Abstract
Silkworm pathogens have been heavily impeding the development of sericultural industry and play important roles in lepidopteran ecology, and some of which are used as biological insecticides. Rapid advances in studies on the omics of silkworm pathogens have produced a large amount of data, which need to be brought together centrally in a coherent and systematic manner. This will facilitate the reuse of these data for further analysis. We have collected genomic data for 86 silkworm pathogens from 4 taxa (fungi, microsporidia, bacteria and viruses) and from 4 lepidopteran hosts, and developed the open-access Silkworm Pathogen Database (SilkPathDB) to make this information readily available. The implementation of SilkPathDB involves integrating Drupal and GBrowse as a graphic interface for a Chado relational database which houses all of the datasets involved. The genomes have been assembled and annotated for comparative purposes and allow the search and analysis of homologous sequences, transposable elements, protein subcellular locations, including secreted proteins, and gene ontology. We believe that the SilkPathDB will aid researchers in the identification of silkworm parasites, understanding the mechanisms of silkworm infections, and the developmental ecology of silkworm parasites (gene expression) and their hosts.
Database URL: http://silkpathdb.swu.edu.cn
Introduction
The silkworm, Bombyx mori, has been domesticated for >5000 years. Today over 178 000 metric tons of silk are produced annually world-wide (http://inserco.org/en/?q=statistics), the biggest producers being China and India (http://sericulturecouncil.com/world-raw-silk-production/). Silkworm diseases, including pébrine (caused by microsporidian Nosema bombycis) which was first described in France in 1856 (1, 2), were one of the factors contributing to the decline of the silk industry in Europe. Silkworm diseases result in losses of over 100 millions of US dollars each year in China alone. In addition to the domestic silkworm, B. mori, other lepidopteran species are also used in silk production. These ‘wild’ silk moths include Antheraea pernyi, Antheraea assamensis, Antheraea yamamai, Philosamia cynthiaricini, Eriogyna pyretoum, Caligula japonica, Samia cynthia, Attacus atlas and Actias selene.
We have recently developed the Silkworm Pathogen Database (SilkPathDB) which we update regularly. This database is an open-access, user-friendly comprehensive resource providing annotation and tools for the analysis of genomic information from various silkworm species and their parasites. The database will enable scientists to easily access and analyze their data, improve experimental design, and facilitate systematic studies, such as functional genomics, genome evolution and systems biology.
We have integrated previously acquired data generated in our laboratories during the process of genome, transcriptome and proteome studies on N. bombycis CQ1 (3), N. antheraeae YY (3), Nosema sp. PM-1, Vairimorpha necatrix BM, B.mori nucleopolyhedrovirus CQ1 and A.pernyi nucleopolyhedrovirus H. Also included in the SilkPathDB are transcriptome and proteome data from studies of host response to infection in the silkworm-pathogen system (4–6).
Genomic analyses of pathogens from silkworm species will continue to be added to the SilkPathDB as new information becomes available. This will give us insight into aspects of the genomic ecology of a range of Lepidoptera. The pathogens most frequently reported from silkworms are viruses, microsporidia, fungi and bacteria. In fact these pathogenic taxa infect a wide variety of Lepidoptera and a broad array of insects; members of these four groups are often used or suggested for use as biological control agents (7, 8). A number of silkworm pathogen genomes have already been sequenced and are publicly available on the SilkPathDB (9, 10).
Datasets and methods
System implementation
The SilkPathDB is built on a platform composed of a Linux Ubuntu Server 14.04, Apache 2.4, MySQL 5.5, PHP 5.5, PostgreSQL 9.3 and BioPerl 1.6. The implementation of SilkPathDB integrates and harnesses three complementary, mature and well supported technologies; Chado (11), GBrowse (12) and Drupal (http://www.drupal.org). The architecture of the SilkPathDB consists of four layers. The core layer employs the Chado schema for storing all datasets and is managed by the PostgreSQL. Outside the core is a configurable layer that controls the communications between the core layer and the outer layers. The third layer is composed of background analysis tools that process and respond to queries, and the Drupal CMS that manages website contents. The outermost layer is an interactive user interface that transmits information between the user and the database.
