Abstract
Drug treatments for hepatocellular carcinoma (HCC) often fail because of multidrug resistance (MDR). The mechanisms of MDR are complex but cancer stem cells (CSCs), which are able to self‐renew and differentiate, have recently been shown to be involved. The deubiquitinating enzyme ubiquitin‐specific protease 22 (USP22) is a marker for CSCs. This study aimed to elucidate the role of USP22 in MDR of HCC and the underlying mechanisms. Using in vitro and in vivo assays, we found that modified USP22 levels were responsible for the altered drug‐resistant phenotype of BEL7402 and BEL/FU cells. Downregulation of USP22 dramatically inhibited the expression of ABCC1 (encoding MRP1) but weakly influenced ABCB1 (encoding P‐glycoprotein). Sirtuin 1 (SIRT1) was reported previously as a functional mediator of USP22 that could promote HCC cell proliferation and enhance resistance to chemotherapy. In this study, USP22 directly interacted with SIRT1 and positively regulated SIRT1 protein expression. Regulation of the expression of both USP22 and SIRT1 markedly affected the AKT pathway and MRP1 expression. Inhibition of the AKT pathway by its specific inhibitor LY294002 resulted in downregulation of MRP1. USP22 and MRP1 expression was detected in 168 clinical HCC samples by immunohistochemical staining, and a firm relationship between USP22 and MRP1 was identified. Together, these results indicate that USP22 could promote the MDR in HCC cells by activating the SIRT1/AKT/MRP1 pathway. USP22 might be a potential target, through which the MDR of HCC in clinical setting could be reversed.
Keywords: hepatocellular carcinoma, MRP1, multidrug resistance, SIRT1, USP22
Abbreviations
- CSC
cancer stem cell
- HCC
hepatocellular carcinoma
- MDR
multidrug resistance
- MRP1
resistance‐associated protein 1
- P‐gp
P‐glycoprotein
- SIRT1
sirtuin 1
- USP22
ubiquitin‐specific protease 22
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies and results in approximately 80 000 patient deaths worldwide per year, representing the second leading cause of cancer death (Torre et al., 2015). The front‐line treatment for HCC is surgical resection and liver transplantation. However, most HCCs are inoperable because patients typically present at advanced stages. Even after surgical resection, the prognosis of HCC remains poor given the high recurrence rate. For advanced‐stage HCCs, systematic therapies are adopted as palliative methods, such as traditional chemotherapy and molecular targeted therapy, but most of them failed to demonstrate significant efficacy in increasing survival rates (Chan et al., 2016; Lin et al., 1997). The response rates of traditional cytotoxic chemotherapy agents, such as adriamycin, fluorouracil, cisplatin, and mitomycin to HCC, are less than 10% (Ge and Huang, 2015; Qin et al., 2013; Zaanan et al., 2013). To date, although sorafenib is the only small‐molecule inhibitor that has been approved by the U.S. Food and Drug Administration for HCC treatment, the survival benefit of the sorafenib treatment arm was modest (Cheng et al., 2009). Multidrug resistance (MDR) is the main reason for HCC drug therapy failure, and complex mechanisms are involved. Extensive studies have revealed that MDR is associated with the epithelial/mesenchymal transition (van Zijl et al., 2011), overexpression of drug efflux pumps, DNA damage repair, the hypoxia‐inducible factor 1‐α signaling pathway, and epigenetic regulation. Recent studies indicate that cancer stem cells (CSCs), which are able to self‐renew and differentiate, are involved in the mechanism of chemoresistance (Chan et al., 2015).
Ubiquitin‐specific protease 22 (USP22), which is described as a CSC marker (Glinsky, 2006; Zhang et al., 2008), belongs to the ubiquitin‐specific processing proteases (USPs), the largest subfamily of deubiquitinating enzymes (DUBs) (Nijman et al., 2005). USP22 is a key subunit of the Spt‐Ada‐Gcn5 acetyl transferase complex (SAGA), which removes ubiquitin from target proteins to regulate the transcription of downstream genes (Reyes‐Turcu et al., 2009). USP22 plays a significant role both in pathology and in physiology via interactions with different substrates (Melo‐Cardenas et al., 2016). USP22 could bind to TRF1 and regulate telomere length (Atanassov et al., 2009). USP22 deubiquitinates cyclin B1, stabilizes cyclin B1 by antagonizing proteasome‐mediated degradation, and promotes it accumulation in the nucleus (Lin et al., 2015; Melo‐Cardenas et al., 2016). Recently, USP22 was found to cooperate with GSK3β to stabilize KDM1A and further promote glioblastoma tumorigenesis (Zhou et al., 2016). Clinically, USP22 has been confirmed to predict tumor recurrence, metastasis, and poor survival after cancer diagnosis in several types of cancer (Glinsky et al., 2005; He et al., 2015). Our group first ensured that knockdown of USP22 can induce cell cycle arrest and inhibit cell growth in the HCC cell line HepG2 (Ling et al., 2012a).
Notably, sirtuin 1 (SIRT1), a member of the sirtuin family of nicotinamide adenine dinucleotide (NAD+)‐dependent class III histone deacetylases that are human homologues of yeast silent information regulator 2, controls a variety of biologic processes ranging from metabolic homeostasis, neurodegenerative diseases, and aging to cancer (Armour et al., 2013). Chen et al. (2012) demonstrated that SIRT1 was overexpressed in HCC and plays an oncogenic role in HCC by enhancing cell proliferation and resistance to chemotherapy. Interestingly, SIRT1 was identified as a mediator of acetylation of USP22 and the SAGA coactivator complex (Armour et al., 2013). USP22 may antagonize p53 through Sirt1 stabilization to suppress cell apoptosis under DNA damage and during embryonic development (Lin et al., 2012). To date, studies exploring the mechanism of USP22 influencing drug resistance of HCC is limited. In this study, we hypothesize that the CSC marker USP22 influences drug sensitivity via regulating SIRT1, which will shed new insights into the mechanisms of MDR in HCC.
2. Materials and methods
2.1. Cell cultures
The human HCC cell lines Bel7402 and Bel7402/5‐fluorouracil (BEL/FU) were purchased from KeyGen Biotech Co., Ltd (Nanjing, China). The Bel‐7402 cells were cultured in RPMI‐1640 (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco) and 100 μg·mL−1 each of penicillin and streptomycin (Invitrogen, Carlsbad, CA, USA) in 5% CO2 at 37 °C. The BEL/FU cells were cultured as described previously (Cheng et al., 2013).
2.2. Reagents and antibodies
5‐Fluorouracil (2,4‐dihydroxy‐5‐fluoropyrimidine), methotrexate (MTX), and ADR (doxorubicin) were purchased from Sigma‐Aldrich (St. Louis, MO, USA). LY294002 was purchased from Selleck Chemicals (Houston, TX, USA). The following antibodies were used in western blot analysis and/or immunohistochemical staining. β‐Actin was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). USP22 and SIRT1 were from Abcam (Cambridge, MA, USA). AKT, phosphorylated AKT (phospho‐Ser473), phosphorylated GSK‐3β (phospho‐Ser9), cleaved PARP, and cleaved caspase‐3 were from Cell Signaling Technology, Inc. (Danvers, MA, USA). P‐glycoprotein (P‐gp) and resistance‐associated protein 1 (MRP1) were from Thermo Fisher Scientific, Inc. (Rockford, IL, USA). Goat anti‐rabbit and goat anti‐mouse IgG peroxidase‐conjugated secondary antibodies were from Thermo‐Pierce (Rockford, IL, USA). The Cell Counting Kit‐8 (CCK‐8) and the Annexin V‐FITC Apoptosis Detection Kit were purchased from KeyGen Biotech. The HistostainTM‐Plus Kits (IgG/Bio, S‐A/HRP, DAB) were purchased from Zhongshan Golden Bridge Co., Ltd. (Beijing, China).
