Abstract
Enrichment cultures were conducted using bismuth subsalicylate as the sole source of carbon and activated sludge as the inoculum. A pure culture was obtained and identified as a Fusarium sp. based on spore morphology and partial sequences of 18S rRNA, translation elongation factor 1-α, and β-tubulin genes. The isolate, named Fusarium sp. strain BI, grew to equivalent densities when using salicylate or bismuth subsalicylate as carbon sources. Bismuth nitrate at concentrations of up to 200 μM did not limit growth of this organism on glucose. The concentration of soluble bismuth in suspensions of bismuth subsalicylate decreased during growth of Fusarium sp. strain BI. Transmission electron microscopy and energy-dispersive spectroscopy revealed that the accumulated bismuth was localized in phosphorus-rich granules distributed in the cytoplasm and vacuoles. Long-chain polyphosphates were extracted from fresh biomass grown on bismuth subsalicylate, and inductively coupled plasma optical emission spectrometry showed that these fractions also contained high concentrations of bismuth. Enzyme activity assays of crude extracts of Fusarium sp. strain BI showed that salicylate hydroxylase and catechol 1,2-dioxygenase were induced during growth on salicylate, indicating that this organism degrades salicylate by conversion of salicylate to catechol, followed by ortho cleavage of the aromatic ring. Catechol 2,3-dioxygenase activity was not detected. Fusarium sp. strain BI grew with several other aromatic acids as carbon sources: benzoate, 3-hydroxybenzoate, 4-hydroxybenzoate, gentisate, d-mandelate, l-phenylalanine, l-tyrosine, phenylacetate, 3-hydroxyphenylacetate, 4-hydroxyphenylacetate, and phenylpropionate.
Bismuth subsalicylate is the active ingredient in Pepto-Bismol. It is one of a class of bismuth-containing medicinals used to treat ulcers and other gastrointestinal disorders. The use of bismuth compounds for this purpose dates back to the 18th century (38). The mechanism of action of bismuth subsalicylate and related compounds was not understood for most of the duration of their medicinal usage.
In general, microbial interactions with bismuth salts and organobismuth compounds are not well known, but there are insights largely derived from studies on sensitivity and resistance to bismuth. Bismuth sulfite has been used in growth medium for the selective isolation of Salmonella strains from foods by inhibiting other bacteria (46). Organobismuth compounds and salts have been shown to inhibit growth of Helicobacter pylori (28), other bacteria implicated in gastrointestinal disorders (27), and some fungi (8, 32).
Even less is known about the metabolic disposition of inorganic bismuth by microbes. Putative bismuth resistance genes were mapped on penicillin resistance plasmids in Staphylococcus aureus to regions containing the cadmium/zinc and mercury resistance genes, but no specific genes were correlated with bismuth resistance (35). Bismuth ions induce bacterial cadmium and arsenical/antimonial resistance operons (12, 19) and synthesis of metal binding peptides in yeast (16), but these phenomena have not been specifically linked to a bismuth resistance phenotype. Bacteria exposed to bismuth salts and organobismuth compounds have been shown to accumulate bismuth by unknown mechanisms onto cell surfaces, in cell walls, and in the cytoplasm (26, 33, 42). In anaerobic ecosystems, trimethylbismuth gas effluent has been detected and attributed to microbial action (4). Several bacterial species have been reported to form reflective “bismuth mirrors” when growing around filter disks soaked with bismuth salts on solid media (28, 33). Although bismuth mirrors have been suggested to arise from bismuth reduction, the lack of analytical procedures has left this issue unresolved.
To better understand resistance and metabolic responses to bismuth by microbes, an enrichment culture approach was used to isolate microbes that could grow using bismuth subsalicylate as the sole carbon source. A filamentous fungus identified as a Fusarium species was isolated. Fusarium sp. strain BI accumulated bismuth intracellularly during growth on bismuth subsalicylate, and the accumulated bismuth was concentrated in phosphorus-rich inclusions in specimens prepared for electron microscopy. The metabolic pathway through which Fusarium sp. strain BI degraded bismuth subsalicylate was determined by assaying the activities of known enzymes involved in salicylate catabolism in crude extracts of biomass grown on salicylate and glucose. The ability of Fusarium sp. strain BI to grow using a variety of other organic acids as carbon sources was also evaluated.
MATERIALS AND METHODS
Enrichment culture.
The minimal medium used for the enrichment culture consisted of a 0.015 M potassium phosphate buffer (pH 7.0) with salts of essential metals and trace elements added to give the following final concentrations: 5 mM NH4Cl, 80 μM MgSO4, 1.8 μM CaCl2, 80 nM Na2MoO4, 50 nM MnSO4, 20 nM Fe(II)SO4, 10 nM CoSO4, 10 nM ZnSO4, 10 nM Fe2(III)(SO4)3, 10 nM Al(SO4)3, 10 nM NiSO4, 10 nM CuSO4, 10 nM H3BO3. Vitamins were added to the following final concentrations: 0.15 μM thiamine, 82 nM biotin, 45 nM folic acid, 0.49 μM pyridoxine, and 0.82 μM nicotinamide. A 15 mM suspension of bismuth subsalicylate (Sigma-Aldrich, St. Louis, Mo.) was prepared in 50 ml of minimal medium, inoculated with 1 ml of activated sewage sludge (Metro Wastewater Treatment Plant, St. Paul, Minn.), and incubated at room temperature on a rotary shaker. After 14 days, 5 ml of the initial enrichment culture was transferred to a fresh 15 mM suspension of bismuth subsalicylate in minimal medium and incubated as described for 4 days, and then a second subculture was started by adding 5 ml of this culture to another fresh 15 mM suspension of bismuth subsalicylate. After incubation for 6 days, the final subculture was streaked for isolated colonies onto a 0.25× Luria-Bertani (LB) agar plate and incubated at 30°C for 72 h. Different colony types were then isolated as pure cultures on additional 0.25× LB agar plates.