Chado is a relational database schema originally designed to store previously reported Drosophila data in a fully integrated manner to include genomic sequence, bibliographic, genetic, phenotypic and molecular data. Chado is now being used as a relational database schema to manage data from a wide variety of organisms (11). One aim is to develop a software system with a broad degree of interoperability. The SilkPathDB data are stored in a Chado-compliant PostgreSQL database using the Generic Model Organism Database (GMOD, http://gmod.org). Chado supports the management and storage of genome annotations and is easily extensible to new data types. All formatted genome features are loaded into the database in batch using a built-in Perl script of Chado, the gmod_bulk_load_gff3.pl.
GBrowse (12, 13) is a combination database and interactive web page for manipulating and displaying annotations of genomes and is a core component of the GMOD. Gene models and annotations are retrieved from the Chado database using the Bio::Chado::Schema Perl API, and loaded into MySQL databases using the GMOD Perl API to provide content utilized in the SilkPathDB GBrowse page.
Drupal is a content management, open source software system. It supports custom modules, themes, content types and functions, and has been widely used to provide the web-based portal for biological databases (14). To manage the content of the five taxa, an ‘Organism’ content type was created. The Bootstrap framework (http://getbootstrap.com) is employed to make a responsive, clean and intuitive user interface that supports both desktop and mobile devices. PHP combined with BioPerl scripts were developed for generating feature index, gene overview and search result pages by retrieving data regarding specific genes and organisms from the Chado database. JQWidgets (http://www.jqwidgets.com) is applied to manage and display the feature index, search result, BLAST output and host-responsive data.
Data and processing
The majority of the data in our database were downloaded from the GenBank (http://www.ncbi.nlm.nih.gov/genbank/) or the Silkworm Database (SilkDB) (http://www.silkdb.org) (15). At present the SilkPathDB contains data from 90 organisms from 5 taxonomic groups (5 microsporidia, 3 fungi, 49 bacteria, 29 viruses and 4 insect hosts) (Table 1).
Table 1.
Summary of organisms and major data types available in SilkPathDB
| No. of records |
||||||
|---|---|---|---|---|---|---|
| Microsporidia | Fungi | Bacteria | Virus | Host | Total | |
| Species | 5 | 3 | 49 | 29 | 4 | 90 |
| Assembly | 2457 | 359 | 49 | 59 | 52 004 | 54 928 |
| Gene | 12 995 | 31 703 | 277 369 | 3012 | 60 637 | 385 716 |
| mRNA | 12 995 | 31 703 | 271 267 | 3012 | 65 890 | 384 867 |
| Protein | 12 995 | 31 703 | 271 267 | 3012 | 65 890 | 384 867 |
| Transposon | 12 570 | 10 127 | 15 821 | 28 | 431 743 | 470 289 |
| Responsive gene | 4069 | 4069 | ||||
| GO | ||||||
| Molecular function | 331 | 951 | 7510 | 34 | 1048 | 9874 |
| Biological process | 275 | 764 | 25 047 | 24 | 919 | 27 029 |
| Cellular component | 106 | 248 | 3011 | 5 | 319 | 3689 |
Newly sequenced genomes which have not been annotated or further characterized are processed for SilkPathDB using a number of procedures. Eukaryotic protein-coding genes are predicted with the programs AUGUSTUS (16) and GeneMark-ES (17). Prokaryotic and viral protein-coding genes are predicted using GeneMarkS (18) and Glimmer (19). Transfer RNA genes are predicted by tRNAscan-SE (20). Ribosomal RNA genes are predicted with RNAmmer (21), INFERNAL (22) and homologous searches. Gene predictions obtained by different methods are combined using GLENA (http://sourceforge.net/projects/glean-gene) to make consensus data sets. DNA and protein features are then formatted into the Generic Feature Format version 3 (https://github.com/The-Sequence-Ontology/Specifications/blob/master/gff3.md) using Sequence Ontology (23).
All predicted proteins are annotated using a BLASTP (24) search against the Swiss-Prot and TrEMBL databases in UniProt (25). To obtain information on protein domains and gene ontology (GO), scores with E-values of 1e-5 or less are aligned against the InterPro (26) database (InterProScan5) (27). At present, 274 361 functional proteins have been annotated against the UniProt database. SilkPathDB GO annotation data comprises over 204 075 automatically assigned annotations using 40 592 unique GO terms, including 9874 Molecular Function terms, 27 029 Biological Process terms and 3689 Cellular Component terms (Table 1).
Transposable elements (TEs) are identified using RepeatModeler version open-1.0.8 (http://www.repeatmasker.org/RepeatModeler.html), which implements RepeatScout (28) and RECON (29). The consensus sequences from RepeatModeler are then mapped across each genome with RepeatMasker version open-4.0.6 (http://www.repeatmasker.org) against Repbase Update 20.11 (30). To date 470 289 TEs have been annotated in the SilkPathDB (Table 1).
