neighborhood of the P/R locus, while others are unlinked (Fig. 3A). Three distinctive features of these Ste3-like receptors are seen in comparison to the mating-type-specific receptors: (i) they present longer C-terminal cytoplasmic regions, (ii) lack pheromone genes in close association, and (iii) show lower levels of intraspecific polymorphism. Non-mating-type-specific receptors have been documented in C. cinerea (41), Phanerochaete chrysosporium (97), L. bicolor (98), Postia placenta (99), and several species of the order Polyporales (100), and many more can be expected to be identified from whole-genome sequence data. For example, up to 29 Ste3-like receptors were identified in the genome of Serpula lacrymans (Fig. 2). A few theoretical possibilities have been proposed for their function (41). One such possibility considers that they may play a role during monokaryotic fruiting, in a way similar to that described for the C. neoformans non-mating-type receptor gene CPR2, whose overexpression elicits homothallic reproduction (101). Another hypothesis suggests that these genes may function in vegetative communication within a species or in conspecific recognition avoiding hybridization (41). Experimental studies addressing the function of these non-mating-type-specific receptors are thus critically needed.
Comparative genomics revealed that gene content and organization (synteny) are generally conserved around the HD locus in Agaricomycetes. Two genes usually border this locus: one encodes a mitochondrial intermediate peptidase (MIP), which has served as a valuable marker for the isolation of HD loci from nonmodel species, and the other is known as a beta-flanking gene (β-fg) for an unknown protein (Fig. 3A) (94, 100, 102). Although this configuration is found in most species analyzed, exceptions have been reported. In S. commune, for example, two large rearrangements at the HD locus have taken place causing the separation of the HD locus into two clearly distinct subloci that now lie more than 500 kb apart and frequently recombine during meiosis (Fig. 3A) (103, 104). A similar situation is observed in Flammulina velutipes (winter mushroom or enoki), but the distance between the two HD subloci is about 70 kb. Interestingly, whereas synteny analysis of the surrounding regions indicates that the separation of the two HD subloci in S. commune emerged via transposition of the two gene clusters, the extant arrangement in F. velutipes was driven by two large inversions (95). Other exceptions to the typical MIP–HD1/HD2–β-fg gene arrangement were observed in the cacao pathogen Moniliophthora roreri, where both MIP and β-fg are located 40 and 60 kb upstream of HD1/HD2 (105), respectively, and in the Shiitake mushroom, Lentinula edodes, where MIP is distant from the HD locus (106). Gene order conservation at the P/R locus is also observed among Agaricomycetes, although to a lesser extent compared to the HD locus. The P/R locus in L. bicolor was shown to be in a chromosomal region more prone to gene duplication, translocations, and accumulation of transposable elements (98), and in the wood-decay basidiomycete Botryobasidium botryosum (order Cantharelalles; Fig. 2), genome inspection revealed the presence of transposons in the immediate vicinity of pheromone and receptor genes (Fig. 3A; this report). Another illustrative example is the position of STE20 and STE12 genes, which varies among the species shown in Fig. 3A.
Tetrapolarity with multiple alleles at both MAT loci is often suggested to promote outcrossing (107, 108). It is true that tetrapolarity is less favorable to selfing than to outcrossing, because the chances that any two haploid siblings from the same diploid parent are compatible is at most 25% for tetrapolarity, compared to 50% from a bipolar cross (107). However, tetrapolarity also yields only 25% compatibility under outcrossing when both mating-type genes are biallelic, so that tetrapolarity in itself does not favor outcrossing compared to selfing under biallelism. It is multiallelism that may restrict inbreeding, however, and the evolution of high numbers of mating types is suspected to be primarily driven by selection for increased compatibility at syngamy under an already established outcrossing mating system. An evolutionary scenario could be one with ancestral mating-type determinism being biallelic at the HD locus and where the high level of gamete dispersal evolves, leading to widespread outcrossing, which in turn selects for rare alleles, yielding multiallelism at the HD locus. Recruiting a biallelic P/R locus in mating-type determinism would not favor outcrossing, because again, compatibility odds will be similar under outcrossing compared to selfing. Different scenarios for explaining the incorporation of the P/R locus in the mating-type determinism system are possible, such as its role in syngamy or mate attraction, or again, evolution first of multiallelism (e.g., due to population differentiation). Studies addressing the pattern of mating in nature, depending on gamete dispersal rates and polymorphism levels at the mating-type genes, will be fundamental to test some of these predictions.
Bipolar Breeding Systems: Genomic Changes and Evolutionary Considerations
Early studies indicated that, although the tetrapolar mating system predominates in the Basidiomycota, a considerable number of species are bipolar. For example, roughly a quarter of the mushroom species are bipolar (1, 13) and, significantly, they occur phylogenetically interspersed with tetrapolar relatives (100). Owing to the occurrence of multiple bipolar species within the basidiomycetes, the origin of tetrapolarity as the ancestral state to this clade is still debatable. However, several lines of evidence argue strongly in favor of this hypothesis. First, the tetrapolar system is architecturally complex, making it unlikely to have arisen multiple times (23, 109). Second, the same mating-type-determining genes (at both P/R and HD loci) are involved in mating-type determinism in species of the three major basidiomycete lineages, suggesting a common origin (41, 42, 108). Third, all bipolar species investigated so far display genomic signatures, suggesting that they derived secondarily from a tetrapolar system (detailed below). Based on these observations, the prevailing hypothesis is that the tetrapolar system evolved in a basidiomycete ancestor following the acquisition of a separate locus (the P/R locus) for MAT determination that segregated independently of the HD locus, and with multiallelism evolving in many lineages (17, 41).
As initially envisaged by John Raper (1), and more recently supported by solid molecular evidence (detailed below), bipolarity in the Basidiomycota appears to be the result of simple genetic changes. There are two general ways this can be achieved: (i) by linkage of the P/R and HD loci into a single, nonrecombining MAT region with often only two alleles of each locus being retained or (ii) by loss of function of the P/R locus in mating-type specificity, conceivably through mutations that cause constitutive expression of the P/R-regulated pathway or that produce a self-compatible allele at the P/R locus.
Transition to bipolarity in smuts and Malassezia through linkage of P/R and HD loci
There are now several direct examples that support the formation of a bipolar system through a chromosomal translocation that places both P/R and HD loci in tight linkage. In the Ustilaginomycotina, a prime example is provided by Ustilago hordei, a bipolar smut pathogen closely related to tetrapolar U. maydis (Fig. 2) (110). With the molecular analysis of the MAT loci, initially in U. maydis (26, 59, 111, 112), the genomic basis of the bipolar mating behavior of U. hordei was subsequently revealed: both P/R and HD complexes were physically linked on the same chromosome, 430 and 500 kb apart on the largest chromosome representing the respective MAT-2 and MAT-1 loci (Fig. 3B), and the presence of inversions in between these complexes was proposed to cause the almost complete lack of recombination of the complexes (113–115), likely resulting in a subsequent accumulation of transposable elements. Detailed analyses have since then been published on mating types in several Ustilaginomycotina spp.
Complete genomes of five species of closely related genera varying in mating-type organization are available: tetrapolar U. maydis (116) and S. reilianum (117) and the bipolar species U. hordei, with the HD and P/R loci 430 to 500 kb apart (118), Sporisorium scitamineum, with the HD and P/R loci 59 kb apart (119, 120) (Fig. 3B), and Ustilago bromivora, with the HD and P/R loci 183 kb apart (121). Based on comparison of chromosomal rearrangements between U. maydis, U. hordei, and S. reilianum, it was suggested that the tetrapolar organization in the S. reilianum lineage was ancestral and that part of S. reilianum chromosome 1 harboring the HD locus was translocated to its chromosome 20 harboring the P/R locus, to generate the bipolar organization in U. hordei (118). This potential scenario was strengthened in the recently published bipolar S. scitamineum genome: homologous parts of S. reilianum chromosomes 1 and 2 seem rearranged to give chromosome 2 in S. scitamineum (120). In all these cases, the presence of many repetitive and transposable elements at the breakpoints suggested they may be functionally involved (122). As stated above, the resulting spacing between the HD and P/R loci in these species varied between 59 and 500 kb, and this was likely a consequence of increasing numbers of such elements due to decreased recombination.
A similar MAT loci configuration has also been recently identified in the human skin commensal yeast Malassezia spp. complex that is responsible for dandruff and other skin pathologies. These include Malassezia sympodialis, Malassezia globosa, and Malassezia yamatoensis (123, 124) (Fig. 2). Similar to U. hordei, the P/R and HD loci in these three Malassezia spp. are also located on the same chromosome and are separated by large chromosomal regions that are >100 kb in size. Interestingly, comparison of the chromosomal regions encompassing the P/R and HD loci in these species also suggests that the linkage between the two MAT loci are the result of two independent events, with one giving rise to the linkage between the two MAT loci in M. sympodialis and M. globosa, while the other rearrangement led to the P/R and HD linkage in M. yamatoensis. Additionally, analyses of natural isolates of M. sympodialis suggest that recombination is still occurring between the P/R and HD loci in natural populations, indicating that linkage between the two MAT loci is not tight. In agreement with this, the 170-kb MAT region of M. globosa does not contain accumulations of repetitive elements as found at the MAT-1 locus of U. hordei (125), as is expected in regions of suppressed recombination. Moreover, a preliminary examination of the P/R-containing region in the genomes of two M. sympodialis strains of opposite mating types revealed conserved gene order in the immediate vicinities of the P/R gene pair, which are found adjacent but inverted in both mating types (Fig. 3B; this report). The breeding system in Malassezia species is thus considered “pseudo-bipolar,” a term initially proposed for the breeding system of the red-pigmented yeast S. salmonicolor to reflect a situation where multiallelism might occur despite the apparent loose linkage of the two MAT regions (45); linkage is expected to be selected for in selfing mating systems, in which rare alleles have no advantage, leading to gradual loss of alleles but two by genetic drift.
Bipolarity in pathogenic Cryptococcus spp. complex and in Trichosporon
Outside the Ustilaginomycotina, deviations from the classic tetrapolar system have also been observed in a variety of species. For example, all the species within the human pathogenic Cryptococcus spp. complex (Tremellomycetes, Agaricomycotina; Fig 2) (126) have bipolar mating systems. In these species, there are two mating types, MATa and MATα, defined by two alleles (a and α) at a single large MAT locus (Fig. 3A). The MAT locus in pathogenic Cryptococcus spp. is ∼120 kb in size and contains about 20 genes, including those encoding mating pheromones and their cognate receptors and homeodomain transcription factors (HD1 or SXI1 and HD2 or SXI2). There is, however, a distinctive feature regarding the normally two-gene homeodomain function in this group of species: the HD1 (or SXI1) gene is unique to MATα strains, whereas HD2 (or SXI2) is specific to the MATa strains (Fig. 3A). This is in contrast to other basidiomycetes in which a divergently transcribed HD1/HD2 gene pair is present in each mating type.
