Abstract
The diversity of fungi in permanently frozen soil from northeastern Siberia was studied by culture-independent PCR amplification of diverse environmental 18S rRNA genes. Elaborate protocols to avoid contamination during drilling, sampling, and amplification were used. A broad diversity of eukaryotic DNA sequences that were 510 bp long, including sequences of various fungi, plants, and invertebrates, could be obtained reproducibly from samples that were up to 300,000 to 400,000 years old. The sequences revealed that ancient fungal communities included a diversity of cold-adapted yeasts, dark-pigmented fungi, plant-parasitic fungi, and lichen mycobionts. DNA traces of tree-associated macrofungi in a modern tundra sample indicated that there was a shift in fungal diversity following the last ice age and supported recent results showing that there was a severe change in the plant composition in northeastern Siberia during this period. Interestingly, DNA sequences with high homology to sequences of coprophilic and keratinophilic fungi indicated that feces, hair, skin, and nails could have been sources of ancient megafauna DNA recently reported to be present in small amounts of Siberian permafrost sediments.
Following death of an organism, the DNA is degraded and extensively modified by temperature-dependent hydrolytic and oxidative damaging processes, which results in accumulation of damaged sites and short DNA fragments (9, 10, 12). Thus, permafrost is considered to be ideal for long-term preservation of biomolecules due to the constant low temperatures, neutral pH, and anaerobic conditions (29).
Culture-independent PCR screening of composite samples enables scientists to describe ecological communities in minute samples (3, 16). This technique was used in recent studies of paleocommunities in Siberian sediments to amplify 16S rRNA genes, chloroplast DNA, and mitochondrial DNA from bacteria, plants, and herbivorous mammals, respectively (27). Numerous copies of these genes, as well as nuclear ribosomal genes, are present in each cell, and these genes are more likely to survive age-dependent degradation than single-copy genes (9). Ancient amplifiable fungal DNA has been shown to survive for at least several thousand years in glacial ice (3, 16) and glacier-associated remains (19) and are therefore likely to be present in fossil permafrost as well.
While microscopic evidence suggests that plant sedimentary chloroplast DNA largely originates from fine rootlets and seeds, there is no morphological evidence of mammalian organic compounds in the sediments. Dung and urine have been suggested as possible DNA sources (11, 27).
Importantly, studies of ancient microbial DNA are highly prone to modern contamination, and strict criteria, established and reported on previously (3, 16), must be fulfilled for authentication; these criteria include independent reproducibility of results by different laboratories.
The purpose of this work was to monitor and characterize the fungal composition of ancient permafrost. We categorized 18S rRNA genes amplified directly from natural communities in order to obtain important novel information on the northeastern Siberian paleoenvironment.
MATERIALS AND METHODS
Samples, DNA extraction, and PCR amplification.
Soil samples were obtained in western Beringia along 1,200 km of the Arctic coast (Table 1) under strict drilling conditions; a column coring method without drill fluids was used along with spiking with recognizable bacterial strains prior to drilling to test for contamination penetrating the cores. The seasonal thawing zone was within the top 0.5 m, and the thickness of the permafrost was calculated to be 800 to 1,000 m (7, 8, 27, 30).
TABLE 1.
Number of retrieved clones, stratigraphic age, core number, and drilling depth (meters below the surface) for the four sedimentary samples
| Sample layer | Age (103 yr) | Core no.a | Depth (m) | No. of clones
|
|||
|---|---|---|---|---|---|---|---|
| 125 bp | 210 bp | 510 bp | Total | ||||
| Modern | 0 | 1 | 0.5 | 25 | 9 | 33 | 67 |
| Holocene | 10 | 1 | 4.0 | —b | 21 | 22 | 43 |
| Late Pleistocene | 20 | 2 | 4.8 | 34 | 19/25c | 30 | 108 |
| Pleistocene | 300-400 | 4 | 9.2 | 25/18c | 35 | 36/22c | 136 |
See reference 27 for core numbers.
Amplification of 125 bp from the 10 ky sample failed. Spiking of samples with Humicula insolens genomic DNA, prior to cycling, revealed inhibition of the PCR in the 10 ky sample (results not shown).
The number after the shill is the number of clones independently obtained in Oxford.