Protein subcellular localizations are predicted via EuSecPred (http://silkpathdb.swu.edu.cn/eusecpred) and ProSecPred (http://silkpathdb.swu.edu.cn/prosecpred), which have been developed for identifying subcellular localizations of eukaryotic and prokaryotic proteins, respectively. Proteins containing signal motifs which are not allocated to cellular components (nucleus, endoplasmic reticulum, cytoplasm, Golgi apparatus, cell membrane) are considered to be secreted proteins. The SilkPathDB now includes 275 164 automatically assigned subcellular localizations including 8705 secreted pathogen proteins.
Silkworm transcriptome and proteome data obtained from silkworms infected with bacteria, viruses and microsporidia have been extracted from published studies and placed in the SilkPathDB. At present the database contains 4069 unique silkworm genes and proteins expressed in response to infection (Table 1), including 47 proteins from Gram-negative Escherichia coli and Gram-positive Bacillus bombyseptieus (5), 2435 genes from B.bombyseptieus (6) and 2521 genes from the microsporidian N. bombycis CQ1 (4).
Results
Data download and search
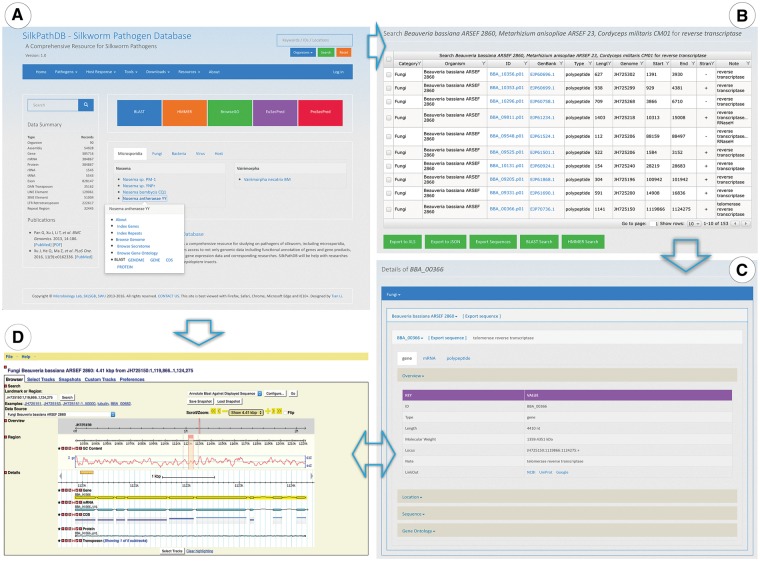
Datasets contained in the SilkPathDB can be downloaded in bulk via the ‘Download Datasets’ which is a hyperlink contained in the ‘Downloads’ drop down menu at the top of the home page (Figure 1A). The available datasets are grouped into the five taxonomic categories (Microsporidia, Fungi, Bacteria, Virus and Host Insects). Each of these categories contains the following data types: genome, transcriptome and proteome.
Figure 1.
Highlights of the Homepage, Search and Gene Overview pages of SilkPathDB. (A) The SilkPathDB homepage with direct access to search, analysis tools and data for each organism. (B) An example of search results presented in a table that can be sorted, filtered and further analyzed. (C) Feature details including overview, location, GO and subcellular localization. (D) The SilkPathDB genome browser, from which feature details can be graphically viewed.
Genome features of each pathogen can be accessed directly via the Pathogen tab at the top of the SilkPathDB homepage (Figure 1A). A drop down menu shows the four pathogen taxa in the database. Clicking on a taxon reveals the species available in the SilkPathDB for that taxon. Clicking on a particular species brings you to the database page for that species. Using the menu tabs, users can access all genes and repeats, browse genomes, secretomes and GO, and download and do a BLAST search of sequences for each species. The GO browser (SilkPathGO) allows users to list genes and export sequences based on GO terms.
A search tool is available on every SilkPathDB page (Figure 1A, top right). This tool allows users to perform queries on single or multiple species. Search results are listed in a table, each column of which can be sorted and filtered for further searches (Figure 1B). Rows of the table can be selected and exported to XLS and JSON files. Selected sequences can be exported in FASTA format. Furthermore, the format supports analysis by BLAST and HMMER. Feature details can be accessed via the hyperlink contained in the ID column heading. The Feature Details page provides major information about a feature, including ID, type, length, location, sequence, GO and subcellular localization (Figure 1C).
