Abstract
Fecal wastes from a variety of farmed livestock were inoculated with livestock isolates of Escherichia coli O157, Listeria monocytogenes, Salmonella, Campylobacter jejuni, and Cryptosporidium parvum oocysts at levels representative of the levels found in naturally contaminated wastes. The wastes were subsequently spread onto a grass pasture, and the decline of each of the zoonotic agents was monitored over time. There were no significant differences among the decimal reduction times for the bacterial pathogens. The mean bacterial decimal reduction time was 1.94 days. A range of times between 8 and 31 days for a 1-log reduction in C. parvum levels was obtained, demonstrating that the protozoans were significantly more hardy than the bacteria. Oocyst recovery was more efficient from wastes with lower dry matter contents. The levels of most of the zoonotic agents had declined to below detectable levels by 64 days. However, for some waste types, 128 days was required for the complete decline of L. monocytogenes levels. We were unable to find significant differences between the rates of pathogen decline in liquid (slurry) and solid (farmyard manure) wastes, although concerns have been raised that increased slurry generation as a consequence of more intensive farming practices could lead to increased survival of zoonotic agents in the environment.
The transfer of pathogens to food through the application of animal manures to agricultural land is well described (6, 17, 25). In addition, pathogen recycling through livestock populations can result from the use of contaminated feces as fertilizer for crops that are subsequently used as animal feed (4). Livestock farming in Europe has become more intensive since the end of the Second World War (31). Productivity increases have been achieved by raising animal stocking densities, which in turn have required that fundamental changes be made in livestock waste management (31). In particular, there has been a shift away from the generation of solid manures to slurry-based production systems over the last 3 decades (14, 31). Solid manures contain bedding materials, such as straw and sawdust, which helps keep the wastes aerated, whereas liquid slurry wastes, which lack bedding, are more anaerobic (31). Thus, although spreading onto land is a traditional method for the disposal of livestock wastes, the nature of the manures being spread has changed relatively recently.
The implications of the change from solid to liquid wastes on the survival of indigenous zoonotic agents are unclear and require further study (14, 31) because, during the warmer months, a large proportion of cattle in temperate climates is turned out to graze pasture. In the United States and Europe grassland is likely to have been fertilized with waste generated from a range of potentially different livestock (30). To date, there has not been a comprehensive assessment of the decline of a range of pathogens in both liquid and solid livestock-derived wastes. In this study we began to address this shortfall in our knowledge by examining the decline of pathogens which had been inoculated into solid and liquid wastes generated on commercial farms. The pathogens studied were animal isolates of Escherichia coli O157, Listeria monocytogenes, Campylobacter jejuni, various Salmonella serotypes, and Cryptosporidium parvum. Contaminated manures were subsequently spread onto fescue pasture planted over a clay soil, and pathogen decline was monitored. These zoonotic agents were studied because they are commonly encountered in manures (10) and collectively cause the majority of food-borne illness in the United Kingdom (1, 2, 14, 25).
MATERIALS AND METHODS
Microorganisms and culture conditions.
The zoonotic agents used for these studies were all recent livestock waste isolates. Salmonella enterica serotype Typhimurium, C. jejuni, L. monocytogenes, and a non-verotoxin-producing E. coli O157 strain have been described previously (15). The bacteria were cultured in broth media supplemented with 3% (wt/vol) ammonium chloride and 1% sodium chloride at 37°C (or 25°C for L. monocytogenes) (8). The headspace in Campylobacter culture vessels was filled with a custom-formulated mixture containing 8% (vol/vol) carbon dioxide, 7% (vol/vol) oxygen, and 85% (vol/vol) nitrogen (British Oxygen Company, Guilford, United Kingdom).
Viable C. parvum oocysts were purchased initially from the Moredun Institute (Edinburgh, United Kingdom) and had been propagated in an ovine host. Oocysts were further propagated in 14-day-old dairy calves. Infection was by feeding milk for three consecutive days, and the total dose was 5 × 106 oocysts per day. Infected feces were collected and analyzed as described below to determine the initial oocyst levels and viability.