2.3. In vitro drug cytotoxic assay
The cytotoxicity of chemotherapeutic drugs was determined by the CCK‐8 assay. Cells (5 × 103) were plated in 100 mL of medium/well in 96‐well plates. After 24 h of incubation, 5‐Fu (0, 100, 200, 500, and 1000 mg·mL−1), MTX (0, 5, 10, 20, and 50 mg·mL−1), and ADR (0, 5, 10, 20, and 50 mg·mL−1) were added, and the CCK‐8 assay was performed 48 h later. Then, the medium was replaced with 100 μL of RPMI‐1640 and 10 μL of CCK‐8 reagent. The plates were gently shaken, and the absorbance was measured at 450 nm with an EnSpire™ 2300 Multilabel Reader (PerkinElmer, Waltham, MA, USA). Five replicates were prepared for each condition, using wells without cells as blanks. The IC50 values were calculated to observe the cytotoxicity of drugs.
2.4. Flow cytometric analysis for cell apoptosis
To evaluate the effects on induction of apoptosis, the cells were examined using the Annexin V‐APC Apoptosis Detection Kit according to the manufacturer's protocols. BEL7402 and BEL/5‐FU cells were seeded into six‐well plates (2 × 105 well). The cells were trypsinized, washed with cold PBS, and suspended in PBS. Then, the cells were stained using 5 μL of Annexin V‐APC and 5 μL of propidium iodide and were incubated at 37 °C for 30 min in the dark. The stained cells were analyzed using an Accuri C6 flow cytometer (Accuri Cytometers Inc., Ann Arbor, MI, USA).
2.5. RT‐PCR and real‐time PCR array analysis
For RT‐PCR, the RNA isolation, PCR, and primers for amplification were as described before (Ling et al., 2012a). For the real‐time PCR array analysis, the Human Cancer Drug Resistance PCR Array (PAHS‐004Z) was purchased from QIAGEN (Valencia, CA, USA). Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) and then reverse‐transcribed into first‐strand cDNA using the QuantiTect Reverse Transcription Kit (QIAGEN) according to the manufacturer's protocol. Real‐time PCR was performed on an ABI Prism 7500 fast real‐time PCR system (Applied Biosystems, Foster City, CA, USA) using the QuantiTect SYBR Green PCR Kit (QIAGEN). The data were analyzed in http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php. The relative expression of the genes was normalized to β‐actin. The primer sequences of MRP1 (ABCC1) and β‐actin could be found in one of our previous studies (Ling et al., 2014b).
2.6. Immunoprecipitation and western blot analysis
An immunoprecipitation kit from Proteintech (Wuhan, China) was used in this study. The main procedure was conducted according to the manufacturer's protocol. Briefly, cell lysates were immunoprecipitated with an antibody against USP22. Precipitated proteins (10 μg) were prepared for western blot analysis, and the detailed procedure has been described previously (Ling et al., 2014a). Primary antibodies (described in the section 2.2.) were incubated at 4 °C overnight. The bands were visualized by chemiluminescence, imaged using a ChemiDoc XRS, and analyzed using Image Lab (both from Bio‐Rad, Hercules, CA, USA).
2.7. Intracellular ADR measurement
Cells were treated with 10 μm of ADR for 1 h, and then, the cells were cultured in drug‐free RPMI‐1640 for an additional 1 h. Next, the cells were trypsinized and harvested. Fluorescence intensity of intracellular ADR was determined by flow cytometry. The wavelengths of excitation light and emission light were 488 and 575 nm. Geomean was calculated and compared among different groups.
2.8. Gene knockdown using shRNA in BEL/FU cells
Transfection was performed in a six‐well plate with 50% confluent cultures. Cells were maintained in 2 mL of complete medium with 5 mg·mL−1 Polybrene per well and were treated with 0.5 μm control, USP22‐ or SIRT1‐specific shRNA lentiviral particles (obtained from Shandong Vigenebio Co., Ltd., Jinan, China) overnight. Then, the medium in each well was replaced with 2 mL of complete medium (without Polybrene). Next, stable clones expressing control, USP22‐ or SIRT1‐specific shRNA were selected using puromycin dihydrochloride. Then, cells were collected for gene expression assays.
2.9. Stable overexpression of USP22 or SIRT1 in BEL‐7402 cells
Lentiviral particles containing empty vector, USP22 vector, or SIRT1 vector were obtained from Shandong Vigenebio Co., Ltd. Cells were plated in a six‐well plate at 50% confluent cultures. Media were removed from plate wells and replaced with media supplemented with Polybrene at a final concentration of 5 μg·mL−1. Next, the cells were infected by adding lentiviral particles to the culture. Stable clones expressing empty vector, USP22, or SIRT1 were selected using puromycin dihydrochloride. Then, cells were collected for gene expression assays.
2.10. In vivo chemosensitivity assay
To investigate whether USP22 is related to drug sensitivity of tumor cells in vivo, the chemosensitivity of 5‐FU was examined in nude mice bearing tumor cell xenografts. Five‐week‐old male athymic nude mice were obtained from the Animal Facility of Zhejiang University. The mice were maintained under pathogen‐free conditions and were provided with sterilized food and water. Briefly, 5 × 106 cells were injected subcutaneously into the right flank of each nude mouse. When mice exhibited palpable tumors (the tumor volume was approximately 100 mm3), BEL7402/mock, BEL7402/USP22, BEL/FU control shRNA, and BEL/FU‐USP22 shRNA tumor‐bearing mice were randomly divided into control and treatment groups (n = 6 mouse per group). The treatment groups received 30 mg·kg−1 5‐FU (intraperitoneal injection) three times per week for 3 weeks, and the control groups received DMSO diluted in physiological saline. The mice were sacrificed, and the tumors were isolated. The tumor volume was detected and calculated using the following formula: volume = 1/2(length × width2).
2.11. Patient samples
Tumor samples from a total of 168 HCC patients who underwent operations at our hospital (First Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang, China) between 2010 and 2014 were used in this study. This study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhejiang University, and informed consent was obtained from all patients. These patients were diagnosed with HCC either before or after surgery. No treatment was administered to the patients before surgery. The diagnosis was confirmed by histopathological examination.
2.12. Immunohistochemical staining
The tumors isolated from the mice were paraffin‐embedded and cut into 10‐μm sections using a microtome cryostat (HM 500 OM; Carl Zeiss, Jena, Germany). Immunohistochemical staining was conducted according to the manufacturer's protocols for the HistostainTM‐Plus Kits. Primary antibodies (as described in the section 2.2.) were incubated at 4 °C overnight. The images were captured with a light microscope (Axiolab; Carl Zeiss), and five images/sample were prepared. The image‐pro plus 4.5 (Media Cybernetics, Silver Spring, MD, USA) software was used to analyze the staining data.
For the tumor samples from HCC patients, a tissue microarray was created from the 168 HCC samples. After histopathological examination, every sample was excised in one core with a 1.0 mm diameter on one tissue array. Immunohistochemical staining was conducted according to the manufacturer's protocols for the HistostainTM‐Plus Kits. The results were scanned by Pannoramic MIDI (3DHISTECH Ltd., Budapest, Hungary), quantified, and analyzed by Pannoramic viewer and quant center software. For each epitope, the staining score for tumor cells was recorded separately. An H‐score, which reflects the expression level of certain protein, was calculated based on the staining intensity as previously described (Azim et al., 2015).