Subsamples (approximately 1 g [wet weight]) were taken from the interior of the frozen core samples (27), and DNA was extracted by using established procedures (27). 18S rRNA genes were amplified by using the following primer pairs. With primers BMBC-R (5′-GTACACACCGCCCGTCG-3′) and NS8 (5′-TCCGCAGGTTCACCTACGGA-3′) (26) amplifying 125 bp of eukaryotic DNA, the initial denaturation at 94°C for 4 min was followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 68°C for 1 min and a final extension at 68°C for 7 min. With primers KW3 (5′-TCCAGCTCCAATAGCGTATA-3′) and SL43 (5′-GAACCACACGTCCTATTC-3′) (15) amplifying 210 bp of ascomycetous DNA, denaturation at 94°C for 2 min was followed by six cycles of 94°C for 45 s, 55°C for 45 s, and 68°C for 90 s, 34 cycles of 94°C for 45 s, 50°C for 45 s, and 68°C for 90 s, and finally 68°C for 7 min. Finally, with primers SR7R (5′-AGTTAAAAAGCTCGTAGTTG-3′) and SR5 (5′-GTGCCCTTCCGTCAATT-3′) (www.biology.duke.edu/fungi/mycolab/primers/htm) amplifying 510 bp of eukaryotic DNA, denaturation at 94°C for 2 min was followed by six cycles of 94°C for 45 s, 50°C for 45 s, and 68°C for 90 s, 34 cycles of 94°C for 45 s, 45°C for 45 s, and 68°C for 90 s, and finally 68°C for 7 min. The final concentrations of the different reagents in the PCR mixtures were as follows: each deoxynucleoside triphosphate, 0.2 mM; 1× High Fidelity buffer; MgSO4, 2 mM; each primer, 1 μM; and Platinum Taq High Fidelity polymerase(Invitrogen), 1.0 U. Autoclaved double-distilled UV-treated water was added to bring the final volume to 50 μl.
DNA from PCR was purified from the PCR mixtures by using a GFX PCR DNA and a gel band purification kit (Amersham Biosciences). Separate reaction mixtures were pooled after purification. Cloning was done by ligating PCR products into the pCR4Blunt-TOPO vector (Zero Blunt TOPO PCR cloning kit for sequencing; Invitrogen) by using a ratio of insert to ligation vector of 1:1 to help ensure a more comprehensive recovery of all PCR products. Sequencing was carried out with a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems).
Precautions and controls.
DNA extractions were carried out at the Ancient DNA Facility at the University of Copenhagen and, independently, at the University of Oxford in fully equipped and physically isolated clean laboratories solely dedicated to ancient DNA work with separate ventilation systems, nightly exposure of surfaces to UV irradiation, and weekly cleaning of surfaces with bleach. The PCR setup was in a positive-flow hood (Holten Laminair HB 2448) exposed to UV light during the night. Both extraction and pre-PCR work were carried out with their own sets of lab clothes, lab tools, and unopened reagents. Tools for pre-PCR work were washed in 5% sodium hypochlorite (28) and exposed to UV light overnight. Full-body suits, facemasks, and sterile surgical gloves were used. Double-distilled, UV-treated, and autoclaved water, as well as PCR buffer, the deoxynucleoside triphosphate mixture, and MgSO4 were centrifuged through 30-kDa filter units (Microcon YM-30; Millipore) that have been reported to retain fragments longer than 45 bp, and the primer solutions were centrifuged through 50-kDa filter units (Microcon YM-50, Millipore) that retained fragments longer than 75 bp (28). Mock extraction and PCR controls were made to test for false-positive results. Independent extraction, PCR, and cloning with its own set of laboratory reagents, including primers, were carried out at University of Oxford to confirm results obtained in Copenhagen.
Sequence analysis.
Phylogenetic profiles for all DNA sequences were generated by using the PhylPro software to investigate evidence of genetic recombination among homologous sequences (25). Recombinant sequences were omitted from further analysis, as were clones that aligned only partially with GenBank hits.