Wastes and inoculation of bacterial pathogens.
Solid farmyard manure (FYM) and/or liquid wastes (slurries or dirty water) from dairy cattle, laying chickens, poultry broilers, or breeder pigs were investigated as part of this study (Table 1). All wastes were fresh (<72 h postdeposition) and were collected from commercial farms. Cultures of bacterial pathogens were introduced directly into the wastes and distributed through the material either by stirring (slurries and dirty water) or by tumbling in a concrete mixer. The initial level of each bacterial pathogen was approximately 1 × 106 CFU g−1. This was representative of the upper limit of pathogen levels found previously in an on-farm survey of fresh livestock wastes (10). C. parvum was introduced into wastes at a level of 5 × 108 oocysts per plot. The mass of waste used for each 3-m2 field plot (25 to 40 kg) was calculated by using an average nitrogen content for each livestock waste type (derived from analyses performed in our laboratory between 1995 and 2000 [n = 16,322] [results not shown]), taking into account a target concentration of 200 kg of total nitrogen Ha−1.
TABLE 1.
Length of time required for a 1-log10 decrease (D value) in the levels of zoonotic agents present in livestock wastes spread onto grass pasture in summera
| Animal source | Waste type | D values (days)a
|
|||
|---|---|---|---|---|---|
| Salmonellae | E. coli O157 | L. monocytogenes | C. jejuni | ||
| Dairy cattle | FYM | 1.79 | 1.47 | 2.97 | 2.31 |
| Dairy cattle | Slurry | 1.61 | 1.63 | 2.01 | 2.65 |
| Dairy cattle | Dirty water | 1.34 | 1.31 | 1.82 | 2.33 |
| Beef cattle | FYM | 1.62 | 1.49 | 1.72 | 3.05 |
| Beef cattle | Slurry | 1.45 | 1.56 | 1.86 | 1.85 |
| Pig | FYM | 1.81 | 1.45 | 3.20 | 1.87 |
| Pig | Slurry | 1.86 | 1.70 | 2.80 | 1.86 |
| Poultry | Broiler litter | 1.77 | 1.63 | 2.13 | 2.53 |
| Sheep | FYM | 1.58 | 1.58 | 2.24 | 2.17 |
The values are means calculated from replicate experiments over 2 years (n = 6). The values were calculated from the initial decline over the 32 days immediately after waste spreading. The correlation coefficient is >0.65 for all times. Dirty water was parlor washings composed of dairy cattle fecal material, milk, and detergents.
Three replicate field plots were used for each waste type. The wastes were left unincorporated on top of the pasture. Controls onto which no manure or pathogen was spread were included for each treatment. Pathogen declines were measured in studies performed over 3 years between 2000 and 2003. During the first 2 years, the experiments commenced in late spring (referred to as summer decline experiments). The year 3 studies commenced in autumn 2002 (winter decline experiments). Full chemical analyses of plots were performed only for the first summer decline experiment. C. parvum decline was monitored only for the second-year summer experiments.
Sample collection and transit.
Samples for analysis were collected from each replicate field plot over a 6-month period. Each sample comprised a minimum of 20 combined subsamples collected to a depth of 15 cm with sterile soil augers. Samples were refrigerated at 2°C and transported from the farm site to laboratories, where analyses were begun within 16 h of sampling.
Microbiological methods.
Bacteria in the samples of waste were enumerated by using filter methods which allow resuscitation of injured cells, as described previously (8). Colony counts were converted to CFU per gram of waste according to the criteria specified by ISO 4833 (15a).
Antibody capture was used to determine the levels of C. parvum by the method described by Pepperell et al. (26), with minor modifications. Briefly, plot subsamples (10 to 20 g) were agitated with 10 volumes of 2 M NaCl containing 0.01% (vol/vol) Tween 20, and 10 ml of the sedimented (30 min) solution was overlaid onto 40 ml of a 1.09-g ml−1 sucrose solution. Each sample was centrifuged at 5,000 × g for 10 min without braking. The top 25 ml of the supernatant was mixed with 25 ml of deionized water and recentrifuged as described above. C. parvum oocysts were enumerated from the pellet by using a GC-combo immunomagnetic separation kit (Dynal Biotech, Wirral, United Kingdom) according to manufacturer's instructions. Viability was assessed by staining with 4′,6′-diamidino-2-phenylindole (DAPI) and propidium iodide and by epifluorescence microscopy by using the method described by Olson et al. (23).
