Abstract
Since the emergence of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrom Coronavirus (MERS-CoV) it has become increasingly clear that bats are important reservoirs of CoVs. Despite this, only 6% of all CoV sequences in GenBank are from bats. The remaining 94% largely consist of known pathogens of public health or agricultural significance, indicating that current research effort is heavily biased towards describing known diseases rather than the ‘pre-emergent’ diversity in bats. Our study addresses this critical gap, and focuses on resource poor countries where the risk of zoonotic emergence is believed to be highest. We surveyed the diversity of CoVs in multiple host taxa from twenty countries to explore the factors driving viral diversity at a global scale. We identified sequences representing 100 discrete phylogenetic clusters, ninety-one of which were found in bats, and used ecological and epidemiologic analyses to show that patterns of CoV diversity correlate with those of bat diversity. This cements bats as the major evolutionary reservoirs and ecological drivers of CoV diversity. Co-phylogenetic reconciliation analysis was also used to show that host switching has contributed to CoV evolution, and a preliminary analysis suggests that regional variation exists in the dynamics of this process. Overall our study represents a model for exploring global viral diversity and advances our fundamental understanding of CoV biodiversity and the potential risk factors associated with zoonotic emergence.
Keywords: coronavirus, viral ecology, bat, evolution
1. Introduction
In 2002/3, SARS-Coronavirus (SARS-CoV) emerged in the Guangdong province of southern China (Drosten et al. 2003; Ksiazek et al. 2003). It quickly spread to twenty-seven countries, infecting 8,098 people with 774 deaths, and was declared the first global pandemic of the 21st century. Bats were identified as the reservoir (Lau et al. 2005; Li et al. 2005) and probable source (Ge et al. 2013) of the outbreak. In 2012, the SARS pandemic was followed by MERS-Coronavirus (MERS-CoV), which emerged in the Middle East (Zaki et al. 2012) with 1,782 confirmed human cases and 640 deaths (as of September 2016). Camels were identified as the likely source of human infections (Reusken et al. 2013; Azhar et al. 2014); however, bats were again found to host closely related (MERS-like) viruses and are therefore assumed to be the original evolutionary source (Woo et al. 2006; Anthony et al. 2012; Corman et al. 2014a,b; Wacharapluesadee et al. 2015; Anthony et al. 2017).
Together, these outbreaks have cemented the coronaviridae as a family of zoonotic concern, and stimulated a surge in viral discovery efforts in bats [reviewed by Drexler et al. (2014)]. These efforts appear to show that almost all human CoVs have zoonotic origins or otherwise circulate in animals, including human 229E [bats (Pfefferle et al. 2009); camels (Sabir et al. 2016)], NL63 [bats (Donaldson et al. 2010; Huynh et al. 2012)], and OC43 [cattle (Vijgen et al. 2005)]. Even non-human CoVs such as porcine epidemic diarrhea virus (PEDV) may have emerged by host switching from other animals [bats (Tang et al. 2006; Huang et al. 2013)]. Overall, it seems that CoV diversity in bats is substantial (Drexler et al. 2014), that these viruses are prone to host switching (Woo et al. 2009), and that they are a current, historic, and future threat to public health.
Despite recent efforts, there are several notable gaps in our understanding of CoV diversity. Foremost, our knowledge of CoVs in resource-limited countries is poor (Drexler et al. 2014). This is particularly problematic given that many of these same areas are predicted to be hotspots of disease emergence (Jones et al. 2008). Second, there has been little effort to understand the evolutionary and ecological drivers of CoV diversity on a global scale, or to evaluate host and regional variation in the factors that contribute to the risk of emergence (e.g. host switching). Indeed, most studies to date have been somewhat limited in their geographic scope and have included only small sample sizes with little epidemiologic or ecological context. In short, there is a critical need for a more global perspective on CoV diversity.
In 2009, the PREDICT project was established, in part, to address this need. Focused on strengthening capacity and identifying viruses in wildlife at high-risk interfaces (PREDICT_Consortium 2014), this USAID Emerging Pandemic Threats (EPT) initiative worked with local partners in twenty countries across Latin America, Africa, and Asia over 5 years to better understand the current diversity of CoVs (as well as other viruses) and evaluate the factors that drive this diversity at different scales. Herein, we report a large diversity of CoV sequences (mostly from bats), show that the biogeography of bats has shaped the diversity of CoVs globally, and provide evidence to suggest there could be regional variation in host switching and the risk for zoonotic emergence.
