Abstract
Most heterotrophic bacteria assimilate CO2 in various carboxylation reactions during biosynthesis. In this study, assimilation of 14CO2 by heterotrophic bacteria was used for isotope labeling of active microorganisms in pure cultures and environmental samples. Labeled cells were visualized by microautoradiography (MAR) combined with fluorescence in situ hybridization (FISH) to obtain simultaneous information about activity and identity. Cultures of Escherichia coli and Pseudomonas putida assimilated sufficient 14CO2 during growth on various organic substrates to obtain positive MAR signals. The MAR signals were comparable with the traditional MAR approach based on uptake of 14C-labeled organic substrates. Experiments with E. coli showed that 14CO2 was assimilated during both fermentation and aerobic and anaerobic respiration. The new MAR approach, HetCO2-MAR, was evaluated by targeting metabolic active filamentous bacteria, including “Candidatus Microthrix parvicella” in activated sludge. “Ca. Microthrix parvicella” was able to take up oleic acid under anaerobic conditions, as shown by the traditional MAR approach with [14C]oleic acid. However, the new HetCO2-MAR approach indicated that “Ca. Microthrix parvicella,” did not significantly grow on oleic acid under anaerobic conditions with or without addition of NO2−, whereas the addition of O2 or NO3− initiated growth, as indicated by detectable 14CO2 assimilation. This is a metabolic feature that has not been described previously for filamentous bacteria. Such information could not have been derived by using the traditional MAR procedure, whereas the new HetCO2-MAR approach differentiates better between substrate uptake and substrate metabolism that result in growth. The HetCO2-MAR results were supported by stable isotope analysis of 13C-labeled phospholipid fatty acids from activated sludge incubated under aerobic and anaerobic conditions in the presence of 13CO2. In conclusion, the novel HetCO2-MAR approach expands the possibility for studies of the ecophysiology of uncultivated microorganisms.
Several isotope-based methods have been introduced in recent years for cultivation-independent characterization of active microorganisms in environmental samples. The novel methodologies include direct isotope analysis of extracted biomarkers, including amino acids, fatty acids, and nucleic acids (9, 10, 24, 32, 36), stable isotope probing (SIP) of DNA or RNA (25, 33), and a new isotope microarray (1). Microautoradiography (MAR) in combination with fluorescence in situ hybridization (FISH) has also been developed for cultivation-independent identification of active bacteria in environmental matrices (3, 21, 29, 30). When targeting heterotrophic bacteria, the traditional MAR approach has been based on the addition of typically 14C- or 3H-labeled organic substrates to environmental samples under defined incubation conditions. Labeled substrate and/or labeled degradation products are then taken up by active heterotrophs and often assimilated into various biomass components. MAR based on inorganic 14CO2 as the labeled precursor has been used successfully for years to target autotrophic organisms, e.g., chemolithotrophic nitrifiers from activated sludge (21) and autotrophic Achromatium cells from freshwater sediments (18). Combined with FISH, the current MAR approach often provides excellent information about activity and identity at the single-cell level in complex environments.
Advances in isotope labeling strategies may further expand the potential applications of the MAR approach. For example, isotope labeling of metabolic active heterotrophic bacteria may be improved by using 14CO2 as isotope source. This suggestion is based on the old observation that most, if not all, heterotrophic organisms assimilate CO2 during biosynthesis in various carboxylation reactions induced by enzymes such as pyruvate carboxylase, phosphoenolpyruvate carboxylase, coenzyme A carboxylase, etc. (5, 13, 41). This phenomenon, often described as “heterotrophic CO2 assimilation,” has been used previously for quantification of microbial activity in environmental samples (35, 38, 39), as a measurer of perturbations by xenobiotic compounds (20), and for autoradiographic detection of growing bacteria (34). The majority of these studies on heterotrophic CO2 assimilation have been inspired by Romanenko (35). However, heterotrophic CO2 assimilation has received somewhat less attention recently in microbial ecology.
Assimilation of 14CO2 in heterotrophic bacteria was visualized by autoradiography as early as 1961, but the scale of autoradiography at that time was reported to range between 0.1 and 1 mm (34). With the tools available today, the resolution of MAR is around a single cell (ca. 1 μm). Hence, 14CO2 labeling combined with visualization by MAR-FISH may provide new insights regarding the function and identity of uncultivated heterotrophs.
Some filamentous bacteria in activated sludge, including “Candidatus Microthrix parvicella” are extremely difficult to isolate (7, 40). “Ca. Microthrix parvicella” is common in activated sludge wastewater treatment systems, where it causes serious foam problems (4, 7). Unfortunately, the organism is hard to grow and the physiology is poorly understood which has made it difficult to establish efficient control measures. Different in situ techniques including the traditional MAR approach has shown that “Ca. Microthrix parvicella” grows mainly (or only) on long-chain fatty acids such as oleic acid (4, 28). Furthermore, “Ca. Microthrix parvicella” can accumulate oleic acid and presumably form storage compounds under anaerobic conditions without nitrite and nitrate present. Under subsequent aerobic conditions, these storage compounds may be used to support growth. However, it remains unclear whether “Ca. Microthrix parvicella” can use nitrite or nitrate as electron acceptor or to what extent oleic acid is used as a growth substrate under anaerobic conditions in the absence of nitrite or nitrate. These are questions of potential great importance for understanding the competitiveness of the organism in activated sludge systems. However, these questions are difficult to address by using current in situ techniques, including the traditional MAR-method.
In the present study, we examined assimilation of 14CO2 by Escherichia coli and Pseudomonas putida in order to optimize conditions for single-cell detection by FISH combined with MAR (HetCO2-MAR). Subsequently, the HetCO2-MAR approach was used for studying the physiology of “Ca. Microthrix parvicella” in activated sludge under different substrate and electron acceptor regimes.
MATERIALS AND METHODS
Isotope labeling of pure cultures.
Pure cultures of E. coli ATCC 25922 and P. putida R1 (26) were grown in liquid mineral medium (LM medium) modified from MacDonald and Spokes (23) with the following composition (per liter): (NH4)2SO4, 0.13 g; KH2PO4, 0.2 g; CaCl2 · 2H2O, 20 mg; MgSO4 · 7H2O, 40 mg; FeNaEDTA, 3.8 mg; HEPES buffer, 4.8 g; and trace element solution (14), 1 ml. The medium was adjusted to pH 7.5 with 10 M NaOH and autoclaved. Various different electron donors and electron acceptors were added from sterile filtered stock solutions before inoculation.