Strain identification.
Wet mounts of spores from a 14-day plate culture were examined by light microscopy. To obtain template DNA for PCR amplification and sequencing, a loop of aerial hyphae from a plate culture was added to 50 μl of sterile distilled H2O (dH2O) in a 1.7-ml microcentrifuge tube. The tube was placed in a boiling water bath for 10 min and then centrifuged. The supernatant (template DNA) was decanted into a sterile microcentrifuge tube. PCR amplification reaction mixtures had 10 μl of 10× PCR buffer, 10 mM each deoxynucleoside triphosphate, 50 mM MgCl2, 2 μl of template DNA, 5 U of Taq DNA polymerase, and MilliQ dH2O to 100 μl. The same cycling regime was used for each set of primers (21). Amplification was verified by agarose gel electrophoresis. PCR product was purified for sequencing using a QIAquick gel extraction kit (QIAGEN, Valencia, Calif.). Primers S1 and S3 (21) and FF1100 and FR1 (45) were used to amplify sections of the 18S rRNA gene, and primers S1, FF1100, and FF700 (45) were used for sequencing. Primers EF-1 and EF-2 were used to amplify a section of the translation elongation factor 1-α gene, and EF-1 was used for sequencing (36). Primers Bt2a and Bt2b (14) were used to amplify and sequence a section of the β-tubulin gene. Sequences were aligned against all DNA sequences deposited in GenBank using BLAST (1).
Photomicroscopy.
Photomicrographs were taken with Portra 100T tungsten film (Kodak, Rochester, N.Y.) and an FX-35 (Nikon, Melville, N.Y.) camera mounted on a Zeiss (Thornwood, N.Y.) standard universal microscope.
Assay for total soluble bismuth.
Total soluble bismuth in cultures and extracts was determined with an Optima 3000 DV inductively coupled plasma optical emission spectrometer (Perkin Elmer, Boston, Mass.). Suspensions of 1.5 mM bismuth subsalicylate were prepared in 250 ml of minimal medium and incubated on a shaker at 30°C. After 24 h, 10-ml aliquots were removed using a sterile pipette and filtered with sterile 25-mm-diameter 0.2-μm-pore-size mesh polyethersulfone syringe filters (Nalgene, Rochester, N.Y.). Experimental flasks were then inoculated with 100 μl of 48-h cultures grown on 1.5 mM salicylate (Sigma-Aldrich) in minimal medium. Ten-milliliter aliquots were then collected from experimental and uninoculated control flasks every 24 h and filtered as described above. Prior to analysis by inductively coupled plasma optical emission spectrometry (ICP-OES), concentrated nitric acid was added to a final concentration of 1% and samples were spiked with 1 mg of yttrium/liter, which served as an internal standard. Plasma parameters were as follows: radio frequency power was 1,350 W, nebulizer gas flow was 0.70 liters/min, auxiliary gas flow was 0.40 liters/min, plasma coolant flow was 15.0 liters/min, and the sample introduction rate was 1.5 liters/min. Bismuth was observed at 223.061 nm, and yttrium was observed at 371.030 nm.
Bismuth toxicity and tolerance.
To determine total dry biomass yield in the presence and absence of Bi3+, 100-ml aliquots (prepared in triplicate) of minimal medium with 2% (wt/vol) glucose containing 0, 2, 20, or 200 μM bismuth nitrate (Sigma-Aldrich) were inoculated with 100 μl of a 48-h culture of Fusarium sp. strain BI (grown on 2% glucose in minimal medium) and incubated on a shaker at 30°C. After 72 h of incubation, biomass was harvested by vacuum filtration on preweighed 0.2-μm-pore-size mesh 47-mm-diameter nylon membranes (Millipore, Billerica, Mass.) and rinsed three times with minimal medium. Filters and collected biomass were dried at 100°C in a drying oven for 48 h, cooled to room temperature in a desiccator for 1 h, and then weighed. To plot growth curves of Fusarium sp. strain BI growing on salicylate and bismuth subsalicylate, 2,000-ml aliquots of 1.5 mM salicylate and bismuth subsalicylate in minimal medium were inoculated with 10 ml of a 48-h culture of Fusarium sp. strain BI grown on 1.5 mM salicylate. Cultures were incubated on a shaker at 30°C. Aliquots of 1.0 ml were collected from the cultures periodically with sterile glass pipettes. Aliquots from the bismuth subsalicylate culture were diluted 1:1 with 1 M disodium tartaric acid (Acros Organics B.V.B.A, Geel, Belgium) to solubilize undissolved bismuth subsalicylate (41). The optical density at 600 nm (OD600) of each aliquot was determined immediately after collection, and readings were plotted versus time.
Electron microscopy and elemental analysis.