2. Methods
2.1 Animals and samples
Bats (n = 12,333), rodents (n = 3,387), and non-human primates (n = 3,470) were humanely sampled (capture and release) from twenty ‘hotspot’ countries (Jones et al. 2008), representing central Africa (Cameroon, Gabon, Democratic Republic of Congo, Republic of Congo, Rwanda, Tanzania, Uganda), Latin America (Peru, Bolivia, Brazil, Mexico), and Asia (Bangladesh, Cambodia, China, Indonesia, Laos, Malaysia, Nepal, Thailand, Viet Nam). Samples were also collected from humans (n = 1,124) in seven countries in central Africa and Asia as a pilot to begin to explore the propensity for viral sharing with wildlife. In all twenty countries, wildlife samples were collected from ‘high risk’ interfaces, where direct or indirect contact with humans might promote zoonotic viral transmission. The selected sampling sites included areas of land-use change (deforestation, conversion to agriculture); sites in and around human dwellings; foci of ecotourism; markets restaurants and farms along the animal value chain; and areas where occupational exposure was likely (animal sanctuaries, agricultural activities). When possible, individuals were identified to lowest taxonomic order (genus and species) and assigned to an age class (adult, subadult, neonate) by the field teams. All samples from swabs (e.g. oral, urine, rectal), fluids (e.g. saliva), and tissues were collected into (1) NucliSens® Lysis Buffer (bioMérieux, Inc., Marcy-I’Étoile, France) and (2) viral transport media, and then frozen in the field in liquid nitrogen and transferred to the laboratory for storage at −80 °C. All animal sampling activities were conducted with permissions from local authorities and under the Institutional Animal Care and Use Committee at the University of California, Davis (protocol number: 16048). All human activities were reviewed and approved by the UC Davis Institutional Review Board (IRB), under protocols: 215253 and 432330.
2.2 Viral discovery
RNA was extracted from all samples, and cDNA prepared using superscript III (Invitrogen). Two broadly reactive consensus PCR assays targeting non-overlapping fragments of the orf1ab were used to detect both known and novel CoVs (Quan et al. 2010; Watanabe et al. 2010). The first [the ‘Watanabe’ assay (Watanabe et al. 2010)] amplified a ∼434 bp fragment of the RNA-dependent RNA polymerase (RdRp) corresponding to nucleotides (NTs) 15,156–15,589 in the human CoV OC43 genome (NC_005147), while the second [the ‘Quan’ assay (Quan et al. 2010)] amplified a ∼332 bp fragment of a different peptide downstream of the RdRp, corresponding to NTs 18,323–18,654. Amplified products of the expected size were cloned and sequenced (traditional Sanger dideoxy sequencing) according to standard protocols, and sequences edited manually in Geneious Pro (ver 9.1.3, Biomatters, Auckland, NZ). We note that this approach was adopted to facilitate viral discovery in resource-poor settings, which are the target of pandemic preparedness initiatives such as USAID PREDICT, and where techniques such as high throughput sequencing are largely unavailable.
2.3 Phylogenetics
Coronavirus sequences from the Quan and Watanabe datasets were aligned with reference sequences collected from GenBank using MUSCLE (Edgar 2004), followed by manual alignment in Se-Al v2.0a11 (http://tree.bio.ed.ac.uk). The best-fitting model of nucleotide substitution for each dataset was selected by Akaike’s Information Criterion (AIC) using jModeltest v2.1.5 (Darriba et al. 2012). For both alignments, the general time reversible model with a gamma distribution of rate heterogeneity was the best-fitting model. Maximum likelihood trees were constructed from each alignment using MEGA v6.06 (Tamura et al. 2013). Histograms of all pairwise sequence identities (%) were plotted, as described previously for hantaviruses (Maes et al. 2009), and used to define cut-offs between viral sequence clusters for analysis (akin to operating taxonomic units).
2.4 Diversity measures
The Earth’s surface was divided into grid cells, each 10° latitude by 10° longitude. Thirty-four of these grid cells included areas from which bats were sampled (bats were identified down to the species level in thirty cells), and twenty-seven cells had samples testing positive for CoVs. If the coordinates listed for a particular bat were on the line between grid cells, it was arbitrarily included in the grid cell with higher latitude/longitude. Viral and host species richness for each cell was calculated by determining the total number of unique virus and bat species within each cell. Alpha diversity was calculated using the Shannon index (Jost et al. 2011), which was chosen over the Gini–Simpson index because it does not place disproportionate weight on dominant species. Since we do not know how representative our sampling was, the Shannon index was deemed the best approach. The effective number of viruses and bats in each cell was then correlated using Kendall’s tau, since the values were not normally distributed. Sensitivity analyses were performed by shifting the grid cells in 1° increments and re-calculating richness and effective species numbers.
Matrices of beta diversity between each combination of grid cells were developed using the Jaccard index (Jost et al. 2011), and Mantel tests were used to determine whether the distance between grids was associated with the dissimilarity in virus and bat species. Geographic distance matrices were calculated by finding the centroid of each grid cell and calculating the geodesic distance between each pair using the R package ‘geosphere’(Geosphere 2015) and tests were performed using Spearman’s rank correlation. All analyses of diversity were conducted in R version 2.3.2.
2.5 Network model
A presence/absence matrix was constructed to show the distribution of viral sequence clusters across all species. Using the Python package NetworkX (Hagberg et al. 2008), a network model was constructed, connecting bat species to all viral clusters that were identified within that species in our data. The network is made up of thirty-one connected components, eighty-two viral clusters, eighty-five bat species, and 159 edges. Networks were plotted using Gephi using the force-directed algorithm ForceAtlas2 (Jacomy et al. 2014). Specifically, we plotted two bipartite graphs, one where hosts are colored by region and one in which they are colored by family.
