Abstract
Purpose of review
The goal of this review was to compare and contrast the results and implications from several recent transcriptomic studies that analyzed the expression of lncRNAs in breast cancer. How many lncRNAs are dysregulated in breast cancer? Do dysregulated lncRNAs contribute to breast cancer etiology? Are lncRNAs viable biomarkers in breast cancer?
Recent findings
Transcriptomic profiling of breast cancer tissues, mostly from The Cancer Genome Atlas, identified thousands of long noncoding RNAs that are expressed and dysregulated in breast cancer. The expression of lncRNAs alone can divide patients into molecular subtypes. Subsequent functional studies demonstrated that several of these lncRNAs have important roles in breast cancer cell biology.
Summary
Thousands of lncRNAs are dysregulated in breast cancer that can be developed as biomarkers for prognostic or therapeutic purposes. The reviewed reports provide a roadmap to guide functional studies to discover lncRNAs with critical biological functions relating to breast cancer development and progression.
Keywords: long noncoding RNAs, breast cancer, biomarkers, The Cancer Genome Atlas, transcriptomics
I. Introduction
Breast cancer is the mostly commonly diagnosed cancer in women throughout the world. Nearly 1.7 million are diagnosed and more than 500,000 women die each year from the heterogeneous genetic disease [1]. There are two main histological subtypes—Invasive Ductal Carcinoma (IDC) and Invasive Lobular Carcinoma (ILC) and five main molecular subtypes—Luminal A, Luminal B, Basal-like, HER2-enriched, and normal-like [2–4]. The five molecular subtypes are defined by the PAM50 gene expression signature [5, 6] that largely correlates with the expression of pathological markers. Luminal A tumors typically express both the Estrogen (ER) and Progesterone Receptors (PR), Luminal B are generally ER+/PR-, HER2-enriched mostly contain amplified ERBB2/HER2 growth factor receptor, and basal-like largely do not express ER, PR, or HER2 (triple-negative or TNBC). Targeted therapies have dramatically improved the clinical outcomes of ER+ and HER2+ breast cancer patients, but resistance to these therapies and metastasis still remain deadly killers that are not fully understood. Targeted therapies do not exist for basal-like tumors, nor are there biomarkers to subdivide this group.
Long noncoding RNAs (lncRNAs) are RNAs longer than 200 nucleotides that have no apparent coding potential. This loose definition incorporates RNAs of varying genomic structures, expression patterns, and functions. LncRNAs perform a wide-variety of biological functions including regulation of transcription, translation, proliferation, apoptosis, etc. They perform these functions largely through their interactions—regulated by their structure—to DNA, RNA, and proteins by acting as guides, scaffolds, and decoys [7–9]. During the past three decades, the expressions, functions, and dysregulation of several lncRNAs were described for all phases of breast cancer initiation and progression. Comprehensive reviews detail the proposed roles of these lncRNAs [7, 10, 11]. It is not our intention to rehash those reviews. Instead, Table 1 and the following short section summarize very briefly and broadly the functions of selected lncRNAs in breast cancer biology that were identified through standard molecular biology and genetic approaches, before reviewing more carefully the insights from recent global analyses of lncRNAs in breast cancer. For comprehensive reviews on the functions of lncRNAs in breast cancer, please refer to [7, 10, 11].
Table 1.
Dysregulated lncRNAs in breast cancer
| LncRNA | Expression in Breast Cancer | Biology |
|---|---|---|
| H19 | Upregulated | Promotes anchorage-independent growth, expression correlates with tumor grade, may lead to drug resistance. [17–19, 104–110] |
| GAS5 | Downregulated | Induces apoptosis, low expression associates with poor prognosis. [50–53, 111, 112] |
| LSINCT5 | Upregulated | Enhances proliferation. [38] |
| HOTAIR | Upregulated | Enhances proliferation and metastasis, activates ligand-independent ER activation and possibly contributes to tamoxifen resistance. [45, 113–116] |
| XIST | Downregulated | Inhibits cell viability and regulates X chromosome inactivation. [13, 14, 22, 23] |
| NEAT1 | Upregulated | Enhances Proliferation and Induces Apoptosis[35–37] |
| MALAT1 | Upregulated | Promotes proliferation, invasion, migration, tumor progression, metastasis, and elevated expression associates with poor survival of ER-/lymph node negative patients and tamoxifen treated ER+ patients. [8, 39–44] |
| lncRNA-ARA | Upregulated in Adriamycin resistant cell lines | Induces proliferation, decreases apoptosis, expression inversely correlates with Adriamycin sensitivity[117] |
| BC200 | Upregulated | Prognostic indicator of tumor progression and enhances cell growth [18, 118] |
| SOX2OT | Upregulated | Inhibits proliferation, enhances anchorage-independent growth, and may be involved in in maintaining a stem cell phenotype [119] |
| TreRNA | Upregulated | Enhances invasion and metastasis. [120] |
| UCA1 | Upregulated | Enhances proliferation, inhibits apoptosis, and possibly involved in tamoxifen resistance. [121, 122] |
| NKILA | Downregulated | Low expression associated with breast cancer metastasis and poor patient prognosis. [123] |
| ZFAS1 | Downregulated | Inhibits proliferation and differentiation. [55, 56] |
| lncRNA-ATB | Upregulated in trastuzumab resistant tumors and cell lines | Promotes trastuzumab resistance, enhances invasion, metastasis, and EMT. [124] |
| BCAR4 | Upregulated | Promotes proliferation, tumorigenesis, metastasis, potentially involved in tamoxifen resistance, and expression associates with poor survival. [125–129] |
| lncRNA-HIT | Upregulated | Promotes migration, invasion, EMT, and metastasis. [130] |
| LINK-A | Upregulated in TNBC | Promotes tumorigenesis and glycolysis. [132] |
| JPX | Downregulated | Limits cell viability, activates XIST. [133] |
| PANDA | Upregulated | Prevents apoptosis. [134] |
| SRA | Upregulated | Enhances migration and tumorigenesis. [28–32] |
| LINC00511 | Upregulated | Enriched in TNBC. [33] |
| 91H | Upregulated | Promotes cell proliferation and tumorigenesis. [24–26] |
| LINC00520 | Upregulated | Enhances migration and invasion[46] |
| Linc-ROR | Upregulated | Enriched in TNBC. Promotes EMT, invasion, metastasis, and multidrug resistance. [47–49] |
II. lncRNAs in breast cancer – early evidence of dysregulation and function
Overview
In the early 1990s, the identification and functions of lncRNAs emerged through traditional molecular biology and genetic techniques. Two of the first described—H19 [12] and XIST [13, 14]—have roles in mammary gland development and breast cancer formation. H19 is a classically imprinted gene that is hormonally-regulated [15], expressed highly during pregnancy[16], overexpressed in breast cancer tumors, promotes anchorage-independent growth[17], significantly correlates with tumor grade [18], and may lead to drug resistance[19]. XIST induces X chromosome inactivation in female mammals by recruiting chromatin-modifying complexes to the targeted X chromosome resulting in histone modification and heterochromatin formation [15, 20, 21]. The expression of XIST is commonly reduced in breast cancer tumor tissues and cell lines with a corresponding increase in phosphorylated-AKT levels [22, 23], suggesting that XIST negatively regulates cell viability through inhibition of AKT. Over the past decade several other lncRNAs were shown to be dysregulated in breast cancer and have important biological functions. The next three subsections briefly discuss potential oncogenic and tumor suppressor lncRNAs involved in breast cancer formation or progression.