Genome browser
Genomes of pathogens and hosts can be viewed with the SilkPathDB genome browser (Figure 1D), which is driven by GMOD GBrowse (12) and supports the visualization of feature position, sequence, G + C content, functional annotation and relationship among features. By clicking the glyph of each feature, details can be viewed in SilkPathDB and GenBank.
Analysis
Homologs from pathogen and host genomic data can be can be obtained through BLAST and HMMER sequence searches. The SilkPathDB BLAST is built with NCBI BLAST+ 2.5.0 (24), NCBI BLAST 2.2.26 (31) and AB-BLAST 2.2.6 (32) and supports multiple database searches. The NCBI BLAST+ is an optimized and strongly recommended version of the former NCBI BLAST, which is not supported by the NCBI. The AB-BLAST is a fast and sensitive BLAST suit improved from the NCBI BLAST. The results give standard and tabular outputs from which both matched targets and matched regions can be exported. The implementation of HMMER in the SilkPathDB is designed to find homologs using the Hidden Markov model (33). The results are then presented in a table that can be sorted, filtered and exported.
Conclusion and future perspective
Genome database has become one of the most useful resources and platforms during scientific research. Except some global and comprehensive databases like GenBank (34) and UniProt (35), there are several databases that provide specific and effective functions for the study of microbial pathogens. Among those related to silkworm pathogens, the Fungal and Oomycete Genomics Resources (FungiDB) (36) and Microsporidia Genomics Resource (MicrosporidiaDB) (37) are two databases that belong to the Eukaryotic Pathogen Database Resources (EuPathDB) (38), which provide abundant data and a sophisticated search strategy system enabling complex interrogations of underlying data. However, we have not found data of fungal pathogens that infect silkworm in the FungiDB, and only found a silkworm microsporidium, N. bombycis CQ1, in the MicrosporidiaDB. Besides, the Virus Pathogen Resource (39) is an integrated repository of data and analysis tools for multiple virus families, including silkworm Cypovirus, but has not provided data of Baculoviridae and Parvoviridae Family. Meanwhile, we have not found genome databases specific for pathogenic bacteria related to silkworm. Therefore, there is no database that provides data and functions of all natural and microbial pathogens of silkworm yet.
The SilkPathDB is a comprehensive resource and platform that provides people with general knowledge, omics data, host-responsive data of silkworm pathogens, as well tools and pipelines for searching and analyzing public and personal data. With these functions, SilkPathDB can be used to aid researchers in the identification of silkworm parasites, understanding the mechanisms of silkworm infections, and elucidating the developmental ecology of silkworm parasites and their hosts.
We continue to incorporate additional data from both pathogens and their hosts into the SilkPathDB. In addition, we will include new datasets and analyses enhancing information on interactions between these pathogens and their hosts. We are also developing tools for further analysis and data mining, such as genomic variation among pathogen isolates, gene transfers between pathogen and host, and gene expression analysis using high throughput sequencing data. With high-throughput sequencing, genomic data is being produced at an enormous rate, creating specialized genomic databases is increasingly important. Making these data accessible through enhanced interfaces and by seamless links between the SilkPathDB and other public databases will be important for understanding interactions between these important domestic insects and their pathogens.
Acknowledgements
The authors thank all people's contribution on this work. They would like to thank the anonymous reviewers for their valuable comments and suggestions.
Funding
This work was supported by the Natural Science Foundation of China (31472151, 31272504 and 31001036), the National Basic Research Program of China (2012CB114604), the Fundamental Research Funds for the Central Universities (XDJK2015A010).
Conflict of interest. None declared.