2.6 Cophylogeny
The Jane (Conow et al. 2010) software tool was used for cophylogenetic reconstructions. This approach applies an a priori ‘cost scheme’ to different evolutionary events in order to test the degree of congruence between two trees (virus and host). These ‘events’ include cospeciation, duplication, host switching, and failure to diverge (or sorting) (Charleston 1998; Conow et al. 2010), which are used to create a minimal-cost reconstruction of the evolutionary history between the viruses and hosts. Another cophylogeny program, CoRe-PA (Merkle et al. 2010), uses a parameter-adaptive approach to estimate an appropriate cost scheme without any prior value assignment. For our analysis, we used Jane with the default pre-determined cost scheme where cospeciation is the null hypothesis and, therefore, the ‘cheapest’ event, with the assumption that host switching would theoretically be more costly than genetic drift within a species to which a virus is already adapted. Analysis was repeated with varied cost values in Jane to test for sensitivity to our cost scheme, as well as in CoRe-PA for robustness, and event types and costs from each reconstruction were compared between the two programs. As these changes ultimately did not significantly influence the outcome of the analysis, exact event assignments used in the final analysis were inferred solely from Jane with the default cost scheme.
The analyses were performed using all unique associations between viruses and their respective hosts (limited to bats). Alpha-CoV sequences and beta-CoV sequences were analyzed separately, since each appears to have diversified independently within bats. This gave us a total of four reconstructions (alpha-Quan, alpha-Watanabe, beta-Quan, beta-Watanabe). Only one instance of each virus–host association was included, even if multiple instances of the same association were observed (i.e. our analysis does not account for the frequency of detection). Associations were excluded if the host was not identified to the species level or if the cytochrome B sequence did not exist in GenBank or could not be amplified by PCR. The topology of the virus tree was inferred from the topology of the full tree for consistency, since subsets of sequences often produce different arrangements.
For each reconstruction, the most recent evolutionary event leading to a virus–host association was recorded. Given that sorting is particularly sensitive to missing data (it indicates a loss on the tree when in fact it may simply be an artifact of under-sampling), we have excluded this event from our analysis. We also distinguished between two potential types of host switching, based on an assumption that there could be differences in zoonotic risk between viruses that only move between closely related bats (e.g. those within the same genus) and those that are able to make more substantive jumps into distantly related species (those in other host genera or families). We therefore use ‘host switching’ to refer specifically to viral lineages that have moved into a host belonging to another genus or family, and ‘sharing’ to indicate a viral lineage that has moved into a different host species of the same genus (without speciating). We selected host genus as the cut-off to be consistent with current conventions of taxonomic hierarchy, and verified that the number of species per genus is relatively consistent between regions. We further verified that the mean genetic distance between host species is consistent between regions (i.e. that the taxonomic distance a virus has to navigate when moving between any two species is roughly similar in all three regions).
A binary regression model with logistic link function was used to investigate whether the degree of host switching varied by region. The dependent variable was host switching, and either cospeciation or sharing was the reference group. Region was the independent variable. We used the generalized estimating equations (GEE) (Liang and Zeger 1986) method in order to account for multiple event types within one family and derive a robust estimate of the standard errors. Event data for all four reconstructions were aggregated for the analysis. Where host–virus associations were duplicated, one was removed. If the duplicated event types did not agree [which can occur given the sensitivity of these methods to variation in topology and number of taxa in each tree (Conow et al. 2010)], we randomly selected one of the two possibilities and repeated the randomization 100 times. The GEE estimates from the 100 random iterations were then combined using rules established by Rubin (1987). The analysis was repeated to explore variation based on bat family (independent variable). Some families only possessed one type of event, which would introduce quasi-complete separation into the GEE model, so they were excluded from the corresponding models. However, the numbers of bat species within these excluded families were very small (<5) compared to the overall number of bat species for the whole study, so we felt this would not impact any overall trends.
2.7 Viral discovery effort
The relationship between sampling effort and viral discovery was evaluated using a log-link Poisson regression model, fitted with the count of viruses for each species as the dependent variable and the number of animals sampled as the independent variable. This model is universally applied with count data. The coefficient of the sample number variable is the log of the ratio of the expected number of viruses with (n + 1) samples collected to the expected number of viruses with n samples collected, where n can be any arbitrary positive integer. Using this estimated coefficient, we calculated the estimated numbers of viruses (viral sequence clusters) that can be found with respect to the sampling effort (i.e. all possible numbers of collected animals within a species), and they were then used to plot the estimated curve together with its 95% confidence band. We clarify that we use the term ‘expected’ to indicate the statistical expectation based on the estimate of the regression model. We acknowledge that the model fit is not as good as the model with an additional quadratic term of the independent variable, nonetheless it avoids the problem of over-fitting and realistically reflects the relationship between sampling effort and viral discovery when the number of animals collected is relatively lower.