Cells applied for isotope labeling were harvested from fresh cultures grown in LM medium (25°C, 150 rpm in the dark) with the same energy substrate as added during isotope labeling. After being harvested, the cells were resuspended in fresh LM medium to a final optical density measured at 600 nm of 0.5 (equivalent to ca. 2 × 108 cells ml−1 derived from microscopically enumeration) and then incubated on the bench for 1 h prior to the simultaneous addition of isotope, electron acceptor, and electron donor.
NaH14CO3 (58 mCi mmol−1; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) was added to 2 ml of cell suspension (described above) in 9.5-ml glass vials (25 μCi ml−1; 0.4 mM H14CO3−). Immediately after addition of H14CO3−, the growth was initiated by the addition of electron donor (3 mM glucose, 6 mM pyruvate, 9 mM acetate, or 0.5 g of yeast extract liter−1) and electron acceptor (10 mM NO3−). Glass vials were sealed with gas-tight thick rubber stoppers, and finally the oxygen regime was adjusted. Anaerobic conditions were obtained by repeated evacuation of headspace prior to isotope addition and subsequent injection of oxygen free N2 (99.999% purity). In parallel, cells were labeled with [14C]glucose (310 mCi mmol−1; Amersham Pharmacia Biotech), [14C]pyruvate (20 mCi mmol−1; American Radiolabeled Chemicals, Inc., St. Louis, Mo.), and [14C]acetate (57 mCi mmol−1; Amersham Pharmacia Biotech), with the same total substrate concentration as in the samples with radiolabeled H14CO3−, except that the isotope concentration was only 5 μCi ml−1. Incorporation of radioactive material during incubation was monitored by using filter count (described below).
Isotope labeling of activated sludge samples.
Activated sludge and foam (mainly filamentous bacteria) was collected at the Mou wastewater treatment plant, located 20 km east of Aalborg, Denmark, and stored overnight at 4°C. The activated sludge was diluted and mixed with foam to at final content of suspended solids of 2 g liter−1. The mixture contained many filamentous organisms, and it was almost exclusively “Ca. Microthrix parvicella” (>90%) as determined by FISH according to the method of Erhart et al. (16). In some experiments anaerobic conditions were applied, here defined as the absence of oxygen, nitrite, and nitrate. If any nitrite or nitrate was present, the sample was incubated without oxygen until nitrite and nitrate disappeared (monitored with test stickers from Merck, Darmstadt, Germany). MAR experiments were incubated with 0.17 μCi of [1-14C]oleic acid ml−1 (57 mCi mmol−1; Amersham-Pharmacia Biotech) or 27.5 μCi of H14CO3− ml−1 (Amersham-Pharmacia Biotech), which is equivalent to 0.5 mM H14CO3−. Unlabeled oleic (1 mM) was added to all samples. The background concentration of unlabeled bicarbonate in the activated sludge was ca. 4 mM. All incubations were conducted at 21°C on a rotary table at 150 rpm. All incubations were carried out in 9-ml serum bottles using a final volume of 2.0 ml. Anaerobic conditions were obtained by repeated evacuation of headspace and subsequent injection of N2 (99.999% purity). In some experiments electron acceptor (2 mM NO3−, 1 mM NO2−, or atmospheric O2) was added after 3 h of incubation. Pasteurized sample (70°C, 10 min) was applied as a negative control. Incorporation of radioactive material was monitored by using filter count as described below.
After 6 h of isotope labeling, all samples were fixed in 4% paraformaldehyde and stored at −20°C in 50% ethanol and 50% phosphate-buffered saline for later MAR analysis as described previously (21).
Parallel samples were incubated with NaH13CO3 (99 atom% 13C; Cambridge Isotope Laboratories, Andover, MS) or [18-13C]oleic acid (99 atom% 13C; Sigma-Aldrich, Milwaukee, Wis.) with the same additions of electron acceptors as described above for the radiolabeled compounds. In addition, samples were labeled with 13CO2 in the presence of 5 mg of allylthiourea (ATU) liter−1. ATU is an inhibitor of autotrophic ammonia oxidation (6). All samples labeled with stable isotopes were fixed in methanol and dichloromethane for later extraction and analysis of 13C-labeled phospholipid fatty acids (PLFAs; see below).
Quantification of radioactive incorporation (filter count).
To avoid loss of radioactive CO2, all samples were taken through gas-tight rubber stoppers with syringes. Subsamples of suspensions (100 μl) from radioactive incubations (pure culture or activated sludge) were filtered through 0.2-μm-pore-size mixed cellulose filter (Advantec MFS, Inc., Pleasanton, Calif.). Subsequently, 5 ml of 0.1 N HCl was added to the filtration unit. After 3 min of acidification, the acid was washed through the filter, and the filter was immediately transferred to a scintillation vial (20 ml) and dissolved in 10 ml of scintillation fluid (Filter-Count; Packard, Groningen, The Netherlands).
Specific radioactivity in the inorganic carbon pool.
For selected samples, an index of specific radioactivity in the inorganic carbon pool was calculated as the ratio between 14CO2 and the total CO2 in the headspace after acidification. The total concentration of inorganic carbon in the samples (LM medium or activated sludge) was determined by headspace gas chromatography after acidification as described previously (11). The radioactive 14CO2 was quantified in headspace samples after acidification by trapping CO2 in ethyleneglycolmonomethylether-ethanolamine (7:1). Trapped CO2 was transferred to 20-ml polyethylene scintillation vials (Packard). The radioactivity was quantified after the addition of 10 ml of scintillation cocktail (Ultima Gold XR; Packard) by liquid scintillation counting (Packard 1600 TR; Packard).
MAR-FISH.
MAR and FISH were carried out as previously described (3, 21) with minor modifications. Briefly, the fixed samples were washed thoroughly in 0.1 N HCl and distilled water. Prior to hybridization, small subsamples were transferred to gelatin-coated coverslips (24 by 60 mm) and immobilized by drying them at 50°C. The samples were briefly rinsed with distilled water to remove precipitates and hybridized with a mixture of fluorescently labeled (Cy3) oligonucleotide probes (Thermo Hybaid, Ulm, Germany) targeting all known Bacteria (12) as described previously (2, 12). In some samples the MAR signal from filamentous bacteria was quantified manually by enumeration of silver grains as described by Nielsen et al. (27).