Chemical methods.
The pH and conductivity of liquid and solid wastes were determined directly with samples without dilution (slurry) and with decimal dilution (FYM) by using methods described previously (10). The dry matter and ammonia concentrations in the waste samples were determined after drying and chemical titration with 0.05 M sulfuric acid, respectively, as described previously (10).
Environmental temperatures and rainfall.
Soil temperature was recorded at a depth of 5 cm by using a Squirrel data logger (Grant Instruments, Cambridge, United Kingdom). Precipitation was collected in a rain gauge located in the buffer strip surrounding the plots and was recorded daily.
Analyses of results.
Log averages and associated standard deviations from each set of three replicates were calculated for each sample. R2 values were determined by the least-squares method, and coefficients of variation (CV) were calculated by dividing the mean by the standard deviation for each sample time. Groups of CVs were compared by using the Mann-Whitney U test for nonparametric data (P < 0.05; SPSS 11.5; SPSS Inc., Chicago, Ill.). The numbers of days required for a 1-log decline in bacterial numbers (D values) were calculated from data generated during the first 16 days immediately after waste spreading. Groups of D values were compared by a one-way analysis of variance with Tukey's post hoc test (SPSS).
RESULTS AND DISCUSSION
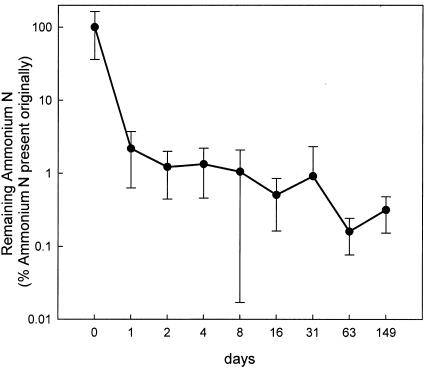
Within 24 h of spreading the fresh wastes onto pasture, ammonia volatilization lowered the crop-available nitrogen (5) (Fig. 1) by >98%. This result is particularly noteworthy because it shows that fresh livestock wastes are a poor source of nitrogen for grassland if the material is broadcast spread onto the surface of a pasture.
FIG. 1.
Loss of ammonium N from experimental plots over time. The data are averages derived from a range of fresh manure types as described in Materials and Methods and are the percentages of ammonium N remaining, as calculated from the amount applied initially. The error bars indicate the standard deviations of the means obtained from 27 experimental plots.
Since the mid-1970s, compositional differences in manures have been shown to have an influence on the bacterial populations in wastes (19). Since in this study we used different batches of wastes over several years, it was important to characterize the wastes used. Chemical analyses (pH, ammonium N, conductivity, and dry matter) were therefore performed for each waste at the start of each experiment. Three sets of chemical analyses were performed each year for each waste type. There were no significant differences (P > 0.05, as determined by analysis of variance) between years when the chemical results were compared. For the year 1 studies, chemical analyses were performed for each sample collected throughout the decline experiments. There were no significant differences (P > 0.05) between time intervals and any of the waste types for each chemical determination 24 h after wastes were spread onto land (data not shown). The result was likely due to the fact that the samples contained a high proportion of field soil. Field soil would buffer pH and normalize the original manure-specific differences in dry matter and conductivity. Since there were no significant differences between plots, chemical analyses were performed only for the initial and final samples in subsequent experiments.