2.13. Statistical analysis
spss 21.0 statistical software (Chicago, IL, USA) was used for the statistical analysis. Values are presented as the mean ± SD. Statistical analyses were performed using Student's t‐test. The analysis of multiple groups was performed by ANOVA with an appropriate post hoc test. Correlation analyses were performed using Pearson's test for quantitative data.
3. Results
3.1. USP22 is highly expressed in the HCC MDR cell line BEL/FU
Our PCR and western blot assay results revealed drastically increased USP22 mRNA and protein expression in BEL/FU cells compared with parental BEL‐7402 cells (Fig. 1). Given that USP22 is a CSC marker, these results implied that USP22 might play a significant role in MDR in human HCC cells.
Figure 1.
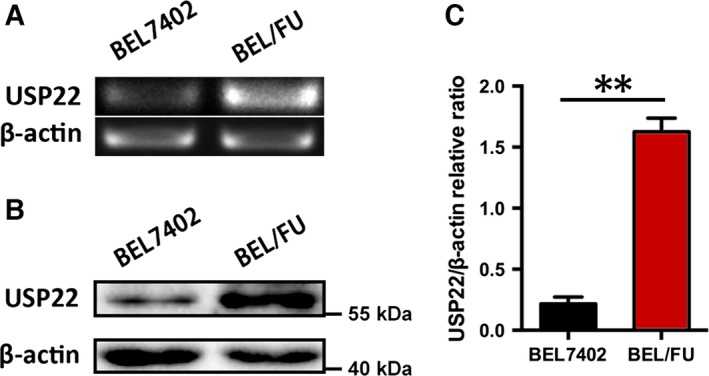
Expression of USP22 in BEL7402 and BEL/FU cells. (A) The USP22 mRNA levels were quantified by qPCR in BEL7402 and BEL/FU cells. (B) The USP22 expression was determined using western blot in BEL7402 and BEL/FU cells. (C) Band intensities are presented after semiquantification using image lab 5.0 software (Bio‐Rad) and normalized with β‐actin. These values are represented as the means ± SD from three independent experiments (**P < 0.01).
3.2. Modulation of USP22 effectively regulates MDR in BEL‐7402 and BEL/FU cells in vitro and in vivo
Given the significantly increased expression of USP22 in BEL/FU cells (Fig. 1), we silenced the USP22 gene by shRNA to elucidate the implication of USP22 in the chemosensitivity of BEL/FU cells in vitro and in vivo. As shown in Fig. 2A,B, the USP22 expression level was significantly reduced in BEL/FU cells treated with USP22 shRNA. The IC50 was calculated using the CCK‐8 assay, and silencing of USP22 significantly increased the chemosensitivity of BEL/FU cells to 5‐FU, MTX, and ADR. We also used a nude mouse model bearing BEL/FU control shRNA cell and BEL/FU control USP22 shRNA cell xenografts to elucidate the implication of USP22 in the chemosensitivity of BEL/FU cells to 5‐FU. As noted in Fig. 2E, no significant difference between BEL/FU control shRNA cells treated with DMSO and BEL/FU USP22 shRNA treated with DMSO (1627 ± 623 mm3 vs 1467 ± 329 mm3, P > 0.05) was observed. Remarkably, silencing USP22 reduced the tumor volume of BEL/FU cells treated with 5‐FU (BEL/FU control shRNA cells treated with 5‐FU vs BEL/FU USP22 shRNA treated with 5‐FU, 945 ± 545 mm3 vs 372 ± 228 mm3, P < 0.05). The in vivo assay results were consistent with the in vitro assay results.
Figure 2.
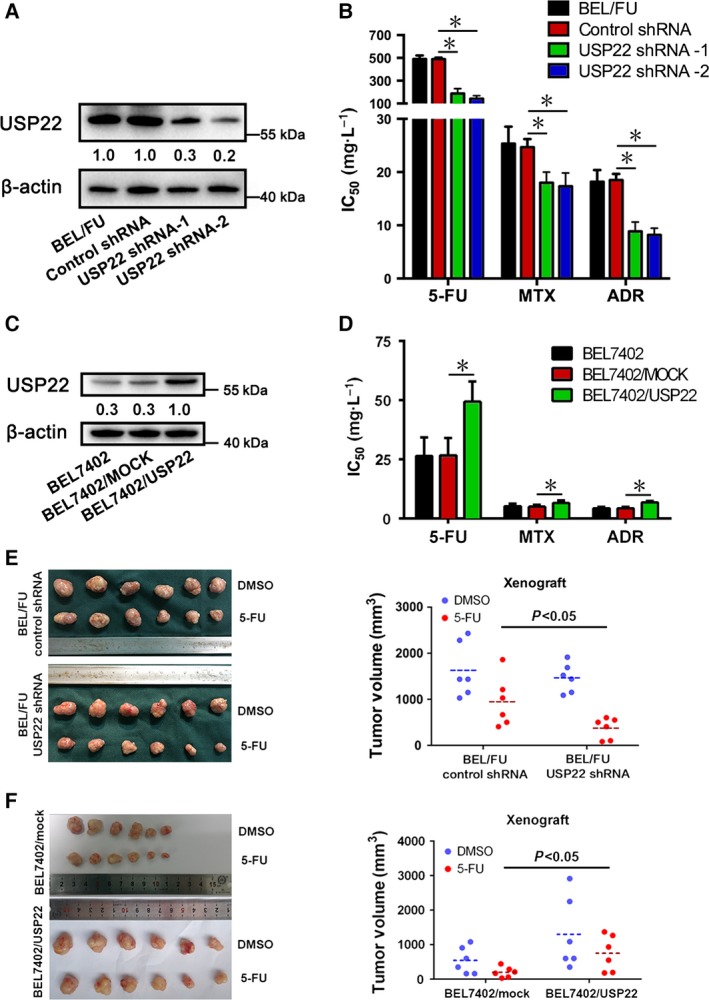
Effect of USP22 on the chemosensitivity of BEL7402 and BEL/FU cells in vitro and in vivo. (A,C) USP22 expression was monitored using western blot analysis in BEL7402 and BEL/FU cells after modulation of USP22. Band intensities were semiquantified using image lab 5.0 software and normalized with AKT or β‐actin. Values are presented as the means under the bands. (B,D) The IC50 values from the CCK‐8 assay were calculated to assess the sensitivity to 5‐FU, MTX, and ADR. These values are presented as means ± SD from three independent experiments (*P < 0.05). (E,F) The xenograft tumor (derived from BEL/FU control shRNA, BEL/FU USP22 shRNA, BEL7402/mock, or BEL7402/USP22 cells) volumes were measured, and the data were quantified and presented (*P < 0.05).
Moreover, lentiviral vector‐mediated overexpression of USP22 protein was performed in BEL‐7402 cells. As shown in Fig. 2C,D, overexpression of USP22 significantly decreased the chemosensitivity of BEL‐7402 cells to 5‐FU, MTX, and ADR. Similarly, the in vivo assay results supported the in vitro results (Fig. 2F). These results supported the role of USP22 as a positive modulator of chemosensitivity in HCC cells.