Upregulated lncRNAs in breast cancer promote proliferation and tumorigenesis
Several lncRNAs that are overexpressed in breast tumors regulate breast cancer biology (Table 1). The antisense compliment of H19—91H—is highly expressed in a subset of breast tumors. 91H promotes cell proliferation and tumorigenesis [24, 25] by preventing histone and DNA methylation on the maternal H19/IGF2 allele leading to increased expression of those two oncogenes [26]. The lncRNA SRA also regulates gene expression, but as a coactivator of ER and PR [27]. At least 470 genes are regulated by hormones in a SRA-dependent manner[28–30] and SRA is necessary for full ERα activity [31]. Overexpression of SRA in the mammary gland of mice leads to epithelial hyperplasia [32]. Interestingly, while it seems SRA functions in breast cancer as a coactivator to ER, it is necessary for full migration of a TNBC cell line, suggesting it also functions independently of ER. A different nuclear receptor, RORγ, regulates LINC00511, which is a highly expressed lncRNA in breast cancer. LINC00511 is enriched in TNBC cell lines and acts as an oncogene in lung cancer [33, 34]. It is unsurprising that lncRNAs involved in breast cancer biology interact with or are regulated by nuclear receptors and other breast cancer players. Another example is NEAT1, which is repressed by BRCA1. NEAT1 localizes to the paraspeckles in the nucleus, is upregulated by hypoxia, induces proliferation, inhibits apoptosis, and high expression correlates with poor survival in breast cancer tumors [35–37]. For other lncRNAs there is relatively little known, like LSINCT5, which is overexpressed in breast and ovarian cancer [38] and enhances proliferation through unknown mechanisms.
Upregulated lncRNAs in breast cancer promote migration, invasion, and tumor progression
MALAT1 is a conserved and alternatively spiced lncRNA [39] that regulates gene expression by altering transcription and post-transcriptional pre-mRNA processing of a large number of genes [40]. MALAT1 is upregulated in breast cancer and promotes invasion and migration of breast cancer cells [41]. The expression of MALAT1 is higher in ER+ and HER2+ tumors, but MALAT1 induces proliferation, tumor progression, and metastasis of TNBC cells, implying a role in all breast cancer subtypes. Further supporting this hypothesis, elevated expression of MALAT1 is associated with poor disease-specific survival of ER- and lymph node negative patients [40] and poor recurrence-free survival in tamoxifen treated ER+ patients [42]. Thus, MALAT1 might serve as an important biomarker or target of therapy for these groups. In fact, antisense oligonucleotides targeting MALAT1 subcutaneously injected in mice with established mammary tumors resulted in cystic and poorly metastasizing tumors implying targeting MALAT1 is a potential therapy to limit the spread of disease [43, 44]. HOTAIR is another lncRNA that enhances breast cancer metastasis. HOTAIR represses HOXD—a metastatic suppressor—by inducing H3K27me3 through the recruitment of PRC2 (25,26). Much like other breast cancer related lncRNAs, HOTAIR is repressed directly by ER. When HOTAIR is upregulated it promotes ligand-independent ER activation and contributes to tamoxifen resistance [45].
Other oncogenes also regulate lncRNAs in breast cancer cells. LINC00520 is controlled by SRC, PIK3CA, and STAT3 and enhances migration and invasion [46]. LINC00520 is highly elevated in breast cancer cells with highest expression in basal-like subtype. Likewise, Linc-ROR is a lncRNA upregulated in breast tumors that is enriched in TNBC. Linc-ROR promotes epithelial-mesenchymal transition (EMT), invasion, metastasis, and multidrug resistance in breast cancer in part by acting as a miRNA sponge [47, 48]. Additionally, estrogen metabolites downregulate NRF2 expression resulting in an upregulation of linc-ROR expression suggesting Linc-ROR may contribute to estrogen-mediated breast tumorigenesis as well as TNBC [49].
Downregulated lncRNAs in breast cancer regulate proliferation, apoptosis, and differentiation
In addition to XIST there are other tumor suppressor-like lncRNAs with downregulated expression in breast cancer (Table 1). Gas5 is a pro-apoptotic snoRNA host gene with reduced expression in breast cancer samples [50]. Gas5 induces apoptosis by acting as a mimic to the glucocorticoid receptor [51] and contains a stem-loop sequence that serves as hormone response element mimic in breast cancer [52]. Low expression of Gas5 is associated with poor prognosis in breast cancer likely due to reduced apoptosis stimulated by chemotherapeutic agents [53, 54]. Zfas1 is another snoRNA host gene that acts as a tumor suppressor. Zfas1 is highly expressed and regulated during mammary gland development of the mouse, and the human orthologue is down-regulated in breast tumors [55]. Reduction of Zfas1 expression enhanced proliferation and differentiation of the HC11 mammary epithelial cell line [55] potentially through interaction with ribosomes and by regulating RPS6 phosphorylation [56].
III. Global analyses reveal widespread dysregulation of lncRNAs in breast cancer tissues and cells
Overview
Early studies demonstrated that lncRNAs are critical to the biology of breast cancer cells and that other undiscovered lncRNAs likely play equally critical roles in breast cancer development and progression. The advent of global expression analyses, especially those applying next-generation sequencing, unleashed a plethora of information about the expression, dysregulation, and function of novel lncRNAs in breast cancer initiation and development. However, discrepancies in specific results exist between studies due to unstandardized analyses and annotations. RNAseq data can be analyzed to allow novel isoform/gene discovery or can simply quantify the expression of RNAs to predefined gene annotations. While there is overlap between annotations from different databases (RefSeq, GENCODE, BodyMap) there are numerous differences due to how they were generated and curation requirements. This section details the early evidence of pervasive transcription through recent studies utilizing novel and publically available datasets. We comment on the similarities and differences in the approaches and how they affect the discoveries of these studies. Table 2 summarizes key prognostic and functional lncRNAs identified through the global analyses.
Table 2.