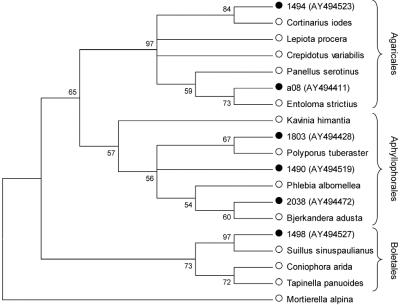
To deduce the taxonomic affiliation, we used the sequences to search publicly available DNA sequence databases. The consensus taxon and the species with the highest levels of sequence similarity were recorded (see supplemental table) from the gapped BLAST record as described in reference 28. Clone sequences and the top hit sequences identified by BLAST searches were aligned with additional 18S rRNA gene GenBank sequences, and the alignments were subjected to phylogenetic analyses in an attempt to assign the sequences to classes and orders (Fig. 1). Neighbor-joining trees were constructed by using the Kimura 2-parameter substitution model with 1,000 bootstrap replications in the MEGA software program.
FIG. 1.
Method for phylogenetic identification. In this analysis 510-bp permafrost homobasidiomycete-related clone sequences (•), top BLAST hit sequences, and taxonomically related GenBank sequences (○) were used to construct a neighbor-joining tree with 1,000 bootstrap replications. All phylogenetic reconstructions can be obtained from the authors upon request.
RESULTS
Culture-independent analysis of 18S rRNA genes from permafrost.
Amplification followed by cloning and sequencing of 125-, 210-, and 510-bp 18S rRNA gene fragments from four different sedimentary samples yielded 356 clones (Table 1). The 210-bp fragments were amplified with ascomycete-specific primers, whereas both the 125- and 510-bp fragments were amplified with universal eukaryotic primers. The Siberian tundra samples ranged in age from a modern, seasonally frozen layer to three prehistoric deposits that were dated to approximately 10,000, 20,000, and 300,000 to 400,000 years ago (0 ky, 10 ky, 20 ky, and 300-400 ky samples, respectively) (Table 1). The highest numbers of clones were retrieved with the 510-bp primers; 510-bp amplicons were successfully obtained from all ancient samples. The data support the hypothesis that DNA of this length from all three major kingdoms (plants, fungi, and animals) is preserved in prehistoric samples.
Fungal diversity.
In this study a broad spectrum of organisms from the Siberian tundra was discovered. Ancient and previously unreported fungal DNA from Ascomycetes, Basidiomycetes, and Zygomycetes were found in the soil and represented fungi belonging to 12 different classes and 10 different orders (Table 2). Amplicons retrieved from the ancient samples were assigned to the following phylogenetically, taxonomically, ecologically, and physiologically broad fungal groups: Ascomycetes (Leotiomycetes, Dothidiomycetes, Dothidiomycetes et Chaetotyriomycetes, Sordariomycetes, Chaetotyriomycetes, Saccharomycetes, Eurotiomycetes, and Lecanoromycetes), Basidiomycetes (Urediniomycetes, Homobasidiomycetes, and Heterobasidiomycetes), and Zygomycetes (Zygomycetes).
TABLE 2.
Consensus rank and top BLAST genera for 18S rRNA gene sequences
| Sample | Kingdom | Phyluma | Class or subclassb | Order | Top BLAST genera (>96%) |
|---|---|---|---|---|---|
| Modern (seasonally frozen active layer) | Viridiplantae | Magnoliophyta | Rosids | Casuarina, Flacourtia, Idesia | |
| Asterids | Pterospora, Vaccinium | ||||
| Bryophyta | Bryopsida | Aulacomnium, Palustriella | |||
| Fungi | Basidiomycota | Urediniomycetes | Uredinales | Bensingtonia | |
| Homobasidiomycetes | Boletales | Suillus | |||
| Agaricales | Cortinarius, Entoloma | ||||
| Aphyllophorales | Limnoperdon, Phlebia | ||||