References
- 1. James J., Becnel T.G.A. (1999) Microsporidia in insects In: Murray Wittner L.M.W. (ed). The Microsporidia and Microsporidiosis. ASM Press, Washington, DC, pp. 447–501. [Google Scholar]
- 2. Jean-Marie Legay G.C. (2004) La phase pastorienne de la sériciculture. La crise de la pébrine et ses conséquences. Nat. Sci. Soc., 12, 413–417. [Google Scholar]
- 3. Pan G., Xu J., Li T. et al. (2013) Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation. BMC Genomics, 14, 186.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma Z., Li C., Pan G. et al. (2013) Genome-wide transcriptional response of silkworm (Bombyx mori) to infection by the microsporidian Nosema bombycis. PLoS One, 8, e84137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhong X.W., Zhao P., Zou Y. et al. (2012) Proteomic analysis of the immune response of the silkworm infected by Escherichia coli and Bacillus bombyseptieus. Insect Sci., 19, 559–569. [Google Scholar]
- 6. Huang L., Cheng T., Xu P. et al. (2009) A genome-wide survey for host response of silkworm, Bombyx mori during pathogen Bacillus bombyseptieus infection. PLoS One, 4, e8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. St Leger R.J., Wang C. (2010) Genetic engineering of fungal biocontrol agents to achieve greater efficacy against insect pests. Appl. Microbiol. Biotechnol., 85, 901–907. [DOI] [PubMed] [Google Scholar]
- 8. Lomer C.J., Bateman R.P., Johnson D.L. et al. (2001) Biological control of locusts and grasshoppers. Annu. Rev. Entomol., 46, 667–702. [DOI] [PubMed] [Google Scholar]
- 9. Xiao G., Ying S.H., Zheng P. et al. (2012) Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep., 2, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao Q., Jin K., Ying S.H. et al. (2011) Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet., 7, e1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mungall C.J., Emmert D.B., FlyBase C. (2007) A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics, 23, i337–i346. [DOI] [PubMed] [Google Scholar]
- 12. Stein L.D., Mungall C., Shu S. et al. (2002) The generic genome browser: a building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stein L.D. (2013) Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief. Bioinformatics, 14, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ficklin S.P., Sanderson L.A., Cheng C.H. et al. (2011) Tripal: a construction toolkit for online genome databases. Database (Oxford), 2011, bar044.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J., Xia Q., He X. et al. (2005) SilkDB: a knowledgebase for silkworm biology and genomics. Nucleic Acids Res., 33, D399–D402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stanke M., Schoffmann O., Morgenstern B. et al. (2006) Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics, 7, 62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borodovsky M., Lomsadze A. (2011) Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr. Protoc. Bioinformatics, Chapter 4, Unit 4 6 1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Besemer J., Lomsadze A., Borodovsky M. (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res., 29, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delcher A.L., Harmon D., Kasif S. et al. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lowe T.M., Eddy S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lagesen K., Hallin P., Rodland E.A. et al. (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res., 35, 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nawrocki E.P., Kolbe D.L., Eddy S.R. (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics, 25, 1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eilbeck K., Lewis S.E., Mungall C.J. et al. (2005) The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol., 6, R44.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Camacho C., Coulouris G., Avagyan V. et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. UniProt C. (2015) UniProt: a hub for protein information. Nucleic Acids Res., 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mitchell A., Chang H.Y., Daugherty L. et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res., 43, D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mulder N., Apweiler R. (2007) InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol. Biol., 396, 59–70. [DOI] [PubMed] [Google Scholar]
- 28. Price A.L., Jones N.C., Pevzner P.A. (2005) De novo identification of repeat families in large genomes. Bioinformatics, 21, i351–i358. [DOI] [PubMed] [Google Scholar]
- 29. Quesneville H., Bergman C.M., Andrieu O. et al. (2005) Combined evidence annotation of transposable elements in genome sequences. PLoS Comput. Biol., 1, 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bao W., Kojima K.K., Kohany O. (2015) Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA, 6, 11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Altschul S.F., Madden T.L., Schaffer A.A. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gish W. (1996–2009) http://blast.advbiocomp.com/.
- 33. Eddy S.R. (2009) A new generation of homology search tools based on probabilistic inference. Genome Inform., 23, 205–211. [PubMed] [Google Scholar]
- 34. Benson D.A., Clark K., Karsch-Mizrachi I. et al. (2015) GenBank. Nucleic Acids Res., 43, D30–D35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. The UniProt, C. (2016) UniProt: the universal protein knowledgebase. Nucleic Acids Res, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stajich J.E., Harris T., Brunk B.P. et al. (2012) FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res., 40, D675–6D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aurrecoechea C., Barreto A., Brestelli J. et al. (2011) AmoebaDB and MicrosporidiaDB: functional genomic resources for Amoebozoa and Microsporidia species. Nucleic Acids Res., 39, D612–D619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aurrecoechea C., Barreto A., Basenko E.Y. et al. (2016) EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res, 45, D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pickett B.E., Sadat E.L., Zhang Y. et al. (2012) ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res., 40, D593–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]