Additional dysregulated lncRNAs in breast cancer identified through recent transcriptomic screens
| lncRNAs | Notes |
|---|---|
| LINC00657, LINC00346, LINC00654, HCG11 | High expression of all four predicted poorer overall survival. Knockdown of LINC00657 decreased proliferation. |
| LINC00705, LINC00310, LINC00704, LINC00574, FAM74A3, UMODL1-AS1, ARRDC1-AS1, HAR1A, LINC00323 | High expression of all nine associated with poor disease free survival. |
| HOTAIRM1, LINC00340 | Enriched in basal breast tumors. |
| HOTAIR | Enriched in HER2 subtype |
| TOPORS-AS1, RP11-35G9.3 | High expression is associated with good patient outcome |
| BCAL8 | Enhances proliferation, anchorage-independent growth, and tumor growth in nude mice. |
| CTD-2015G9.2, CTD2527121.15, LINC00393, LINC001198, RP11-10A14.5, RP11-19E11.1 | Enriched in basal-like breast cancer. |
| LINC01297 RP11-303E16.2 | In ER+ tumors, LINC01297 is highly expressed and RP11-303E16.2 is downregulated. Together their expression can divide patients into ER+ and ER-. |
| CYTOR | Correlates with poor survival and is critical for proliferation and migration |
| HIF1A-AS2, AK124454 | Part of an mRNA-lncRNA signature that predicts recurrence-free survival and pathologic complete remission after taxane-based neoadjuvant chemotherapy. Both lncRNAs promote proliferation, invasion, and paclitaxel resistance. |
| BRCAT49 | Breast cancer and lineage-associated lncRNA located near a SNP associated to breast cancer in several GWASs. |
| DCCAM-AS1 | Highly expressed in breast cancer tissues. Strongly induced by estrogen. Enhances proliferation, invasion, anchorage-independent growth of cell lines and xenograft growth and metastasis in mice. |
| lncRNA152, lncRNA67 | Estrogen regulated lncRNAs that are overexpressed in breast cancer tumors. Both are necessary and sufficient for proliferation of certain breast cancer cell lines. |
| LIMT | Prevents migration, invasion, and metastasis. Depleted in basal-like and HER2 positive tumors. Loss of LIMT is correlated with a decrease in survival. |
Evidence of pervasive transcription
Commonly used gene expression microarrays do not contain probes for most lncRNAs—an obvious problem for using previously published data for lncRNA discovery. However, the use of genome tiling arrays provided early hints of pervasive transcription of the human genome. Bertone et al. constructed a series of 134 high-density tiling arrays that represented 1.5 Gb of non-repetitive sequence of the human genome and discovered 10,595 transcripts from poly-A selected liver RNA not detected by other methods [57]; early evidence that novel transcripts were awaiting discovery.
Next generation sequencing of total RNA (RNAseq) does not rely on predesigned probes and thus allows for easier discovery of novel RNAs. With this technology several groups demonstrated that over 70% of the genome is transcribed even though only 1.5% of the transcripts encode for proteins [58–61]. But the questions remained if the novel transcripts were real and functional or if they were the product of a leaky transcriptional system or imperfect detection and computational technologies. To address this, Guttman et. al used a multi-omic approach to define a conservative cohort of 1,675 large intergenic noncoding RNAs (lincRNAs) in mouse cells that are 1) expressed, 2) multi-exonic, 3) in areas of regulatory histone marks (H3K4me3 and H3K36me), and 4) do not overlap protein coding or miRNA genes [62]. Regulatory histone marks and splicing of transcripts imply that these lincRNAs are functional and are not merely transcriptional noise; however, it does not preclude single-exon RNAs or those without active histone marks from being functional. The ENCODE Consortium used RNAseq data from 15 different cell lines without the conservative requirements of Guttman et al. They reported that 74.7% of the human genome is covered by primary transcripts, but that no individual cell line expressed more than 56.7% of the known transcripts. Work from ENCODE is continuously updated and the latest version of Human GENCODE (v.25) as of December 2016 (https://www.gencodegenes.org/stats/current.html) [58] has 15,767 lncRNA genes and another 14,650 pseudogenes. This means the number of long noncoding genes is nearly double that of protein-coding genes, thus ushering in the excitement of new candidates to understand, diagnose, and treat human disease including breast cancer.
Insights from The Cancer Genome Atlas
Concurrent to ENCODE, The Cancer Genome Atlas (TCGA) was sequencing numerous cancers with the goal of understanding the molecular changes leading to tumor formation. TCGA is a collaborative that published multilevel-omic data on thousands of tumors in over 33 different cancer types. For breast cancer alone, there are publically available RNAseq expression data from 1098 tumors. The processed expression data that are publically available were calculated using MapSplice followed by RSEM with a generic annotation file to hg19 that is similar to the UCSC Gene standard track (December 2009 version) [63, 64]. This pipeline does not allow for novel gene discovery and includes the expression of 20,532 total genes and 73,671 transcripts. This data was mined and reanalyzed by several groups to determine the expression and function of lncRNAs in breast cancer biology and their potential utility as biomarkers, molecular classifiers, or therapeutic targets.
Through the cBioPortal [65] Liu et al.[66] examined the 2730 annotated lncRNAs in the fully processed TCGA data. They found that 577 of these lncRNAs have altered RNA expression, copy number variations, or mutations in at least 1% of the breast cancer tumors. The overexpression of four lncRNAs—LINC00657, LINC00346, LINC00654, and HCG11— predicts a poorer overall survival and the combined overexpression of another 9 lncRNAs— LINC00705, LINC00310, LINC00704, LINC00574, FAM74A3, UMODL1-AS1, ARRDC1-AS1, HAR1A, LINC00323— is associated with poorer disease free survival, suggesting that lncRNAs are viable biomarkers in breast cancer. However, because TCGA data is relatively new and the five-year relative survival rate of women diagnosed with localized breast cancer is 98.5%[67] associations with disease outcomes based on TCGA data may still be premature. To examine the functional significance of a lncRNA in the new signature, LINC00657 was knocked out with CRISPR/Cas9 in MCF7 and MDA-MB-231 breast cancer cell lines. The knock-out cell lines proliferated slowly and formed smaller and fewer colonies in a clonogenic assay. This indicates that LINC00657 is an oncogene that is overexpressed in a subset of breast cancer tumors.
Another comprehensive analysis of lncRNAs in breast cancer using TCGA data was performed on 658 non-normal-like IDC samples with defined molecular subtypes by PAM50 scores [68]. Using aligned data from TCGA, ‘raw’ sequencing reads were obtained by back converting alignment to FASTQ files. The reads were then realigned with MOSAIK[69] and transcripts were quantified with HTSeq and DESeq [70] using GENCODEv.15 that contained 13,159 annotated lncRNAs. After filtering for lncRNAs with expression near or below levels of detection (FPKM greater than or equal to 1 in at least 10% of samples), 1,623 lncRNAs were examined further. GREAT analysis [71] demonstrated that the expressed lncRNAs were genomically positioned near protein coding genes (PCGs) with established roles in breast cancer such as ESR1, GATA3, and FOXA1. Additionally, 937 of these lncRNAs have histone marks associated with enhancer regions (H3K37ac and H3K4me2) in human mammary epithelial cell lines, suggesting the lncRNAs are cis-acting regulators of expression of genes that drive breast carcinogenesis. Unsupervised hierarchical clustering of breast cancer samples by the expression of lncRNAs highly correlated with PAM50 scores and defined molecular subtypes of breast cancer. Notably, there is a clear distinction between basal and ER+ subtypes with 122 lncRNAs, including HOTAIRM1 and LINC00340, overexpressed (FC>=2; FDR<0.05) in the basal subgroup. HOTAIR and 57 other lncRNAs were overexpressed in the HER2-enriched subtype and 96 lncRNAs were upregulated in the luminal subtypes. In contrast to the previous study only 6 lncRNAs were associated with patient outcome (FDR < 0.05) including two— TOPORS-AS1 and RP11-35G9.3—that are associated with good outcome [68].