| Cantarellales | Hydnum | ||||
| Stereales | Acanthophysium | ||||
| Heterobasidiomycetes | Filobasidiales | Cryptococcus, Filobasidium | |||
| Ascomycota | Leotiomycetes | Helotiales | Chalara, Cyathicula, Neobulgaria, Phialocephala | ||
| Sordariomycetes | Lecythophora | ||||
| Zygomycota | Zygomycetes | Mucorates | Umbelopsis | ||
| Mortierellales | Mortierella | ||||
| Holocene (10 ky) | Viridiplantae | Magnoliophyta | Rosids | Flacourtia | |
| Chlorophyta | Chlorophyceae | Haematococcus | |||
| Fungi | Basidiomycota | Homobasidiomycetes | Aphyllophorales | Amylocystis | |
| Heterobasidiomycetes | Filobasidiales | Cryptococcus | |||
| Cystofilobasidiales | Mrakia | ||||
| Tremellales | Cryptococcus | ||||
| Ascomycota | Dothidiomycetes | Pleosporales | Delitschia | ||
| Dothidiomycetes et Chaetotyriomycetes | Raciborskiomyces | ||||
| Sordariomycetes | Sordariales | Sordaria | |||
| Late Pleistocene (20 ky) | Viridiplantae | Magnoliophyta | Rosids | Flacourtia, Platytheca, Cneorum | |
| Liliopsida | Oryza | ||||
| Bryophyta | Polytrichopsida | Polytrichum | |||
| Chlorophyta | Trebouxiphyceae | Microthamniales | Trebouxia | ||
| Fungi | Basidiomycota | Urediniomycetes | Uredinales | Melamspora | |
| Homobasidiomycetes | Aphyllophorales | Phanerochaete | |||
| Heterobasidiomycetes | Filobasidiales | Cryptococcus | |||
| Tremellales | Cryptococcus, Bullera | ||||
| Ascomycota | Leotiomycetes | Lachnum, Stromatinia, Blumeria, Monilinia | |||
| Saccharomycetes | Candida | ||||
| Dothidiomycetes | Westerdykella, Leptosphaeria | ||||
| Sordariomycetes | Phialophora | ||||
| Dothidiomycetes et Chaetotyriomycetes | Hormonema, Raciborskiomyces, Aureobasidium, Mycosphaerella | ||||
| Eurotiomycetes | Warcupiella | ||||
| Zygomycota | Zygomycetes | Mortierellates | Mortierella | ||
| Pleistocene (300-400 ky) | Viridiplantae | Magnoliophyta | Rosids | Flacourtia | |
| Liliopsida | Poales | Oryza, Festuca, Rhynchospora, Triticum | |||
| Bryophyta | Bryopsida | Diphyscium, Cinclidium | |||
| Chlorophyta | Trebouxiphyceae | Microthamniales | Trebouxia | ||
| Prasiolales | Chlorella | ||||
| Fungi | Basidiomycota | Urediniomycetes | Sporobolomyces | ||
| Homobasidiomycetes | Agaricales | Heliocybe, Panellus, Entoloma | |||
| Heterobasidiomycetes | Cystofilobasidiales | Cryptococcus | |||
| Tremefalles | Bullera, Cryptococcus | ||||
| Ascomycota | Leotiomycetes | Cadophora, Monilinia, Stromatinia, Rhytisma, Rhexocercosporidium | |||
| Dothidiomycetes | Herpotrichia, Aureobasidium | ||||
| Sordariomycetes | Phialophora | ||||
| Chaetotyriomycetes | Exophiala | ||||
| Lecanoromycetes | Lecanorales | Cladonia, Cetraria, Pilophorus, Physcia, Umbilicaria |
Bryophyta is not a phylum but a nonranked taxonomic level between kingdom and class.
Eudicotylidon subclasses Rosids and Asterids are dominated by shrubs and trees, whereas Liliopsida comprises monocotyledonous herbs.
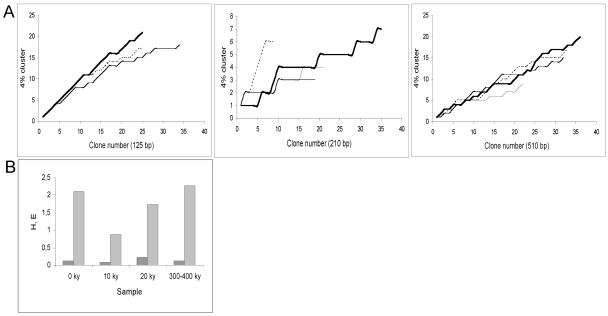
Representatives of six classes and four orders of the Viridiplantae groups of flowering plants, mosses, and algae were also found (Table 2). In addition to plants and fungi, two classes and one order of Alveolata, two classes and two orders of Metazoa, and some Cercozoan DNA were found. Diversity curves were constructed for all samples and primer sets to investigate the level of redundancy (Fig. 2). The curves show no evidence of reaching saturation and hence indicate that the diversity spectrum is not complete.