One-milliliter aliquots of 96-h cultures grown in 1.5 mM bismuth subsalicylate and 1.5 mM salicylate (Sigma-Aldrich) were concentrated by centrifugation, rinsed with minimal medium, and pelleted again. Pellets were fixed with 2% glutaraldehyde, postfixed with 1% OsO4, and dehydrated in an ethanol series. Fixed biomass was then infiltrated with EMbed 812 epoxy resin (Electron Microscopy Sciences, Hatfield, Pa.) in steps of 30, 60, 90, and 100% resin in absolute ethanol. After infiltration, fixed biomass was embedded in flat molds with 100% resin at 60°C for 48 h. Ultrathin (60 nm) sections were cut with a diamond knife on a microtome and collected on nylon mesh grids. Sections were viewed using a CM12 transmission electron microscope (TEM) (Philips Electron Optics, Eindhoven, Netherlands), and elemental composition of discrete regions in sections was determined by energy-dispersive spectroscopy (EDS) with a Power MX ultrathin window EDS system (EDAX, Mahwah, N.J.).
Enzyme activity assays.
Cultures for preparation of crude extracts were grown in 2,000-ml batches on a shaker at 30°C in minimal medium. Ten milliliters of a 48-h culture grown on the corresponding substrate was used as inoculum. Substrate (30 mmol total) was added in stages as follows: 50% at time of inoculation, 25% after 24 h, and 25% after 48 h. Biomass was harvested 52 h after inoculation by vacuum filtration and rinsed as described above. Crude extracts were prepared by macerating the collected biomass in a chilled (4°C) mortar and pestle with 2× (vol/wt) 0.033 mM potassium phosphate buffer (pH 7.0, 4°C) and 2× (wt/wt) Empore Filter Aid 400 high-density glass beads (3M, St. Paul, Minn.). The resulting slurry was dispensed into microcentrifuge tubes and centrifuged at 4°C for 15 min. Supernatants (crude extracts) were decanted from each tube and pooled; the pellets were discarded. Total protein content of the crude extract was estimated using the Bio-Rad Protein Assay (Hercules, Calif.) (7), and a standard curve was prepared using bovine serum albumin. All activity assays were conducted using a Beckman DU 740 spectrophotometer (Fullerton, Calif.). Crude extracts were assayed for salicylate hydroxylase activity (47), catechol 1,2-dioxygenase activity (20), catechol 2,3-dioxygenase activity (2), gentisate 1,2-dioxygenase activity (30), and hexokinase activity (5) using previously published procedures.
Polyphosphate extraction and analysis.
Biomass from 72-h cultures grown on 1.5 mM salicylate or bismuth subsalicylate was collected by vacuum filtration as described above. Wet biomass (0.4 g) was ground in 1.2 ml of 2% (wt/vol) trichloroacetic acid (Fisher Scientific, Hampton, N.H.) in a chilled mortar and pestle with 0.2 g of ground glass beads and then centrifuged for 5 min. Polyphosphate of different chain lengths was extracted from the supernatant using the methods of Clark et al. (10). Polyphosphate concentration was determined in the fractions as orthophosphate using the colorimetric assay of Saheki et al. (39). Prior to analysis, polyphosphate in the fractions was converted to orthophosphate by acid hydrolysis, and orthophosphate released from polyphosphates was differentiated from orthophosphate from other sources by previously published methods (25). For ICP-OES analysis, samples were acidified with 1 ml of concentrated nitric acid, diluted to 12 ml with MilliQ dH2O, and filtered with 0.2 μm mesh syringe filters as described above.
Growth of Fusarium sp. strain BI on other substrates.
Net Fusarium sp. strain BI biomass yield was determined from growth on 0.25 mmol of various organic substrates [bismuth subsalicylate, salicylic acid, catechol (Sigma-Aldrich), gentisic acid (Sigma-Aldrich), benzoic acid (Mallinckrodt, Hazelwood, Mo.), 3-hydroxybenzoic acid (Sigma-Aldrich), 4-hydroxybenzoic acid (Eastman, Kingsport, Tenn.), phenylacetic acid (Eastman), 3-hydroxyphenylacetic acid (Sigma-Aldrich), 4-hydroxyphenylacetatic acid, 3-phenylpropionic (hydrocinnamic) acid (Sigma-Aldrich), d-(R)-mandelic acid (Sigma-Aldrich), l-(S)-mandelic acid (Sigma-Aldrich), l-tyrosine (Sigma-Aldrich), l-phenylalanine (Sigma-Aldrich), d-glucose, and cyanuric acid (Sigma-Aldrich)]. One-hundred-milliliter aliquots of 2.5 mM substrate in minimal medium (prepared in triplicate) were inoculated from 5-day plate cultures of Fusarium sp. grown on 0.25× LB plates and incubated on a shaker at 30°C. After 96 h, biomass was collected by vacuum filtration and rinsed as described above. Undissolved bismuth subsalicylate was solubilized as described above. Biomass was collected on filters and dried and weighed as described above.
Nucleotide sequence accession numbers.
The partial 18S rRNA, translation elongation factor 1-α, and β-tubulin gene sequences have been deposited in GenBank under accession numbers AY725264, AY725265, and AY25266, respectively.
RESULTS AND DISCUSSION
Enrichment culture and strain identification.