13C-PLFA extraction and analysis.
Microbial lipids from activated sludge were analyzed for the abundance of 13C after incubation with NaH13CO3 or [13C]oleic acid. Lipids were extracted by using a mixture of dichloromethane and methanol as described previously (37). Phospholipids (polar lipids) were separated from other extractable lipids by silicic acid column chromatography and then subjected to mild alkaline methanolysis to form fatty acid methyl esters (37). 13C-PLFA methyl esters were then analyzed on a Finnigan Delta Plus XL gas chromatograph combustion isotope ratio mass spectrometer (ThermoQuest, Bremen, Germany). The gas chromatograph (Hewlett-Packard 6890) was equipped with a HP-5MS column (60 m by 0.25 mm [inner diameter]), and a GC/C III combustion interface. He was used as the carrier gas. Fatty acids were identified and named as described previously (19). δ13C values were determined based on authentic standards certified relative to the international standard PeeDee Belemnite.
RESULTS
Isotope labeling of test cultures.
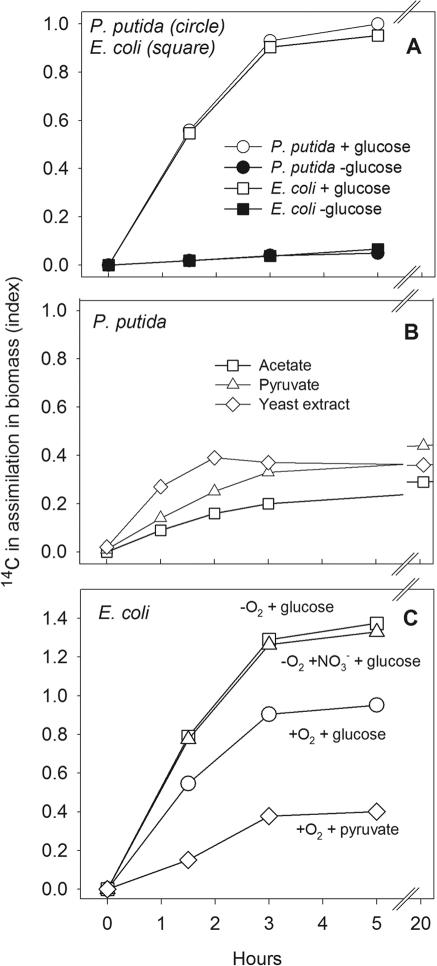
Assimilation of 14CO2 by E. coli and P. putida was measured under different growth conditions to examine the effects of selected electron donors and organic substrates on CO2 assimilation (Fig. 1). To facilitate comparisons of strains, substrates, and electron acceptors, the maximum assimilation observed when growing P. putida aerobically on glucose (3 mM) is defined as index value 1. The absolute amount of 14CO2 assimilated by P. putida at index 1 was equivalent to assimilation of ca. 8% of the added 14CO2 in 5 h (2 μCi ml−1). With an initial cell density of 2 × 108 cells ml−1, the average cell specific isotope labeling was ∼10−14 Ci cell−1 (considering equal assimilation of isotope among cells). From our experiences this is more than 1 order of magnitude above the experimental detection limit for MAR on single cells with an exposure time of 3 to 4 days.
FIG. 1.
Isotope labeling of P. putida and E. coli with 14CO2 in the presence of different substrates and oxygen regimes. P. putida after 5 h with glucose is defined as index 1.
E. coli and P. putida assimilated nearly the same amount of 14CO2 when grown under comparable conditions with 3 mM glucose (Fig. 1A). There was hardly any 14CO2 assimilation in parallel samples incubated without glucose (less than index 0.06). 14CO2 assimilation by P. putida varied slightly depending on which organic substrate was used for growth (Fig. 1B). Concentrations of acetate, pyruvate, and yeast extract were adjusted to obtain approximately the same concentration of organic carbon as in the glucose treatments shown in Fig. 1A (see Materials and Methods for details). All organic substrates supported 14CO2 assimilation, although the activity was lower than observed for glucose (less than index 0.4). No significant changes in net 14CO2 assimilation were observed by extending the incubation period from 3 to 20 h (Fig. 1B). The observed 14CO2 assimilation was comparable or greater than in parallel samples incubated without 14CO2 in the presence of [14C]glucose, [14C]acetate, or [14C]pyruvate. In these experiments, the maximum isotope labeling was index 0.6 after 9 h of incubation with [14C]glucose (data not shown).
When E. coli was grown anaerobically on glucose under fermentative conditions or with NO3− as electron acceptor (Fig. 1C), significant amounts of 14CO2 was also assimilated. No difference was seen between anaerobic samples grown with or without addition of NO3− as electron acceptor. The amount of 14CO2 incorporated under anaerobic conditions with glucose as substrate was greater than under aerobic conditions (Fig. 1C). E. coli was also grown aerobically with pyruvate (Fig. 1C), and the assimilation of 14CO2 after 3 h was close to index 0.4, which is comparable to what was observed for P. putida (Fig. 1B).
Differences in growth rates, CO2 assimilation activity, and isotope dilution resulted in variations in cell-specific 14CO2 incorporation under the different incubation conditions. Incubation for 3 h with glucose and 14CO2 resulted in a cell-specific radioactivity of ca. 0.6 × 10−14, 1.0 × 10−14, or 1.2 × 10−14 Ci cell−1 for cells incubated under aerobic conditions, anaerobic conditions with nitrate as electron acceptor, or fermentative conditions, respectively. Nonlinear dilution of 14CO2 by 12CO2 produced during mineralization of the added organic substrates made it very difficult to quantify exactly the amount of total CO2 incorporated during the different incubation conditions. For example, the isotope was diluted to 15 and 17% of the initial specific activity during aerobic incubation of P. putida and E. coli, respectively, for 3 h with glucose as the substrate. Anaerobic incubation of E. coli with glucose and NO3− resulted in a reduction to 25% of the specific activity, whereas incubation under fermentative conditions diluted the isotope to 38% of the initial specific activity. Regardless, the amounts of 14CO2 assimilated into bacterial cells during the experiments (10−15 to 10−14 Ci cell−1) were sufficient to allow clear visualization by MAR with an exposure time of 3 to 5 days (see below).