3.3. Modulation of USP22 in BEL/FU and BEL‐7402 cells regulates 5‐FU‐induced cell apoptosis and intracellular ADR concentrations
The number of apoptotic cells was examined after 48 h of 5‐FU or DMSO incubation by flow cytometric analysis. A significant increase in apoptosis was observed in BEL/FU USP22 shRNA cells with 5‐FU treatment compared with control cells, whereas no noticeable difference in the number of apoptotic cells incubated with DMSO was observed (Fig. 3A). Cleaved caspase‐3 and cleaved parp, two markers of apoptosis, were monitored to confirm 5‐FU‐induced apoptosis by western blot analysis. The levels of cleaved caspase‐3 and cleaved parp were upregulated upon inhibition of USP22 (Fig. 3B). Concomitantly, the number of apoptotic BEL‐7402/USP22 cells was reduced compared with BEL‐7402/mock cells upon treatment with 5‐FU (Fig. 3C). Cleaved caspase‐3 and cleaved parp were downregulated (Fig. 3D). To further confirm the effect of USP22 on the regulation of MDR, the fluorescence intensity of intracellular ADR was determined by flow cytometry. As shown in Fig. 3E, the concentration of intracellular ADR was increased in BEL/FU USP22 shRNA cells. In addition, overexpression of USP22 also reduced the intracellular ADR concentration in BEL7402 cells. Conclusively, these results suggested that overexpression of USP22 inhibited 5‐FU‐induced cell apoptosis and promoted ADR efflux in HCC cells.
Figure 3.
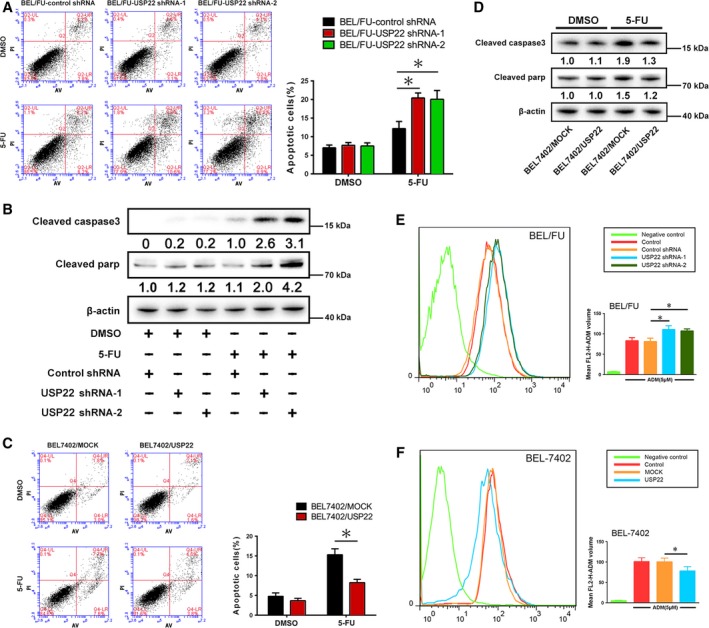
USP22 regulates cell apoptosis and intracellular ADR in BEL7402 and BEL/FU cells. (A,C) After knockdown or overexpression of USP22 in BEL/FU or BEL7402 cells, cells were examined using Annexin V/PI staining. Then, the distribution of apoptotic cells was measured by flow cytometric analysis. The percentages of apoptotic cells are presented in the bar graph (*P < 0.05). (B,D) Cleaved caspase‐3 and cleaved parp were monitored using western blot analysis in BEL7402 and BEL/FU cells. Band intensities were semiquantified using image lab 5.0 software and normalized with AKT or β‐actin. Values are represented as the means under the bands. (E,F) Intracellular ADR was measured by flow cytometric analysis. Mean FL2‐H‐ADM volume is presented in the bar graph as means ± SD from three independent experiments (*P < 0.05).
3.4. PCR array results imply that downregulation of USP22 in BEL/FU cells decreases the expression of MRP1, but not P‐gp
As we previously described, USP22 silencing increased the concentration of intracellular ADR. We suggest that USP22 may have an influence on MDR proteins. A cancer drug resistance PCR array revealed that downregulation of USP22 dramatically inhibited the expression of ABCC1 (encoding MRP1) but weakly influenced ABCB1 (encoding P‐gp), as presented in Fig. S1A. The expression of P‐gp was verified by western blot analysis (Fig. S1B).
3.5. USP22 activates the AKT/MRP1 pathway depending on SIRT1 in HCC cells
As shown in Fig. 4A, knockdown of USP22 by shRNA inhibited the AKT/MRP1 pathway compared with control shRNA cells and wild‐type cells. We observed remarkably decreased expression of phosphorylated AKT and phosphorylated GSK‐3β in BEL/FU USP22 shRNA cells. Similarly, overexpression of USP22 in BEL/7402 cells activated the AKT/MRP1 pathway (Fig. 4B). The downregulation of SIRT1 induced suppression of the AKT/MRP1 pathway in BEL/FU cells (Figs 4C and S2A), and the overexpression of SIRT1 activated the AKT/MRP1 pathway in BEL7402 cells (Fig. 4D). SIRT1 expression was positively regulated by USP22 (Fig. 4A,B), whereas SIRT1 knockdown or overexpression did not influence USP22 expression (Fig. 4C,D). Moreover, SIRT1 deficiency attenuated USP22‐induced activation of the AKT/MRP1 pathway in BEL7402 cells, suggesting that USP22 regulated the AKT/MRP1 pathway in a SIRT1‐dependent manner (Fig. 5A). USP22 directly interacts with SIRT1 based on the co‐immunoprecipitation assay (Fig. 5B).
Figure 4.
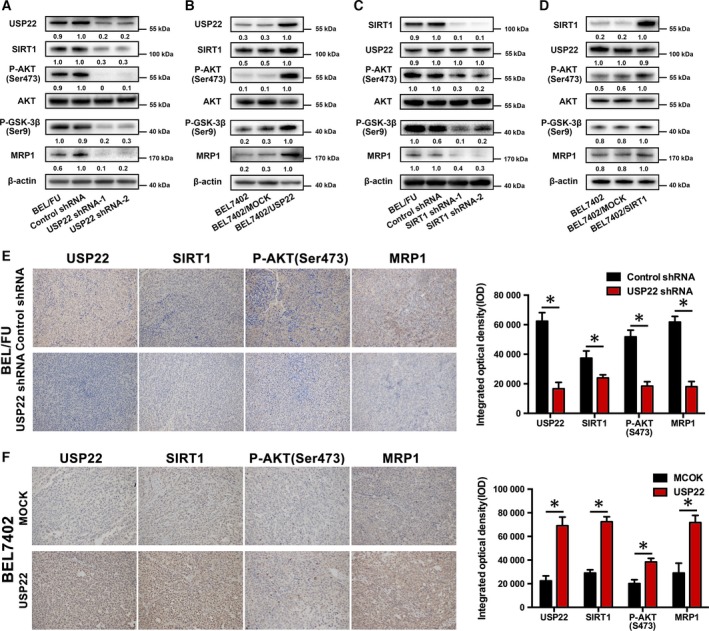
Regulation among USP22, SIRT1, and the AKT/MRP1 pathway. (A,B) After knockdown or overexpression of USP22 in BEL/FU or BEL7402 cells, USP22, SIRT1, phosphorylated AKT, AKT, phosphorylated GSK‐3β, and MRP1 were monitored using western blot analysis. (C,D) After knockdown or overexpression of SIRT1 in BEL/FU or BEL7402 cells, USP22, SIRT1, phosphorylated AKT, AKT, phosphorylated GSK‐3β, and MRP1 were monitored using western blot analysis. Band intensities were semiquantified using image lab 5.0 software and normalized with AKT or β‐actin. Values are represented as the means under the bands. (E,F) The expression of USP22, SIRT1, phosphorylated AKT, and MRP1 in xenograft tumors (derived from BEL/FU control shRNA, BEL/FU USP22 shRNA, BEL7402/mock, or BEL7402/USP22 cells) was detected by IHC. The data were quantified and are represented as the means ± SD (*P < 0.05).