Accordingly, each mating type in pathogenic Cryptococcus spp. has been reduced to the minimal number of HD genes required to function upon fusion of compatible cells (127). Recessive losses of genes from one mating type could likely occur because each is sheltered in a permanent hemizygous state due to recombination suppression within the MAT locus, associated with extensive chromosomal rearrangements. However, gene conversion does occur within these regions and at a frequency that is at least comparable to the crossover frequencies in other chromosomal regions (71). Additionally, it has been shown that recombination hotspots associated with the presence of GC-rich motifs are located at the regions that flank the MAT locus in Cryptococcus (128, 129), which could potentially function as repressors of crossover within the MAT locus. Comparison of the MAT loci of closely related tetrapolar species such as C. amylolentus (71) and Cryptococcus heveanensis (70) suggests that the single MAT locus in the pathogenic Cryptococcus spp. is the result of fusion of ancestral P/R and HD loci, possibly mediated by repetitive sequences and transposable elements that may act as substrates for nonhomologous recombination (130, 131).
A recent genomic study found that in the oil-accumulating yeast Trichosporon oleaginosus (recently renamed Cutaneotrichosporon oleaginosus) (Fig. 2 and Fig. 3A), as well as in some of its sister species, the mating-type regions show significant structural differences from those of other Tremellomycetes that possess tetrapolar breeding systems. Specifically, no homologs of the HD2 transcription factor could be identified in the genomes of T. oleaginosus and Trichosporon asahii, suggesting that this transcription factor might have been lost during the evolution of the Trichosporon spp. complex (132). Alternatively, this gene is absent from only one mating type like the situation in the Cryptococcus spp. complex, a possibility which cannot be ruled out since the three sequenced strains carried the same alleles of P/R and HD genes. However, if the latter hypothesis ends up being true, then such gene loss would represent a case of parallel evolution in Trichosporon and the human pathogenic Cryptococcus spp. complex given that the last common ancestor of the two lineages most likely contained the standard HD1/HD2 gene arrangement. It would be of interest in this scenario to test whether the HD gene loss is associated with established genetic linkage between the genomic regions that contain the P/R and HD genes (Fig. 3A). Unfortunately, the fragmented nature of the available genome assembly of T. oleaginosus (132) and the lack of descriptions of a sexual cycle for any Trichosporon species prevent a more definitive answer at this time.
Expansion of the mating-type locus in Microbotryum following linkage of P/R and HD loci
The findings in the aforementioned bipolar species support the hypothesis of a tendency toward expansion of the regions of recombination suppression in mating-type regions in fungi (133), although it is not yet clear what underlying factors have been driving this trend, e.g., whether it is simply the linkage of the two mating-type loci. In line with this, a more extreme case of the evolution of bipolarity from tetrapolarity by linkage between the two MAT loci is observed in the anther-smut fungus Microbotryum lychnidis-dioicae. All species belonging to the genus Microbotryum are castrating floral smut pathogens that parasitize plants in the Caryophyllaceae family (Fig. 1B), forming a complex of sibling species highly specialized on different plant species (61, 134, 135). The most studied species is M. lychnidis-dioicae, which parasitizes Silene latifolia (136). This fungal species is bipolar, with only one MAT locus and two alleles, called a1 and a2 (137). This is among the first fungal species for which bipolar mating types have been described (138) and also the first fungal species for which size-dimorphic mating-type chromosomes were revealed (139). A sex event is required before each new plant infection, and M. lychnidis-dioicae displays a highly selfing mating system under heterothallism, performing mostly intratetrad mating (a.k.a. automixis) (140–142). The populations are therefore highly homozygous throughout much of the genome (143, 144).
A recent high-quality assembly of the genome of M. lychnidis-dioicae has revealed that bipolarity and mating-type chromosome dimorphism are due to the linkage of the P/R and HD loci in a very large region of suppressed recombination (145, 146). The centromere is also located in the nonrecombining region (145), which leads to the segregation of mating types in the first meiotic division. The mating-type chromosomes of M. lychnidis-dioicae thus do not recombine across 90% of their lengths (a1, 3.5 mega-base pairs [Mbp]; a2, 4.0 Mbp), with only small pseudo-autosomal regions at both edges that recombine. The nonrecombining region is highly rearranged, with at least 210 inversion events. As expected due to the less efficient selection in genomic regions without recombination (147), the cessation of recombination has led to genomic degeneration, with accumulation of transposable elements, nonsynonymous substitutions, biased gene expression, and the hemizygous loss of hundreds of genes (145, 148).
Most other anther-smut fungi are bipolar with dimorphic mating-type chromosomes that also show degeneration (148); one clade has recently been found to have a tetrapolar breeding system: in particular, Microbotryum saponariae and Microbotryum lagerheimii, which parasitize, respectively, Saponaria officinalis and Silene vulgaris (73). In these closely related species, the two MAT loci segregate independently, being located on different chromosomes, but are linked to their respective centromeres (73). This situation may have resulted from a reversion from bipolarity to tetrapolarity or from the independent evolution of the linkage of MAT loci under automixis in the other lineages. Indeed, both bipolarity with linkage to the centromere, as in M. lychnidis-dioicae, and linkage of both MAT loci to their respective centromeres on different chromosomes, as in M. saponariae, are equally favorable under automixis in anther-smut fungi (73, 145).
Transition to bipolarity through loss of function of the P/R genes in mating-type determination
It is not surprising that changes in the configuration of the MAT loci can alter the mating behavior of individuals, sometimes resulting in transitions between different breeding systems. For example, in tetrapolar heterothallic species, the HD locus encodes genes that heterodimerize during mating to form active transcription regulators, ensuring the proper development following syngamy. However, the heterodimer can only form between HD1 and HD2 proteins derived from different alleles. Thus, if recombination occurs between the HD1 and HD2 genes resulting in bringing together compatible HD1 and HD2 alleles on the same chromosome, a haploid individual bearing this novel HD genotype pair would transition from compatibility determined by two sequential compatibility checkpoints (P/R and HD) to a single mechanism (P/R) inherited in a bipolar manner. This newly formed HD allele could also be the first step in a transition from heterothallism to homothallism provided that compatible pheromones and receptors are brought together at the P/R locus in a subsequent event. Examples of these types of transition have been reported in C. cinerea, where fusion of the HD1 and HD2 genes from different subunits results in self-compatibility and constitutive sexual development (149). Alternatively, mutations in a pheromone receptor gene in C. cinerea resulted in a receptor that was either constitutively activated (150) or that responded erroneously to a normally incompatible pheromone encoded within the same allele (151), removing its function in mating-type determination.
Studies of several other Agaricomycetes, including Coprinellus disseminatus (152), Pholiota microspora (153, 154), P. chrysosporium (94), and Heterobasidion irregulare (155), have shown that a transition from a tetrapolar to a bipolar breeding system was indeed due to the uncoupling of the P/R from mating-type-determining functions. These bipolar species retained nonetheless a multiallelic HD locus, with the number of alleles defining the number of mating types (e.g., ∼123 mating types are estimated for C. disseminatus) (16, 152). Therefore, a new mating type will arise as soon as a novel HD allele is generated. However, evolving novel HD alleles that are still able to heterodimerize with extant alleles, while restricting self-activation, will become increasingly challenging (156). Compared to tetrapolar multiallelic species such as C. cinerea and S. commune, and assuming that HD alleles are found at equal frequency in the population, the probability of randomly chosen individuals being compatible in an outcrossing event is theoretically greater in bipolar multiallelic species: only a single locus needs to be compatible, rather than two in the tetrapolar situation (16). Furthermore, considering that mating in mushroom-forming fungi is preceded by basidiospore (meiospore) dispersal, outcrossing rather than selfing is more likely to occur (16, 61). Therefore, and because mating-type compatibility is not discriminated prior to syngamy, the costs of landing on an incompatible mate are in this case very high. It can thus be easily envisaged that mutations at the P/R locus causing loss of function in mating-type determination may have been selected for, in some circumstances, because the chance of compatibility would be greatly increased, and there is no selective pressure anymore to restrict inbreeding, which is already avoided by gamete dispersal.
Homothallism in the Basidiomycota
In some fungal species, haploid cells are universally compatible for mating, including among clonemates (a.k.a. intrahaploid mating, same-clone mating, or haploid selfing) (15, 79, 156). We call these species homothallic, and several proximal mechanisms have been proposed to account for such mating behavior: (i) the presence of compatible sets of MAT genes, either fused or unlinked, in one haploid genome (primary homothallism), (ii) mating-type switching (either bidirectional or unidirectional), and (iii) the lack of any gene-based mating-type discrimination (also known as the ability for unisexual reproduction or same-sex mating) (reviewed in 157, 158). Some other fungal species, referred to as pseudohomothallic, also have the ability to complete the sexual cycle without the apparent need for syngamy (159–161). However, pseudohomothallism is achieved by packaging two nuclei of compatible mating types derived from the same meiosis into a single spore. This process is thus more similar to diploid selfing via automixis or intratetrad mating and has different genetic consequences compared to intrahaploid mating (79, 162).
Homothallism is present in three main fungal lineages (the phyla Mucoromycota, Ascomycota, and Basidiomycota), indicating that this form of sexual behavior may provide a selective advantage under certain conditions, for instance, by promoting universal compatibility (79, 162). Compared to other phyla, however, homothallic basidiomycete species are less common. Whether this relates to the complex genetic underpinnings of the mating system in this phylum, as described above, or is because they can more easily evolve multiple alleles, allowing high frequencies of mating compatibility, is still an unresolved matter. Primary homothallism has been proposed for a handful of species in the Basidiomycota, most notably Sistotrema brinkmannii (163), in which homothallic and bipolar heterothallic individuals are found in the same population, and for the cacao pathogen Moniliophtora perniciosa (164). A partial genome sequence is only available for the latter species, which upon examination revealed various genes for putative pheromones and receptors and one or possibly two HD1 genes (165), but their genomic organization as well as their function during the sexual cycle remain undetermined. We present below details of two basidiomycete species for which there has been progress in understanding the genetic basis of homothallism.