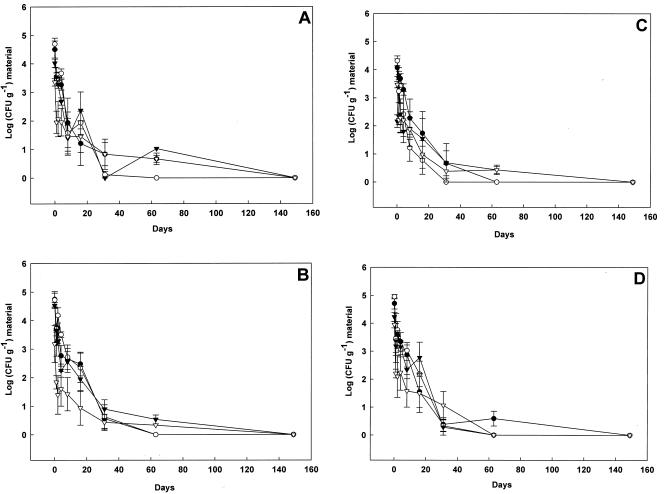
The declines of bacterial zoonotic agents in cattle wastes, which were typical of the declines observed for each of the waste types, are shown as Fig. 2. In general, the declines were immediate, and there was no evidence of initial bacterial proliferation. The data in Fig. 2 are the means for two independent trials performed in consecutive summers, and three independently analyzed replicates were used for each year. Although figures of this sort are a useful visual indication of decline, they are difficult to interpret and do not easily allow comparisons to be made between different experiments. Thus, in keeping with other studies of this type (12, 13, 16), D values were calculated from the initial linear summer declines, and they are summarized in Table 1. The average D values ranged from 1.31 to 3.20 days.
FIG. 2.
Decline of Salmonella (•), Listeria (▿), Campylobacter (▾), and E. coli O157 (O) in cattle wastes. Solid wastes (A and C) and liquid wastes (B and D) were obtained from beef cattle (A and B) and dairy cattle (C and D).
Although D values are useful for comparing initial linear rates of pathogen decline, the rate falls as the least hardy organisms are removed from the population and leave behind only those organisms that are better equipped for survival. Since the decline is not fully linear (Fig. 2), it is not possible to extrapolate pathogen survival from initial levels of zoonotic agents. Table 2 shows the longest interval of time that was observed before the levels of zoonotic agents declined below the limits of detection. The data in Table 2 were derived from the two summer experiments and the single winter experiment for bacteria and from a single summer experiment for the protozoan. L. monocytogenes was the hardiest zoonotic agent studied and survived for 128 days before the concentration declined to below detectable levels. The genus Listeria can be routinely isolated from soil and is therefore likely to be well-adapted to this niche.
TABLE 2.
Longest recorded lengths of time until zoonotic agents could no longer be isolated from the surface of grass pasture sampled as described in Materials and Methodsa
| Waste type | Longest recorded time (days) for:
|
||||
|---|---|---|---|---|---|
| Salmonellae | E. coli O157 | L. monocytogenes | C. jejuni | C. parvum | |
| Dairy cattle FYM | 42 | 16 | 128 | 42 | 30 |
| Dairy cattle slurry | 63 | 32 | 42 | 63 | 30 |
| Beef cattle FYMb | 63 | 32 | 63 | 63 | 30 |
| Beef cattle slurryb | 32 | 32 | 63 | 32 | 30 |
| Pig FYM | 32 | 32 | 63 | 32 | 30 |
| Pig Slurry | 16 | 32 | 63 | 16 | 63 |
| Poultry FYM | 63 | 32 | 42 | 42 | 30 |
| Sheep FYM | 16 | 63 | 128 | 16 | 30 |
| Dirty water | 42 | 32 | 128 | 32 | 30 |
Zoonotic agents were introduced into livestock wastes which were subsequently spread onto grassland without incorporation. The majority of the data are the longest times observed for any of the three decline experiments which were performed separately during two summers and a single winter season.
Data were obtained from summer decline experiments only.