Figure 5.
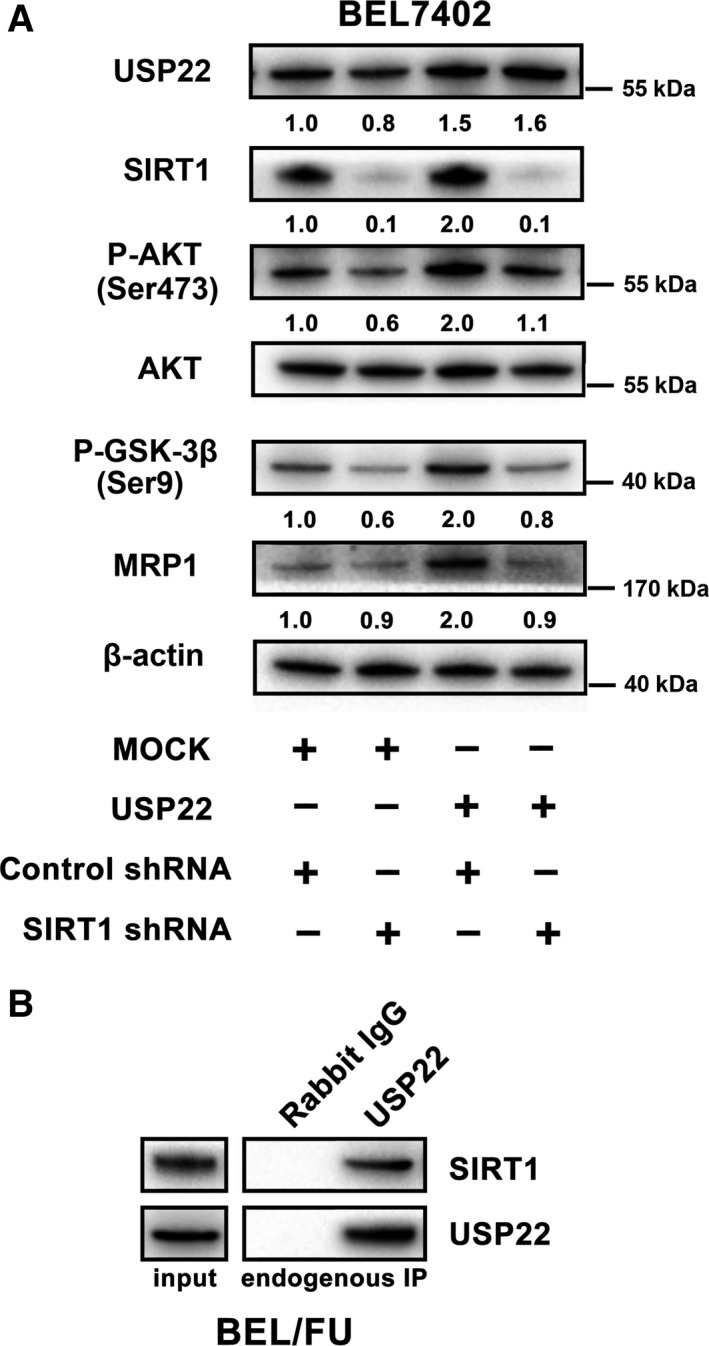
Deficiency of SIRT1 partially attenuates USP22‐induced activation of the AKT/MRP1 pathway in BEL7402 cells. (A) AKT/MRP1 pathway expression was monitored by western blot analysis in BEL7402 cells with USP22 overexpression and/or SIRT1 knockdown by shRNA. Values are represented as the means under the bands. (B) Co‐immunoprecipitation assays were used to verify the interaction between USP22 and SIRT1.
In addition, we also evaluated the variation in SIRT1 and the AKT/MRP1 pathway in the xenografts of the nude mouse. Consistent with the in vitro results, SIRT1 and the AKT/MRP1 pathway were inhibited in BEL/FU USP22 shRNA cell‐derived xenografts and positively regulated in BEL/USP22 cell‐derived xenografts compared with control (Fig. 4E,F).
3.6. Modulation of SIRT1 in BEL/FU and BEL‐7402 cells regulates 5‐FU‐induced cell apoptosis and intracellular ADR concentrations
We next tested the effect of SIRT1 on 5‐FU‐induced cell apoptosis and intracellular ADR concentration. Constitutively suppressed SIRT1 caused a significant increase in apoptotic cells after 5‐FU treatment compared with the control cells in BEL/FU SIRT1 shRNA cells (Fig. 6A). Inhibition of SIRT1 stimulated the expression of cleaved caspase‐3 and cleaved parp (Fig. 6B) in BEL/FU cells treated with 5‐FU. Similarly, the apoptotic BEL‐7402/SIRT1 cells decreased upon treatment with 5‐FU (Fig. 6C). Cleaved caspase‐3 and cleaved parp were downregulated in BEL7402/SIRT1 cells (Fig. 6D). As shown in Fig. 6E,F, the concentration of intracellular ADR was increased in BEL/FU SIRT1 shRNA cells, and overexpression of SIRT1 reduced the intracellular ADR concentration in BEL7402 cells.
Figure 6.
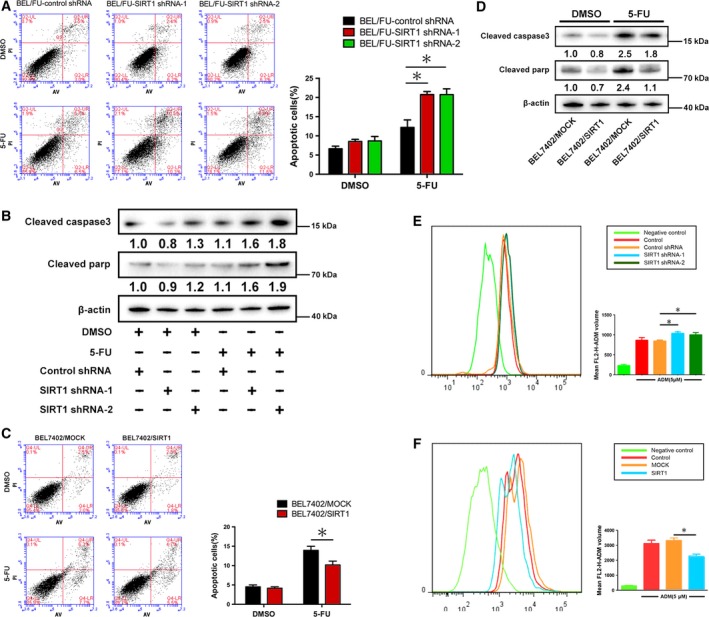
SIRT1 regulates cell apoptosis and intracellular ADR in BEL7402 and BEL/FU cells. (A,C) After knockdown or overexpression of SIRT1 in BEL/FU or BEL7402 cells, cells were examined using Annexin V/PI staining. Then, the distribution of apoptotic cells was measured by flow cytometric analysis. Apoptotic cell percentages are presented in the bar graph (*P < 0.05). (B,D) Cleaved caspase‐3 and cleaved parp were monitored using western blot analysis in BEL7402 and BEL/FU cells. Band intensities were semiquantified using image lab 5.0 software and normalized with AKT or β‐actin. Values are represented as the means under the bands. (E,F) Intracellular ADR was measured by flow cytometric analysis. Mean FL2‐H‐ADM volumes are presented in the bar graph as means ± SD from three independent experiments (*P < 0.05).