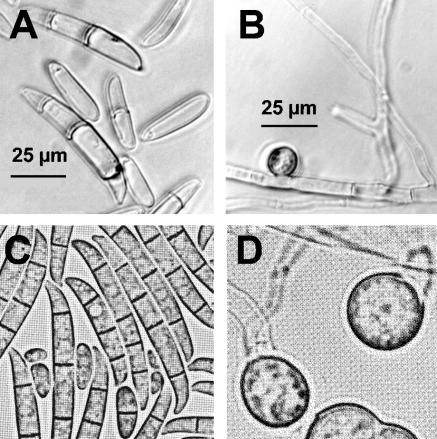
Growth was evident in the initial enrichment culture after 14 days. After subculturing the enrichment culture to verify growth on bismuth subsalicylate, a filamentous fungus was isolated that was identified as Fusarium sp. based on spore type and morphology (Fig. 1). Microconidia, macroconidia, and chlamydospores were all present (Fig. 1A and B). To confirm the genus of the isolate, partial sequences of 18S rRNA, translation elongation factor 1-α, and β-tubulin genes from the isolate were aligned against sequences in GenBank. 18S rRNA gene sequence from the isolate was most similar (1,048 of 1,051 nucleotides [nt]) to 18S rRNA gene sequences from Fusarium culmorum strain UPSC 1981 (GenBank accession number AF48073) and an uncultured ascomycete (GenBank accession number AB074659). Translation elongation factor 1-α gene sequence from the isolate had the highest similarity (656 of 673 nt) to corresponding sequences from Fusarium solani f. sp. batatas strains NRRL22402 and NRRl22400 (GenBank accession numbers AF178344 and AF178343, respectively). Sequence from the β-tubulin gene of the isolate was most similar (267 of 290 nt) to β-tubulin gene sequence from Gibberella zeae PH-1 (teleomorph of Fusarium graminearum) (GenBank accession number XM389706). Due to the varied results from the sequence alignments and lack of a globally accepted Fusarium taxonomy (34), identification of the isolate to the species level was not considered possible. The isolate was designated Fusarium sp. strain BI.
FIG. 1.
Light micrographs (A and B) (magnification, ×1,300) showing types and morphology of Fusarium sp. strain BI spores. Macroconidia and microconidia (A) and chlamydospores (B) were all present in this isolate. For comparison, light micrographs previously published by Toussoun and Nelson (44) of macroconidia and microconidia (C) (magnification, ×950) and chlamydospores (D) (magnification, ×1,660) from a Fusarium solani strain are shown (reproduced with permission of The Pennsylvania State University Press).
Depletion of bismuth from the culture medium and bismuth tolerance.
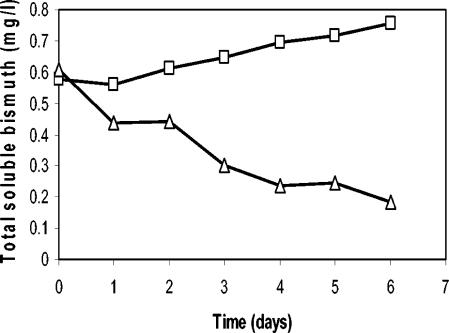
While organobismuth compounds have been shown to inhibit growth of fusaria (8, 29), mechanisms of bismuth tolerance and toxicity in fusaria and other fungi have not previously been explored in depth. Evaluation of bismuth reduction by Fusarium sp. strain BI was hampered by the lack of both a commercially available stable bismuth isotope and a practical assay to distinguish bismuth oxidation states. However, changes in bismuth concentration in the culture medium and cellular localization of bismuth were determined in the present study. ICP-OES was used to monitor the concentration of soluble bismuth during growth of Fusarium sp. strain BI in a 1.5 mM bismuth subsalicylate suspension in minimal medium (Fig. 2). Total soluble bismuth in inoculated suspensions decreased by ∼0.4 mg/liter (∼75% of initial concentration) after 6 days, while total soluble bismuth in the uninoculated control increased by ∼0.2 mg/liter (Fig. 2) during the same period. The increase in soluble bismuth in the control is likely due to slow solubilization of bismuth subsalicylate, which is relatively insoluble in aqueous media. Therefore, the net decrease in soluble bismuth during growth of the culture was closer to 0.6 mg/liter.
FIG. 2.
Plot of total soluble bismuth versus time in 1.5 mM bismuth subsalicylate suspensions in minimal medium that was uninoculated (□) or inoculated with Fusarium sp. strain BI (Δ). Bismuth concentrations were determined by ICP-OES.
Independent experiments were conducted to test bismuth tolerance of Fusarium sp. strain BI. No discernible difference in biomass production was observed in cultures grown in the presence of 0 to 200 μM bismuth nitrate in minimal medium with glucose as the sole carbon source. Optical density data showed that cultures grown on salicylate reached stationary phase ∼24 h sooner than cultures grown on bismuth subsalicylate. However, the cultures achieved equivalent optical densities at stationary phase (data not shown).
Bismuth ions have a high affinity for thiolate ligands and are proposed to be a hazard in the cytoplasmic environment (38). Regulating cytoplasmic levels of free essential and nonessential metal ions is central to fungal metal tolerance (13). Because intracellular bismuth accumulation did not reduce the growth yield of Fusarium sp. strain BI, this organism appears to have a mechanism that either maintains low levels of intracellular bismuth or renders bismuth nontoxic inside the cell.
Localization of accumulated bismuth.