Yan et al. used a different computational pipeline to perform a comprehensive characterization of lncRNAs of 5,037 tumors across 13 cancer types—including breast cancer— in TCGA. They used Partek to calculate expression levels from the downloaded alignment (.bam) files to GENCODE V.18 that contains 13,562 lncRNA genes and 14,181 pseudogenes. All lncRNAs were considered to be expressed if their 50th percentile RPKM was greater than 0 or if the 90th-percentile RPKM value was more than 0.1. Using this criteria they state that 4,329 lncRNAs are detectable in breast cancer—nearly 3 times more than the aforementioned studies using the same data—and that 1,150 are differentially expressed between the 990 breast cancer tumors vs. the 106 normal samples examined [72]. Their results agree with ENCODE data that lncRNAs are expressed or dysregulated in more cell type [60]—or in this case cancer type—specific manner than PCGs. They further found that dysregulated lncRNAs are located in somatic copy number variable regions, their promoters have altered DNA methylation patterns, and they contain cancer-associated SNPs. A cluster analysis of expressed lncRNAs from 817 breast tumors highly correlated with molecular subtypes based on PAM scores—much like the previous studies—and had a high correlation with clinical subtypes [72]. Consistent with the Su et al.[68] report the basal-like subtype formed the strongest cluster apart from all other subtypes. Using an entropy based metric they demonstrated that the expression of lncRNAs are more specific than PCGs for defining subtypes [72]. Because lncRNAs are expressed and dysregulated in a cancer and subtype specific manner and because they are highly structured and potentially more stable in blood and urine, it is possible that lncRNAs will be better biomarkers for diagnosis and prognosis than PCGs. The authors argued that cancer driver lncRNAs can be found by using their comprehensive characterization of lncRNAs with a functional screen. As a proof of concept they designed a siRNA screen of 19 lncRNAs that had cancer specific genomic or epigenomic alterations and that correlated with patient survival. The screen and subsequent assays revealed that reduction of BCAL8 reduces proliferation of breast cancer cell lines, anchorage-independent growth, and tumor growth when injected into nude mice. Using a Guilt-by-association (GBA) analysis [73] they showed that BCAL8 is co-expressed with cell cycle genes at the RNA and protein level [72]. Additional lncRNAs were not examined, but the data and pipeline are a straightforward approach to discovering other lncRNAs that regulate important biological phenotypes of breast cancer cells.
Beyond TCGA
Open data allows for increasing the power of studies through the combination of datasets. TCGA breast cancer data are a resource that have been integrated with other novel and publically-available datasets. Iyer et al. curated 7,256 RNAseq libraries (including 5,847 from TCGA) from tumors, normal tissues, and cell lines and used an ab initio assembly methodology [74–76]—that allows for novel gene discovery—to define the MiTranscriptome compendium including 58,648 expressed lncRNAs, 79% of which were previously unannotated [59]. In breast cancer alone they discovered 1,115 and 134 lineage and cancer specific transcripts respectively. Consistent with other studies, the newly discovered lncRNAs were expressed at lower levels than PCGs and overlapped disease-associated SNPs. BRCAT49 is a previously uncharacterized breast cancer and lineage specific lncRNA that is near a SNP associated to breast cancer in several GWASs [77–81]. This suggests that analyses allowing for novel lncRNA detection have more power to identify functional players in breast cancer biology than analysis techniques that simply align to previously annotated genes. Extending on this, Niknafs et al. identified 63 potentially estrogen-regulated, breast cancer lncRNAs by intersecting lncRNAs upregulated in tumor vs. normal and ER+ vs. ER-tumors [82]. This screen identified DCCAM-AS1 as an estrogen regulated lncRNA that is overexpressed in breast cancer. Silenced expression of DCCAM-AS1 decreased proliferation, invasion, anchorage-independent growth of tested breast cancer cell lines and reduced xenograft growth and metastasis when injected into mice. Overexpression of DCCAM-AS1 promoted growth in the absence of estrogen indicating a possible role in tumor progression and tamoxifen resistance [82]. DDCAM-AS1 was found by another group to be the most abundant lncRNA of 133 that are regulated by ligand-independent ERα activity, further suggesting a role in tamoxifen resistance [83].
Bradford et al. combined TCGA data with RNA-Seq data from 69 breast cancer patients that they generated at Huntsman Cancer Hospital [84]. For consistency, they reanalyzed the TCGA data through the same pipeline they applied for their new samples—StarAlign followed by Cufflinks [74–76] using the GRCh38 genome build with GENCODEv22 gene annotation. They only analyzed a set of 6062 lncRNAs that did not overlap other genes. This is critical as TCGA RNAseq data is not stranded, meaning individual reads can align perfectly to lncRNAs and PCGs when they are orientated in an antisense fashion (Figure 1a). User decisions and algorithm design decide how to treat ambiguous reads. They can be discarded, assigned completely to one gene, or divided in a variety of ways. This choice is often not listed in the methods, but determines the final expression of both antisense genes. If reads are divided, it is possible that RNAseq data will incorrectly state that both antisense genes are expressed.
Figure 1. Difficulties associated with studying lncRNAs.
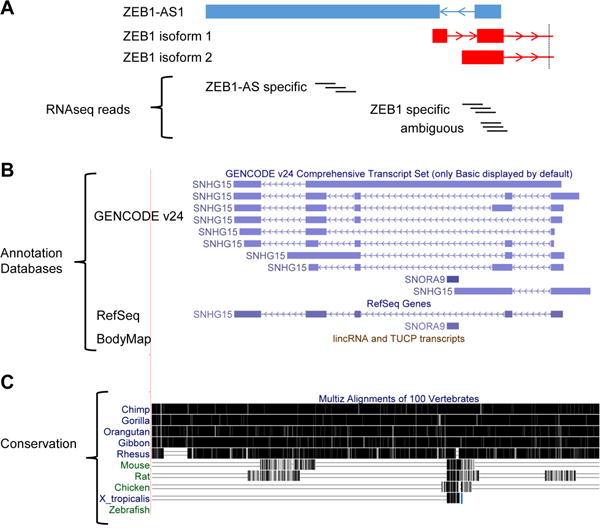
(A) Unstranded RNAseq reads can be ambiguous when mapping to antisense lncRNAs. (Top) Annotations of lncRNA ZEB1-AS1 (blue) and two ZEB1 PCG isoforms (red; cropped for simplicity). Arrows indicate the direction of transcription. (Bottom) RNAseq reads are accurately assigned when they align to specific exons of the antisense pair, but are ambiguous when they map to the shared exons. (B) Annotations between databases vary. Annotation of SNHG15 from three commonly used databases— GENCODE, RefSeq, and BodyMap—are shown. BodyMap does not include an annotation for SNHG15. (C) LncRNAs are not highly conserved. The conservation of SNHG15 is shown with black bars representing sequence conservation between different species. Note that SNHG15 is only conserved among primates.