MAR of pure cultures.
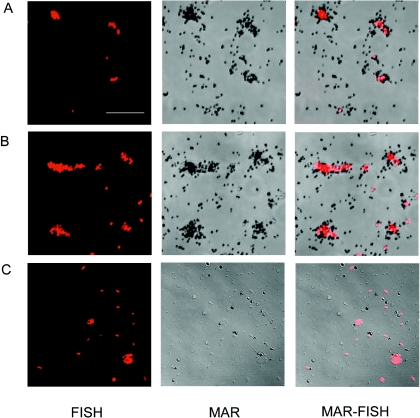
An example of MAR-positive cells is shown in Fig. 2A, where P. putida was grown aerobically on glucose in the presence of 14CO2. As a relative strong beta-emitter, the incorporated 14C produces silver grains several microns from the labeled cells, but most silver grains could be detected on top or in the very proximity of individual cells. P. putida was also MAR positive when grown on yeast extract (Fig. 2B), pyruvate, or acetate (images not shown). Furthermore, the absence of organic substrate severely reduced 14CO2 assimilation, and no MAR-positive cells were observed (Fig. 2C).
FIG. 2.
FISH, MAR, and superimposed MAR-FISH of P. putida after incubation with 14CO2. Glucose (A) and yeast extract (B) were used as substrates. (C) Negative control without any organic substrate addition. All samples were incubated at aerobic conditions. Scale bar, 10 μm.
E. coli was MAR positive after both aerobic (Fig. 3A) and anaerobic (Fig. 3B) growth on glucose in the presence of 14CO2. It is clearly seen that the MAR signal from E. coli labeled under anaerobic fermenting conditions was at least as strong as the signal from cells labeled under aerobic conditions. This is in agreement with the biomass labeling results shown in Fig. 1C. Positive MAR signals were also seen for E. coli grown with pyruvate and 14CO2 (image not shown). E. coli was MAR-negative in the absence of added organic substrates (Fig. 3C).
FIG. 3.
MAR and FISH and superimposed MAR-FISH of E. coli after incubation with 14CO2. Glucose was used as energy substrate in the presence (A) or absence (B) of oxygen. (C) Negative control without any organic substrates added. Scale bars, 10 μm.
14CO2 assimilation by activated sludge.
Activated sludge with a high occurrence of foam-forming filamentous “Ca. Microthrix parvicella” was investigated for 14CO2 assimilation under different growth conditions to identify factors important for the metabolic activity of the bacterial population. Oleic acid (1 mM) was added to the sludge, together with 14CO2. Under aerobic conditions, a significant assimilation was observed, indicating heterotrophic CO2 assimilation taking place in the activated sludge (evaluated by filter counts). After 7 h, the isotope labeling of biomass in the activated sludge was quantified to 0.43 mCi g of suspended solids−1. This corresponded to an assimilation of 3.1% of the added 14CO2 in the presence of 1 mM oleic acid. In contrast, very little 14CO2 was assimilated under anaerobic conditions (3 h), indicating insignificant heterotrophic CO2 assimilation in the presence of oleic acid (0.04 mCi g of suspended solids−1). However, when the anaerobic activated sludge was subsequently exposed to either oxygen or nitrate, substantial amounts of 14CO2 were assimilated into biomass (0.36 and 0.42 mCi g of suspended solids−1, respectively). The addition of NO2− after 3 h under anaerobic conditions induced assimilation of only 0.11 mCi g of suspended solids−1. Negative control with pasteurized sludge assimilated no detectable 14CO2 (<0.001 mCi g of suspended solids−1).
MAR of activated sludge.
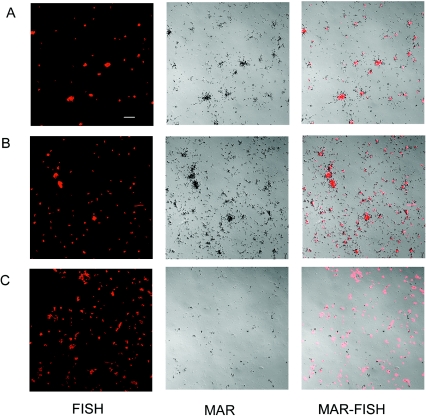
Two types of MAR approaches were conducted in order to reveal different physiological features of “Ca. Microthrix parvicella.” In the first experiment, a traditional MAR approach was carried out with [14C]oleic acid as the radiolabeled substrate. [14C]oleic acid was added to activated sludge for 4 h under aerobic and anaerobic conditions. Under both conditions, filamentous “Ca. Microthrix parvicella” were MAR positive (data not shown), indicating an active uptake of [14C]oleic acid under both aerobic and anaerobic conditions. However, a different result appeared when the HetCO2-MAR approach with a combination of 14CO2 and unlabeled oleic acid was used. “Ca. Microthrix parvicella” filaments were MAR positive under aerobic conditions (Fig. 4A) but MAR negative under anaerobic conditions (Fig. 4B). The MAR signal was quantified by counting the silver grain density along the filaments. In the presence of oxygen, 1.46 silver grains μm−1 were observed along the MAR-positive filaments (Table 1), whereas the numbers of silver grains along the filaments incubated in the absence of oxygen (Fig. 4B) were not significantly different from the background (Table 1). In addition to the filamentous “Ca. Microthrix parvicella,” some nonfilamentous bacteria also assimilated sufficient 14CO2 to be MAR positive under aerobic conditions (Fig. 4A).
FIG. 4.
MAR of filamentous “Ca. Microthrix parvicella” in activated sludge after incubation with 14CO2 in the presence of unlabeled oleic acid under aerobic (A) and anaerobic (B) conditions. Scale bars, 10 μm.
TABLE 1.
Quantification of MAR signal from “Ca. Microthrix parvicella” in activated sludge after 14CO2 labeling in the presence of oleic acida
| Treatment | No. of silver grains μm−1 ± SD |
|---|---|
| O2 (7 h) | 1.46 ± 0.36 |
| No O2 (3 h) → O2 (4 h) | 1.27 ± 0.22 |
| No O2 (3 h) → No O2 + NO3− (4 h) | 1.34 ± 0.46 |
| No O2 (3 h) → No O2 + NO2− (4 h) | 0.23 ± 0.20 |
| No O2 (7 h) | 0.04 ± 0.10 |
Expressed as described previously by Nielsen et al. (27). SD indicates the standard deviation between enumerated filaments.