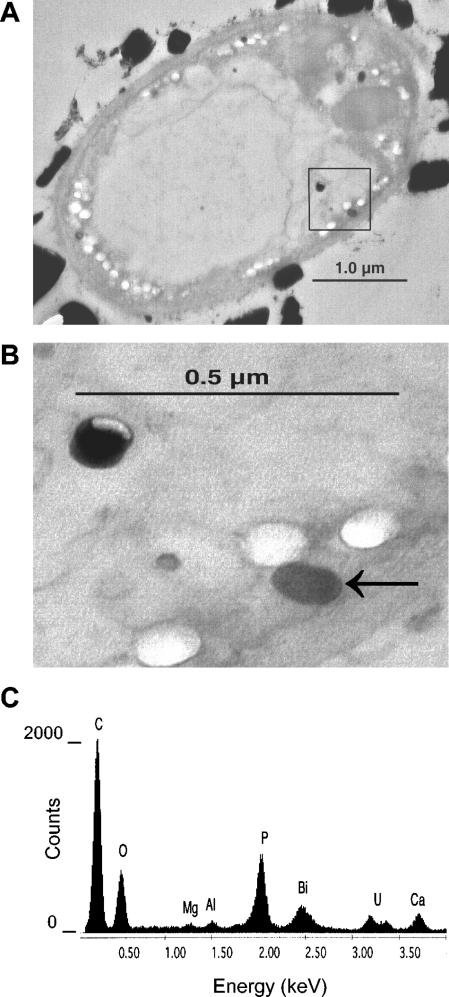
TEM-EDS analysis of ultrathin (60 nm) sections of Fusarium sp. strain BI was used to investigate if soluble bismuth depleted from the medium during growth was accumulated by the organism. In cross-sections of hyphal filaments grown on bismuth subsalicylate as a sole carbon source, discrete electron-dense regions were observed in the cytoplasm and central vacuole (Fig. 3A and B). Electron-transparent regions of similar size and shape to the electron-dense regions (Fig. 3A and B) likely represent the locations of electron-dense granules that were torn from or fell out of the sections. EDS analysis of the electron-dense regions revealed the presence of bismuth, phosphorus, calcium, and sometimes also magnesium and/or aluminum (Fig. 3C). Uranium was present (Fig. 3C) because the sections were stained with uranyl acetate; aluminum is a component of the culture medium and could have been accumulated by the organism during growth. The spot size of the beam used for EDS analysis was slightly larger than the diameter of the granules, and material surrounding the granule is included in the spectrum (Fig. 3C). Separate EDS analysis of areas surrounding the electron-dense granules showed that no phosphorus, bismuth, or other metals were present in these regions (data not shown). No electron-dense granules were found located in the cell walls. EDS analysis of electron-dense regions outside the cell wall showed that bismuth was present in higher concentrations and that phosphorus was present in lower concentrations in these regions than in the intracellular granules (Fig. 3A, spectrum not shown). These regions are assumed to be undissolved bismuth subsalicylate crystals, and the presence of phosphorus can be attributed to phosphate in the growth medium. In control samples grown on salicylate, intracellular electron-dense regions were also present (data not shown). EDS analysis (data not shown) of these regions revealed the presence of phosphorus and low levels of calcium and aluminum (bismuth was not detected).
FIG. 3.
TEM (A and B) and EDS (C) analysis of Fusarium sp. strain BI grown in a 1.5 mM bismuth subsalicylate suspension in minimal medium. Intracellular electron-dense granules were visible in cross-sections of hyphal filaments (A) (magnification, ×28,000). A magnified view (B) (magnification, ×45,000) of the boxed area in panel A shows that the electron-dense and electron-transparent regions are of similar size and shape. The spectrum (C) obtained from EDS analysis of the electron-dense region indicated by the arrow in panel B revealed the presence of bismuth, phosphorus, calcium, and traces of magnesium and aluminum in the granule. Uranium appears in the spectrum because the section was stained with uranyl acetate. The carbon and oxygen peaks may represent both the composition of the granule and fixed biological material around the granule, as the beam spot size was slightly larger than the granule. keV, kilo-electron volts.
Polyphosphate isolation and association with bismuth.
Inorganic polyphosphate has been identified in a wide range of organisms (23), including several species of Fusarium (11, 15, 31). Among other functions, polyphosphate is known to chelate metal ions in some biological systems (22). Polyphosphate was extracted from cells grown on bismuth subsalicylate or salicylate and was fractionated into three separate fractions: (i) short-chain soluble, (ii) long-chain soluble, and (iii) granular (10). Polyphosphate content of each fraction was determined as orthophosphate after acid hydrolysis via a colorimetric assay (39, 25).
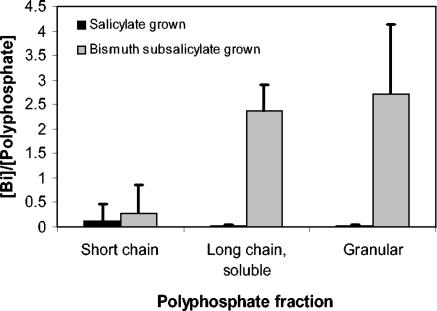
Short-chain polyphosphates were present in low concentrations, and long-chain polyphosphates were present in relatively high concentrations, which is in accordance with previous studies of fusaria (31). Each fraction was analyzed for bismuth content by ICP-OES, and the ratio of bismuth to phosphate from polyphosphate was calculated (Fig. 4). The negative control culture grown on salicylate showed only background levels of bismuth in all fractions, but polyphosphate levels were equivalent to those in fractions from biomass grown on bismuth subsalicylate. The long-chain-soluble and granular fractions from bismuth subsalicylate-grown biomass had the highest bismuth:phosphate ratios (Fig. 4). Average bismuth concentrations in these fractions were ∼50 times higher than the average bismuth concentrations in the short-chain fractions. In biomass grown on bismuth subsalicylate, the bismuth concentrations in the short-chain polyphosphate fractions ranged from 0.01 to 0.03 mM (average, 0.02 mM), bismuth concentrations in the long-chain-soluble polyphosphate fractions ranged from 0.91 to 1.15 mM (average, 1.03 mM), and bismuth concentrations in the granular polyphosphate fractions ranged from 0.77 to 1.31 mM (average, 1.04 mM).