2.8 Factors associated with CoV positivity
Due to the small numbers of positive individuals obtained for rodents, non-human primates, and humans, only bat data were analyzed for factors associated with positivity. A total of 12,333 bats were tested. This included 5,624 males and 5,767 females (922 were not sexed). Among aged bats 7,385 were adults, 1,029 were sub-adults, and 8 were neonates (3,911 were not aged). Binary logistic regression models were used to evaluate significant variables, restricting analyses to species for which ≥50 individuals were tested. Model selection was first performed in R (version 3.2.5) (R Foundation for Statistical Computing, 2008) using the R package ‘gmulti’ and the step function, and the best model determined by AIC. A robust estimation of standard error was calculated by clustering by individual Animal Identification number to account for non-independence of multiple tests from the same animal using STATA 13.1, SE (College Station, TX, USA). The independent variables included in analyses were Specimen Type (blood, feces/rectal swabs, guano, oral/rectal sample, oral/nasal swabs, tissue, urine/urogenital swabs), Host Taxon (family, sub-family, genus), Host Age (adult, sub-adult, neonate), Season (wet, dry), and Interface (animal use, human activity, land use, pristine area). Season was designated as ‘wet’ or ‘dry’ according to month and proximity to the equator for each country. Interfaces were broadly grouped according to the type and intensity of animal contact with people, specifically (1) Land Use (animals sampled in areas with crops, extractive industries, livestock activities), (2) Animal Use (hunting, markets, restaurants, trade, wild animal farms, wildlife managements, zoos/sanctuaries, handling by veterinarian/researchers), (3) Human Activities (ecotourism, in and around human dwellings), and (4) Pristine (where animal human contact was not likely). The model fit for each region was evaluated using the Hosmer–Lemeshow goodness-of-fit test and pseudo R2 values.
2.9 Virus ‘Hotspot’ Maps
The number of viral sequence clusters (richness) in each sub-clade was summarized by host family (Supplementary Table S1), and chi square tests used to evaluate whether viral sub-clade and host family were independent of each other. Bat species richness maps were then created to infer hotspots of CoV diversity. For each viral clade, spatial distribution data on all bat species belonging to associated families was obtained from the International Union for Conservation of Nature (IUCN). Using ArcGIS version 10 (ESRI 2011), bat species were quantified by counting overlapping polygons. The resulting count data were then converted to a raster and plotted. All sampled bats, as well as all identified sequence clusters within the clade in question, were then plotted over the raster data.
3. Results
3.1 Global diversity of CoVs
A total of 19,192 animals and humans were assayed for the presence of CoV by consensus PCR (cPCR) (Table 1). The majority were bats (n = 12,333), representing 282 species from twelve families. Overall, the proportion of CoV positive individuals was 8.6% in bats (n = 1,065/12,333) and 0.2% in non-bats (n = 17/6,859). In other words, over 98% of all positive individuals were bats.
Table 1.
Summary of individuals tested and number positive for at least one coronavirus by host taxa.
| Host taxa tested | No. individuals tested | No. individuals positive | No. distinct viruses detected |
|---|---|---|---|
| Bats | 12,333 | 1,065 | 91 |
| Non-Human Primates | 3,470 | 4 | 2 |
| Rodents and Shrews | 3,387 | 11 | 7 |
| Humans | 1,124 | 2 | 2 |
| Total | 19,192 | 1,082 | 100a |
aNote: Numbers do not total as two viruses were detected in two taxa.
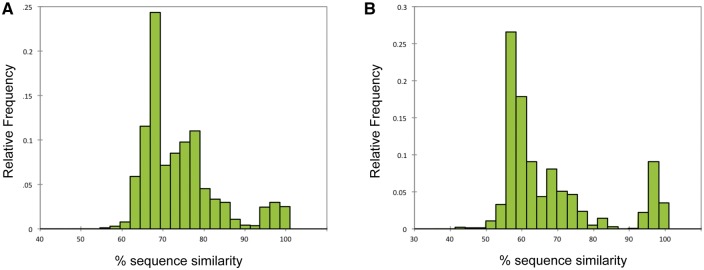
Partial sequences were obtained from two non-overlapping fragments of the orf1ab gene, yielding 654 sequences for the ‘Quan’ region and 950 for the ‘Watanabe’ region (see ‘Methods’). There was a 27% overlap, where sequences were obtained for both regions from the same sample. The distribution of pairwise sequence identities defined a 90% cut-off between taxa (Fig. 1), and the resulting monophyletic groups (in which all sequences had ≥90% identity) used as our operating taxonomic units. Groups that shared less than 90% identity to a known sequence were labeled sequentially as PREDICT_CoV-1, -2, -3 etc; while groups sharing ≥90% identity to a sequence already in GenBank were considered to be strains of a known virus and assigned the same name as the matching sequence (e.g. Kenya_bat_CoV_KY33 or HKU-9). Sequences that shared >90% identity but were found in different hosts were considered part of the same taxonomic unit. Based on these criteria, 100 discrete viral taxa were identified, ninety-one of which were found in bats (Table 1; Fig. 2). Importantly, we make no claims that these groups correspond to ‘species’ as full orf1ab replicase sequences would be required for that (King et al. 2012). Instead we clarify that we are using these partial fragments to cluster sequences into ‘operational taxonomic units’ (analogous to OTU clustering in microbiome research). We also stress that our cut-off was determined based on two discrete regions within the orf1ab separated by >3,000 nucleotides, and that these regions correspond to unique peptides post-translational cleavage.