Like others, Bradford et al. demonstrated that highly and variably expressed lncRNAs were sufficient to separate breast cancer tumors into their molecular subtypes. Further, they defined a list of six lncRNAs that are significantly overexpressed in basal-like compared to other breast cancer subtypes—CTD-2015G9.2, CTD2527121.15, LINC00393, LINC001198, RP11-10A14.5, RP11-19E11.1[84]. Note the lack of overlap of enriched lncRNAs between studies. This is likely due to the different annotations and methods of analysis used by the independent groups. Interestingly, using a GBA method and integration with cell line expression data in the Cancer Cell Line Encyclopedia [85] they discovered that a high proportion of a specific cell type in the tumor microenvironment caused some tumors to not cluster well into their corresponding subtypes [84]. This suggests that the expression of lncRNAs can potentially be used to study tumor heterogeneity and define cell-type composition. Accordingly, they defined certain lncRNAs as putatively associated with breast cancer stromal and immune cells.
TCGA data has also been used as a validation set. Grembergen et al. [86] repurposed Affymetrix Human Genome U133 Plus 2.0 array data from 823 breast tumors and 172 normal samples across a collection of 7 data sets downloaded from Gene Expression Omnibus (GEO) [87] by BLASTing probes against the LNCipedia database v.2.1 [88]. Unfortunately, repurposing probes potentially introduces the same bias as using unstranded RNAseq for calling expression of antisense lncRNAs. Nevertheless, 3053 annotated lncRNAs had corresponding probes on the array. Of those, 215 were aberrantly expressed in at least 10% of the tumors. The expression of this set of lncRNAs clearly distinguished breast tumors from normal samples and the signature was validated with TCGA expression data. In agreement with all other studies, the expression of lncRNAs separated ER+ (high expression of LINC01297 and low expression of RP11-303E16.2) and ER-tumors and correlated well with molecular subtypes defined by PAM50. With functional assays, they demonstrated that high expression of CYTOR—one of the dysregulated lncRNAs—correlated with poor survival and is critical for proliferation and migration [86].
Two other studies used repurposed microarray data to define lncRNA prognostic signatures. Jiang et al. performed a prospective observational study that demonstrated an integrated mRNA-lncRNA signature had better prognostic value to predict recurrence-free survival and pathologic complete remission after taxane-based neoadjuvant chemotherapy than clinicopathologic parameters in a cohort of 275 TNBC patients [89]. However, this signature was limited by the number of probes covering lncRNA loci (195 lncRNAs were differentially expressed between normal and tumor) on the Affymetrix Human Transcriptome Array 2.0 implying that this signature can be improved if developed with RNAseq data. Functionally, they confirmed that the two lncRNAs in the signature—HIF1A-AS2 and AK124454—promote proliferation, invasion, and paclitaxel resistance [89].
The other was a retrospective study of 164 banked frozen tumors that used repurposed Agilent microarray data (2811 lncRNAs covered) and showed that a lncRNA expression profile could predict metastasis independently of traditional prognostic markers [90]. It is unclear if the lncRNA expression signature is better than a PCG or lncRNA-PCG hybrid classifier, but it is further evidence that lncRNAs are viable biomarkers to define molecular subtypes of breast cancer and to predict recurrence.
Novel lncRNAs defined by regulation by breast cancer driver genes
Other novel breast cancer associated lncRNAs have been discovered through analysis of cell lines treated with estrogen [91] and EGF [92]—a hormone and ligand respectively that drive subsets of breast tumors. One study identified 1,888 lncRNAs in MCF7 cells, 700 of which were not previously annotated in current databases (GENCODE, UCSC, RefSeq, BodyMap, MiTranscriptome). They combined RNAseq data following 3 hours of estrogen treatment with global run-on sequencing (GRO-seq) [93] data at several time points after treatment [91]. This “lncM” set only includes lncRNAs with FPKM > 1 that have evidence of a primary transcript, as determined by GRO-seq. Therefore, this is a conservative estimate of the number of lncRNAs expressed in MCF7 cells. Certainly expanding the time points following estrogen treatment or repeating in other breast cancer cell lines will aid in the discovery of additional breast cancer lncRNAs. On average the lncM lncRNAs were more nuclear and less stable than other lncRNAs or PCGs, which is likely how GRO-seq improved detection. Estrogen regulated the expression of 531 lncM genes indicating they may be important for breast cancer cell biology at least in ER+ tumors. Like all previous studies, the lncM genes were able to cluster normal from cancerous samples and were as effective as PCGs in determining the molecular subtypes. They characterized two lncRNAs—lncRNA152 and lncRNA67—that are regulated by estrogen, have elevated expression in breast tumors, and have expression that is correlated to PCGs associated with cell-cycle regulation. They demonstrated that both of these lncRNAs are necessary and sufficient for proliferation of certain breast cancer cell lines and knockdown of either altered the expression of many cell cycle genes [91]. Interestingly, estrogen signaling downregulates lncRNA152 even though both estrogen and lncRNA152 drive proliferation, suggesting a negative feedback loop that was not explored further.
The EGF study analyzed expression of REFseq annotated lncRNAs with probes on the Agilent SurePrint G3 Human microarray following EGF treatment of the immortalized but non-transformed MCF10a breast epithelial cell line [92]. They reported 346 lncRNAs were significantly (greater than 1.5 FC) altered at a minimum of one time point. Eleven of these lncRNAs correlated with shorter overall and relapse-free patient survival in the METABRIC dataset, suggesting they are involved in breast cancer etiology or are preferentially expressed in HER2+ or basal-like subtypes—as these subtypes have poorer prognosis. Loss of function and gain of function analyses demonstrated that one of the clinically correlated lncRNAs—LIMT (LINC01089)—prevents migration, invasion, and metastasis. LIMT is depleted in basal-like and HER2+ tumors and loss of expression correlates with decreased survival [94]. Finally, another study reported 1,382 lncRNAs were differentially expressed (FC> 2.0; FDR<0.05) in 7 pairs of HER2+ tumors versus adjacent normal tissue [95]. It is unclear what annotation was used, but it would be interesting to determine if these lncRNAs are enriched in HER2+ tumors from TCGA or are regulated by EGF. Likewise, examining lncRNA expression following activation of other breast cancer driver genes or mutations such as PIK3CA or IGF1R will certainly identify more breast cancer associated lncRNAs.