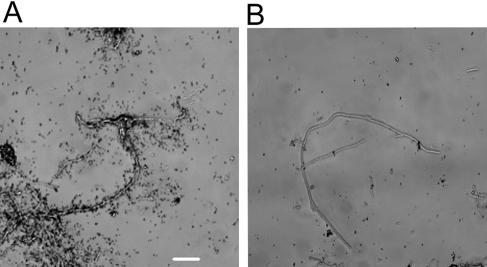
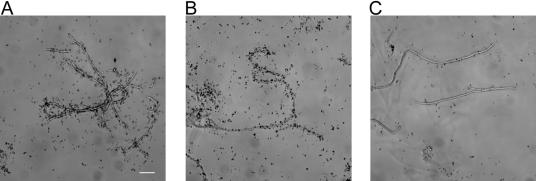
To examine whether “Ca. Microthrix parvicella” take up oleic acid under anaerobic conditions for storage purposes, which may subsequently be used for growth under aerobic conditions, assimilation of 14CO2 was investigated after incubation of sludge samples with oleic acid under anaerobic conditions for 3 h, followed by a shift to aerobic conditions for 4 h (Fig. 5A). Parallel experiments with [14C]oleic acid showed that all dissolved [14C]oleic acid (measured as radioactivity in filtrated samples) was assimilated after 0.5 to 1 h (data not shown). With 14CO2 as the isotope substrate, MAR-positive filaments were clearly observed after the anaerobic-aerobic transition, suggesting that the oleic acid taken up under anaerobic conditions stimulated metabolic activity and potentially growth under aerobic conditions. In order to see whether 14CO2 assimilation could also be induced by nitrite or nitrate addition, samples incubated for 3 h under anaerobic conditions with oleic acid were subsequently exposed to nitrate-reducing (Fig. 5B) or nitrite-reducing conditions (Fig. 5C). MAR-positive filaments were observed with nitrate as electron acceptor but hardly any with nitrite (Fig. 5B and C). Quantification of the MAR signals (Table 1) showed that a comparable amount of 14CO2 was assimilated after a transition from anaerobic to aerobic or nitrate-reducing conditions. The amount of 14CO2 assimilated after the transition was comparable to what was observed for filaments incubated under aerobic conditions for 7 h without transitions (Student t test, P > 0.05 [not significant]). In contrast, transition from anaerobic to nitrite-reducing conditions induced significant less assimilation of 14CO2 (Fig. 5C and Table 1), which was nevertheless above the MAR signal observed for samples incubated for 7 h in the absence of oxygen (Student t test, P < 0.01). Thus, the results show that oleic acid was most likely taken up and stored under anaerobic conditions without induction of significant growth as indicated by heterotrophic CO2 fixation. Stored oleic acid or oleic acid derivatives (e.g., lipids) were then able to stimulate metabolic activity and likely growth when oxygen or nitrate was supplied as electron acceptors.
FIG. 5.
MAR of filamentous “Ca. Microthrix parvicella” in activated sludge labeled with 14CO2 in the presence of oleic acid under anaerobic conditions (3 h), followed by the addition of oxygen (A), nitrate (B), or nitrite (C). Scale bar, 10 μm.
13C-PLFA profiles.
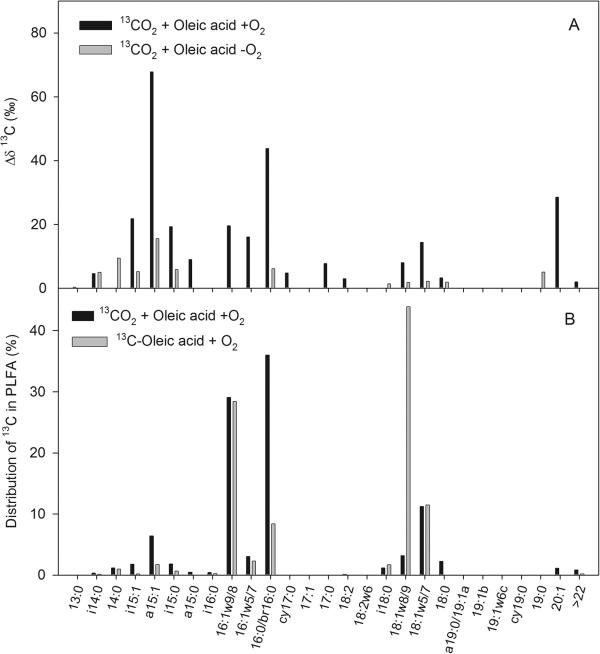
Activated sludge samples with filamentous “Ca. Microthrix parvicella” were incubated with 13CO2 and oleic acid to investigate the assimilation of CO2 into microbial biomass. 13C-PLFA profiles confirmed that 13CO2 was incorporated into cell macromolecules such as phospholipids (Fig. 6). Incubation of sludge samples in the presence of 13CO2 and ATU (an inhibitor of autotrophic ammonia-oxidizing bacteria), had little effect on the resulting 13C-PLFA profiles. Only three fatty acid groups representing 16:0 and 16:1 species were less labeled in the presence of ATU than without ATU (data not shown). These fatty acids represent PLFAs that are also produced by some autotrophic ammonia-oxidizing bacteria (8). Since the majority of 13C-enriched PLFAs were not affected by ATU inhibition, it may be concluded that heterotrophic rather than chemo-litho-autotrophic CO2 assimilation appeared to dominate in the sludge samples. This may be due partly to the low initial concentration in the sludge of energy sources for typical autotrophs such as ammonia oxidizers (i.e., an NH4+ concentration of <0.01 mM).
FIG. 6.
[13C]PLFA profiles from activated sludge with “Ca. Microthrix parvicella” incubated with oleic acid. (A) Comparison of 13C-PLFA profiles after incubation with oleic acid and 13CO2 in the presence or absence of oxygen. Enrichment in 13C is expressed as changes in δ13C compared to control samples incubated without 13CO2 (Δδ13C). (B) Comparison of 13C-PLFA profiles after incubation with either 13CO2 and nonlabeled oleic acid or nonlabeled CO2 and [13C]oleic acid. The relative distribution of 13C incorporated into PLFAs is expressed as a percentage.