FIG. 4.
Ratio of bismuth to polyphosphate in extracts of Fusarium sp. strain BI biomass grown on salicylate and bismuth subsalicylate. Biomass was ground and extracted to yield fractions containing polyphosphate of different chain lengths (10). Bismuth concentrations were determined in each fraction by ICP-OES. Polyphosphate concentrations were determined colorimetrically (39) as orthophosphate released after acid hydrolysis of polyphosphate in each fraction (25). Error bars represent the standard deviations of three replicates.
The TEM-EDS analysis showed that accumulated bismuth was localized in phosphate-rich granules located primarily in the cytoplasm (Fig. 3). This result, coupled with the high concentrations of bismuth found in the long-chain and granular polyphosphate fractions from biomass grown on bismuth subsalicylate, suggests that at least some of the bismuth ions accumulated by Fusarium sp. strain BI during degradation of bismuth subsalicylate are complexed with polyphosphate. This interaction may have a role in bismuth tolerance in this organism. Experiments conducted in vitro have shown that Bi3+ can complex with polyphosphate (43). To our knowledge, no previous reports of bismuth accumulation in polyphosphate granules in a biological system have been published.
Degradation of the salicylate moiety.
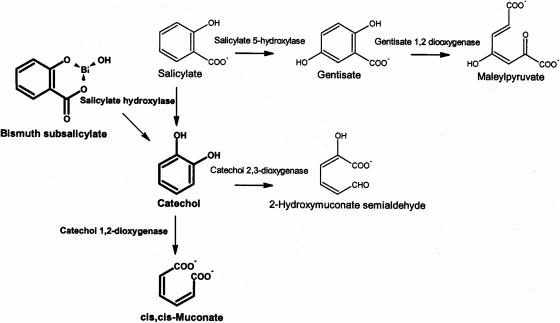
Bismuth subsalicylate is composed of a Bi3+ ion chelated by a salicylate molecule (Fig. 5). An organism utilizing this compound as a carbon source would be expected to degrade it through one or more of the known salicylate catabolic pathways (Fig. 5). Fusarium sp. strain BI grew with salicylate, gentisate, or catechol as a sole carbon source (Fig. 6). During growth of Fusarium sp. strain BI on bismuth subsalicylate or salicylate, no degradative intermediates accumulated in the culture medium as detected by UV/visible light spectrophotometry. Crude extracts of Fusarium sp. strain BI grown on salicylate and glucose were assayed for specific activities of the enzymes in the predicted pathways, with hexokinase activity assayed as a control (Table 1). Catechol 1,2-dioxygenase activity was induced eightfold in cultures grown on salicylate compared to cultures grown on glucose (Table 1). Catechol 2,3-dioxygenase activity was not detected in either preparation (Table 1). Gentisate 1,2-dioxygenase activity was equivalent in extracts from cultures grown on salicylate or glucose (Table 1). These results suggest that Fusarium sp. strain BI degrades salicylate primarily through a catechol intermediate, with ring cleavage catalyzed by a catechol 1,2-dioxygenase (Fig. 5). By extension, this is proposed as the pathway through which Fusarium sp. strain BI degrades bismuth subsalicylate (Fig. 5).
FIG. 5.
Known pathways for salicylate catabolism in fungi and/or bacteria. Intermediates in the proposed primary pathway of bismuth subsalicylate degradation by Fusarium sp. strain BI are shown in boldface.
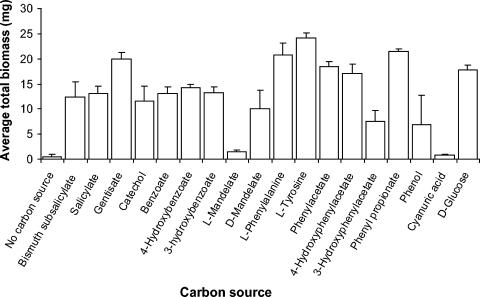
FIG. 6.
Total dry biomass of Fusarium sp. strain BI grown for 96 h on 0.25 mmol of various substrates as sole carbon sources. Error bars represent the standard deviation of three replicates.
TABLE 1.
Specific activities of enzymes in crude extracts of Fusarium sp. strain B1 grown on different substratesa
| Growth substrate | Sp act (nmol min−1 mg protein−1)
|
||||
|---|---|---|---|---|---|
| Salicylate hydroxylase | Catechol 1,2- dioxygenase | Catechol 2,3- dioxygenase | Gentisate 1,2- dioxygenase | Hexo- kinase | |
| Salicylate | 108 ± 15 | 228 ± 43 | ND | 58 ± 20 | 8 ± 2 |
| Glucose | ND | 29 ± 14 | ND | 33 ± 43 | 192 ± 28 |
Activity was measured by NADH oxidation (decrease in A340 nm) for salicylate hydroxylase, production of cis,cis muconate (increase in A260 nm) for catechol 1,2-dioxygenase, production of maleylpyruvate (increase in A375 nm) for catechol 2,3-dioxygenase, substrate depletion (decrease in A321 nm) for gentisate 1,2-dioxygenase, and NADP+ reduction (increase in A340 nm) for hexokinase. Values are means (± standard deviation) of three to five replicates from single extracts. ND, no activity detected.