Homothallic breeding system of Phaffia rhodozyma
Recently, inspection of the genomes of three strains of the homothallic yeast P. rhodozyma, belonging to the order Cystofilobasidiales (Fig. 2) (166), revealed the presence of pheromone precursor and receptor genes organized in two gene clusters, located ∼5 kb apart. Each cluster was composed of one pheromone and one pheromone receptor gene (either MFA1/STE3-1 or MFA2/STE3-2; Fig. 3A) (167). In contrast, a single HD1/HD2 gene pair was found in the genome that was not linked to the P/R locus (Fig. 2) (167). In elucidating the molecular interactions involved in mating compatibility in P. rhodozyma, David-Palma et al. (168) pursued targeted gene replacement of the MAT genes and genetic crosses using single, double, and triple mutants (168). They determined that both P/R gene clusters produced intercompatible gene products, enabling self-signaling, and each pheromone only interacted with the receptor of the other cluster (i.e., Mfa1 activates Ste3-2 and Mfa2 activates Ste3-1; Fig. 3A). Additionally, the single pair of HD1 and HD2 homeodomain proteins was also shown to be required and apparently cooperated for normal completion of the sexual cycle (168).
Even though the genetic makeup of MAT loci in P. rhodozyma is suggestive of primary homothallism, David-Palma et al. (168) pointed out why this configuration departs in several aspects from what would be expected from simply gathering in a haploid genome the intercompatible alleles of MAT genes. First, although the two pheromone receptor genes exhibited considerable sequence similarity (∼50% amino acid identity), phylogenetic analyses indicated that they were more closely related to each other than to receptor genes in other species, and both seemed to derive from the MATα/A1-related allele lineage (Fig. 4). This indicates that the two pheromone receptor genes in Phaffia have diverged much more recently than in most basidiomycetes (possibly coalescing within the genus) (M. David-Palma, personal communication), and the extant arrangement may be the result of fusion of two mating-type variants. The second aspect concerns the HD genes, in that only one HD1/HD2 pair is present, but both proteins are required for sexual development. Based on evidence of a very weak interaction between the two HD proteins in a yeast two-hybrid assay (168), it was proposed that at the generation of a compatible HD1/HD2 pair within the haploid genome there would need to be one or more mutations relaxing the structural hindrance that normally prevents interactions between HD proteins encoded by the same locus in heterothallic species (168). Similar investigations are currently under way in other species of the Cystofilobasidiales to reconstruct the evolutionary transitions in breeding systems in this lineage (M. David-Palma, personal communication).
Homothallism in C. neoformans
While most homothallic species can initiate sexual reproduction through fusion with any other haploid cell, including clonemates, they do so through the presence of two intercompatible mating alleles within a haploid genome. In the human pathogenic fungus C. neoformans, the proximal mechanism for homothallism is different because it concerns haploid cells with a single mating-type allele in their genome (169). Specifically, while C. neoformans has a bipolar breeding system in which mating typically occurs between MATa and MATα cells, it has been found that, in response to nutrient limitation, MATα cells can also complete the sexual cycle in the absence of MATa cells (169). The α-α diploid state can be established through either (i) cellular and nuclear fusion of two clonemates, (ii) nuclear fusion between mother and daughter nuclei, (iii) endoreplication of the entire genome of a single cell resulting in a diploid nucleus, or (iv) fusion of cells of the same mating type but of different genetic lineages (169). While the first three pathways represent different ways to achieve same-clone mating (haploid selfing), the latter enables outcrossing and thus the possibility to admix genetic diversity despite identical cell-type identity of the mating partners. In all cases, the resulting diploid cell undergoes monokaryotic filamentation and subsequent basidium formation, where meiosis takes place to produce the haploid spores as in the heterothallic mating cycle (Fig. 1C).
Importantly, recent studies using this model basidiomycete have shown that α-α mating between genetically identical cells can generate phenotypic and genotypic diversity de novo, including single-nucleotide polymorphisms (less frequently), chromosomal translocations, and aneuploidy (more frequently) which, although usually likely deleterious, may rarely provide a beneficial novelty (170). Moreover, crossovers have also been detected within the MAT locus during meiosis of genotypes resulting from α-α mating (129), presumably because the two MATα alleles are colinear, unlike a-α combinations (130). It should be pointed out that among the C. neoformans natural isolates, the vast majority (up to >99%) are of the α mating type (171). Thus, α-α mating could be beneficial when a compatible mating partner for a heterothallic cross is hard to find in nature. In turn, it is also possible that α-α mating played a role in generating the highly imbalanced mating-type ratio observed among natural isolates. Recently, this type of homothallism determinism has also been identified in the human pathogen Candida albicans (172, 173) as well as in the tree wound-infecting fungus Huntiella moniliformis (174), suggesting that it could be more prevalent among fungal species than is currently appreciated.
Evolutionary considerations on the costs of homothallism
Although it has been traditionally accepted among mycologists that homothallism evolved to promote same-clone mating (170, 175), it is important to point out that the potential for same-clone mating of a species as assessed in a laboratory setting does not imply that this is the prevalent mating system of the species in nature. Moreover, the sexual cycle involving clonemates does not result in actual recombination, the supposedly main advantage of sexual reproduction (176), while still incurring some costs (e.g., slower reproduction and maintenance of the meiotic machinery). Same-clone mating is less efficient at purging deleterious mutations: homologous chromosomes carrying different deleterious mutations that occurred in a given large clonal lineage can of course be recombined and produce a chromosome free of deleterious mutations (177), but this constitutes more outcrossing (i.e., mating between two different genotypes) than same-clone mating per se. Besides, in a population undergoing only same-clone mating, different deleterious mutations that are fixed by genetic drift in distinct clonal lineages cannot be eliminated from the populations, and Muller’s ratchet will inexorably lower population fitness (133). Furthermore, same-clone mating cannot bring together different beneficial mutations that evolved in distinct clonal lineages (Hill-Robertson interference). A transition to asexuality would bypass some of the costs of sex while producing haploid progeny that are genetically the same as the participants in same-clone mating.
Homothallism has been suggested instead to have evolved due to the selective advantage of increasing the number of available mates, because it can be seen as a universal compatibility (15, 79). The frequency of outcrossing in homothallic species under experimental conditions of mate choice has rarely been explored, but outcrossing has still been shown to occur in some homothallic fungi (178, 179). Homothallism would therefore be advantageous through universal compatibility under outcrossing. However, given that same-clone mating incurs costs of sex without some of the major benefits of recombination, heterothallism may be favored by selection for increasing the chance to produce recombinant offspring when there is limited haploid dispersal (15, 79). Nevertheless, same-clone mating may allow a benefit from indirect advantages of the sexual cycle without breaking apart beneficial allelic combinations (180, 181). Such advantages include the production of resistant sexual spores as a life history constraint, changes in ploidy level and physiology, virus elimination, and genome defenses that depend on the sexual cycle, such as repeat-induced point mutation (182) or meiotic silencing of unpaired DNA (183). Recent studies also showed that same-clone mating can generate de novo mutations and new beneficial phenotypes (170).
New Genome Sequencing Projects: What Can They Tell Us about MAT Gene Structure and Evolution?
With the exponential growth of data resulting from several large-scale genomics initiatives, such as the 1000 Fungal Genomes Project at the Joint Genome Institute (184) and the Fungal Genome Initiative at the Broad Institute (185), it is now possible to identify and compare the genomic organization of MAT genes across many fungal species. For example, genome surveys allowed the detection of P/R and HD genes in important plant pathogens with complex life cycles, such as the rust fungi (Pucciniales, Pucciniomycotina; Fig. 3B) (40, 186). Indeed, because many rust species are macrocyclic (i.e., completing their sexual stage on a different and sometimes obscure or unknown alternative host plant) and sometimes unculturable, sexual compatibility has been very difficult to study. Some reports have shed light on possible breeding systems: several Puccinia and Uromyces spp. may have a simple bipolar system (187), whereas a more complicated tetrapolar system with multiple allelic specificities may be present in Melampsora lini (188) and the related oat crown rust pathogen, Puccinia coronata (189). Whether these and other species have bi- or multiallelic MAT loci remains unknown, but new insight is now arising from different fungal genome sequencing initiatives.
In a recent study, several new wheat rust Puccinia spp. genomes were analyzed for the presence of mating-type genes (186). In three cereal-infecting rust fungi (Puccinia triticina [Pt], Puccinia graminis f.sp. tritici, and Puccinia striiformis f.sp. tritici), one divergently transcribed HD1/HD2 gene pair was found, reminiscent of the organization in other basidiomycetes; two alleles of each were present in the dikaryotic urediniospores, which is the cell type produced upon mating. Preliminary analyses of multiple genome sequences allude to the presence of more than two alleles in nature (G. B., unpublished). Three homologs for the pheromone receptor (Ste3) were identified in the dikaryotic urediniospores. Phylogenetic analysis, comparing Ste3 proteins from various rusts and other basidiomycetes, revealed that one STE3 gene (PtSTE3.2-1) was present in two allelic forms, whereas the other two (PtSTE3.2-2 and PtSTE3.2-3) likely represented the two mating specificities, one in each haplotype genome (186) (Fig. 3B and Fig. 4). This was substantiated by revealing many more single-nucleotide polymorphisms in STE3.2-1 homologs in P. triticina, P. graminis f.sp. tritici, and P. striiformis f.sp. tritici among several sequenced genomes; the conclusion, also based on differential expression in various life cycle stages, is that this gene may represent a novel function (186), suggesting that MAT pathway components may be repurposed. In further support, a pheromone precursor gene (PtMFA2) was identified 650 bp from PtSTE3.2-2, divergently transcribed as found, e.g., in the P/R a2 locus of U. maydis. However, the current rather fragmented Puccinia genomes did not allow for further conclusions about the possible physical linkage of the HD and P/R gene complexes to constitute a bipolar arrangement, but based on the current assembly and some mapping data in P. triticina, these loci are at least 216 kb apart (186). Current developments of single-molecule sequencing by Pacific Biosciences, focusing on generating very long kb-sized reads, are expected to improve these genome assemblies in the near future.
CONCLUSIONS
The basidiomycete fungi have been found to undertake nearly every permutation of reproductive compatibility mechanisms, and the recent literature reviewed here illustrates that the high diversity at mating compatibility loci has remained for several decades “the most challenging problem in the control of [their] sexuality” (190). The advent of inexpensive molecular tools now provides access to genetic information in a broader range of nonmodel species, including at the population-genomic level, so that more comprehensive databases are being assembled. Such surveys shed light on the generalities of what does happen in nature, beyond our glimpses and speculations into what can happen in exceptional cases (vis-à-vis reference 191). Just as importantly, the large-scale comparative approaches that derive from improved resolution of phylogenomic techniques (192) may provide new understanding of the ecological and evolutionary forces responsible for transitions in breeding systems and underlying genetic determinants. Fungi thus provide rich opportunities for the match of tractable study systems with questions pertaining to sexual life histories as a defining characteristic retained among diverse eukaryotic lineages.
FIGURE 3b.