Figure 1.
Histogram of the relative frequency of pairwise sequence identities used to define the cutoff between operating taxonomic units for all CoV sequences detected. A bimodal distribution was observed and a cutoff of 90% sequence identity used to separate sequences from both the Watanabe (Panel A) and Quan (Panel B) assays into discrete viral taxa.
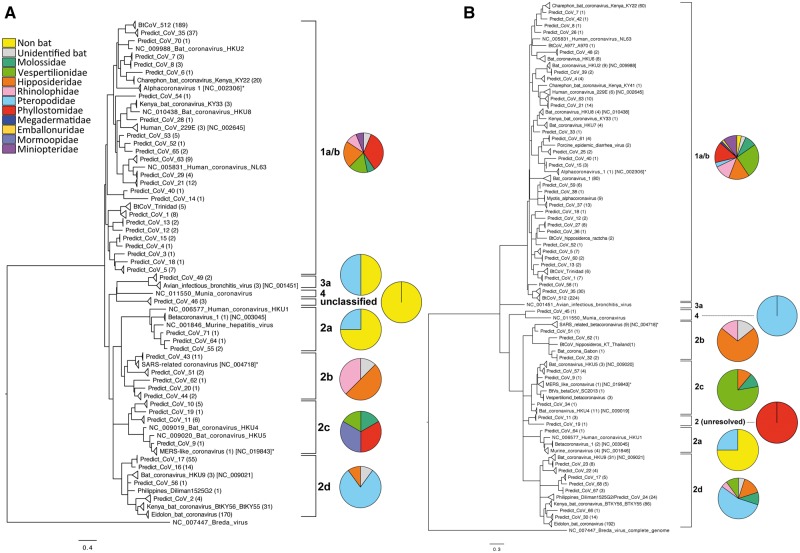
Figure 2.
Maximum likelihood phylogenetic reconstructions for all partial CoV RdRp fragments for both the Quan (Panel A) and Watanabe (Panel B) assays. Sequences are collapsed into clades, representing our operating taxonomic units (sequences sharing ≥90% identity) and number of sequences for each taxon is indicated in parentheses. Representative published sequences from GenBank have been included for comparison (GenBank accession number indicated). Both trees were rooted using the related Breda virus (NC_007447) and all nodes have ≥60% bootstrap support. Pie charts indicate the distribution of viral taxa by host (bat) family, in each virus sub-clade.
Of note was the detection of subgroup 2a CoV sequences in bats, humans and non-human primates from four different countries. This sub-clade is largely considered the rodent sub-clade (Lau et al. 2015; Wang et al. 2015; Tsoleridis et al. 2016), but here we detected sequences corresponding to the known virus betacoronavirus-1 in Pteropus medius from Bangladesh, Pteropus alecto from Indonesia, and from humans in China, and sequences corresponding to murine coronavirus in non-human primates from Nepal (these sequences were detected by four different laboratories, and no rodent samples were processed at the same time) (Fig. 2). In addition, we report the discovery of several sub-clade 2b and 2c CoV sequence clusters (the SARS and MERS sub-clades, respectively) (Fig. 2), and the detection of human coronavirus 229E-like sequences in hipposideros and rhinolophus bats sampled in ROC, Uganda, Cameroon and Gabon, supporting previous suggestions that 229E has zoonotic origins (Pfefferle et al. 2009; Hu et al. 2015). Finally, we highlight the detection of avian-associated infectious bronchitis virus-like sequences (IBV) in bats and a closely related virus (PREDICT_CoV-49) in non-human primates from Bangladesh, as well as porcine epidemic diarrhea virus (PEDV) in bats (Fig. 2). Again, we confirm that no livestock samples were processed in any of these laboratories that might have acted as a source of contamination.
3.2 Factors driving viral diversity
Viral α-diversity (the ‘effective number’ of species) was significantly correlated with bat α-diversity (tau = 0.325, P = 0.022) (Fig. 3), suggesting that more viruses will be found in regions where bat diversity is higher. This association was maintained when viral richness, rather than effective number of species, was used as the diversity index (tau = 0.421, P < 0.001). Viral and bat β-diversity were also significantly correlated (Mantel test; rho = 0.575, P < 0.001). In both cases, diversity differentiated almost entirely into three discrete communities by region (Fig. 3). Only Africa and Asia shared any viral sequence clusters (PREDICT_CoV-35 and HKU9; Fig. 4), demonstrating that bats have had a strong biogeographic influence on the global ecology and evolution of CoVs.
Figure 3.