The pathway for salicylate degradation by Fusarium sp. strain BI described above follows previously described pathways of salicylate degradation by yeasts and filamentous fungi: conversion of salicylate to catechol by salicylate hydroxylase and cleavage of the aromatic ring by catechol 1,2-dioxygenase (Fig. 5) (2, 40). This pathway links to the Krebs cycle via β-ketoadipate (2, 40). Catechol 2,3-dioxygenase activity is well documented in bacteria (24), but we found only one report of this activity in a fungus (37). Conversion of salicylate to gentisate by salicylate 5-hydroxylase has been described for bacteria (17), but to our knowledge it has not been described for fungi. We did not find previously published descriptions of salicylate degradation by fusaria. Cain et al. reported a Fusarium oxysporum strain that did not grow on salicylate but grew well on catechol (9). Catechol and gentisate are intermediates in other catabolic pathways that do not include salicylate (18), and catechol 1,2-dioxygenase activity and gentisate 1,2-dioxygenase activity have been detected in fusaria grown on other aromatic substrates (6).
Growth on other aromatic compounds.
Fusarium sp. strain BI could grow using a variety of aromatic compounds, primarily aromatic acids, as sole carbon sources. Measurable growth was detected on 12 of 14 compounds tested (in addition to bismuth subsalicylate, salicylate, catechol, and gentisate), with negative results only on l-mandelate and cyanuric acid (Fig. 6). Fusaria have previously been reported to grow on benzoic acid, 3-hydroxybenzoic acid, 4-hydroxybenzoic acid, phenol, phenylacetic acid, and 3-phenylacetic acid (3, 6).
Acknowledgments
We thank Gib Ahlstrand for TEM-EDS operation and image processing, Russell Anderson for performing ICP-OES analysis, James Groth for interpretation of spore morphology, Corby Kistler for gene sequencing recommendations, and Mike Sadowsky for technical advice and assistance.
This work was supported by Department of Energy grant no. DE-FG02-01ER63268 and National Institutes of Health National Institute of General Medical Sciences Biotechnology Training Grant no. GM08347.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. J., and S. Dagley. 1980. Catabolism of aromatic acids in Trichosporon cutaneum. J. Bacteriol. 141:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barz, W. 1971. Über den abbau aromatischer verbindungen durch Fusarium oxysporum schlecht. Arch. Mikrobiol. 78:341-352. [PubMed] [Google Scholar]
- 4.Bentley, R., and T. G. Chasteen. 2002. Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol. Mol. Biol. Rev. 66:250-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmeyer, H. U., M. Grassl, and H.-E. Walter. 1983. Reagents for enzymatic analysis, p. 222-223. In H. U. Bergmeyer, J. Bergmeyer, and M. Grassl (ed.), Methods of enzymatic analysis, 3rd ed., vol. 2. Verlag Chemie, Weinheim, Germany.
- 6.Boominathan, K., and A. Mahadevan. 1989. Dissimilation of aromatic substances by fungi. Zentralbl. Mikrobiol. 144:37-45. [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Burrell, R. E., C. T. Corke, and R. G. Goel. 1983. Fungitoxicity of organoantimony and organobismuth compounds. J. Agric. Food Chem. 31:85-88. [Google Scholar]
- 9.Cain, R. B., R. F. Bilton, and J. A. Darrah. 1968. The metabolism of aromatic acids by micro-organisms. Metabolic pathways in the fungi. Biochem. J. 108:797-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, J. E., H. Beegen, and H. G. Wood. 1986. Isolation of intact chains of polyphosphate from “Propionibacterium shermanii” grown on glucose or lactate. J. Bacteriol. 168:1212-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebel, J. P., and S. Muller. 1958. Cytochemical investigations of the inorganic polyphosphates present in living organisms. III. Presence of polyphosphates in some lower organisms. Exp. Cell Res. Suppl. 15:36-42. [DOI] [PubMed] [Google Scholar]
- 12.Endo, G., and S. Silver. 1995. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 177:4437-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadd, G. M. 1993. Tansley review No. 47. Interactions of fungi with toxic metals. New Phytol. 124:25-60. [Google Scholar]
- 14.Glass, N. L., and G. C. Donaldson. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61:1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbik, L. T., and I. A. Ellanskaya. 1981. Metachromatin inclusions in conidia and hyphae cells of Fusarium lk. ex fr. species. Mikrobiol. Zh. 43:342-346. [PubMed] [Google Scholar]
- 16.Grill, E., E.-L. Winnacker, and M. H. Zenk. 1986. Synthesis of seven different homologous phytochelatins in metal-exposed Schizosaccharomyces pombe cells. FEBS Lett. 197:115-120. [Google Scholar]
- 17.Grund, E., B. Denecke, and R. Eichenlaub. 1992. Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4. Appl. Environ. Microbiol. 58:1874-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harper, D. B. 1977. Fungal degradation of aromatic nitriles. Enzymology of C-N cleavage by Fusarium solani. Biochem. J. 167:685-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji, G., and S. Silver. 1992. Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI1258. J. Bacteriol. 174:3684-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kacchy, A. N., and V. V. Modi. 1976. Catechol metabolism in Pseudomonas aeruginosa: regulation of meta-fission pathway. Ind. J. Exp. Biol. 14:163-165. [PubMed] [Google Scholar]