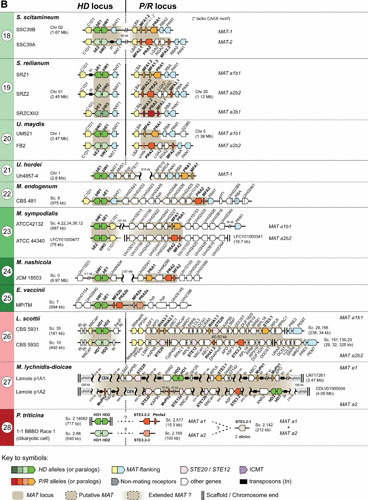
Schematic showing the genomic structure and diversity of MAT loci in representative basidiomycete lineages. The genomic organization of the homeodomain (HD) and pheromone/receptor (P/R) MAT loci is shown for selected species of (A) the Agaricomycotina and (B) the Ustilaginomycotina and Pucciniomycotina. Arrows indicate genes and their direction of transcription. Putative MAT loci are shaded in light brown, and MAT genes are colored as indicated in the key with different color grades representing different alleles (or paralogs). When known, conserved genes flanking MAT loci (colored light yellow or light blue) are shown within each lineage. Genes that encode components of the pheromone response pathway are shown in pink and are in many cases within the MAT locus. Putative homologs of a protein required for posttranslational modification of pheromone precursors (isoprenyl cysteine methyltransferase [ICMT]) are colored purple and appear near the P/R locus in some species. P/R loci no longer determining mating-type specificity in bipolar Agaricomycetes are depicted with a gray background. In M. lychnidis-dioicae, the two mating-type chromosomes are highly rearranged and enriched in transposable elements, so that only a small number of genes is depicted. Of note, whereas in M. nashicola P/R and HD genes are far apart on the same chromosome, in E. vaccinii and M. endogenum the two sets of genes are closer together. Other genes or genomic features are colored or represented as given in the key. For citations and additional details, see text. Gene names or their associated protein accession numbers are shown as they appear in their respective genome databases, except in species of Ustilaginomycotina and Tremellomycetes, where names were given based on sequence identity to the closest homolog in U. maydis and C. neoformans, respectively.
GLOSSARY OF TERMS
Anisogamy: Situation in a species with two classes of gamete sizes—one class of large gametes (female function) and one class of small gametes (male function); a single genotype can produce both types of gametes in most anisogamous fungi, so that there are no different sexes.
Automixis: A form of diploid selfing resulting from the fusion among products of a single meiosis (i.e., intratetrad mating).
Bipolarity: Breeding system with a single locus controlling mating type; it is often associated with only two alleles in fungi.
Breeding system: Mechanism determining reproductive compatibility, i.e., with no mating type (homothallism) or mating types determined by one locus (bipolar) or with two independent loci (tetrapolar); it is important to distinguish the breeding system from the mating system, because, even if they likely coevolved, the understanding of their relationships and their evolutionary implications can only be precisely achieved by recognizing that they refer to different concepts (15, 17).
Clonemates: Cells with identical genotypes resulting from clonal division of a mother cell.
Diploid selfing: Mating among meiotic products from a single diploid individual.
Haploid selfing (or same-clone mating, intrahaploid mating): Mating among haploid clonemates.
Heterothallism: Situation in which a given haploid genotype can only mate with another haploid genotype carrying a different allele at the mating-type locus (loci). It is important to distinguish the terms “heterothallism” and “self-sterility,” the latter usually being used to refer to the inability of diploid selfing rather than haploid selfing in the scientific literature, which have very different evolutionary consequences. Even in fungi, many heterothallic species can undergo selfing (e.g., Microbotryum anther smuts; Fig. 1).
Hill-Robertson interference (named after the two scientists who first proposed the concept): A phenomenon referring to less efficient selection because of linkage disequilibria (generated either by drift or in a clonal population); for example, in the case of beneficial mutations appearing in different individuals at two different genes in linkage disequilibrium, selection can only fix one of the two.
Homeodomain protein: Protein that harbors a characteristic approximately 60-amino acid helical protein-folding structure, highly conserved among eukaryotes, that binds DNA at so-called conserved homeobox sequences of about 180 bp and often regulates transcription of other sets of genes; in basidiomycetes, these transcription factors control a set of genes involved in the mating process, including morphological changes. The term “homeo” refers to the term “homeosis designing,” the replacement of a body part by another body part in animals, as can occur when transcription factors are affected by mutations.
Homothallism: Situation in which a haploid genotype can mate with any other haploid genotype in the population, including its clonemates; homothallism is typically assessed by the ability of a haploid genotype to produce sexual structures alone in a petri dish; however, it is important to recognize that it does not inform on the mating system actually occurring in natural populations. It is also important to distinguish the terms “homothallism” and “self-fertility,” the latter usually being used in the scientific literature to refer to the ability of diploid selfing rather than haploid selfing, which have very different evolutionary consequences. Using “self-fertility” for “homothallism” can only lead to confusion in the actual phenomenon that is referred to and therefore in the understanding of concepts and evolution and breeding and mating systems.
Inbreeding: Mating occurring among related individuals.
Isogamy: Situation in a species in which all gametes have similar sizes.
Karyogamy: Fusion of haploid nuclei following mating.
Mating system: The pairing of mating participants in nature, considering their genetic relatedness, i.e., outcrossing versus inbreeding or selfing (and either haploid or diploid selfing).
Mating types: Different types of individuals or gametes in the population, with gametes of similar sizes, but mating compatibility being usually determined by the exchange of molecular signals and being possible only between different mating types. It is important to distinguish sexes from mating types because the evolutionary causes of their evolution and their evolutionary consequences are very different (15).
Muller’s ratchet (proposed by Hermann Joseph Muller): A process in which the chromosomes of asexual populations accumulate inexorably deleterious mutations where the lack of recombination prevents regenerating chromosomes free of deleterious mutation from two homologous chromosomes carrying different deleterious mutations.
Outcrossing: Mating among different genotypes, usually unrelated individuals.
Selfing: Mating among meiotic products from the individual; used in the broader literature to mean “diploid selfing.” See Diploid selfing, Haploid selfing.
Sexes: Different types of individuals in the population, one type producing large gametes (females) and the other type producing small gametes (males).
Syngamy: Haploid cell fusion during mating.
Tetrapolarity: Breeding system with two unlinked loci controlling mating type; it is often associated with multiple alleles in fungi.
ACKNOWLEDGMENTS
M.A.C. acknowledges postdoctoral grant SFRH/BPD/79198/2011 from Fundação para a Ciência e a Tecnologia, Portugal, and the Unidade de Ciências Biomoleculares Aplicadas – UCIBIO, which is cofinanced by FCT/MEC (UID/Multi/04378/2013) and by the ERDF under the PT2020 Partnership Agreement (POCI-01-0145-FEDER-007728). M.E.H. acknowledges support from the National Institutes of Health (award R15GM119092), and T.G. acknowledges ERC starting grant GenomeFun 309403 and grant ANR-09-0064-01.
We acknowledge the Broad Institute, the U.S. Department of Energy Joint Genome Institute, the RIKEN Center for Life Science Technologies for providing free access to many Basidiomycota genomes, and the many researchers and funding agencies that have contributed to these vast and ever-increasing public resources. We apologize to those whose work we may have failed to cite.
REFERENCES
- 1.Raper JR. 1966. Genetics of Sexuality in Higher Fungi. Roland Press, New York, NY. [PubMed] [Google Scholar]
- 2.Morrow CA, Fraser JA. 2009. Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res 9:161–177 10.1111/j.1567-1364.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 3.Hibbett DS, et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycol Res 111:509–547 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Kirk MP, Cannon PF, Minter DW, Stalpers JA. 2008. Dictionary of the Fungi, 10th ed. CABI, Oxon, United Kingdom. [Google Scholar]
- 5.Hibbett DS. 2006. A phylogenetic overview of the Agaricomycotina. Mycologia 98:917–925 10.3852/mycologia.98.6.917. [DOI] [PubMed] [Google Scholar]
- 6.Begerow D, Bauer R, Boekhout T. 2000. Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol Res 104:53–60 10.1017/S0953756299001161. [DOI] [Google Scholar]
- 7.Begerow D, Schäfer AM, Kellner R, Yurkov A, Kemler M, Oberwinkler F, Bauer R. 2014. Ustilaginomycotina, p 295–329. In McLaughlin DJ, Spatafora JW (ed), Systematics and Evolution: Part A. Springer, Heidelberg, Germany. 10.1007/978-3-642-55318-9_11 [DOI] [Google Scholar]
- 8.Aime MC, Toome M, McLaughlin DJ. 2014. Pucciniomycotina, p 271–294. In McLaughlin DJ, Spatafora JW (ed), Systematics and Evolution: Part A. Springer. Heidelberg, Germany. 10.1007/978-3-642-55318-9_10 [DOI] [Google Scholar]
- 9.Aime MC, Matheny PB, Henk DA, Frieders EM, Nilsson RH, Piepenbring M, McLaughlin DJ, Szabo LJ, Begerow D, Sampaio JP, Bauer R, Weiss M, Oberwinkler F, Hibbett D. 2006. An overview of the higher level classification of Pucciniomycotina based on combined analyses of nuclear large and small subunit rDNA sequences. Mycologia 98:896–905 10.3852/mycologia.98.6.896. [DOI] [PubMed] [Google Scholar]
- 10.Bary A, Balfour BI, Garnser HEF. 1887. Comparative Morphology and Biology of the Fungi, Mycetozoa and Bacteria. Clarendon Press, Oxford, United Kingdom. 10.5962/bhl.title.56861 [DOI] [Google Scholar]
- 11.Kniep H. 1928. Die Sexualität der Niederen Pflanzen. G. Fischer, Jena, Germany. [Google Scholar]
- 12.Buller AHR. 1930. The biological significance of conjugate nuclei in Coprinus lagopus and other hymenomycetes. Nature 126:686–689 10.1038/126686a0. [DOI] [Google Scholar]
- 13.Whitehouse HLK. 1949. Multiple-allelomorph heterothallism in the fungi. New Phytol 48:212–244 10.1111/j.1469-8137.1949.tb05120.x. [DOI] [Google Scholar]