Assimilation of 13CO2 into microbial PLFAs was stimulated by the addition of oleic acid to sludge samples (Fig. 6A). However, more 13CO2 was incorporated into PLFAs under aerobic compared to anaerobic conditions, which is in agreement with the MAR results shown in Fig. 4. Comparison with control samples incubated with 13CO2 but without added oleic acid revealed that especially PLFAs representing 16:1ω8/9, 16:0, 18:1ω9/8, and 18:1ω5/7 were labeled in the presence of oleic acid. This was confirmed to a large extent by incubating aerobic sludge samples directly with [13C]oleic acid (Fig. 6B). Comparison of 13C-PLFA profiles from samples incubated with 13CO2 and unlabeled oleic acid with profiles from samples incubated with [13C]oleic acid and unlabeled CO2 revealed differences in the relative enrichments of some PLFAs but relatively few differences among which PLFAs were labeled (Fig. 6B). Thus, the majority of the labeled PLFAs in Fig. 6B likely originated from microorganisms including “Ca. Microthrix parvicella” that are associated with oleic acid metabolism in the activated sludge. Some of these microorganisms produced long-chain PLFAs representing unknown C20-C22 compounds (Fig. 6). MAR investigations showed that “Ca. Microthrix parvicella” was a dominant consumer of oleic acid in aerobic sludge, but also other bacteria consumed oleic acid under aerobic conditions. As a result, it is not known to what extent the unusual microbial fatty acids (C20-C22) can be linked directly to oleic acid consumers such as “Ca. Microthrix parvicella.”
DISCUSSION
In this study we have shown that CO2 assimilation occurred during active metabolism of different organic substrates by several heterotrophic bacteria grown under various electron acceptor conditions. This 14CO2 incorporation was sufficient for single cell detection by MAR (HetCO2-MAR).
The traditional MAR approach (when targeting heterotrophic organisms) relies on the application of radioactively labeled organic compounds supplied as substrate. In the work presented here, HetCO2-MAR combined with traditional MAR enabled differentiation between assimilation of substrate (traditional MAR) and active metabolism (HetCO2-MAR) by the target organism “Ca. Microthrix parvicella.” Hence, HetCO2-MAR used as supplement to the traditional MAR is proposed as a potential powerful tool for future studies on the ecophysiology of heterotrophic microorganisms. In addition, the HetCO2-MAR approach has a number of potential advantages compared to the traditional MAR. The traditional MAR approach is inherently limited by the availability of isotope-labeled substrates. Homogeneously isotope-labeled complex substrates is normally not commercially available, and it is often difficult and expensive to label active organisms, which only respond to complex organic substrates. In the present study, we showed that complex organic substrates (exemplified by yeast extract) induced significant assimilation of 14CO2, sufficient for MAR visualization on a single-cell level (Fig. 2B). In combination with FISH, the HetCO2-MAR approach opens for numerous future applications focusing on 14CO2 assimilation by metabolically active heterotrophic microorganisms using a range of nonlabeled organic substrates. The HetCO2-MAR approach also minimizes problems associated with surface adhesive or hydrophobic compounds, which may attach to bacterial surface and induce false MAR-positive cells when the traditional MAR approach is used. In addition, the HetCO2-MAR approach will be very cost-effective when screening the metabolism (but not possible assimilation without further metabolic activity) of many different single compounds in order to elucidate substrate preferences for known or unknown organisms in environmental systems.
A successful MAR experiment relies on sufficient incorporation of the radiotracer into bacterial cells. We have used an exposure time of 3 to 5 days, which requires an incorporation of ca. 10−15 Ci cell−1. To get this amount of 14CO2 into heterotrophic cells, an experimental design with optimized concentrations of unlabeled and labeled bicarbonate, incubation time, and biomass concentration is required. In experiments with E. coli and P. putida, we used relatively high biomass concentrations, which caused some isotope dilution due to CO2 production during respiration (see below). This problem can be eliminated if much lower biomass concentrations are used. In the study of activated sludge with “Ca. Microthrix parvicella”, we used very high concentrations of 14CO2 to overcome the high background concentration of bicarbonate in the activated sludge. Thus, HetCO2-MAR can be made at least as sensitive as the traditional MAR if conditions are optimized for a specific system. HetCO2-MAR may particularly increase the sensitivity (as compared to traditional MAR) in cases where large organic molecules are only available with one or a few 14C atoms.
Our results support previous suggestions that assimilation of 14CO2 is a general phenomenon in metabolically active heterotrophic microorganisms (17, 38). Heterotrophic CO2 assimilation induced by the presence of organic substrates was confirmed by filter count, MAR visualization, and analysis of PLFAs. It has been suggested previously that heterotrophic bacteria assimilate relative constant quantities of inorganic carbon during growth (35), and recently we reported an assimilation of 1.4% ± 0.7% of cell carbon produced by P. putida (38). The assimilation of CO2 has been reported partly to depend on the presence of electron acceptor (15) and to be related to variations in the concentration of external CO2 (22). In the present study we did not determine the total assimilation of CO2 relative to the biomass production. However, with an average carbon content of 1.2 × 10−14 mol of C cell−1, the observed assimilation of 14CO2 in aerobically grown P. putida with glucose (ca. 10−14 Ci cell−1) corresponds to a total CO2 assimilation of at least 3% of biomass C based on the initial specific activity (14C/12C) of 14CO2. In addition, we have clearly shown that different incorporations of the added 14CO2 in our experiments could primarily be explained by different dilutions of the added isotope, dependent on the production of unlabeled CO2 during the incubation. As an example, after 3 h of incubation of E. coli, the specific activity (14C/12C) of 14CO2 in anaerobic samples was more than twice the specific activity in aerobic samples due to less production of unlabeled CO2 during fermentative metabolization of glucose. Hence, the observed stimulated assimilation of 14CO2 by E. coli under anaerobic conditions (Fig. 1C) can be explained mainly by less dilution of 14CO2, since fermentative metabolism of glucose produces less CO2 than aerobic metabolism.