FIG. 2.
Diversity curves (A) and statistical analysis of diversity indices (B). (A) Clusters (4%; 2% DNA damage plus 2% sequence heterogeneity) (28) were plotted against clone number by following the order of sampling. The amplicon length is indicated for each graph. Dotted line, modern sample; thin line,10 ky sample; medium line, 20 ky sample; thick line, 300-400 ky sample. (B) Shannon's diversity index (H) (light grey bars) and Shannon's equitability (E) (dark grey bars) were calculated for fungus-related sequence communities obtained with 510-bp primers by using the 4% clusters for species definition.
Quality control.
Independent DNA extraction and replication at the Ancient Biomolecules Centre at the University of Oxford for the 20 ky and 300-400 ky samples yielded 65 clones (Table 1). PCR controls, which were used at both Oxford and Copenhagen to test for the presence of extraneous DNA, were negative in all analyses. All replicated sequence data were subjected to a bootstrap test for independent reproducibility (27, 30). This test confirmed that sequence populations obtained in Copenhagen and Oxford represent the same underlying distribution. Of the 15 classes identified in the 300-400 ky sample, 12 were found in analyses performed in Oxford and Copenhagen. The authenticity of the data was further supported by sequences showing high homology to common arctic lichen fungi (Cladonia metaminiata and Cetraria islandica), soil-inhabiting anaerobic earth mites, and protists, as well as fungal psychrotolerant species of Cryptococcus, Mortierella, Geomyces, and Mrakia previously found in polar regions (6, 23).
DISCUSSION
510-bp amplicons retrieved from the ancient permafrost.
PCR-amplified 18S rRNA gene fragments that were 510 bp long and were from organisms trapped in permafrost for 300,000 to 400,000 years represent the longest independently authenticated nuclear eukaryote DNA sequences of this age. Predictions of DNA decay based on in vitro observations have been used to argue that amplification of ancient DNA fragments longer than a few hundred base pairs should be impossible (12, 17). The amplification of eukaryotic 18S rRNA genes, together with the previously reported amplification of 600-bp 16S prokaryotic rRNA genes in 400,000- to 600,000-year-old permafrost samples (30) and the amplification of approximately 1-kb mitochondrial DNA sequences from Holocene permafrost-preserved penguin bone (14), indicates that the maximum amplifiable length of ancient DNA is somewhat greater. There have been several interesting reports on revival of ancient anabiotic or cryobiotic microorganisms from permafrost in Siberia and Antarctica (1, 21, 24). However, the observations have not been reproduced yet, and it seems unlikely that the ability to transform into such cryopreserved resting forms alone can explain the broad diversity of DNA obtained in this study. A more plausible explanation is the exceptional preservation conditions for DNA offered by the Beringian permafrost (13, 29). These include constant subzero temperatures, an almost neutral pH, and an anaerobic environment. All of these conditions are factors that reduce the rates of chemical DNA degradation and oxidative damage (29).
Possible biases in data.
Sampling of cloned PCR products did not exhaust the diversity present in the samples investigated (Fig. 2A), and continued sampling would likely reveal even more taxa, further expanding our knowledge of ancient Siberian ecosystems. Therefore, we remain cautious about comparing diversity indices for the different time periods examined in the present study (Fig. 2B). Furthermore, caution may be necessary if the observed fraction was derived from species with better protected DNA and/or more easily extracted DNA than other species not identified in this analysis.
Fungal life forms in prehistoric Siberia.
During previous thermo- and cryochrones Beringia served as an ice-free refugee, which allowed the presence and growth of cold-climate-adapted plants, animals, and fungi (4). As a consequence, the biodiversity of Beringia was reported to be relatively rich compared to that of polar ice caps (27), and the ratio of indigenous DNA to exogenous DNA was expected to be relatively high in the present study.