- 14.Meinhardt F, Esser K, Lemke PA. 1990. Sex determination and sexual differentiation in filamentous fungi. Crit Rev Plant Sci 9:329–341 10.1080/07352689009382294. [DOI] [Google Scholar]
- 15.Billiard S, López-Villavicencio M, Devier B, Hood ME, Fairhead C, Giraud T. 2011. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev Camb Philos Soc 86:421–442 10.1111/j.1469-185X.2010.00153.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.James TY. 2015. Why mushrooms have evolved to be so promiscuous: insights from evolutionary and ecological patterns. Fungal Biol Rev 29:167–178 10.1016/j.fbr.2015.10.002. [DOI] [Google Scholar]
- 17.Nieuwenhuis BPS, Billiard S, Vuilleumier S, Petit E, Hood ME, Giraud T. 2013. Evolution of uni- and bifactorial sexual compatibility systems in fungi. Hered (Edinb) 111:445–455 10.1038/hdy.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casselton LA, Olesnicky NS. 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev 62:55–70. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SC, Ni M, Li W, Shertz C, Heitman J. 2010. The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev 74:298–340 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kües U. 2015. From two to many: multiple mating types in Basidiomycetes. Fungal Biol Rev 29:126–166 10.1016/j.fbr.2015.11.001. [DOI] [Google Scholar]
- 21.Kües U, Casselton LA. 1992. Fungal mating type genes: regulators of sexual development. Mycol Res 96:993–1006 10.1016/S0953-7562(09)80107-X. [DOI] [Google Scholar]
- 22.Kothe E. 1996. Tetrapolar fungal mating types: sexes by the thousands. FEMS Microbiol Rev 18:65–87 10.1111/j.1574-6976.1996.tb00227.x. [DOI] [PubMed] [Google Scholar]
- 23.Raper J, Flexer A. 1971. Mating systems and evolution of the Basidiomycetes, p 149–167. In Petersen RH (ed), Evolution in the Higher Basidiomycetes. University of Tennessee Press, Knoxville, TN. [Google Scholar]
- 24.Whitehouse HLK. 1951. A survey of heterothallism in the Ustilaginales. Trans Br Mycol Soc 34:340–355 10.1016/S0007-1536(51)80061-5. [DOI] [Google Scholar]
- 25.Buller AHR. 1950. Researches on Fungi: the Sexual Process in the Uredinales. University of Toronto Press, Toronto, Canada. [Google Scholar]
- 26.Bölker M, Urban M, Kahmann R. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441–450 10.1016/0092-8674(92)90182-C. [DOI] [PubMed] [Google Scholar]
- 27.Manolaridis I, Kulkarni K, Dodd RB, Ogasawara S, Zhang Z, Bineva G, O’Reilly N, Hanrahan SJ, Thompson AJ, Cronin N, Iwata S, Barford D. 2013. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature 504:301–305 10.1038/nature12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raudaskoski M, Kothe E. 2010. Basidiomycete mating type genes and pheromone signaling. Eukaryot Cell 9:847–859 10.1128/EC.00319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue C, Hsueh YP, Heitman J. 2008. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev 32:1010–1032 10.1111/j.1574-6976.2008.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamiya Y, Sakurai A, Tamura S, Takahashi N, Abe K, Tsuchiya E, Fukui S, Kitada C, Fujino M. 1978. Structure of rhodotorucine A, a novel lipopeptide, inducing mating tube formation in Rhodosporidium toruloides. Biochem Biophys Res Commun 83:1077–1083 10.1016/0006-291X(78)91505-X. [DOI] [PubMed] [Google Scholar]
- 31.Kamiya Y, Sakurai A, Tamura S, Takahashi N, Abe K, Tsuchiya E, Fukui S. 1978. Isolation of rhodotorucine A, a peptidyl factor inducing the mating tube formation in Rhodosporidium toruloides. Agric Biol Chem 42:1239–1243. [Google Scholar]
- 32.Miyakawa T, Tabata M, Tsuchiya E, Fukui S. 1985. Biosynthesis and secretion of tremerogen A-10, a polyisoprenyl peptide mating pheromone of Tremella mesenterica. Eur J Biochem 147:489–493 10.1111/j.0014-2956.1985.00489.x. [DOI] [PubMed] [Google Scholar]
- 33.Spellig T, Bölker M, Lottspeich F, Frank RW, Kahmann R. 1994. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J 13:1620–1627. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore TD, Edman JC. 1993. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol 13:1962–1970 10.1128/MCB.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendland J, Vaillancourt LJ, Hegner J, Lengeler KB, Laddison KJ, Specht CA, Raper CA, Kothe E. 1995. The mating-type locus B α 1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J 14:5271–5278. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Shea SF, Chaure PT, Halsall JR, Olesnicky NS, Leibbrandt A, Connerton IF, Casselton LA. 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148:1081–1090. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caldwell GA, Naider F, Becker JM. 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol Rev 59:406–422. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaelis S, Barrowman J. 2012. Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease. Microbiol Mol Biol Rev 76:626–651 10.1128/MMBR.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coelho MA, Rosa A, Rodrigues N, Fonseca A, Gonçalves P. 2008. Identification of mating type genes in the bipolar basidiomycetous yeast Rhodosporidium toruloides: first insight into the MAT locus structure of the Sporidiobolales. Eukaryot Cell 7:1053–1061 10.1128/EC.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duplessis S, Cuomo CA, Lin YC, Aerts A, Tisserant E, Veneault-Fourrey C, Joly DL, Hacquard S, Amselem J, Cantarel BL, Chiu R, Coutinho PM, Feau N, Field M, Frey P, Gelhaye E, Goldberg J, Grabherr MG, Kodira CD, Kohler A, Kües U, Lindquist EA, Lucas SM, Mago R, Mauceli E, Morin E, Murat C, Pangilinan JL, Park R, Pearson M, Quesneville H, Rouhier N, Sakthikumar S, Salamov AA, Schmutz J, Selles B, Shapiro H, Tanguay P, Tuskan GA, Henrissat B, Van de Peer Y, Rouzé P, Ellis JG, Dodds PN, Schein JE, Zhong S, Hamelin RC, Grigoriev IV, Szabo LJ, Martin F. 2011. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci USA 108:9166–9171 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kües U, James TY, Heitman J. 2011. Mating type in basidiomycetes: unipolar, bipolar, and tetrapolar patterns of sexuality, p 97–160. In Pöggeler S, Wöstemeyer J (ed), Evolution of Fungi and Fungal-Like Organisms, vol 6. Springer, Heidelberg, Germany. 10.1007/978-3-642-19974-5_6 [DOI] [Google Scholar]
- 42.Maia TM, Lopes ST, Almeida JM, Rosa LH, Sampaio JP, Gonçalves P, Coelho MA. 2015. Evolution of mating systems in basidiomycetes and the genetic architecture underlying mating-type determination in the yeast Leucosporidium scottii. Genetics 201:75–89 10.1534/genetics.115.177717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L, Petit E, Hood ME. 2016. Variation in mate-recognition pheromones of the fungal genus Microbotryum. Hered (Edinb) 116:44–51 10.1038/hdy.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akada R, Minomi K, Kai J, Yamashita I, Miyakawa T, Fukui S. 1989. Multiple genes coding for precursors of rhodotorucine A, a farnesyl peptide mating pheromone of the basidiomycetous yeast Rhodosporidium toruloides. Mol Cell Biol 9:3491–3498 10.1128/MCB.9.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coelho MA, Sampaio JP, Gonçalves P. 2010. A deviation from the bipolar-tetrapolar mating paradigm in an early diverged basidiomycete. PLoS Genet 6:e1001052 10.1371/journal.pgen.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abe K, Kusaka I, Fukui S. 1975. Morphological change in the early stages of the mating process of Rhodosporidium toruloides. J Bacteriol 122:710–718. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Day AW. 1976. Communication through fimbriae during conjugation in a fungus. Nature 262:583–584 10.1038/262583a0. [DOI] [PubMed] [Google Scholar]
- 48.Feldbrügge M, Kämper J, Steinberg G, Kahmann R. 2004. Regulation of mating and pathogenic development in Ustilago maydis. Curr Opin Microbiol 7:666–672 10.1016/j.mib.2004.10.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Zarnack K, Eichhorn H, Kahmann R, Feldbrügge M. 2008. Pheromone-regulated target genes respond differentially to MAPK phosphorylation of transcription factor Prf1. Mol Microbiol 69:1041–1053. [DOI] [PubMed] [Google Scholar]
- 50.Nadal M, García-Pedrajas MD, Gold SE. 2008. Dimorphism in fungal plant pathogens. FEMS Microbiol Lett 284:127–134 10.1111/j.1574-6968.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- 51.Hartmann HA, Kahmann R, Bölker M. 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J 15:1632–1641. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 52.Urban M, Kahmann R, Bölker M. 1996. Identification of the pheromone response element in Ustilago maydis. Mol Gen Genet 251:31–37. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Banham AH, Asante-Owusu RN, Göttgens B, Thompson S, Kingsnorth CS, Mellor E, Casselton LA. 1995. An N-terminal dimerization domain permits homeodomain proteins to choose compatible partners and initiate sexual development in the mushroom Coprinus cinereus. Plant Cell 7:773–783 10.1105/tpc.7.6.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kämper J, Reichmann M, Romeis T, Bölker M, Kahmann R. 1995. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81:73–83 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- 55.Kahmann R, Bölker M. 1996. Self/nonself recognition in fungi: old mysteries and simple solutions. Cell 85:145–148 10.1016/S0092-8674(00)81091-0. [DOI] [PubMed] [Google Scholar]
- 56.Badrane H, May G. 1999. The divergence-homogenization duality in the evolution of the b1 mating type gene of Coprinus cinereus. Mol Biol Evol 16:975–986 10.1093/oxfordjournals.molbev.a026187. [DOI] [PubMed] [Google Scholar]
- 57.Yee AR, Kronstad JW. 1993. Construction of chimeric alleles with altered specificity at the b incompatibility locus of Ustilago maydis. Proc Natl Acad Sci USA 90:664–668 10.1073/pnas.90.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yee AR, Kronstad JW. 1998. Dual sets of chimeric alleles identify specificity sequences for the bE and bW mating and pathogenicity genes of Ustilago maydis. Mol Cell Biol 18:221–232 10.1128/MCB.18.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, Herskowitz I, Kahmann R. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295–306 10.1016/0092-8674(90)90744-Y. [DOI] [PubMed] [Google Scholar]
- 60.Wahl R, Zahiri A, Kämper J. 2010. The Ustilago maydis b mating type locus controls hyphal proliferation and expression of secreted virulence factors in planta. Mol Microbiol 75:208–220 10.1111/j.1365-2958.2009.06984.x. [DOI] [PubMed] [Google Scholar]