IV. Conclusions
The vast majority of the genome is transcribed even though less than 2% of it encodes for proteins. In fact, there are as many lncRNAs as there are PCGs, but the functions of most are unknown. Over 80% of GWAS SNPs associated with disease are in noncoding regions [96] and early studies revealed that lncRNAs like H19, XIST, HOTAIR, and MALAT1 (Table 1) are critical regulators of breast cancer cell biology. Recent transcriptomic analyses followed by targeted functional validations demonstrate that a large number of novel lncRNAs are dysregulated in breast cancer tissues and play important roles in breast cancer cell biology. Clearly the choice of computational pipeline and annotation database used caused discordant results, but all studies agree that the expression of lncRNAs can be used as biomarkers to define breast cancer subtypes and potentially predict recurrence and survival. lncRNAs are exciting potential biomarkers in breast cancer because 1) they are more cell-type and cancer-type specific than PCGs, 2) the functional unit is easily and directly quantifiable, 3) they are highly structured and stable allowing for potential detection in the blood and urine, 4) they are dysregulated in breast cancer subtypes, and 5) the lncRNA PCAS1 is FDA approved as a biomarker for prostate cancer–thus precedent is set [97].
However, there are many obstacles to studying and utilizing lncRNAs as biomarkers or targets of therapy. 1) Most lncRNAs were recently identified in large transcriptomic studies, but have not been functionally characterized. Large siRNA and CRISPR screens [98] are beginning to demonstrate that many lncRNAs have critical biological roles, but more direct functional studies and mouse models like those of Sauvageau et al. [99] are necessary to prescribe functions to the vast majority of lncRNAs. 2) The annotations between the large databases are inconsistent (Figure 1b) largely because most are annotated based on short RNAseq reads and not on full length transcripts. New sequencing technologies such as single molecule, real-time sequencing [100](Pacific Biosciences) that use long reads have the potential to improve annotations if they become cheap enough to be widely used. 3) Understanding mutations and SNPs in lncRNAs is more difficult than PCGs because we cannot infer function based on reading frames. However, new techniques such as PARIS [101] and SHAPE [102] are defining secondary structures of lncRNAs at a global level. It is possible that once common secondary structures are defined and prescribed functions then we will be able to stratify the significance of a mutation based on disruption of these structures. 4) Because they were recently identified and were not on microarray chips, there are few publically available datasets to mine. Even TCGA data that measured gene expression with RNAseq was not stranded, meaning most antisense lncRNAs cannot be accurately measured because single reads map equally well to the gene on either strand (Figure 1a). 5) Many lncRNAs are unique to primates [103] making functional genetic studies in lower organisms nearly impossible (Figure 1c).
Despite the challenges, new technologies have exponentially increased our understanding of lncRNA expression, dysregulation, and function in breast cancer initiation and progression. With more sequencing under different experimental and developmental conditions new lncRNAs will be discovered and with them excitement will grow for the use of lncRNAs in precision medicine to diagnose and treat breast cancer patients.
Acknowledgments
We acknowledge Sreeroopa Som for her comments during the planning of the manuscript. We also thank our funding sources. DB was supported in part by a Susan G. Komen Foundation Fellowship. AW was supported in part through the Computer Science, Biology, and Biomedical Informatics and the University of Pittsburgh Cancer Institute Academies, which are supported through grants from the Doris Duke Foundation-Clinical Research Experiences for High School Students at the University of Pittsburgh (grant #2014154), the National Cancer Institute CURE Program (3P30CA047904-27S2), and support from the Jack Kent Cook Foundation and donations from UPMC and grateful parents and patients. Additionally, we would like to thank the team from the Department of Biomedical Informatics National Library of Medicine Training Program Grant in Biomedical Informatics (T15 LM007059), the University of Pittsburgh Cancer Institute (UPCI) Cancer Center Support Grant for the Cancer Bioinformatics Service (P30 CA47904), the Clinical and Translational Science Institute Biomedical Informatics Core (UL1 RR024153).
Abbreviations
- lncRNA
long noncoding RNA
- PCG
Protein coding gene
- GWAS
Genomewide Association Studies
- RNAseq
RNA sequencing
- TCGA
The Cancer Genome Atlas
- ER
Estrogen Receptor
- PR
Progesterone Receptor
- TNBC
Triple-negative Breast Cancer
- SNP
Single Nucleotide Polymorphism
- EMT
epithelial-mesenchymal transition
Footnotes
Conflict of Interest
Andrew Warburton and David Boone declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors
References
- 1.American Cancer Society. Cancer Facts and Figures 2016 2016 [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Bernard PS, Parker JS, Mullins M, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen TO, Parker JS, Leung S, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs Kevin. Mol Cell. 2012;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansji H, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet. 2014;5:1–15. doi: 10.3389/fgene.2014.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerk S, Schwarzenbacher D, Adiprasito J, Stotz M, Hutterer G, Gerger A, Ling H, Calin G, Pichler M. Current Status of Long Non-Coding RNAs in Human Breast Cancer. Int J Mol Sci. 2016;17:1485. doi: 10.3390/ijms17091485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 14.Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 15.Shore AN, Herschkowitz JI, Rosen JM. Noncoding RNAs involved in mammary gland development and tumorigenesis: There’s a long way to go. J Mammary Gland Biol Neoplasia. 2012;17:43–58. doi: 10.1007/s10911-012-9247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adriaenssens E, Lottin S, Dugimont T, Fauquette W, Coll J, Dupouy JP, Boilly B, Curgy JJ. Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene. 1999;18:4460–4473. doi: 10.1038/sj.onc.1202819. [DOI] [PubMed] [Google Scholar]
- 17.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 18.Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J, Weedon J, Diallo R, Böcker W, Tiedge H. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25:2125–2133. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 19.Doyle LA, Yang W, Rishi AK, Gao Y, Ross DD. H19 gene overexpression in atypical multidrug-resistant cells associated with expression of a 95-kilodalton membrane glycoprotein. Cancer Res. 1996;56:2904–2907. [PubMed] [Google Scholar]
- 20.Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12:815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 21.Brockdorff N. Chromosome silencing mechanisms in X-chromosome inactivation: unknown unknowns. Development. 2011;138:5057–5065. doi: 10.1242/dev.065276. [DOI] [PubMed] [Google Scholar]