Use of 14CO2 for isotope labeling of active heterotrophic microorganisms requires that CO2 assimilation activity correlate with cell metabolic activity. Our results clearly showed that a significant assimilation of CO2 only took place when organic substrate was added to cultures of E. coli or P. putida. A small amount of 14CO2 was assimilated without added substrates (less than index 0.06, Fig. 1), but assimilation was not sufficient to obtain a positive MAR signal with the exposure time applied (Fig. 2C and 3C). A low assimilation of CO2 in the absence of added organics could be due to use of internal storage compounds or organic exudates released from starved or dead cells. Hence, our observations suggest that heterotrophic CO2 assimilation is substrate responsive and that starved or metabolic inactive bacteria only assimilate small amounts of CO2. Nonetheless, we highly recommend including control experiments without the addition of organic substrate in HetCO2-MAR experiments with environmental samples. Background assimilation of CO2 may take place either due to the metabolism of storage products or organic substrates present in the sample or may be linked to autotrophic activity.
In HetCO2-MAR experiments, 14CO2 added to the external medium will diffuse into bacterial cells until isotope equilibrium is approached. Hence, all cells (active and inactive) may contain 14CO2 after the incubation. Gray et al. (18) elegantly showed that thoroughly acidification of the cells before MAR exposure removed dissolved and precipitated CO2 inside and around the cells. Pearl et al. (31) reported that several species of heterotrophic bacteria precipitated carbonates in marine stromatolites (laminated lithified CaCO3) and that these species contributed to the formation of the stromatolites. Thus, it cannot be ruled out that intracellular precipitation of less-soluble [14C]carbonates may lead to positive MAR signals in rare cases. However, we did not observe MAR-positive cells in a range of control experiments with pasteurized samples or in experiments without the addition of energy sources or electron acceptors. These experiments suggested that thorough acidification prior to MAR exposure did remove all inorganic radiocarbon from the cells (including precipitates). Hence, it is expected that intracellular precipitation of carbonates is a minor problem for the HetCO2-MAR approach. This is supported by the observation that metabolically active heterotrophs assimilate isotope labeled CO2 into organic macromolecules such as PLFAs (Fig. 6). Furthermore, this is in agreement with previous studies suggesting that heterotrophs assimilate CO2 into biomass components such as PLFA (38) and RNA (1). These compounds, and other organic macromolecules, will not be affected by an acidification prior to MAR exposure and will lead to visualization of cells that were de facto metabolically active.
The ecophysiological study of the filamentous bacterium “Ca. Microthrix parvicella” in activated sludge showed that this organism could take up oleic acid under anaerobic conditions, but the activity was not associated with a detectable increase in metabolic activity, as indicated by heterotrophic CO2 assimilation. Heterotrophic CO2 assimilation depends on both anabolic and catabolic processes and is stimulated during cell growth (38). Hence, our results support previous suggestions indicating that “Ca. Microthrix parvicella” is able to take up oleic acid under anaerobic conditions and form storage compounds (e.g., lipids) without initiating balanced growth (4, 28). The stored oleic acid or oleic acid derivatives are then able to support cell growth if oxygen becomes available (4, 28). This hypothesis was supported by the combined results from the traditional MAR and HetCO2-MAR experiments, and it was further confirmed by the severely attenuated incorporation of 13CO2 into PLFAs under an aerobic conditions (Fig. 6A). Quantitative MAR results supported these findings and also suggested that nitrate could be used as an efficient electron acceptor in the absence of oxygen (Table 1). This was further supported by nitrate-stimulated 13CO2 incorporation into PLFAs in the absence of oxygen (measured as δ13C in PLFAs [data not shown]). Under nitrite-reducing conditions, however, only a small amount of 14CO2 was incorporated, indicating that “Ca. Microthrix parvicella” was not able to oxidize the storage product with nitrite as an electron acceptor. Quantitative results based on heterotrophic assimilation of CO2 must be interpreted with caution, since changes in metabolism (e.g., electron acceptor) may lead to different cascades of carboxylations in the biosynthesis. However, the observed trends are in agreement with pure culture studies where “Ca. Microthrix parvicella” seem to be able to reduce nitrate to nitrite (40). This metabolic feature (anaerobic storage of long-chain fatty acid, followed by growth with oxygen or nitrate) can probably explain why “Ca. Microthrix parvicella” grows extremely well in nutrient removal plants under alternating conditions with oxygen and nitrate present as an electron acceptor.
In summary, the novel HetCO2-MAR approach made it possible for the first time on a single-cell level to distinguish better between uptake and storage of organic compounds and metabolism that initiates true growth. This was clearly illustrated by the unique information on electron acceptors preferences by “Ca. Microthrix parvicella” that was obtained with the HetCO2-MAR approach. To our knowledge, no other methods available would possibly answer this type of question on a single-cell level. We suggest that the HetCO2-MAR approach will expand the possibilities for studying the ecophysiology of uncultivated heterotrophic microorganisms.
Acknowledgments
This project was financed by Danish Technical Research Council postdoc grant 26-03-0036 and the framework program Activity and Diversity in Complex Microbial Systems. Additional support to M.H. from the R98 Foundation is greatly appreciated.
We thank Marianne Stevenson, Jane Ildal, Kirsten Maagaard, and Elizabeth Andersen for skilled technical assistance.
REFERENCES
- 1.Adamczyk, J., M. Hesselsoe, N. Iversen, A. Lehner, P. H. Nielsen, M. Schloter, P. Roslev, and M. Wagner. 2003. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 69:6875-6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microb. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreasen, K., and P. H. Nielsen. 1997. Application of microautoradiography to the study of substrate uptake by filamentous microorganisms in activated sludge. Appl. Environ. Microbiol. 63:3662-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreasen, K., and P. H. Nielsen. 2000. Growth of Microthrix parvicella in nutrient removal activated sludge plants: studies of in situ physiology. Water Res. 34:1559-1569. [Google Scholar]
- 5.Barker, H. A. 1941. The chemistry and metabolism of bacteria. Annu. Rev. Biochem. 10:553-580. [Google Scholar]
- 6.Bedard, C., and R. Knowles. 1989. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microb. Rev. 53:68-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackall, L. L., H. Stratton, D. Bradford, T. delDot, C. Sjorup, E. M. Seviour, and R. J. Seviour. 1996. “Candidatus Microthrix parvicella,” a filamentous bacterium from activated sludge sewage treatment plants. Int. J. Syst. Bacteriol. 46:344-346. [DOI] [PubMed] [Google Scholar]
- 8.Blumer, M., T. Chase, and S. W. Watson. 1969. Fatty acid in the lipids of marine and terrestrial nitrifying bacteria. J. Bacteriol. 99:366-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodelier, P. L., P. Roslev, T. Henckel, and P. Frenzel. 2000. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature 403:421-424. [DOI] [PubMed] [Google Scholar]
- 10.Boschker, H. T. S., and J. J. Middelburg. 2002. Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol. Ecol. 40:85-95. [DOI] [PubMed] [Google Scholar]
- 11.Chai, X. S., Q. Lou, and J. Y. Zhu. 2001. Analysis of nonvolatile species in a complex matrix by headspace gas chromatography. J. Chromatogr. A. 909:249-257. [DOI] [PubMed] [Google Scholar]
- 12.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 13.Dijkhuizen, L., and W. Harder. 1985. Microbial metabolism of carbon dioxide, p. 409-423. In H. Dalton (ed.), Comprehensive biotechnology, vol 1: the principles of biotechnology. Pergamon Press, Ltd., Oxford, England.