The data generated in this study agree broadly with the data from the few studies that have been performed previously with individual zoonotic agents. Salmonellae were previously shown to survive in slurries which were allowed to dry on pastures for between 2 and 36 weeks (20). More recently, Bolton and colleagues inoculated cattle feces with E. coli O157 and subsequently spread the preparation onto grass pasture (3). After 50 days, the measured reduction in E. coli O157 levels was between 4 and 5 orders of magnitude. E. coli released from the spread waste was still detectable in the soil, without enrichment, after 99 days. A laboratory-based study in which grass-topped soil cores and a constant incubation temperature of 18°C were used resulted in a D value of roughly 60 days for E. coli O157 (22). Laboratory-based studies tend to show better survival for pathogens because the organisms are not subject to diurnal temperature fluctuations, UV irradiation from sunlight, and the drying effects of moving air (14).
The bacterial levels for the winter spreading studies showed much more variation than the bacterial levels for the summer experiments. Comparisons of coefficients of variation (standard deviation/mean) (data not shown) for all of the bacteria between the two seasons showed that the effect was significant (P < 0.05, as determined by the paired t test). The reasons for the seasonal differences in the variabilities of the bacterial levels recovered from the replicate plots are unclear. There were no obvious differences in the setup between the winter experiment and the summer studies performed in the previous 2 years. Nonetheless, for many of the winter studies, the variation and error were so large that we were unable to perform regression reliably (r2 < 0.65) on the bacterial declines and thus calculate D values. The temperatures were much lower and, at times, below freezing during the winter. It is possible that partial freezing of the waste plots may have contributed to the variation, although the waste was spread at as uniform a thickness as possible. Furthermore, low-dry-matter slurries were easier to apply evenly to plots than solid wastes. If partial freezing were the cause of the variation, we would have expected differences between the CVs for the slurries and FYM, but there were no significant differences between these waste types.
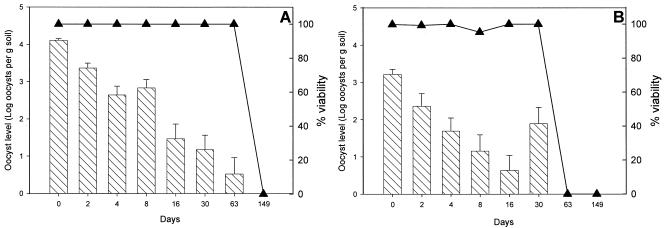
The recovery of oocysts from plots was poor, and, typically, less than 0.005% of the oocysts inoculated was actually recovered over the course of the experiments. Typical declines of C. parvum are shown in Fig. 3. Due to the difficulties in recovering oocysts, declines were measured for a single summer experiment only during the first year. Although identical numbers of oocysts were used to inoculate each waste type, the levels of recovery of C. parvum from manures with higher dry matter contents (>10%, wt/vol) were typically 1 order of magnitude lower than those from slurries. A possible explanation for these differences is that it is more likely that oocysts encounter and attach to organic matter in higher-dry-matter-content wastes and thus they are more difficult to recover (21).
FIG. 3.
Environmental decline of C. parvum spread onto grass. Oocysts were inoculated into livestock wastes (pig slurry [A] and pig FYM [B]), which were subsequently spread onto grass pasture planted in a clay-loam soil. The bars and the y axis on the left indicate the mean levels of oocysts recovered from each sample. The error bars indicate the standard deviations of the means obtained from three independently analyzed experimental plots. The solid triangles and the y axis on the right show the mean viability of the oocysts isolated from each plot on which waste was spread.
The numbers of oocysts recovered from the plots declined with time, and D values were calculated from the log declines of oocyst numbers. As expected, these data indicated that the oocysts were hardier than the bacteria. The D values calculated over 30 days for oocysts ranged from 31 days (sheep FYM) to 8 days (dirty water).