Comparison of viral and bat diversity. The earth’s surface was divided into grid cells by latitude and longitude (10 × 10 degree units) for diversity calculations (Panel A). Grid cells where bats were sampled are numbered in each region. Alpha diversity (Shannon H) for virus (Panel B) and host (Panel C) were correlated, indicating that areas of high bat diversity also have high viral diversity. Darker cells indicate higher alpha diversity (i.e. more viral or host taxa) in each grid cell. Beta diversity was also correlated between virus (Panel D) and host (Panel E), and differentiated into three discrete communities by region—Latin America (grid cells 1–10), Africa (grid cells 11–20), and Asia (grid cells 21–34). Shading indicates that either viruses (in red) or hosts (in blue) are shared between two corresponding grid cells, with darker cells indicating higher pairwise similarity.
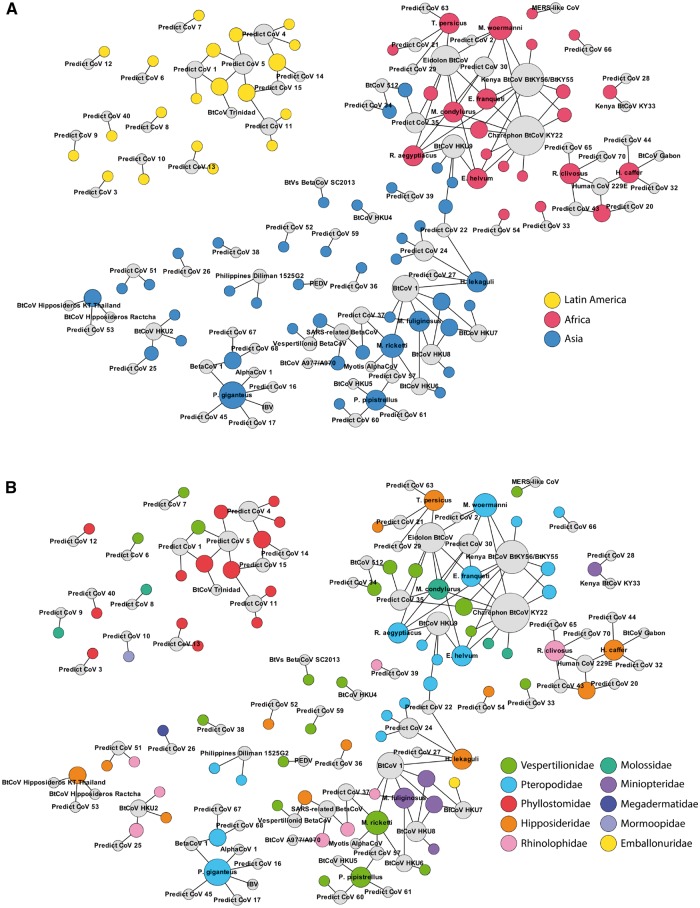
Figure 4.
Network model showing the connection of CoVs and their hosts. Viral sequence clusters (colored grey) are connected to host species, either by region (Panel A) or family (Panel B). Viral and host and communities separate almost entirely by region; only Africa and Asia are connected by two shared viruses (HKU9 and PREDICT_CoV-35) found in species from both continents. Networks also show that viruses appear to be shared by multiple host families in Africa and Asia, while being more restricted to a single family in Latin America.
Co-phylogenetic reconciliation analysis was used to investigate the evolutionary mechanisms driving the virus–host associations observed, for example host switching, viral sharing, or co-speciation (see ‘Methods’). Overall, host switching was the dominant mechanism, followed by co-speciation (Fig. 5). When analyzed by region, host switching (inter-genus transmission) remained dominant in Africa and Asia, but not in Latin America. Despite fewer host-switching events in Latin America, there was a concomitant increase in virus sharing (intra-genus transmission). This suggests that viruses are still switching hosts in Latin America but preferentially move between closely related species, while in Africa and Asia viruses cross between more distantly related species. To assess the sensitivity of our results to our chosen method (i.e. use of a pre-defined cost scheme in the program Jane), we repeated the analysis for the Quan alpha-CoV and beta-CoV reconstructions using CoRePA, a parameter-adaptive approach that estimates an appropriate cost scheme without any prior value assignment. Comparing the results, we noted 91% agreement between Jane and CoRePA in alpha-CoV reconstruction (thirty-four events, of which thirty-one agreed), and 100% agreement in the beta-CoV reconstructions (twenty-six events, all of which agreed). We further note that the cost scheme calculated in CoRePA was 0.7 for host switching and 0.1 for co-speciation—supporting the default cost scheme used in Jane (i.e. that host switching should be considered more ‘costly’ than co-speciation).
Figure 5.
Relative proportion of evolutionary events leading to observed virus:host associations for CoVs in bats. Cophylogenetic reconstructions were used to identify each event (Supplementary Fig. S1: Panels A–D), and significance evaluated by region. Across all regions, host switching was the dominant evolutionary event (Panel A). When separated by region, host switching remained dominant in Africa (Panel B) and Asia (Panel C), but not in Latin America (Panel D).
A bipartite network model connecting viral sequence clusters to their hosts supports these results, illustrating that bat CoVs were connected to several host families in Africa and Asia while being largely restricted to a single bat family in Latin America (Fig. 4, Panel B). Further, host switching was nearly four times more likely than virus sharing in Africa, when compared with Latin America (OR: 3.858; P = 0.040). CoVs in Asia were also more likely to host switch than share when compared with Latin America, however the results were not significant (OR: 2.474; P = 0.143). No particular bat family was associated with increased host switching, but this may reflect small sample sizes when the data are summarized by family.