In conclusion, similar to USP22, SIRT1 has a remarkable influence on 5‐FU‐induced cell apoptosis and intracellular ADR concentration. These results confirmed the role of SIRT1 in USP22‐induced MDR in HCC cells.
3.7. Inhibition of the AKT signaling pathway suppresses the expression of MRP1 and promotes 5‐FU‐induced apoptosis in HCC cells
As aberrant expression of USP22 led to changes in the AKT pathway and MRP1, we tested whether MRP1 expression was regulated by AKT. Inhibition of the AKT pathway by LY294002 (10 μm, 20 μm) efficiently suppressed MRP1 expression in BEL/FU cells in both mRNA and protein levels (Figs 7A and S2B). LY294002 (10 μm) also suppressed USP22‐induced high expression of MRP1 via inhibition of AKT pathway in BEL7402 cells (Fig. 7B). Both of these findings indicated that USP22 upregulated MRP1 depending on the AKT pathway. In addition, cleaved caspase‐3 and cleaved parp were upregulated after BEL/FU cells treated with 5‐FU were incubated with LY294002 (Fig. 7C).
Figure 7.
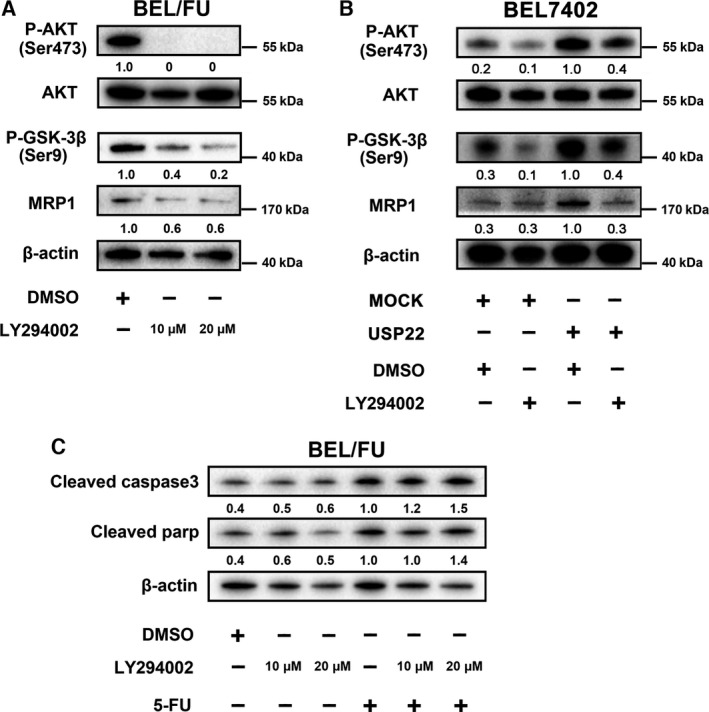
Inhibition of the AKT pathway suppresses MRP1 expression and promotes 5‐FU‐induced apoptosis in BEL/FU cells. (A) LY294002 (10 μm, 20 μm) was used to treat BEL/FU cells, and phosphorylated AKT, AKT, phosphorylated GSK‐3β, and MRP1 were monitored using western blot analysis. (B) AKT/MRP1 pathway expression was monitored by western blot analysis in BEL7402 cells with USP22 overexpression and/or LY294002. (C) Cleaved caspase‐3 and cleaved parp were monitored using western blot analysis in cells treated with LY294002 and/or 5‐FU. Band intensities were semiquantified using image lab 5.0 software and normalized with AKT or β‐actin. Values are represented as the means under the bands.
3.8. USP22 and MRP1 expressions are positively correlated in HCC tissues
Finally, USP22 and MRP1 expressions were detected in 168 HCC samples by immunohistochemical staining. Upon evaluation of H‐scores, a high positive correlation between USP22 and MRP1 was observed (P < 0.001, Pearson's correlation = 0.89) (Fig. 8B), further confirming the relationship between USP22 and MRP1.
Figure 8.
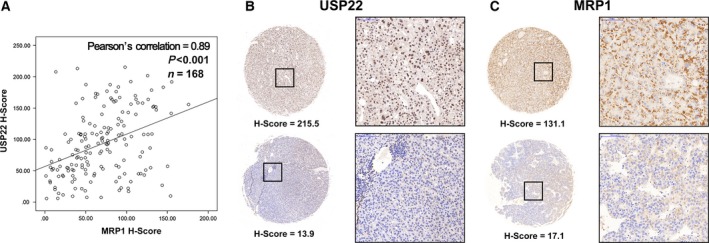
Correlation between USP22 and MRP1 protein expression in 168 HCC samples. (A) Positive correlation between USP22 (y‐axis) and MRP1 (x‐axis) H‐scores by immunohistochemistry (P < 0.001, Pearson's correlation = 0.89). (B) USP22 expression detected by immunohistochemical staining of the primary tumor in two patients diagnosed with HCC. (C) MRP1 expression detected by immunohistochemical staining of the primary tumor in two patients diagnosed with HCC.
4. Discussion
Multidrug resistance is a multifactorial phenomenon and is the major obstacle in the successful and effective chemotherapeutic treatment for HCC. Elucidation of the mechanism of MDR will help to identify effective targets to reverse MDR of HCC in clinical settings. In this study, we demonstrated that USP22 is significant for the MDR of HCC cells in vitro and in vivo and deeply explored the mechanism underlying this phenomenon (Fig. 9).
Figure 9.
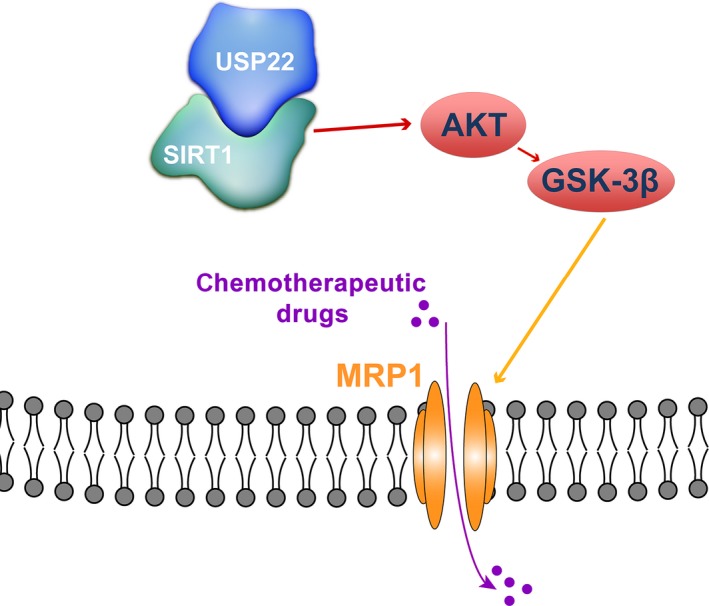
Proposed mechanisms responding to USP22‐mediated MDR in HCC. USP22 directly interacted with SIRT1 and then upregulated MRP1 through the activation of AKT/GSK‐3β pathway, which promoted the efflux of chemotherapeutic drugs of HCC cells.