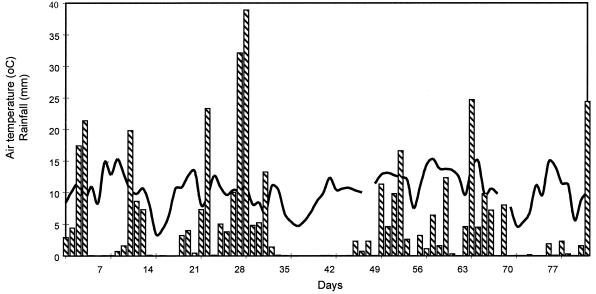
In addition to measuring the levels of oocysts, we used differential DAPI-propidium iodide staining to determine whether the oocysts were actually viable. Although the levels of oocysts declined with time, the viability of the recovered oocysts did not decrease appreciably. This observation is interesting because we expected the viability of the oocysts to decline as the numbers recovered fell. Rapid degradation of dead oocysts can occur, leaving no sporozoites to stain (23). The possibility that we observed decreases in the levels of oocysts because C. parvum was migrating out of the experimental plots also cannot be discounted. Alternatively, the data could mean that the oocysts were not recovered as effectively as viable protozoans. Environmental temperature and rainfall are shown in Fig. 4. The rainfall varied considerably over the course of the experiment but was, on occasion, high enough to generate pools of surface water on the impervious clay pasture during the summer. Thus, it is possible that the C. parvum decline measured was a combination of both oocyst death and washout from the plots. Although in the majority of plots we could not detect oocysts after a single month, the calculated D values suggest that oocysts could survive for a longer time.
FIG. 4.
Rainfall (bars) and temperature (line) for grass pasture plots spread with animal wastes inoculated with zoonotic agents, including C. parvum. The wastes were spread in late spring, and temperature and rainfall were monitored for 12 weeks after spreading. The gaps in the temperature profile were generated on the days that the temperature recorder was taken indoors for downloading.
A previous study (32) showed that Cryptosporidium oocysts placed in bags fashioned from semipermeable membranes on the surface of a grass pasture survived for 2 to 4 weeks in the summer in Scotland. Although radical differences between the experimental setups mean that direct comparisons are difficult, this result is interesting because the survival times are very similar to those that we observed. The use of semipermeable bags restricted oocyst movement in the Scottish study, and hence true decline was measured. Our setup measured decline by true death, as well as movement from plots. Thus, longer survival for the Scottish study would be expected compared with our results. It is possible that the wastes into which our oocysts were inoculated provided protection to the protozoans by preventing them from drying excessively.
Spreading of livestock wastes onto land used for the production of food or animal feeds is widely regarded as the least environmentally damaging disposal method (31). However, the practice is still fraught with pitfalls. It can lead to problems with odor as nitrogen in the form of volatile ammonium N is lost to the atmosphere, and nitrogen converted to nitrate is soluble and a pollutant of watercourses (29). Traditionally, because the environmental aspects of chemical contamination are obvious in the short term, prevention of such pollution was the most important consideration during waste disposal. However, this study and other reports (2, 3, 7, 9, 11, 18, 24, 27, 28) have shown there are also significant microbiological risks which need to be taken into account when animal wastes are spread onto land.
Our observations are that the levels of commonly encountered bacterial zoonotic agents decline rapidly when wastes are applied onto the surface of a grass pasture. A typical D value for the initial decline is 2 to 3 days. L. monocytogenes could, however, be recovered from some plots for up to 128 days. Given that L. monocytogenes is a ubiquitous soil organism, the risks to food safety posed by its extended survival are currently unclear. If a precautionary approach is adopted, the length of time after which it can be assumed that the concentrations of bacterial zoonotic agents in livestock wastes have declined to below detectable levels is 4 months. However, further studies are required to better assess how survival in soil is related to risk to food. In addition to these studies, there have been a number of reports of bacterial survival for significant periods in wastes spread onto land. Thus, it is prudent for waste-spreading guidelines aimed at farmers to take account of the potential microbiological risks to food production and food animals, as well as reductions in environmental pollution and improvements in nutrient recycling.
Acknowledgments
This study was funded by the B17 organic wastes program of the United Kingdom Food Standards Agency.
We gratefully acknowledge laboratory technical assistance by Barbara Rarata, Dawood Sadiq, David Petri, Gemma Simpson, and Gemma Lapworth; Aldwyn Clarke for sample collection; and Rob Davies, Veterinary Laboratories Agency, Weybridge, United Kingdom, for the kind gift of livestock isolates of Salmonella.