- 61.Giraud T, Yockteng R, López-Villavicencio M, Refrégier G, Hood ME. 2008. Mating system of the anther smut fungus Microbotryum violaceum: selfing under heterothallism. Eukaryot Cell 7:765–775 10.1128/EC.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kahmann R, Kämper J. 2004. Ustilago maydis: how its biology relates to pathogenic development. New Phytol 164:31–42 10.1111/j.1469-8137.2004.01156.x. [DOI] [PubMed] [Google Scholar]
- 63.Fedler M, Luh KS, Stelter K, Nieto-Jacobo F, Basse CW. 2009. The a2 mating-type locus genes lga2 and rga2 direct uniparental mitochondrial DNA (mtDNA) inheritance and constrain mtDNA recombination during sexual development of Ustilago maydis. Genetics 181:847–860 10.1534/genetics.108.096859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schirawski J, Heinze B, Wagenknecht M, Kahmann R. 2005. Mating type loci of Sporisorium reilianum: novel pattern with three a and multiple b specificities. Eukaryot Cell 4:1317–1327 10.1128/EC.4.8.1317-1327.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kellner R, Vollmeister E, Feldbrügge M, Begerow D. 2011. Interspecific sex in grass smuts and the genetic diversity of their pheromone-receptor system. PLoS Genet 7:e1002436 10.1371/journal.pgen.1002436. (Erratum, .) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Riess K, Schön ME, Lutz M, Butin H, Oberwinkler F, Garnica S. 2016. On the evolutionary history of Uleiella chilensis, a smut fungus parasite of Araucaria araucana in South America: uleiellales ord. nov. in Ustilaginomycetes. PLoS One 11:e0147107 10.1371/journal.pone.0147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gillissen B, Bergemann J, Sandmann C, Schroeer B, Bölker M, Kahmann R. 1992. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68:647–657 10.1016/0092-8674(92)90141-X. [DOI] [PubMed] [Google Scholar]
- 68.Puhalla JE. 1970. Genetic studies of the b incompatability locus of Ustilago maydis. Genet Res 16:229–232 10.1017/S0016672300002457. [DOI] [Google Scholar]
- 69.Wong GJ, Wells K. 1985. Modified bifactorial incompatibility in Tremella mesenterica. Trans Br Mycol Soc 84:95–109 10.1016/S0007-1536(85)80223-0. [DOI] [Google Scholar]
- 70.Metin B, Findley K, Heitman J. 2010. The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insights into the evolution of sex and sex-determining chromosomal regions in fungi. PLoS Genet 6:e1000961 10.1371/journal.pgen.1000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Findley K, Sun S, Fraser JA, Hsueh Y-P, Averette AF, Li W, Dietrich FS, Heitman J. 2012. Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex. PLoS Genet 8:e1002528 10.1371/journal.pgen.1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guerreiro MA, Springer DJ, Rodrigues JA, Rusche LN, Findley K, Heitman J, Fonseca A. 2013. Molecular and genetic evidence for a tetrapolar mating system in the basidiomycetous yeast Kwoniella mangrovensis and two novel sibling species. Eukaryot Cell 12:746–760 10.1128/EC.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hood ME, Scott M, Hwang M. 2015. Breaking linkage between mating compatibility factors: tetrapolarity in Microbotryum. Evolution 69:2561–2572 10.1111/evo.12765. [DOI] [PubMed] [Google Scholar]
- 74.Yurkov A, Guerreiro MA, Sharma L, Carvalho C, Fonseca Á. 2015. Multigene assessment of the species boundaries and sexual status of the basidiomycetous yeasts Cryptococcus flavescens and C. terrestris (Tremellales). PLoS One 10:e0120400 10.1371/journal.pone.0120400. (Erratum, .) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koh CM, Liu Y, Moehninsi, Du M, Ji L. 2014. Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol 14:50 10.1186/1471-2180-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nichols CB, Fraser JA, Heitman J. 2004. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol Biol Cell 15:4476–4489 10.1091/mbc.E04-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith DG, Garcia-Pedrajas MD, Hong W, Yu Z, Gold SE, Perlin MH. 2004. An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot Cell 3:180–189 10.1128/EC.3.1.180-189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coelho MA, Gonçalves P, Sampaio JP. 2011. Evidence for maintenance of sex determinants but not of sexual stages in red yeasts, a group of early diverged basidiomycetes. BMC Evol Biol 11:249 10.1186/1471-2148-11-249. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Billiard S, López-Villavicencio M, Hood ME, Giraud T. 2012. Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J Evol Biol 25:1020–1038 10.1111/j.1420-9101.2012.02495.x. [DOI] [PubMed] [Google Scholar]
- 80.Riquelme M, Challen MP, Casselton LA, Brown AJ. 2005. The origin of multiple B mating specificities in Coprinus cinereus. Genetics 170:1105–1119 10.1534/genetics.105.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Day PR. 1960. The structure of the A mating type locus in Coprinus lagopus. Genetics 45:641–650. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lukens L, Yicun H, May G. 1996. Correlation of genetic and physical maps at the A mating-type locus of Coprinus cinereus. Genetics 144:1471–1477. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kües U, Casselton LA. 1993. The origin of multiple mating types in mushrooms. J Cell Sci 104:227–230. [Google Scholar]
- 84.Kües U, Richardson WV, Tymon AM, Mutasa ES, Göttgens B, Gaubatz S, Gregoriades A, Casselton LA. 1992. The combination of dissimilar alleles of the A α and A β gene complexes, whose proteins contain homeo domain motifs, determines sexual development in the mushroom Coprinus cinereus. Genes Dev 6:568–577 10.1101/gad.6.4.568. [DOI] [PubMed] [Google Scholar]
- 85.Pardo EH, O’Shea SF, Casselton LA. 1996. Multiple versions of the A mating type locus of Coprinus cinereus are generated by three paralogous pairs of multiallelic homeobox genes. Genetics 144:87–94. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stajich JE, Wilke SK, Ahrén D, Au CH, Birren BW, Borodovsky M, Burns C, Canbäck B, Casselton LA, Cheng CK, Deng J, Dietrich FS, Fargo DC, Farman ML, Gathman AC, Goldberg J, Guigó R, Hoegger PJ, Hooker JB, Huggins A, James TY, Kamada T, Kilaru S, Kodira C, Kües U, Kupfer D, Kwan HS, Lomsadze A, Li W, Lilly WW, Ma LJ, Mackey AJ, Manning G, Martin F, Muraguchi H, Natvig DO, Palmerini H, Ramesh MA, Rehmeyer CJ, Roe BA, Shenoy N, Stanke M, Ter-Hovhannisyan V, Tunlid A, Velagapudi R, Vision TJ, Zeng Q, Zolan ME, Pukkila PJ. 2010. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci USA 107:11889–11894 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fowler TJ, Mitton MF, Rees EI, Raper CA. 2004. Crossing the boundary between the Bα and Bβ mating-type loci in Schizophyllum commune. Fungal Genet Biol 41:89–101 10.1016/j.fgb.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 88.Kües U, Nelson DR, Liu C, Yu G-J, Zhang J, Li J, Wang X-C, Sun H. 2015. Genome analysis of medicinal Ganoderma spp. with plant-pathogenic and saprotrophic life-styles. Phytochemistry 114:18–37 10.1016/j.phytochem.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 89.Kamada T. 2002. Molecular genetics of sexual development in the mushroom Coprinus cinereus. BioEssays 24:449–459 10.1002/bies.10083. [DOI] [PubMed] [Google Scholar]
- 90.Kües U. 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev 64:316–353 10.1128/MMBR.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Casselton LA, Kües U. 2007. The origin of multiple mating types in the model mushrooms Coprinopsis cinerea and Schizophyllum commune, p 283–300. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in Fungi: Molecular Determination and Evolutionary Implications. ASM Press, Washington, DC. 10.1128/9781555815837.ch17 [DOI] [Google Scholar]
- 92.Tymon AM, Kües U, Richardson WV, Casselton LA. 1992. A fungal mating type protein that regulates sexual and asexual development contains a POU-related domain. EMBO J 11:1805–1813. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swiezynski KM, Day PR. 1960. Heterokaryon formation in Coprinus lagopus. Genet Res 1:114–128 10.1017/S0016672300000112. [DOI] [Google Scholar]
- 94.James TY, Lee M, van Diepen LT. 2011. A single mating-type locus composed of homeodomain genes promotes nuclear migration and heterokaryosis in the white-rot fungus Phanerochaete chrysosporium. Eukaryot Cell 10:249–261 10.1128/EC.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Peer AF, Park S-Y, Shin P-G, Jang K-Y, Yoo Y-B, Park Y-J, Lee B-M, Sung G-H, James TY, Kong W-S. 2011. Comparative genomics of the mating-type loci of the mushroom Flammulina velutipes reveals widespread synteny and recent inversions. PLoS One 6:e22249 10.1371/journal.pone.0022249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Freihorst D, Fowler TJ, Bartholomew K, Raudaskoski M, Horton JS, Kothe E. 2016. The mating-type genes of the basidiomycetes, p 329–349. In Wendland J (ed), Growth, Differentiation and Sexuality, 3rd ed, vol 13. Springer International Publishing, Cham, Switzerland. 10.1007/978-3-319-25844-7_13 [DOI] [Google Scholar]
- 97.Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 98.Niculita-Hirzel H, Labbé J, Kohler A, le Tacon F, Martin F, Sanders IR, Kües U. 2008. Gene organization of the mating type regions in the ectomycorrhizal fungus Laccaria bicolor reveals distinct evolution between the two mating type loci. New Phytol 180:329–342 10.1111/j.1469-8137.2008.02525.x. [DOI] [PubMed] [Google Scholar]
- 99.Martinez D, et al. 2009. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA 106:1954–1959 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.James TY, Sun S, Li W, Heitman J, Kuo H-C, Lee Y-H, Asiegbu FO, Olson A. 2013. Polyporales genomes reveal the genetic architecture underlying tetrapolar and bipolar mating systems. Mycologia 105:1374–1390 10.3852/13-162. [DOI] [PubMed] [Google Scholar]
- 101.Hsueh YP, Xue C, Heitman J. 2009. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J 28:1220–1233 10.1038/emboj.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.James TY. 2007. Analysis of mating-type locus organization and synteny in mushroom fungi: beyond model species, p 317–331. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in Fungi: Molecular Determination and Evolutionary Implications. ASM Press, Washington, DC. 10.1128/9781555815837.ch19 [DOI] [Google Scholar]