- 22.Huang YS, Chang CC, Lee SS, Jou YS, Shih HM. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget. 2016 doi: 10.18632/oncotarget.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galupa R, Heard E. X-chromosome inactivation: New insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Berteaux N, Aptel N, Cathala G, et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–6745. doi: 10.1128/MCB.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X, Adriaenssens E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6:29209–23. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vennin C, Spruyt N, Robin Y-M, Chassat T, Le Bourhis X, Adriaenssens E. The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/IGF2 expression through epigenetic modifications. Cancer Lett. 2016 doi: 10.1016/j.canlet.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 27.Lanz RB, McKenna NJ, Onate SA, et al. A Steroid Receptor Coactivator, SRA, Functions as an RNA and Is Present in an SRC-1 Complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Wu HT, Zhu N, Shi YN, Liu Z, Ao BX, Liao DF, Zheng XL, Qin L. Steroid receptor RNA activator: Biologic function and role in disease. Clin Chim Acta. 2016;459:137–146. doi: 10.1016/j.cca.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Foulds CE, Tsimelzon A, Long W, Le A, Tsai SY, Tsai M-J, O’Malley BW. Research resource: expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Mol Endocrinol. 2010;24:1090–105. doi: 10.1210/me.2009-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vicent GP, Nacht AS, Zaurin R, Font-Mateu J, Soronellas D, Le Dily F, Reyes D, Beato M. Unliganded progesterone receptormediated targeting of an RNA-containing repressive complex silences a subset of hormone-inducible genes. Genes Dev. 2013;27:1179–1197. doi: 10.1101/gad.215293.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavarretta ITR, Mukopadhyay R, Lonard DM, Cowsert LM, Bennett CF, O’Malley BW, Smith CL. Reduction of coactivator expression by antisense oligodeoxynucleotides inhibits ERalpha transcriptional activity and MCF-7 proliferation. Mol Endocrinol. 2002;16:253–70. doi: 10.1210/mend.16.2.0770. [DOI] [PubMed] [Google Scholar]
- 32.Lanz RB, Chua SS, Barron N, Söder BM, DeMayo F, O’Malley BW. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol Cell Biol. 2003;23:7163–76. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh TG, Wang SCM, Acharya BR, Goode JM, Graham JD, Clarke CL, Yap AS, Muscat GEO. The Nuclear Receptor, ROR??, Regulates Pathways Necessary for Breast Cancer Metastasis. EBioMedicine. 2016;6:59–72. doi: 10.1016/j.ebiom.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng-Cao Sun, Shu-Jun Li, Guang Li, Rui-Xi Hua, X-HZ, D-JL Long Intergenic Noncoding RNA 00511 Acts as an Oncogene in Non–small-cell Lung Cancer by Binding to EZH2 and Suppressing p57. Mol Ther Acids. 2016;5:e385. doi: 10.1038/mtna.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choudhry H, Albukhari A, Morotti M, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunwoo H, Dinger ME, Wilusz JE. MEN ε / β nuclear retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles MEN ε / β Nuclear Retained Non-Coding RNAs are Up-regulated Upon. Genome Res. 2009;3:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–61. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8:496–505. doi: 10.4161/rna.8.3.14800. [DOI] [PubMed] [Google Scholar]
- 39.Meseure D, Vacher S, Lallemand F, et al. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br J Cancer. 2016:1–10. doi: 10.1038/bjc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jadaliha M, Zong X, Malakar P, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, Zhang Y, Xing Y, Xi T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding MIR-1 with cdc42. Biochem Biophys Res Commun. 2016;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 42.Huang N-S, Chi Y, Xue J, Liu M, Huang S, Mo M, Zhou S, Wu J. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. 2016;7:37957–37965. doi: 10.18632/oncotarget.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendell JT. Targeting a Long Noncoding RNA in Breast Cancer. 2016 doi: 10.1056/NEJMcibr1603785. http://dx.doi.org/10.1056/NEJMcibr1603785. [DOI] [PubMed]
- 45.Xue X, Yang YA, Zhang A, Fong K-W, Kim J, Song B, Li S, Zhao JC, Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2015;35:2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henry WS, Hendrickson DG, Beca F, et al. LINC00520 is induced by Src, STAT3, and PI3K and plays a functional role in breast cancer. 2016;1 doi: 10.18632/oncotarget.11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330–8. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Xia J, Li Q, Yao Y, Eades G, Gernapudi R, Duru N, Kensler TW, Zhou Q. NRF2/Long noncoding RNA ROR signaling regulates mammary stem cell expansion and protects against estrogen genotoxicity. J Biol Chem. 2014;289:31310–31318. doi: 10.1074/jbc.M114.604868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mourtada-Maarabouni M, Pickard M, Hedge V, Farzaneh F, Williams G. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28373:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 51.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pickard MR, Williams GT. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget. 2016;7:10104–10116. doi: 10.18632/oncotarget.7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: Implications for chemotherapy. Breast Cancer Res Treat. 2014;145:359–370. doi: 10.1007/s10549-014-2974-y. [DOI] [PubMed] [Google Scholar]
- 54.Mourtada-Maarabouni M, Pickard M, Hedge V, Farzaneh F, Williams G. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28373:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 55.Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–91. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansji H, Leung EY, Baguley BC, Finlay GJ, Cameron-Smith D, Figueiredo VC, Askarian-Amiri ME. ZFAS1: a long noncoding RNA associated with ribosomes in breast cancer cells. Biol Direct. 2016;11:62. doi: 10.1186/s13062-016-0165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–6. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 58.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. • This was a key study that used ab initio assembly to charachterize 58,648 lncRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koboldt DC, Fulton RS, McLellan MD, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu H, Li J, Koirala P, Ding X, Chen B, Wang Y, Wang Z, Wang C, Xu Z, Mo Y-Y. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.American Cancer Society. Cancer Facts & Figures 2015. Atlanta Am Cancer Soc 2015 [Google Scholar]
- 68.Su X, Malouf GG, Chen Y, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5:9864–76. doi: 10.18632/oncotarget.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hillier LW, Marth GT, Quinlan AR, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 70.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan X, Hu Z, Feng Y, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. • This study provided a comprehensive portrait of lncRNAs across cancers. They describe lncRNAs altered in cancers at transcriptional, genomic, and epigenetic levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steijger T, Abril JF, Engström PG, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10:1177–84. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 78.Li J, Humphreys K, Heikkinen T, et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat. 2011;126:717–727. doi: 10.1007/s10549-010-1172-9. [DOI] [PubMed] [Google Scholar]
- 79.Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michailidou K, Hall P, Gonzalez-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, Prensner JR, Poliakov A, Singhal U, Xiao L, Kregel S, Siebenaler RF, Zhao SG, Uhl M, Gawronski A, Hayes DF, Pierce LJ, Cao X, Collins C, Backofen R, Sahi FF. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7 doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miano V, Ferrero G, Reineri S, Caizzi L, Annaratone L, Ricci L, Cutrupi S, Castellano I, Cordero F, De Bortoli M. Luminal long non-coding RNAs regulated by estrogen receptor alpha in a ligand-independent manner show functional roles in breast cancer. Oncotarget. 2016;7:3201–16. doi: 10.18632/oncotarget.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bradford JR, Cox A, Bernard PCN. Consensus Analysis of Whole Transcriptome Profiles from Two Breast Cancer Patient Cohorts Reveals Long Non-Coding RNAs Assocaited with Intrinsic Subtype and the Tumour Microenvironment. PLoS One. 2016;11:e0163238. doi: 10.1371/journal.pone.0163238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Grembergen O1, Bizet M2, de Bony EJ1, Calonne E1, Putmans P1, Brohée S3, Olsen C4, Guo M5, Bontempi G6, Sotiriou C3, Defrance M7FF. Portraying breast cancers with long noncoding RNAs. Sci Adv. 2016;2:e1600220. doi: 10.1126/sciadv.1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P. LNCipedia: A database for annotated human IncRNA transcript sequences and structures. Nucleic Acids Res. 2013 doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang YZ, Liu YR, Xu XE, Jin X, Hu X, Da Yu K, Shao ZM. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Cancer Res. 2016;76:2105–2114. doi: 10.1158/0008-5472.CAN-15-3284. [DOI] [PubMed] [Google Scholar]