REFERENCES
- 1.Adak, G. K., S. M. Long, and S. J. O'Brien. 2002. Trends in indigenous foodborne disease and deaths, England and Wales: 1992 to 2000. Gut 51:832-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bicudo, J. R., and S. M. Goyal. 2003. Pathogens and manure management systems: a review. Environ. Technol. 24:115-130. [DOI] [PubMed] [Google Scholar]
- 3.Bolton, D. J., C. M. Byrne, J. J. Sheridan, D. A. McDowell, and I. S. Blair. 1999. The survival characteristics of a non-toxigenic strain of Escherichia coli O157:H7. J. Appl. Microbiol. 86:407-411. [DOI] [PubMed] [Google Scholar]
- 4.Brackett, R. E. 1999. Incidence, contributing factors, and control of bacterial pathogens in produce. Postharvest Biol. Technol. 15:305-311. [Google Scholar]
- 5.Chambers, B. G., K. A. Smith, and B. F. Pain. 2000. Strategies to encourage better use of nitrogen in animal manures. Soil Use Manag. 16:157-161. [Google Scholar]
- 6.Cieslak, P. R., T. J. Barrett, P. M. Griffin, K. F. Gensheimer, G. Beckett, J. Buffington, and M. G. Smith. 1993. Escherichia coli O157-H7 infection from a manured garden. Lancet 342:367. [DOI] [PubMed] [Google Scholar]
- 7.Cole, D. J., V. R. Hill, F. J. Humenik, and M. D. Sobsey. 1999. Health, safety, and environmental concerns of farm animal waste. Occup. Med.-State of the Art Rev. 14:423-448. [PubMed] [Google Scholar]
- 8.Driehuis, F., and S. J. W. H. Elferink. 2000. The impact of the quality of silage on animal health and food safety: a review. Vet. Q. 22:212-216. [DOI] [PubMed] [Google Scholar]
- 9.Faust, M. A. 1982. Relationship between land-use practices and fecal bacteria in soils. J. Environ. Qual. 11:141-146. [Google Scholar]
- 10.Fenlon, D. R., and J. Wilson. 1998. The quantitative assessment of Listeria monocytogenes growth in a laboratory ensiling system allowing limited aerobic spoilage. Grass Forage Sci. 53:292-295. [Google Scholar]
- 11.HeinonenTanski, H., P. Leinonen, E. M. Niskanen, M. M. Mielonen, and H. Rasanen. 1998. Aeration improves the hygiene of cattle slurry and the quality of grass forage and silage. Acta Agric. Scand. Sect. B Soil Plant Sci. 48:212-221. [Google Scholar]
- 12.Himathongkham, S., S. Bahari, H. Riemann, and D. O. Cliver. 1999. Survival of Escherichia coli O157:H7 and Salmonella typhimurium in cow manure and cow manure slurry. FEMS Microbiol. Lett. 178:251-257. [DOI] [PubMed] [Google Scholar]
- 13.Himathongkham, S., H. Riemann, S. Bahari, S. Nuanualsuwan, P. Kass, and D. O. Cliver. 2000. Survival of Salmonella typhimurium and Escherichia coil O157:H7 in poultry manure and manure slurry at sublethal temperatures. Avian Dis. 44:853-860. [PubMed] [Google Scholar]
- 14.Hutchison, M. L., F. A. Nicholson, K. Smith, W. C. Keevil, and T. Moore. 2000. A study of on-farm manure applications to agricultural land and an assessment of the risks of pathogen transfer into the food chain. MAFF report FS2526. Ministry of Agriculture Fisheries and Foods, London, United Kingdom. [Online.] http://www.pathogens.org.
- 15.Hutchison, M. L., L. W. Walters, T. Moore, K. M. Crookes, and S. M. Avery. 2004. Effect of length of time before incorporation on the survival of pathogenic bacteria present in livestock wastes applied to agricultural soil. Appl. Environ. Microbiol. 70:5111-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.International Organization for Standardization. 1991. Methods for microbiological examination of food and animal feeding stuffs. Enumeration of micro-organisms. Colony count technique at 30°C. ISO 4833. International Organization for Standardization, Geneva, Switzerland.