- 103.Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker SE, Bartholomew KA, Coutinho PM, Erdmann S, Fowler TJ, Gathman AC, Lombard V, Henrissat B, Knabe N, Kües U, Lilly WW, Lindquist E, Lucas S, Magnuson JK, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze FW, vanKuyk PA, Horton JS, Grigoriev IV, Wösten HA. 2010. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol 28:957–963 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 104.Raper JR, Baxter MG, Ellingboe AH. 1960. The genetic structure of the incompatibility factors of Schizophyllum commune: the A-factor. Proc Natl Acad Sci USA 46:833–842 10.1073/pnas.46.6.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Díaz-Valderrama JR, Aime MC. 2016. The cacao pathogen Moniliophthora roreri (Marasmiaceae) possesses biallelic A and B mating loci but reproduces clonally. Hered (Edinb) 116:491–501 10.1038/hdy.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Au CH, Wong MC, Bao D, Zhang M, Song C, Song W, Law PTW, Kües U, Kwan HS. 2014. The genetic structure of the A mating-type locus of Lentinula edodes. Gene 535:184–190 10.1016/j.gene.2013.11.036. [DOI] [PubMed] [Google Scholar]
- 107.Hsueh YP, Fraser JA, Heitman J. 2008. Transitions in sexuality: recapitulation of an ancestral tri- and tetrapolar mating system in Cryptococcus neoformans. Eukaryot Cell 7:1847–1855 10.1128/EC.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heitman J, Sun S, James TY. 2013. Evolution of fungal sexual reproduction. Mycologia 105:1–27 10.3852/12-253. [DOI] [PubMed] [Google Scholar]
- 109.Hibbett DS, Donoghue MJ. 2001. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst Biol 50:215–242 10.1080/10635150151125879. [DOI] [PubMed] [Google Scholar]
- 110.Wang QM, Begerow D, Groenewald M, Liu XZ, Theelen B, Bai FY, Boekhout T. 2015. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Stud Mycol 81:55–83 10.1016/j.simyco.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Froeliger EH, Leong SA. 1991. The a mating-type alleles of Ustilago maydis are idiomorphs. Gene 100:113–122 10.1016/0378-1119(91)90356-G. [DOI] [PubMed] [Google Scholar]
- 112.Kronstad JW, Leong SA. 1989. Isolation of two alleles of the b locus of Ustilago maydis. Proc Natl Acad Sci USA 86:978–982 10.1073/pnas.86.3.978. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bakkeren G, Kronstad JW. 1994. Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc Natl Acad Sci USA 91:7085–7089 10.1073/pnas.91.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bakkeren G, Kronstad JW. 1996. The pheromone cell signaling components of the Ustilago a mating-type loci determine intercompatibility between species. Genetics 143:1601–1613. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee N, Bakkeren G, Wong K, Sherwood JE, Kronstad JW. 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc Natl Acad Sci USA 96:15026–15031 10.1073/pnas.96.26.15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kämper J, et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 117.Schirawski J, Mannhaupt G, Münch K, Brefort T, Schipper K, Doehlemann G, Di Stasio M, Rössel N, Mendoza-Mendoza A, Pester D, Müller O, Winterberg B, Meyer E, Ghareeb H, Wollenberg T, Münsterkötter M, Wong P, Walter M, Stukenbrock E, Güldener U, Kahmann R. 2010. Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330:1546–1548 10.1126/science.1195330. [DOI] [PubMed] [Google Scholar]
- 118.Laurie JD, Ali S, Linning R, Mannhaupt G, Wong P, Güldener U, Münsterkötter M, Moore R, Kahmann R, Bakkeren G, Schirawski J. 2012. Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 24:1733–1745 10.1105/tpc.112.097261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Que Y, Xu L, Wu Q, Liu Y, Ling H, Liu Y, Zhang Y, Guo J, Su Y, Chen J, Wang S, Zhang C. 2014. Genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut. BMC Genomics 15:996 10.1186/1471-2164-15-996. (Erratum, 16:244, .) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Taniguti LM, Schaker PDC, Benevenuto J, Peters LP, Carvalho G, Palhares A, Quecine MC, Nunes FRS, Kmit MCP, Wai A, Hausner G, Aitken KS, Berkman PJ, Fraser JA, Moolhuijzen PM, Coutinho LL, Creste S, Vieira MLC, Kitajima JP, Monteiro-Vitorello CB. 2015. Complete genome sequence of Sporisorium scitamineum and biotrophic interaction transcriptome with sugarcane. PLoS One 10:e0129318 10.1371/journal.pone.0129318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rabe F, Bosch J, Stirnberg A, Guse T, Bauer L, Seitner D, Rabanal FA, Czedik-Eysenberg A, Uhse S, Bindics J, Genenncher B, Navarrete F, Kellner R, Ekker H, Kumlehn J, Vogel JP, Gordon SP, Marcel TC, Münsterkötter M, Walter MC, Sieber CMK, Mannhaupt G, Güldener U, Kahmann R, Djamei A. 2016. A complete toolset for the study of Ustilago bromivora and Brachypodium sp. as a fungal-temperate grass pathosystem. eLife 5:e20522 10.7554/eLife.20522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gray YH. 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet 16:461–468 10.1016/S0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- 123.Gioti A, Nystedt B, Li W, Xu J, Andersson A, Averette AF, Münch K, Wang X, Kappauf C, Kingsbury JM, Kraak B, Walker LA, Johansson HJ, Holm T, Lehtiö J, Stajich JE, Mieczkowski P, Kahmann R, Kennell JC, Cardenas ME, Lundeberg J, Saunders CW, Boekhout T, Dawson TL, Munro CA, de Groot PW, Butler G, Heitman J, Scheynius A. 2013. Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. MBio 4:e00572-12 10.1128/mBio.00572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, Saunders CW, Reeder NL, Reilman RA, Scheynius A, Sun S, Billmyre BR, Li W, Averette AF, Mieczkowski P, Heitman J, Theelen B, Schröder MS, De Sessions PF, Butler G, Maurer-Stroh S, Boekhout T, Nagarajan N, Dawson TL Jr. 2015. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet 11:e1005614 10.1371/journal.pgen.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, Kronstad JW, Deangelis YM, Reeder NL, Johnstone KR, Leland M, Fieno AM, Begley WM, Sun Y, Lacey MP, Chaudhary T, Keough T, Chu L, Sears R, Yuan B, Dawson TL Jr. 2007. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci USA 104:18730–18735 10.1073/pnas.0706756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, Boekhout T. 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 78:16–48 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 127.Hull CM, Boily MJ, Heitman J. 2005. Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot Cell 4:526–535 10.1128/EC.4.3.526-535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hsueh YP, Idnurm A, Heitman J. 2006. Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet 2:e184 10.1371/journal.pgen.0020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun S, Billmyre RB, Mieczkowski PA, Heitman J. 2014. Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genet 10:e1004849 10.1371/journal.pgen.1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Janbon G, et al. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10:e1004261 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun S, Xu J. 2009. Chromosomal rearrangements between serotype A and D strains in Cryptococcus neoformans. PLoS One 4:e5524 10.1371/journal.pone.0005524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kourist R, Bracharz F, Lorenzen J, Kracht ON, Chovatia M, Daum C, Deshpande S, Lipzen A, Nolan M, Ohm RA, Grigoriev IV, Sun S, Heitman J, Brück T, Nowrousian M. 2015. Genomics and transcriptomics analyses of the oil-accumulating basidiomycete yeast Trichosporon oleaginosus: insights into substrate utilization and alternative evolutionary trajectories of fungal mating systems. MBio 6:e00918-15 10.1128/mBio.00918-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Idnurm A, Hood ME, Johannesson H, Giraud T. 2015. Contrasted patterns in mating-type chromosomes in fungi: hotspots versus coldspots of recombination. Fungal Biol Rev 29:220–229 10.1016/j.fbr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Le Gac M, Hood ME, Fournier E, Giraud T. 2007. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution 61:15–26 10.1111/j.1558-5646.2007.00002.x. [DOI] [PubMed] [Google Scholar]
- 135.Le Gac M, Hood ME, Giraud T. 2007. Evolution of reproductive isolation within a parasitic fungal species complex. Evolution 61:1781–1787 10.1111/j.1558-5646.2007.00144.x. [DOI] [PubMed] [Google Scholar]
- 136.Bernasconi G, Antonovics J, Biere A, Charlesworth D, Delph LF, Filatov D, Giraud T, Hood ME, Marais GA, McCauley D, Pannell JR, Shykoff JA, Vyskot B, Wolfe LM, Widmer A. 2009. Silene as a model system in ecology and evolution. Hered (Edinb) 103:5–14 10.1038/hdy.2009.34. [DOI] [PubMed] [Google Scholar]
- 137.Devier B, Aguileta G, Hood ME, Giraud T. 2009. Ancient trans-specific polymorphism at pheromone receptor genes in basidiomycetes. Genetics 181:209–223 10.1534/genetics.108.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kniep H. 1919. Untersuchungen über den Antherenbrand (Ustilago violacea Pers.). Ein Beitrag zum Sexualitätsproblem. Ztschr Bot 11:257–284. [Google Scholar]
- 139.Hood ME. 2002. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics 160:457–461. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Giraud T, Jonot O, Shykoff JA. 2005. Selfing propensity under choice conditions in a parasitic fungus, Microbotryum violaceum, and parameters influencing infection success in artificial inoculations. Int J Plant Sci 166:649–657 10.1086/430098. [DOI] [Google Scholar]
- 141.Hood ME, Antonovics J. 2000. Intratetrad mating, heterozygosity, and the maintenance of deleterious alleles in Microbotryum violaceum (=Ustilago violacea). Hered (Edinb) 85:231–241 10.1046/j.1365-2540.2000.00748.x. [DOI] [PubMed] [Google Scholar]
- 142.Hood ME, Antonovics J, Koskella B. 2004. Shared forces of sex chromosome evolution in haploid-mating and diploid-mating organisms: Microbotryum violaceum and other model organisms. Genetics 168:141–146 10.1534/genetics.104.029900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Giraud T. 2004. Patterns of within population dispersal and mating of the fungus Microbotryum violaceum parasitising the plant Silene latifolia. Hered (Edinb) 93:559–565 10.1038/sj.hdy.6800554. [DOI] [PubMed] [Google Scholar]
- 144.Gladieux P, Vercken E, Fontaine MC, Hood ME, Jonot O, Couloux A, Giraud T. 2011. Maintenance of fungal pathogen species that are specialized to different hosts: allopatric divergence and introgression through secondary contact. Mol Biol Evol 28:459–471 10.1093/molbev/msq235. [DOI] [PubMed] [Google Scholar]