Amplification of modern and ancient fungus-related 18S rRNA genes, most likely from both vegetative cells and propagules, represents a complex fungal spectrum very similar to the contemporary Arctic and Antarctic fungal spectrum (5, 23). Fungi that were shown by a BLAST homology search to be closely related to the saprophytic genus Panellus (Agaricales) seem to have played an important role in composting of plant material. Nevertheless, BLAST homology searches suggested that there was a predominance of yeasts (Cryptococcus, Bullera, and Candida) and plant-parasitic fungi (Rhytisma, Blumeria, Stromatinia, Sporobolomyces, and Melampsora) in the prehistoric samples (Table 2). Sequences affiliated with encapsulated basidiomycetous yeasts of the Cryptococcus type were found in all ancient samples, possibly due to the ability of the organisms to survive dry freezing and to assimilate a variety of carbon sources (23). Clone sequences similar to the sequences of plant-parasitic fungi in the Leotiomycetes (ascomycetes with fruiting bodies) and Urediniomycetes (rusts) were found in ancient sediments. Recovery of clone sequences with high levels of homology to plant-parasitic fungi was to some extent expected since public databases contain overrepresentations of DNA from such fungi. Nonetheless, it is feasible to see plant parasitism as an adaptation to unfriendly habitats that yields benefits such as plant carbon and moisture, as well as the physical protection offered by the host tissue.
A large number of cloned sequences retrieved from the 20 ky and 300-400 ky sample sediments exhibited high levels of homology (98 to 100%) to small-subunit rRNA genes of black-pigmented Ascomycota associated with modern plants found in polar regions, including Phialophora sp., Aureobasidium pullulans, Stromatinia rapulum, and Rhytisma acerinum (5, 18, 23). The dark pigment melanin offers protection from DNA-damaging UV light and transforms the absorbed radiation to heat, which might be a positive coselective factor in cold environments.
Traces of DNA from lichen symbionts.
The importance of lichens as a food source for mammals in contemporary arctic regions is well established (22). Our finding of lichens in ancient permafrost may explain the ability of the region to sustain diverse populations of both microbes and megafauna throughout the last ice age. Multiple clones identified as Lecanoromycetes in the Pleistocene 300-400 ky sample were identified by BLAST analysis as genetic relatives of the lichenizing mycobiont genera Cladonia, Pilophorus, Cetraria, Physcia, Lecania, Caloplaca, Bunodophoron, and Umbilicaria. Both Cladonia metaminiata and Cetraria islandica, to which we found 100% similarity, are common cold-region inhabitants (2). In the same sample sequences of possible photobionts (Trebouxia and Chlorella homologues) were present among sequences assigned to green algae (Trebouxiophyceae). Trebouxia and Umbilicaria reportedly associate to form lichens capable of maintaining photosynthetic activity at temperatures down to −17°C (20).
Possible source of megafaunal DNA in permafrost.
DNA sequences in the 10 ky sample showed high similarity to the coprophilic fungi Delitschia winteri (Pleosporales) and Sordaria humana (Sordariales). Previous work (27) performed with the 10,000- and 20,000-year-old sediments revealed the dominance of mitochondrial sequences from extinct and extant megafauna. It was proposed that the major contributor of DNA from large herbivorous animals could be endothelial cells present in feces and urine (27). This hypothesis is supported by the presence of DNA from coprophilic fungi in this study. Interestingly, the presence of DNA with high levels of similarity to 18S rRNA gene sequences of the keratinophilic fungi Phialophora sp. and Geomyces pannorum indicate that mammal fur and nails are alternative sources of DNA.
Fungi and flora in the context of climatic changes.
The recovery of rRNA genes from green plants in this study (Table 2) supports the hypothesis that the Beringian vegetation changed dramatically following the last glacial maximum (19,000 years ago) due to climatic changes (4, 27). From a tundra steppe dominated by grasses and herbs, the postglacial period featured development of a tundra dominated by shrubs, which successively developed into modern conditions. Our data indicate that grasses belonging to the order Poales and willow-like shrubs were numerous in the 300-400 ky and 20 ky samples. In the 10 ky and modern samples grasses were absent, and there was a far more diverse population of larger green plants. The observed shift in vegetation is consistent with observed changes in composition of fungal DNA. Yeast-like fungi and fungi that are parasitic on either herbs or grasses seemed to be dominant during the last ice age (10 ky and 20 ky samples), which was followed by an increase in sequences affiliated with homobasidiomycetous root-associated macrofungi (Suillus, Hydnum, and Cortinarius) recovered from the modern sample (0 ky sample). A statistical analysis of the shift in fungal community structures calculated by using Shannon's H value suggested that there was decreasing fungal diversity from the Pleistocene (300-400 ky sample) to the Holocene (10 ky sample), presumably followed by an increase in diversity up to modern times (Fig. 2B). The 10 ky sample, in addition, exhibited the lowest equitability of the four samples, which can be explained by the dominance of a single cluster of Cryptococcus (Basidomycota)-associated sequences.