- 16.Jiang, X. P., J. Morgan, and M. P. Doyle. 2003. Thermal inactivation of Escherichia coli O157:H7 in cow manure compost. J. Food Prot. 66:1771-1777. [DOI] [PubMed] [Google Scholar]
- 17.Jones, D. L. 1999. Potential health risks associated with the persistence of Escherichia coli O157 in agricultural environments. Soil Use Manag. 15:76-83. [Google Scholar]
- 18.Jones, K., S. Howard, and J. S. Wallace. 1999. Intermittent shedding of thermophilic campylobacters by sheep at pasture. J. Appl. Microbiol. 86:531-536. [DOI] [PubMed] [Google Scholar]
- 19.Jones, P. W. 1976. The effect of temperature, solids content and pH on the survival of salmonellas in cattle slurry. Br. Vet. J. 132:284-293. [DOI] [PubMed] [Google Scholar]
- 20.Jones, P. W. 1986. Sewage sludge as a vector of salmonellosis, p. 21-33. In J. C. Block, A. H. Haielaar, and P. L'Hermite (ed.), Epidemiological studies of risks associated with the agricultural use of sewage sludge. Elsevier, London, United Kingdom.
- 21.Kuczynska, E., and D. R. Shelton. 1999. Method for detection and enumeration of Cryptosporidium parvum oocysts in feces, manures, and soils. Appl. Environ. Microbiol. 65:2820-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maule, A. 1996. Survival of the verotoxigenic strain E. coli O157:H7 in laboratory-scale microcosms, p. 61-65. In D. Kay and C. Fricker (ed.), Coliforms and Escherichia coli: problem or solution. Royal Society of Chemistry, Cambridge, United Kingdom.
- 23.Olson, M. E., J. Goh, M. Phillips, N. Guselle, and T. A. McAllister. 1999. Giardia cyst and Cryptosporidium oocyst survival in water, soil, and cattle feces. J. Environ. Qual. 28:1991-1996. [Google Scholar]
- 24.Paiba, G. A., J. W. Wilesmith, and S. J. Evans. 1999. Excretion of VTEC O157 by cattle. Vet. Rec. 144:708. [PubMed] [Google Scholar]
- 25.Pell, A. N. 1997. Manure and microbes: public and animal health problem. J. Dairy Sci. 80:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pepperell, R., J. Massanet-Nicolau, V. M. Allen, and S. Buncic. 2003. Potential for spread of some bacterial and protozoan pathogens via abattoir wastes applied on agricultural land. Food Prot. Trends 23:315-325. [Google Scholar]
- 27.Rahn, K., S. A. Renwick, R. P. Johnson, J. B. Wilson, R. C. Clarke, D. Alves, S. McEwen, H. Lior, and J. Spika. 1997. Persistence of Escherichia coli O157:H7 in dairy cattle and the dairy farm environment. Epidemiol. Infect. 119:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1992. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 58:3494-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmelsgaard, S. E. 1998. The effect of crop, N-level, soil type and drainage on nitrate leaching from Danish soil. Soil Use Manag. 14:30-36. [Google Scholar]
- 30.Smith, K. A., A. J. Brewer, J. Crabb, and A. Dauven. 2001. A survey of the production and use of animal manures in England and Wales. III. Cattle manures. Soil Use Manag. 17:77-87. [Google Scholar]
- 31.Strauch, D., and G. Ballarini. 1994. Hygienic aspects of the production and agricultural use of animal wastes. J. Vet. Med. Ser. B-Zentbl. Veterinaermed. Reihe B Infect. Dis. Vet. Public Health 41:176-228. [DOI] [PubMed] [Google Scholar]
- 32.Svoboda, I., I. Read, J. S. Kemp, S. E. Wright, R. L. Coop, J. L. Mawdsley, R. J. Merry, M. K. Theodoru, B. F. Pain, Z. Bukhari, and H. V. Smith. 1997. Cryptosporidium on cattle farms, p. 3-20. Cryptosporidium in water—the challenge to policy makers and water managers. The Chartered Institution of Water and Environmental Management, Glasgow, United Kingdom.