- 21.Kappe, R., C. Fauser, C. N. Okeke, and M. Maiwald. 1996. Universal fungus-specific primer systems and group-specific hybridization oligonucleotides for 18S rDNA. Mycoses 39:25-30. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg, A. 1995. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J. Bacteriol. 177:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulaev, I. S. 1979. The biochemistry of inorganic polyphosphates, p. 17-21. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 24.Lee, Y. L., and S. Dagley. 1977. Comparison of two dioxygenases from Pseudomonas putida. J. Bacteriol. 131:1016-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenz, B., and H. C. Schröder. 1999. Methods for investigation of inorganic polyphosphates and polyphosphate-metabolizing enzymes. Prog. Mol. Subcell. Biol. 23:217-239. [DOI] [PubMed] [Google Scholar]
- 26.Mahony, D. E., S. Lim-Morrison, L. Bryden, G. Faulkner, P. S. Hoffman, L. Agocs, G. G. Briand, N. Burford, and H. Maguire. 1999. Antimicrobial activities of synthetic bismuth compounds against Clostridium difficile. Antimicrob. Agents Chemother. 43:582-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manhart, M. D. 1990. In vitro antimicrobial activity of bismuth subsalicylate and other bismuth salts. Rev. Infect. Dis. 12(Suppl. 1):S11-S15. [DOI] [PubMed] [Google Scholar]
- 28.Marshall, B. J., J. A. Armstrong, G. J. Francis, N. T. Nokes, and S. H. Wee. 1987. Antibacterial action of bismuth in relation to Campylobacter pyloridis colonization and gastritis. Digestion 37(Suppl. 2):16-30. [DOI] [PubMed] [Google Scholar]
- 29.McClellan, W. D. 1947. Response of Gladiolus to corm treatments. N. Am. Gladiolus Counc. Bull. 41:20. [Google Scholar]
- 30.Middelhoven, W. J. 1993. Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeastlike fungi. Antonie Leeuwenhoek 63:125-144. [DOI] [PubMed] [Google Scholar]
- 31.Muller, S., and J. P. Ebel. 1958. Study of condensed phosphates contained in various fungi. Bull. Soc. Chim. Biol. 40:1153-1161. [PubMed] [Google Scholar]
- 32.Murafuji, T., Y. Miyoshi, M. Ishibashi, A. F. Mustafizur Rahman, Y. Sugihara, I. Miyakawa, and H. Uno. 2004. Antifungal activity of organobismuth compounds against the yeast Saccharomyces cerevisiae: structure-activity relationship. J. Inorg. Biochem. 98:547-552. [DOI] [PubMed] [Google Scholar]
- 33.Nadeau, O. W., D. W. Gump, G. M. Hendricks, and D. H. Meyer. 1992. Deposition of bismuth by Yersinia enterocolitica. Med. Microbiol. Immunol. 181:145-152. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, P. E., M. C. Dignani, and E. J. Anaissie. 1994. Taxonomy, biology, and clinical aspects of Fusarium species. Clin. Microbiol. Rev. 7:479-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick, R. P., and C. Roth. 1968. Plasmid-linked resistance to inorganic salts in Staphylococcus aureus. J. Bacteriol. 95:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Donnell, K., H. C. Kistler, E. Cigelnik, and R. C. Ploetz. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 95:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, T. R., N. Hameed, and A. M. Martin. 1990. Initial steps of phloroglucinol metabolism in Penicillium simplicissimum. Arch. Microbiol. 153:438-443. [Google Scholar]
- 38.Sadler, P. J., H. Li, and H. Sun. 1999. Coordination chemistry of metals in medicine: target sites for bismuth. Coord. Chem. Rev. 186:689-709. [Google Scholar]
- 39.Saheki, S., A. Takeda, and T. Shimazu. 1985. Assay of inorganic phosphate in the mild pH range, suitable for measurement of glycogen phosphorylase activity. Anal. Biochem. 148:277-281. [DOI] [PubMed] [Google Scholar]
- 40.Shailubhai, K., N. N. Rao, and V. V. Modi. 1982. Degradation of benzoate and salicylate by Aspergillus niger. Ind. J. Exp. Biol. 20:166-168. [PubMed] [Google Scholar]
- 41.Sox, T. E., and C. A. Olson. 1989. Binding and killing of bacteria by bismuth subsalicylate. Antimicrob. Agents Chemother. 33:2075-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoltenberg, M., M. Martiny, K. Sorensen, J. Rungby, and K. A. Krogfelt. 2001. Histochemical tracing of bismuth in Helicobacter pylori after in vitro exposure to bismuth citrate. Scand. J. Gastroenterol. 36:144-148. [PubMed] [Google Scholar]
- 43.Thilo, E. 1962. Condensed phosphates and arsenates. Adv. Inorg. Chem. Radiochem. 4:1-75. [Google Scholar]
- 44.Toussoun, T. A., and P. E. Nelson. 1976. Fusarium: a pictorial guide to the identification of Fusarium species, 2nd ed. The Pennsylvania State University Press, University Park, Pa.
- 45.Vainio, E. J., and J. Hantula. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104:927-936. [Google Scholar]
- 46.Wilson, J. W., and E. M. M. Blair. 1926. A combination of bismuth and sodium sulphite affording an enrichment and selective medium for the typhoid-paratyphoid groups of bacteria. J. Pathol. Bacteriol. 29:310-311. [Google Scholar]
- 47.Yamamoto, S., M. Katagiri, H. Maeno, and O. Hayaishi. 1965. Salicylate hydroxylase, a monooxygenase requiring flavin adenine dinucleotide. J. Biol. Chem. 240:3408-3413. [PubMed] [Google Scholar]