- 14.Donaldson, J. M., and G. S. Henderson. 1989. A dilute medium to determine population size of ammonium oxidizers in forest soils. Soil Sci. Soc. Am. J. 53:1608-1611. [Google Scholar]
- 15.Doronina, N. V., and Y. A. Trotsenko. 1984. Levels of carbon dioxide assimilation in bacteria with different pathways of C1 metabolism. Mikrobiologiya 53:885-889. [Google Scholar]
- 16.Erhart, R., D. Bradford, R. J. Seviour, R. Amann, and L. L. Blackall. 1997. Development and use of fluorescent in situ hybridization probes for the detection and identification of “Microthrix parvicella” in activated sludge. Syst. Appl. Microbiol. 20:310-318. [Google Scholar]
- 17.Fuchs, J. A., F. O. Glöckner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray, N. D., R. Howarth, R. W. Pickup, J. G. Jones, and I. M. Head. 1999. Substrate uptake by uncultured bacteria from the genus Achromatium determined by microautoradiography. Appl. Environ. Microbiol. 65:5100-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnsen, A. R., A. Winding, U. Karlson, and P. Roslev. 2002. Linking of microorganisms to phenanthrene metabolism in soil by analysis of 13C-labeled cell lipids. Appl. Environ. Microbiol. 68:6106-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, B. T., and V. I. Romanenko. 1984. Xenobiotic perturbation of microbial growth as measured by CO2 uptake in aquatic heterotrophic bacteria. J. Great Lake Res. 10:245-250. [Google Scholar]
- 21.Lee, N., P. H. Nielsen, K. Andreasen, S. Juretschko, J. L. Nielsen, K. H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analysis in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, W. K. W. 1982. Estimating heterotrophic bacterial productivity by inorganic radiocarbon uptake: importance of establishing time courses of uptake. Mar. Ecol. Prog. Ser. 8:167-172. [Google Scholar]
- 23.MacDonald, R. M., and J. R. Spokes. 1980. A selective diagnostic medium for ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 8:143-145. [Google Scholar]
- 24.MacGregor, B. J., V. Bruchert, S. Fleischer, and R. Amann. 2002. Isolation of small-subunit rRNA for stable isotopic characterization. Environ. Microbiol. 4:451-464. [DOI] [PubMed] [Google Scholar]
- 25.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Møller, S., A. R. Pedersen, L. K. Poulsen, E. Arvin, and S. Molin. 1996. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assayed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl. Environ. Microbiol. 62:4632-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen, P. H., M. A. de Muro, and J. L. Nielsen. 2000. Studies on the in situ physiology of Thiothrix spp. present in activated sludge. Environ. Microbiol. 2:389-398. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen, P. H., P. Roslev, T. E. Dueholm, and J. L. Nielsen. 2002. Microthrix parvicella, a specialized lipid consumer in anaerobic-aerobic activated sludge plants. Water Sci. Technol. 46:73-80. [PubMed] [Google Scholar]
- 29.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paerl, H. W., T. F. Steppe, and R. P. Reid. 2001. Bacterially mediated precipitation in marine stromatolites. Environ. Microbiol. 3:123-130. [DOI] [PubMed] [Google Scholar]
- 32.Pelz, O., L. A. Cifuentes, B. T. Hammer, C. A. Kelley, and R. B. Coffin. 1998. Tracing the assimilation of organic compounds using δ13C analysis of unique amino acids in the bacterial peptidoglycan cell wall. FEMS Microbiol. Ecol. 25:229-240. [Google Scholar]
- 33.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 34.Romanenko, V. I. 1961. Use of the autoradiographic method for a quantitative assay of methane-oxidizing bacteria. Mikrobiologiya 30:292-293. [Google Scholar]
- 35.Romanenko, V. I. 1964. Heterotrophic assimilation of CO2 by bacterial flora of water. Mikrobiologiya 33:610-614. [PubMed] [Google Scholar]
- 36.Roslev, P., and N. Iversen. 1999. Radioactive fingerprinting of microorganisms that oxidize atmospheric methane in different soils. Appl. Environ. Microbiol. 65:4064-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roslev, P., N. Iversen, and K. Henriksen. 1998. Direct fingerprinting of metabolically active bacteria in environmental samples by substrate specific radiolabeling and lipid analysis. J. Microbiol. Methods 31:99-111. [Google Scholar]
- 38.Roslev, P., M. B. Larsen, D. Jørgensen, and M. Hesselsoe. 2004. Use of heterotrophic CO2 assimilation as a measurer of metabolic activity in planktonic and sessile bacteria. J. Microbiol. Methods 59:381-393. [DOI] [PubMed] [Google Scholar]
- 39.Saralov, A. I., I. N. Krylova, and S. I. Kuznetsov. 1984. A modification of Sorokin's method for proportional determination of bacterial chemosynthesis and organotrophic CO2 assimilation in lakes. Mikrobiologiya 53:989-996. [Google Scholar]
- 40.Tandoi, V., S. Rossetti, L. L. Blackall, and M. Majone. 1998. Some physiological properties of an Italian isolate of “Microthrix parvicella.” Water Sci. Technol. 37:1-8. [Google Scholar]
- 41.Werkman, C. H., and H. G. Wood. 1942. On the metabolism of bacteria. Bot. Rev. VIII:1-68. [Google Scholar]