3.3 Estimated number of CoVs in bats
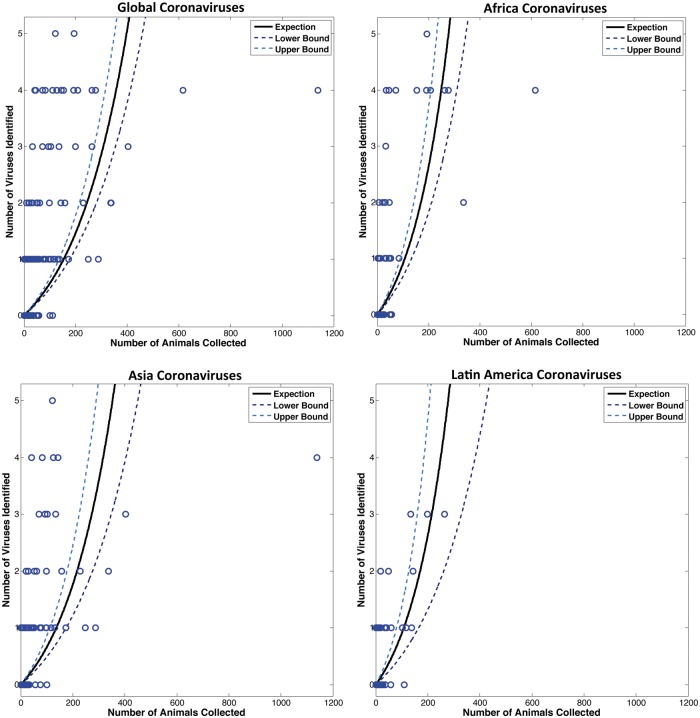
A large number of host (bat) species in our study were negative for CoV (n = 197), however none of these species were sampled extensively (Fig. 6). We found that all species with sample sizes >110 individuals were positive for one or more CoVs, suggesting that we would have detected CoVs in some of the negative species in our study if sampling effort were increased. Due to the high number of these negative species and their effect on the average number of virus sequence clusters per species, they were excluded from our estimates. By retaining only those species for which >110 individuals were sampled (there were twenty-seven species that qualified), we estimated the average number of CoVs per species to be 2.67 (std = 1.38), thus accounting for the likely scenario that some species will have more viruses and others less. We then extrapolated the average to all 1,200 bat species to estimate a total potential richness of 3,204 CoVs (range = 1,200–6,000 CoVs), most of which have yet to be described.
Figure 6.
Relationship between sampling effort and viral detection among all bat species sampled. A Poisson regression model was used to estimate the expected number of viruses based on the number of animals sampled, by species (each circle indicates a separate species). The 95% confidence intervals are indicated.
3.4 Factors predicting CoV positivity
In order to refine future surveillance efforts to find the undiscovered diversity of CoVs in bats, we evaluated the factors predicting CoV positivity. For each region, the best model included specimen type, bat family (or in the case of Latin America, subfamily), season, and animal–human interface (Table 2). Season was not included in the top model for Latin America, but the model with the season included was within two AIC (delta AIC = 1.06) and therefore considered to have as much support as the top model (Burnham and Anderson 2002). Specimen type was highly associated with CoV positivity, and samples containing feces or fecal swabs were significantly more likely to test positive than other specimen types in all regions. Bat family was important in Asia and Africa. Season was also significant, with samples collected during the dry season more likely to test positive in Africa and Asia than those collected during the wet season (albeit with low odds ratios). Age class appears to be important, as sub-adults were more likely to test positive than adults in Africa and Asia. Sex was not related to CoV positivity in Asia or Latin America while males were slightly more likely to be CoV positive in Africa (OR = 1.5, 1.2–1.9 CI, P < 0.001). Broad categories of animal–human interfaces were significantly associated with CoV positivity among bats in all three regions: in Africa and Latin America, bats sampled at the animal use interface category were more likely to be positive, and in Asia, bats sampled at the animal use and human activities interfaces were more likely to be positive than those sampled at other interfaces. The significance of the animal interface in all three regions suggests that practices around animal use may be important for disease transmission.
Table 2.