Absence of ubiquitous highly air-sporulating saprophytes.
Despite the broad diversity of fungal DNA amplified from the samples, the absence of the most common, highly air-sporulating, soil-inhabiting saprophytes Aspergillus, Penicillium, Cladosporium, and Fusarium is striking. It is unlikely that any procedure that broke big, elongate, encapsulated spores (e.g., spores of Cryptococcus) would fail to break the small, spherical spores of the ubiquitous saprophytes (J. W. Taylor, personal communication). The absence of the most numerous fungi in present environments, however, supports the hypothesis that modern-day contamination was successfully avoided in sampling and subsequent amplification of ancient DNA.
Supplementary Material
Acknowledgments
Eske Willerslev was supported by the Royal Society UK.
Expert technical assistance in the laboratory by Sanne R. Jensen and Tina B. Brand was highly appreciated. We also thank Søren Flensted for constructive criticism during the study and John W. Taylor for reviewing the manuscript.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abyzov, S. S. 1993. Microorganisms in the Antarctic ice, p. 265-295. In E. I. Friedmann (ed.), Antarctic microbiology. Wiley-Liss, New York, N.Y.
- 2.Augustin, J., S. Matula, and A. Dandar. 1999. Isolation and purification of lichenan from Iceland moss (Certraria islandica). Biologia 54:289-295. [Google Scholar]
- 3.Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, I., P. Matheus, B. Shapiro, D. Jensen, and A. Cooper. 2002. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science 295:2267-2270. [DOI] [PubMed] [Google Scholar]
- 5.Bergero, R., M. Girlanda, G. C. Varese, D. Intili, and A. M. Luppi. 1999. Psychrooligotrophic fungi from Arctic soils of Franz Joseph Land. Polar Biol. 21:361-368. [Google Scholar]
- 6.DePriest, P. T., N. V. Ivanova, D. Fahselt, V. Alstrup, and A. Gargas. 2000. Sequences of psycrophilic fungi amplified from glacier-preserved ascolichens. Can. J. Bot. Rev. Can. Bot. 78:1450-1459. [Google Scholar]
- 7.Gilichinsky, D. A., G. M. Hlebnikova, D. G. Zvyagintcev, D. G. Fedorov-Davydov, and N. N. K. 1989. Mikrobiologicheskaya harakteristika pri izuchenii osadochnyh porod kriolitozony. Ser. Geologicheskaya 6:103-115. [Google Scholar]
- 8.Gilichinsky, D. A., E. A. Vorobyova, L. G. Erokhina, D. G. Fyordorov-Davydov, N. R. Chaikovskaya, and D. G. Fyordorov-Dayvdov. 1992. Long-term preservation of microbial ecosystems in permafrost. Adv. Space Res. 12:255-263. [DOI] [PubMed] [Google Scholar]
- 9.Greenwood, A. D., C. Capelli, G. Possnert, and S. Paabo. 1999. Nuclear DNA sequences from late Pleistocene megafauna. Mol. Biol. Evol. 16:1466-1473. [DOI] [PubMed] [Google Scholar]
- 10.Handt, O., M. Hoss, M. Krings, and S. Pääbo. 1994. Ancient DNA—methodological challenges. Experientia 50:524-529. [DOI] [PubMed] [Google Scholar]
- 11.Hofreiter, M., J. L. Betancourt, A. P. Sbriller, V. Markgraf, and H. G. McDonald. 2003. Phylogeny, diet, and habitat of an extinct ground sloth from Cuchillo Cura, Neuquen Province, southwest Argentina. Quaternary Res. 59:364-378. [Google Scholar]
- 12.Hofreiter, M., D. Serre, H. N. Poinar, M. Kuch, and S. Pääbo. 2001. Ancient DNA. Nat. Rev. Genet. 2:353-359. [DOI] [PubMed] [Google Scholar]