First, we defined SIRT1 as a significant mediator in USP22‐driven MDR in HCC. Given that SIRT1 is deubiquitinated by USP22 and stabilized at the protein level (Armour et al., 2013; Lin et al., 2012) and previous studies have reported that SIRT1 could negatively influence the chemosensitivity of HCC cells (Chen et al., 2012), our results supported the notion that USP22 increases SIRT1 protein levels in HCC cells. Interestingly, we found that modulation of USP22 or SIRT1 could influence the intracellular ADR concentration, which might partly bridge the induction of MDR by USP22 and SIRT1 in HCC cells. Subsequently, using qPCR array analysis, we found that knockdown of USP22 could drastically inhibit the expression of MRP1, but not P‐gp, in BEL/FU cells. Overexpression of drug efflux transporters belonging to adenosine triphosphate‐dependent binding cassette (ABC) protein family is often responsible for the MDR of cancer (Kathawala et al., 2015; Wolking et al., 2015). The data of PCR array revealed some other interesting findings such as the downregulation of ABCC5 and CYP2D6, which regulates the drug metabolism in cell. We think it is valuable for us to investigate the relationship between USP22 and ABCC5 or CYP2D6. However, both MRP1, also known as ABCC1, and P‐gp, also known as ABCB1, play crucial roles in MDR of HCC (Ling et al., 2012b; Qian et al., 2015). Thus, we demonstrated that silencing of USP22 did not influence the protein level of P‐gp and further explored how USP22 regulated the expression of MRP1 and whether the process was dependent on SIRT1. We will demonstrate the relationship between USP22 and ABCC5 or CYP2D6 in our future studies.
The AKT signaling pathway controls the expression and function of numerous proteins, including MRP1, driving cancer cell MDR (Li et al., 2010; Wang et al., 2016b). Several studies have revealed the relationship between USP22 and the AKT signaling pathway (Cheng et al., 2013; Liu et al., 2012; Zhou et al., 2016; Zhuang et al., 2015). Therefore, the active status of the AKT signaling pathway was detected under the modulation of USP22 in HCC cells in the present study. We observed that USP22 could positively regulate the AKT pathway in a SIRT1‐dependent manner. The PI3K/AKT inhibitor (LY294002) could effectively suppress the expression of MRP1 and promote 5‐FU‐induced apoptosis in BEL/FU cells. Collectively, USP22 might deubiquitinate SIRT1 and subsequently activate the AKT pathway, increasing the expression of MRP1 to induce MDR in HCC cells. Several studies have described the regulation between SIRT1 and the AKT pathway (Pinton et al., 2013; Wang et al., 2016a). SIRT1 could deacetylate AKT and promote its activation (Sundaresan et al., 2011). Our results demonstrated that SIRT1 could promote the AKT activation, but not increased the expression level of AKT in HCC cells, which were consistent with the previous findings. The exact modification of the AKT pathway by SIRT1 in HCC cells needs further demonstration.
Moreover, we also identified a high expression correlation between USP22 and MRP1 in tumor samples from 168 HCC patients, which further confirmed the role of USP22 in the induction of MDR in HCC and supported the molecular mechanism we previously investigated. We will evaluate the clinical significance of combined USP22 and MRP1 detection in HCC tissues in predicting the survival rate or chemotherapy response in our future study.
Taken together, the present study found that USP22 can promote the MDR in HCC cells via activating the SIRT1/AKT/MRP1 pathway. USP22 might be a potential target, through which the MDR of HCC in clinical setting could be reversed. Given that SIRT1 is a promising regulator of metabolism and autophagy (Bellet et al., 2016; Ou et al., 2014), which are also significant for MDR in cancer cells (Bhattacharya et al., 2016), it is important to further explore the mechanism of USP22/SIRT1‐induced MDR in HCC cells. Moreover, an increasing number of USP22 substrates were discovered (Melo‐Cardenas et al., 2016). One of them is cyclin B1 (Lin et al., 2015). Owing to its relationship with the chemotherapeutic resistance of several types of human cancers (Glinsky, 2005), cyclin B1 may also mediate the USP22‐induced MDR in HCC cells, which is valuable for future exploration.
Author contributions
SZ and XX conceived and coordinated the project. SL and JL designed the research study. SL, HX, QS, HD, and LD performed experiments and acquisition of data. SL, XW, PS, LZ, and JL analyzed and interpreted data. SL and JL wrote the paper and critically reviewed the manuscript. All authors read and approved the final version of the manuscript.
Supporting information
Fig. S1. (A) Cancer drug resistance PCR array analysis in BEL/FU cells transfected with control shRNA or USP22 shRNA. A The gene list was presented as BEL/FU USP22 shRNA vs BEL/FU control shRNA. (B) Western blot assay was used to demonstrated that P‐gp (encoded by ABCB1) was not varied in BEL/FU cells with downregulation of USP22.
Fig. S2. MRP‐1 mRNA expression in BEL/FU cells with SIRT1 downregulation and AKT inactivation.
Acknowledgements
This work was supported by the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (Grant No. 81421062), the Major Research Plan of the National Natural Science Foundation of China (No. 91542205), National High‐tech R&D Program of China (863 Program) (No. 2012AA020204), Science and Technology Ministry of Youth Project, Yangtze River Scholar Project, Projects of Medical and Health Technology Program in Zhejiang Province (2015117734).
Contributor Information
Xiao Xu, Email: zjxu@zju.edu.cn.
Shusen Zheng, Email: shusenzheng@zju.edu.cn.
References
- Armour SM, Bennett EJ, Braun CR, Zhang XY, McMahon SB, Gygi SP, Harper JW and Sinclair DA (2013) A high‐confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol 33, 1487–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S and Dent SY (2009) Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell 35, 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim HA Jr, Peccatori FA, Brohee S, Branstetter D, Loi S, Viale G, Piccart M, Dougall WC, Pruneri G and Sotiriou C (2015) RANK‐ligand (RANKL) expression in young breast cancer patients and during pregnancy. Breast Cancer Res 17, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Masri S, Astarita G, Sassone‐Corsi P, Della Fazia MA and Servillo G (2016) Histone deacetylase SIRT1 controls proliferation, circadian rhythm, and lipid metabolism during liver regeneration in mice. J Biol Chem 291, 23318–23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya B, Mohd Omar MF and Soong R (2016) The Warburg effect and drug resistance. Br J Pharmacol 173, 970–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LH, Luk ST and Ma S (2015) Turning hepatic cancer stem cells inside out – a deeper understanding through multiple perspectives. Mol Cells 38, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Wong AM, Lee K, Wong N and Chan AK (2016) Personalized therapy for hepatocellular carcinoma: where are we now? Cancer Treat Rev 45, 77–86. [DOI] [PubMed] [Google Scholar]
- Chen HC, Jeng YM, Yuan RH, Hsu HC and Chen YL (2012) SIRT1 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma and its expression predicts poor prognosis. Ann Surg Oncol 19, 2011–2019. [DOI] [PubMed] [Google Scholar]
- Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS et al (2009) Efficacy and safety of sorafenib in patients in the Asia‐Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double‐blind, placebo‐controlled trial. Lancet Oncol 10, 25–34. [DOI] [PubMed] [Google Scholar]