- 90.Sørensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M, Larsen MJ, Kruse TA. Long non-coding RNA expression profiles predict metastasis in lymph node-negative breast cancer independently of traditional prognostic markers. Breast Cancer Res. 2015;17:55. doi: 10.1186/s13058-015-0557-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, Annotation, and Functional Analysis of Long Noncoding RNAs Controlling Cell-Cycle Gene Expression and Proliferation in Breast Cancer Cells. Mol Cell. 2015;59:698–711. doi: 10.1016/j.molcel.2015.06.023. • This study integrated steady-state RNAseq with GROSeq to define novel lncRNAs in MCF7 cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sas‐Chen A, Aure MR, Leibovich L, et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol Med. 2016;39:e201606198. doi: 10.15252/emmm.201606198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sas-chen A, Aure MR, Leibovich L, et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. 2016:1–13. doi: 10.15252/emmm.201606198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang F, Lyu S, Dong S, Liu Y, Zhang X, Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther. 2016;9:761–772. doi: 10.2147/OTT.S97664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Van Poppel H, Haese A, Graefen M, De La Taille A, Irani J, De Reijke T, Remzi M, Marberger M. The relationship between Prostate CAncer gene 3 (PCA3) and prostate cancer significance. BJU Int. 2012;109:360–366. doi: 10.1111/j.1464-410X.2011.10377.x. [DOI] [PubMed] [Google Scholar]
- 98.Zhu S, Li W, Liu J, et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nat Biotechnol. 2016;34:1279–1286. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sauvageau M, Goff LA, Lodato S, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013 doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–5. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu Z, Zhang QC, Lee B, et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016;165:1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. RNA SHAPE analysis in living cells. Nat Chem Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Juan V, Crain C, Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28:1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. BioEssays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 106.Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280:29625–29636. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 107.Sun H, Wang G, Peng Y, Zeng Y, Zhu QN, Li TL, Cai JQ, Zhou HH, Zhu YS. H19 lncRNA mediates 17$β$-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol Rep. 2015;33:3045–3052. doi: 10.3892/or.2015.3899. [DOI] [PubMed] [Google Scholar]
- 108.Berteaux N, Aptel N, Cathala G, et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–6745. doi: 10.1128/MCB.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, Dugimont T, Curgy JJ. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002;23:1885–1895. doi: 10.1093/carcin/23.11.1885. [DOI] [PubMed] [Google Scholar]
- 110.Dugimont T, Montpellier C, Adriaenssens E, Lottin S, Dumont L, Iotsova V, Lagrou C, Stéhelin D, Coll J, Curgy JJ. The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene. 1998;16:2395–401. doi: 10.1038/sj.onc.1201742. [DOI] [PubMed] [Google Scholar]
- 111.Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: Implications for chemotherapy. Breast Cancer Res Treat. 2014;145:359–370. doi: 10.1007/s10549-014-2974-y. [DOI] [PubMed] [Google Scholar]
- 112.Kino M, Hur DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA Gas5 Is a Growth Arrest and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci Signal. 2010 doi: 10.1126/scisignal.2000568.Noncoding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gupta R a, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:1–10. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang X, Luo E, Liu X, Han B, Yu X, Peng X. Delphinidin-3-glucoside suppresses breast carcinogenesis by inactivating the Akt/HOTAIR signaling pathway. doi: 10.1186/s12885-016-2465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiang M, Huang O, Xie Z, Wu S, Zhang X, Shen A, Hu S, Geng M, Shen K. A novel long non-coding RNA-ARA : Adriamycin Resistance Associated. Biochem Pharmacol. 2014;87:254–283. doi: 10.1016/j.bcp.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 118.Singh R, Gupta SC, Peng W-X, Zhou N, Pochampally R, Atfi A, Watabe K, Lu Z, Mo Y-Y. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7:e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Askarian-amiri ME, Seyfoddin V, Smart CE, Wang J, Kim JE, Hansji H. Emerging Role of Long Non-Coding RNA SOX2OT in SOX2 Regulation in Breast Cancer. 2014;9:1–10. doi: 10.1371/journal.pone.0102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gumireddy K, Li A, Yan J, et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. 2013;32:2672–2684. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu C, Yang M, Ren Y, Wu C, Wang L. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. 2016:4362–4368. [PubMed] [Google Scholar]
- 122.Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo Y-Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu B, Sun L, Liu Q, et al. A Cytoplasmic NF-??B Interacting Long Noncoding RNA Blocks I??B Phosphorylation and Suppresses Breast Cancer Metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 124.Shi S-J, Wang L-J, Yu B, Li Y-H, Jin Y, Bai X-Z. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6 doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xing Z, Park PK, Lin C, Yang L. LncRNA BCAR4 wires up signaling transduction in breast cancer. RNA Biol. 2015;12:681–689. doi: 10.1080/15476286.2015.1053687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Godinho M, Meijer D, Setyono-Han B, Dorssers LCJ, Van Agthoven T. Characterization of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. J Cell Physiol. 2011;226:1741–1749. doi: 10.1002/jcp.22503. [DOI] [PubMed] [Google Scholar]
- 127.Chen F, Mo J, Zhang L. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumor Biol. doi: 10.1007/s13277-016-5256-y. [DOI] [PubMed] [Google Scholar]
- 128.Godinho MFE, Sieuwerts AM, Look MP, Meijer D, Foekens JA, Dorssers LCJ, van Agthoven T. Relevance of BCAR4 in tamoxifen resistance and tumour aggressiveness of human breast cancer. Br J Cancer. 2010;103:1284–91. doi: 10.1038/sj.bjc.6605884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lin GG, Scott JG. NIH Public Access. 2012;100:130–134. [Google Scholar]
- 130.Richards EJ, Zhang G, Li ZP, et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) ??: LncRNA-hit-mediated TGF??-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290:6857–6867. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Van Grembergen O, Bizet M, De Bony EJ, et al. Portraying breast cancers with long noncoding RNAs. 2016:1–16. doi: 10.1126/sciadv.1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:1–12. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X-chromosome inactivation. Cell. 2011;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]