- 13.Hoss, M., P. Jaruga, T. H. Zastawny, M. Dizdaroglu, and S. Pääbo. 1996. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 24:1304-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert, D. M., P. A. Ritchie, C. D. Millar, B. Holland, A. J. Drummond, and C. Baroni. 2002. Rates of evolution in ancient DNA from Adelie penguins. Science 295:2270-2273. [DOI] [PubMed] [Google Scholar]
- 15.Landvik, S., K. N. Egger, and T. Schumacher. 1997. Towards a subordinal classification of the Pezizales (Ascomycota): phylogenetic analyses of SSU rDNA sequences. Nord. J. Bot. 17:403-418. [Google Scholar]
- 16.McInerney, J. O., and M. Mullarkey. 2001. Bacteria and archea: molecular techniques reveal astonishing diversity. Biodiversity 3:3-10. [Google Scholar]
- 17.Pääbo, S., R. G. Higuchi, and A. C. Wilson. 1989. Ancient DNA and the polymerase chain-reaction—the emerging field of molecular archaeology. J. Biol. Chem. 264:9709-9712. [PubMed] [Google Scholar]
- 18.Robinson, C. H. 2001. Cold adaptation in Arctic and Antarctic fungi. New Phytol. 151:341-353. [Google Scholar]
- 19.Rollo, F., M. Ubaldi, L. Ermini, and I. Marota. 2002. Otzi's last meals: DNA analysis of the intestinal content of the Neolithic glacier mummy from the Alps. Proc. Natl. Acad. Sci. USA 99:12594-12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeter, B., G. A. Green, K. Kappen, and R. D. Seppelt. 1994. Carbon dioxide exchange at subzero temperatures. Field measurements on Umbilicaria aprina in Antarctica. Cryptogam. Bot. 4:233-241. [Google Scholar]
- 21.Soina, V. S., E. A. Vorobiova, D. G. Zvyagintsev, and D. A. Gilichinsky. 1995. Preservation of cell structures in permafrost—a model for exobiology. Adv. Space Res. 15:237-242. [DOI] [PubMed] [Google Scholar]
- 22.Storeheier, P. V., S. D. Mathiesen, N. J. C. Tyler, and M. A. Olsen. 2002. Nutritive value of terricolous lichens for reindeer in winter. Lichenologist 34:247-257. [Google Scholar]
- 23.Vishniac, H. S. 1996. Biodiversity of yeasts and filamentous microfungi in terrestrial Antarctic ecosystems. Biodivers. Conserv. 5:1365-1378. [Google Scholar]
- 24.Vorobyova, E., V. Soina, M. Gorlenko, N. Minkovskaya, N. Zalinova, A. Mamukelashvili, D. Gilichinsky, E. Rivkina, and T. Vishnivetskaya. 1997. The deep cold biosphere: facts and hypothesis. FEMS Microbiol. Rev. 20:277-290. [Google Scholar]
- 25.Weiller, G. F. 1998. Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Mol. Biol. Evol. 15:326-335. [DOI] [PubMed] [Google Scholar]
- 26.White, T. J., M. A. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, N.Y.
- 27.Willerslev, E., A. J. Hansen, J. Binladen, T. B. Brand, M. T. P. Gilbert, B. Shapiro, M. Bunce, C. Wiuf, D. A. Gilichinsky, and A. Cooper. 2003. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300:791-795. [DOI] [PubMed] [Google Scholar]
- 28.Willerslev, E., A. J. Hansen, B. Christensen, J. P. Steffensen, and P. Arctander. 1999. Diversity of Holocene life forms in fossil glacier ice. Proc. Natl. Acad. Sci. USA 96:8017-8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willerslev, E., A. J. Hansen, and H. N. Poinar. 2004. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol. Evol. 19:141-147. [DOI] [PubMed] [Google Scholar]
- 30.Willerslev, E., A. J. Hansen, R. Rønn, T. B. Brand, I. Barnes, and C. Wiuf. 2004. Long time persistence of bacterial DNA. Curr. Biol. 14:R9-R10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.