- Cheng L, Luo S, Jin C, Ma H, Zhou H and Jia L (2013) FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis 4, e923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S and Huang D (2015) Systemic therapies for hepatocellular carcinoma. Drug Discov Ther 9, 352–362. [DOI] [PubMed] [Google Scholar]
- Glinsky GV (2005) Death‐from‐cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle 4, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Glinsky GV (2006) Genomic models of metastatic cancer: functional analysis of death‐from‐cancer signature genes reveals aneuploid, anoikis‐resistant, metastasis‐enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle 5, 1208–1216. [DOI] [PubMed] [Google Scholar]
- Glinsky GV, Berezovska O and Glinskii AB (2005) Microarray analysis identifies a death‐from‐cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 115, 1503–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Jin YJ, Zhang YH, Meng HX, Zhao BS, Jiang Y, Zhu JW, Liang GY, Kong D and Jin XM (2015) Ubiquitin‐specific peptidase 22 overexpression may promote cancer progression and poor prognosis in human gastric carcinoma. Transl Res 165, 407–416. [DOI] [PubMed] [Google Scholar]
- Kathawala RJ, Gupta P, Ashby CR Jr and Chen ZS (2015) The modulation of ABC transporter‐mediated multidrug resistance in cancer: a review of the past decade. Drug Resist Updat 18, 1–17. [DOI] [PubMed] [Google Scholar]
- Li L, Wei XH, Pan YP, Li HC, Yang H, He QH, Pang Y, Shan Y, Xiong FX, Shao GZ et al (2010) LAPTM4B: a novel cancer‐associated gene motivates multidrug resistance through efflux and activating PI3K/AKT signaling. Oncogene 29, 5785–5795. [DOI] [PubMed] [Google Scholar]
- Lin DY, Lin SM and Liaw YF (1997) Non‐surgical treatment of hepatocellular carcinoma. J Gastroenterol Hepatol 12, S319–S328. [DOI] [PubMed] [Google Scholar]
- Lin Z, Tan C, Qiu Q, Kong S, Yang H, Zhao F, Liu Z, Li J, Kong Q, Gao B et al (2015) Ubiquitin‐specific protease 22 is a deubiquitinase of CCNB1. Cell Discov 1, pii: 15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD et al (2012) USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell 46, 484–494. [DOI] [PubMed] [Google Scholar]
- Ling S, Feng T, Ke Q, Fan N, Li L, Li Z, Dong C, Wang C, Xu F, Li Y et al (2014a) Metformin inhibits proliferation and enhances chemosensitivity of intrahepatic cholangiocarcinoma cell lines. Oncol Rep 31, 2611–2618. [DOI] [PubMed] [Google Scholar]
- Ling X, He Y, Zhang G, Zhou Y and Yan B (2012b) Increased P‐glycoprotein expression in mitochondria is related to acquired multidrug resistance in human hepatoma cells depleted of mitochondrial DNA. Int J Oncol 40, 109–118. [DOI] [PubMed] [Google Scholar]
- Ling SB, Sun DG, Tang B, Guo C, Zhang Y, Liang R, Wang LM (2012a) Knock‐down of USP22 by small interfering RNA interference inhibits HepG2 cell proliferation and induces cell cycle arrest. Cell Mol Biol (Noisy‐le‐grand) 58(Suppl), OL1803–OL1808. [PubMed] [Google Scholar]
- Ling S, Tian Y, Zhang H, Jia K, Feng T, Sun D, Gao Z, Xu F, Hou Z, Li Y et al (2014b) Metformin reverses multidrug resistance in human hepatocellular carcinoma Bel7402/5‐fluorouracil cells. Mol Med Rep 10, 2891–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Jiang SX, Yang YM, Xu H, Liu JL and Wang XS (2012) USP22 acts as an oncogene by the activation of BMI‐1‐mediated INK4a/ARF pathway and Akt pathway. Cell Biochem Biophys 62, 229–235. [DOI] [PubMed] [Google Scholar]
- Melo‐Cardenas J, Zhang Y, Zhang DD and Fang D (2016) Ubiquitin‐specific peptidase 22 functions and its involvement in disease. Oncotarget 7, 44848–44856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna‐Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK and Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786. [DOI] [PubMed] [Google Scholar]
- Ou X, Lee MR, Huang X, Messina‐Graham S and Broxmeyer HE (2014) SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells 32, 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton G, Manente AG, Murer B, De Marino E, Mutti L and Moro L (2013) PARP1 inhibition affects pleural mesothelioma cell viability and uncouples AKT/mTOR axis via SIRT1. J Cell Mol Med 17, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian JQ, Sun P, Pan ZY and Fang ZZ (2015) Annonaceous acetogenins reverses drug resistance of human hepatocellular carcinoma BEL‐7402/5‐FU and HepG2/ADM cell lines. Int J Clin Exp Pathol 8, 11934–11944. [PMC free article] [PubMed] [Google Scholar]
- Qin S, Bai Y, Lim HY, Thongprasert S, Chao Y, Fan J, Yang TS, Bhudhisawasdi V, Kang WK, Zhou Y et al (2013) Randomized, multicenter, open‐label study of oxaliplatin plus fluorouracil/leucovorin versus doxorubicin as palliative chemotherapy in patients with advanced hepatocellular carcinoma from Asia. J Clin Oncol 31, 3501–3508. [DOI] [PubMed] [Google Scholar]
- Reyes‐Turcu FE, Ventii KH and Wilkinson KD (2009) Regulation and cellular roles of ubiquitin‐specific deubiquitinating enzymes. Annu Rev Biochem 78, 363–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta M, Gupta MP (2011) The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal 4, ra46. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J and Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108. [DOI] [PubMed] [Google Scholar]
- Wang H, Jia XH, Chen JR, Yi YJ, Wang JY, Li YJ, Xie SY (2016b) HOXB4 knockdown reverses multidrug resistance of human myelogenous leukemia K562/ADM cells by downregulating P‐gp, MRP1 and BCRP expression via PI3K/Akt signaling pathway. Int J Oncol 49, 2529–2537. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang D, Yan T and Yuan H (2016a) MiR‐138‐5p promotes TNF‐alpha‐induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp Cell Res 345, 199–205. [DOI] [PubMed] [Google Scholar]
- Wolking S, Schaeffeler E, Lerche H, Schwab M and Nies AT (2015) Impact of genetic polymorphisms of ABCB1 (MDR1, P‐Glycoprotein) on drug disposition and potential clinical implications: update of the literature. Clin Pharmacokinet 54, 709–735. [DOI] [PubMed] [Google Scholar]
- Zaanan A, Williet N, Hebbar M, Dabakuyo TS, Fartoux L, Mansourbakht T, Dubreuil O, Rosmorduc O, Cattan S, Bonnetain F et al (2013) Gemcitabine plus oxaliplatin in advanced hepatocellular carcinoma: a large multicenter AGEO study. J Hepatol 58, 81–88. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL and McMahon SB (2008) The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell‐cycle progression. Mol Cell 29, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Lin K, Zhang S, Chen Y, Zhang N, Xue J, Wang Z, Aldape KD, Xie K, Woodgett JR et al (2016) Nuclear GSK3beta promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat Cell Biol 18, 954–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang YJ, Liao ZW, Yu HW, Song XL, Liu Y, Shi XY, Lin XD and Zhou TC (2015) ShRNA‐mediated silencing of the ubiquitin‐specific protease 22 gene restrained cell progression and affected the Akt pathway in nasopharyngeal carcinoma. Cancer Biol Ther 16, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zijl F, Mall S, Machat G, Pirker C, Zeillinger R, Weinhaeusel A, Bilban M, Berger W and Mikulits W (2011) A human model of epithelial to mesenchymal transition to monitor drug efficacy in hepatocellular carcinoma progression. Mol Cancer Ther 10, 850–860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) Cancer drug resistance PCR array analysis in BEL/FU cells transfected with control shRNA or USP22 shRNA. A The gene list was presented as BEL/FU USP22 shRNA vs BEL/FU control shRNA. (B) Western blot assay was used to demonstrated that P‐gp (encoded by ABCB1) was not varied in BEL/FU cells with downregulation of USP22.
Fig. S2. MRP‐1 mRNA expression in BEL/FU cells with SIRT1 downregulation and AKT inactivation.