- 145.Badouin H, Hood ME, Gouzy J, Aguileta G, Siguenza S, Perlin MH, Cuomo CA, Fairhead C, Branca A, Giraud T. 2015. Chaos of rearrangements in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae. Genetics 200:1275–1284 10.1534/genetics.115.177709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hood ME, Petit E, Giraud T. 2013. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics 193:309–315 10.1534/genetics.112.146266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14:113–124 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fontanillas E, Hood ME, Badouin H, Petit E, Barbe V, Gouzy J, de Vienne DM, Aguileta G, Poulain J, Wincker P, Chen Z, Toh SS, Cuomo CA, Perlin MH, Gladieux P, Giraud T. 2015. Degeneration of the nonrecombining regions in the mating-type chromosomes of the anther-smut fungi. Mol Biol Evol 32:928–943 10.1093/molbev/msu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kües U, Göttgens B, Stratmann R, Richardson WV, O’Shea SF, Casselton LA. 1994. A chimeric homeodomain protein causes self-compatibility and constitutive sexual development in the mushroom Coprinus cinereus. EMBO J 13:4054–4059. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Haylock RW, Economou A, Casselton LA. 1980. Dikaryon formation in Coprinus cinereus: selection and identification of B factor mutants. J Gen Microbiol 121:17–26. [Google Scholar]
- 151.Olesnicky NS, Brown AJ, Honda Y, Dyos SL, Dowell SJ, Casselton LA. 2000. Self-compatible B mutants in coprinus with altered pheromone-receptor specificities. Genetics 156:1025–1033. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.James TY, Srivilai P, Kües U, Vilgalys R. 2006. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics 172:1877–1891 10.1534/genetics.105.051128. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Aimi T, Yoshida R, Ishikawa M, Bao D, Kitamoto Y. 2005. Identification and linkage mapping of the genes for the putative homeodomain protein (hox1) and the putative pheromone receptor protein homologue (rcb1) in a bipolar basidiomycete, Pholiota nameko. Curr Genet 48:184–194 10.1007/s00294-005-0012-7. [DOI] [PubMed] [Google Scholar]
- 154.Yi R, Tachikawa T, Ishikawa M, Mukaiyama H, Bao D, Aimi T. 2009. Genomic structure of the A mating-type locus in a bipolar basidiomycete, Pholiota nameko. Mycol Res 113:240–248 10.1016/j.mycres.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 155.Olson A, et al. 2012. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol 194:1001–1013 10.1111/j.1469-8137.2012.04128.x. [DOI] [PubMed] [Google Scholar]
- 156.Perrin N. 2012. What uses are mating types? The “developmental switch” model. Evolution 66:947–956 10.1111/j.1558-5646.2011.01562.x. [DOI] [PubMed] [Google Scholar]
- 157.Wilson AM, Wilken PM, van der Nest MA, Steenkamp ET, Wingfield MJ, Wingfield BD. 2015. Homothallism: an umbrella term for describing diverse sexual behaviours. IMA Fungus 6:207–214 10.5598/imafungus.2015.06.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Roach KC, Feretzaki M, Sun S, Heitman J. 2014. Unisexual Reproduction, p 255–305. In Friedmann T, Dunlap JC, Goodwin SF (ed), Advances in Genetics, vol 85. Academic Press, San Diego, CA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Raju NB, Perkins DD. 1994. Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora, and Podospora. Dev Genet 15:104–118 10.1002/dvg.1020150111. [DOI] [PubMed] [Google Scholar]
- 160.Merino ST, Nelson MA, Jacobson DJ, Natvig DO. 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143:789–799. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Callac P, Spataro C, Caille A, Imbernon M. 2006. Evidence for outcrossing via the Buller phenomenon in a substrate simultaneously inoculated with spores and mycelium of Agaricus bisporus. Appl Environ Microbiol 72:2366–2372 10.1128/AEM.72.4.2366-2372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin X, Heitman J. 2007. Mechanisms of homothallism in fungi and transitions between heterothallism and homothallism, p 35–57. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in Fungi: Molecular Determination and Evolutionary Implications. ASM Press, Washington, DC. 10.1128/9781555815837.ch3 [DOI] [Google Scholar]
- 163.Ullrich RC, Raper JR. 1975. Primary homothallism-relation to heterothallism in the regulation of sexual morphogenesis in Sistotrema. Genetics 80:311–321. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Griffith GW, Hedger JN. 1994. The breeding biology of biotypes of the witches’ broom pathogen of cocoa, Crinipellis perniciosa. Heredity 72:278–289 10.1038/hdy.1994.38. [DOI] [Google Scholar]
- 165.Kües U, Navarro-González M. 2010. Mating-type orthologous genes in the primarily homothallic Moniliophthora perniciosa, the causal agent of witches’ broom disease in cacao. J Basic Microbiol 50:442–451 10.1002/jobm.201000013. [DOI] [PubMed] [Google Scholar]
- 166.David-Palma M, Libkind D, Sampaio JP. 2014. Global distribution, diversity hot spots and niche transitions of an astaxanthin-producing eukaryotic microbe. Mol Ecol 23:921–932 10.1111/mec.12642. [DOI] [PubMed] [Google Scholar]
- 167.Bellora N, Moline M, David-Palma M, Coelho MA, Hittinger CT, Sampaio JP, Goncalves P, Libkind D. 2016. Comparative genomics provides new insights into the diversity, physiology, and sexuality of the only industrially exploited tremellomycete: Phaffia rhodozyma. BMC Genomics 17:901. 10.1186/s12864-016-3244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.David-Palma M, Sampaio JP, Gonçalves P. 2016. Genetic dissection of sexual reproduction in a primary homothallic basidiomycete. PLoS Genet 12:e1006110 10.1371/journal.pgen.1006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lin X, Hull CM, Heitman J. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 170.Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. 2013. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol 11:e1001653 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. 2011. Cryptococcus: from Human Pathogen to Model Yeast. ASM Press, Washington, DC. [Google Scholar]
- 172.Alby K, Bennett RJ. 2011. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc Natl Acad Sci USA 108:2510–2515 10.1073/pnas.1017234108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Alby K, Schaefer D, Bennett RJ. 2009. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460:890–893 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wilson AM, Godlonton T, van der Nest MA, Wilken PM, Wingfield MJ, Wingfield BD. 2015. Unisexual reproduction in Huntiella moniliformis. Fungal Genet Biol 80:1–9 10.1016/j.fgb.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 175.Whitehouse HLK. 1949. Heterothallism and sex in the fungi. Biol Rev Camb Philos Soc 24:411–447 10.1111/j.1469-185X.1949.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 176.Aanen DK, Hoekstra RF. 2007. Why sex is good: on fungi and beyond, p 527–534. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in Fungi: Molecular Determination and Evolutionary Implications. ASM Press, Washington, DC. 10.1128/9781555815837.ch32 [DOI] [Google Scholar]
- 177.Roach KC, Heitman J. 2014. Unisexual reproduction reverses Muller’s ratchet. Genetics 198:1059–1069 10.1534/genetics.114.170472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Attanayake RN, Tennekoon V, Johnson DA, Porter LD, del Río-Mendoza L, Jiang D, Chen W. 2014. Inferring outcrossing in the homothallic fungus Sclerotinia sclerotiorum using linkage disequilibrium decay. Hered (Edinb) 113:353–363 10.1038/hdy.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Talas F, McDonald BA. 2015. Genome-wide analysis of Fusarium graminearum field populations reveals hotspots of recombination. BMC Genomics 16:996 10.1186/s12864-015-2166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.López-Villavicencio M, Debets AJ, Slakhorst M, Giraud T, Schoustra SE. 2013. Deleterious effects of recombination and possible nonrecombinatorial advantages of sex in a fungal model. J Evol Biol 26:1968–1978 10.1111/jeb.12196. [DOI] [PubMed] [Google Scholar]
- 181.Otto SP. 2009. The evolutionary enigma of sex. Am Nat 174(Suppl 1):S1–S14 10.1086/599084. [DOI] [PubMed] [Google Scholar]
- 182.Selker EU. 1991. Repeat-induced point mutation and DNA methylation, p 258–265. In Bennett JW, Lasure LL (ed), More Gene Manipulations in Fungi. Academic Press, San Diego, CA. 10.1016/B978-0-12-088642-5.50018-1 [DOI] [Google Scholar]
- 183.Shiu PK, Raju NB, Zickler D, Metzenberg RL. 2001. Meiotic silencing by unpaired DNA. Cell 107:905–916 10.1016/S0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 184.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42(D1):D699–D704 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cuomo CA, Birren BW. 2010. The Fungal Genome Initiative and lessons learned from genome sequencing. Methods Enzymol 470:833–855. [DOI] [PubMed] [Google Scholar]
- 186.Cuomo CA, Bakkeren G, Khalil HB, Panwar V, Joly D, Linning R, Sakthikumar S, Song X, Adiconis X, Fan L, Goldberg JM, Levin JZ, Young S, Zeng Q, Anikster Y, Bruce M, Wang M, Yin C, McCallum B, Szabo LJ, Hulbert S, Chen X, Fellers JP. 2017. Comparative analysis highlights variable genome content of wheat rusts and divergence of the mating loci. G3 (Bethesda) 7:361–376 10.1534/g3.116.032797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Anikster Y, Eilam T, Mittelman L, Szabo LJ, Bushnell WR. 1999. Pycnial nectar of rust fungi induces cap formation on pycniospores of opposite mating type. Mycologia 91:858–870 10.2307/3761539. [DOI] [Google Scholar]
- 188.Lawrence GJ. 1980. Multiple mating-type specificities in the flax rust Melampsora lini. Science 209:501–503 10.1126/science.209.4455.501. [DOI] [PubMed] [Google Scholar]
- 189.Narisawa K, Yamaoka Y, Katsuya K. 1994. Mating type of isolates derived from the spermogonial state of Puccinia coronata var. coronata. Mycoscience 35:131–135 10.1007/BF02318489. [DOI] [Google Scholar]
- 190.Raper JR. 1960. The control of sex in fungi. Am J Bot 47:794–808 10.2307/2439117. [DOI] [Google Scholar]
- 191.Travis J. 2006. Is it what we know or who we know? Choice of organism and robustness of inference in ecology and evolutionary biology: (American Society of Naturalists Presidential Address). Am Nat 167:303–314. [DOI] [PubMed] [Google Scholar]
- 192.Wang Z, Nilsson RH, James TY, Dai Y, Townsend JP. 2016. Future perspectives and challenges of fungal systematics in the age of big data, p 25–46. In Li D-W (ed), Biology of Microfungi. Springer International Publishing, Cham, Switzerland. 10.1007/978-3-319-29137-6_3 [DOI] [Google Scholar]
- 193.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Coelho MA, Gonçalves C, Sampaio JP, Gonçalves P. 2013. Extensive intra-kingdom horizontal gene transfer converging on a fungal fructose transporter gene. PLoS Genet 9:e1003587 10.1371/journal.pgen.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bennett RJ, Turgeon BG. 2017. Fungal sex: the Ascomycota. Microbiol Spectrum 4(5):FUNK-0005-2016. 10.1128/microbiolspec.FUNK-0005-2016. [DOI] [PubMed] [Google Scholar]