Variables associated with coronavirus positive bat tests in Africa, Asia, and Latin America. Regional datasets for each logistic regression model consisted of bat species where greater than fifty bats were tested. Age datasets were a subset of the regional datasets as age class was not determined for all individuals. Numbers in bold are statically significant for P < 0.05.
| Odds ratio | P | 95% Confidence intervals | ||||
|---|---|---|---|---|---|---|
| Africa | ||||||
| Sample type | Lower | Upper | ||||
| Feces/rectal swab | 350.98 | <0.001 | 146.64 | 840.07 | ||
| Oral/nasal swab | 5.05 | <0.001 | 2.03 | 12.54 | ||
| Oral/rectal swab | 30.98 | <0.001 | 13.46 | 71.30 | ||
| Blood | 1.65 | 0.540 | 0.33 | 8.09 | ||
| Tissue | 1.00 | |||||
| Host family | ||||||
| Hipposideridae | 1.22 | 0.559 | 0.62 | 2.41 | ||
| Pteropodidae | 3.84 | <0.001 | 2.27 | 6.49 | ||
| Molossidae | 1.00 | |||||
| Season | ||||||
| Dry | 2.42 | <0.001 | 1.79 | 3.25 | ||
| Wet | 1.00 | |||||
| Interface | ||||||
| Animal use | 1.98 | 0.001 | 1.35 | 2.91 | ||
| Pristine area | 1.39 | 0.624 | 0.37 | 5.29 | ||
| Land use | 1.66 | 0.219 | 0.74 | 3.74 | ||
| Human activity | 1.00 | |||||
| Age | ||||||
| Subadult | 5.91 | <0.001 | 4.28 | 8.17 | ||
| Adult | 1.00 | |||||
| Asia | ||||||
| Sample type | ||||||
| Feces/rectal swab | 62.80 | <0.001 | 15.44 | 255.49 | ||
| Oral/nasal swab | 13.25 | <0.001 | 3.11 | 56.38 | ||
| Urine/urogenital swab | 46.92 | <0.001 | 10.85 | 202.80 | ||
| Guano | 22.12 | <0.001 | 3.66 | 133.56 | ||
| Tissue, Oral/rectal swab | 1.00 | |||||
| Host family | ||||||
| Emballonuridae | 0.31 | 0.26 | 0.04 | 2.37 | ||
| Miniopteridae | 9.78 | <0.001 | 5.69 | 16.81 | ||
| Pteropodidae | 2.16 | <0.001 | 1.29 | 3.62 | ||
| Rhinolophidae | 1.08 | 0.82 | 0.57 | 2.03 | ||
| Vespertilionidae | 3.67 | <0.001 | 2.18 | 6.18 | ||
| Hipposideridae | 1.00 | |||||
| Season | ||||||
| Dry | 1.49 | 0.03 | 1.03 | 2.16 | ||
| Wet | 1.00 | |||||
| Interface | ||||||
| Animal use | 3.30 | 0.01 | 1.29 | 8.40 | ||
| Human activity | 3.48 | 0.01 | 1.36 | 8.95 | ||
| Pristine area | 1.81 | 0.35 | 0.52 | 6.35 | ||
| Land use | 1.00 | |||||
| Age | ||||||
| Subadult | 1.84 | 0.00 | 1.26 | 2.67 | ||
| Adult | 1.00 | |||||
| Latin America | ||||||
| Sample type | ||||||
| Feces/rectal swab | 15.66 | 0.000 | 5.02 | 48.79 | ||
| Oral/rectal swab | 27.86 | 0.000 | 6.86 | 113.22 | ||
| Oral/nasal swab | 1.00 | |||||
| Host subfamily | ||||||
| Carolliinae | 6.95 | 0.06 | 0.90 | 53.76 | ||
| Stenodermatinae | 5.04 | 0.12 | 0.67 | 37.89 | ||
| Glossophaginae | 1.00 | |||||
| Interface | ||||||
| Animal use | 5.25 | 0.01 | 1.48 | 18.65 | ||
| Human activity | 2.73 | 0.12 | 0.77 | 9.69 | ||
| Land use | 4.22 | 0.10 | 0.78 | 22.95 | ||
| Pristine area | 1.00 | |||||
| Season | ||||||
| Dry | 1.30 | 0.52 | 0.58 | 2.89 | ||
| Wet | 1.00 | |||||
| Age | ||||||
| Subadult | 1.73 | 0.15 | 0.83 | 3.62 | ||
| Adult | 1.00 | |||||
3.5 Future sampling effort
Based on our study, we were able to estimate the sampling effort that would be required to find all CoVs in bats, based on a Poisson regression model of our data (Fig. 6). With 154 individuals, we estimate that an average of one CoV will be detected (95% CI 136–177). With 397 individuals, we estimate that up to five CoVs will be detected (95% CI 351–458). As we did not observe any species with greater than five viral sequence clusters, we make the assumption that sampling 397 individuals should capture the full diversity of CoVs in each bat species.
3.6 Predicting the distribution of unknown CoVs
Viral sub-clade and host family were not independent of each other (χ2 by clade = P < 0.001; Supplementary Table S1), demonstrating that different clades have significant associations with specific bat families. For example, subgroup 2d CoVs were significantly associated with pteropid bats, and were only identified in regions where pteropid bats exist. Likewise, subgroup 2b CoVs were associated with rhinolophid and hipposiderid bats and 2c with vespertilionid bats. To ‘predict’ the potential distribution of unknown CoVs, we plotted the known distribution of bats belonging to these families and assume that ‘hotspots’ of bat diversity infer hotspots of viral diversity for each sub-clade (Fig. 7). We note that the alphaCoV genus has been considered as one group, since they do not resolve well into sub-clades (de Groot et al. 2009), and that no 2a map was generated given that only one bat virus from this clade was identified.
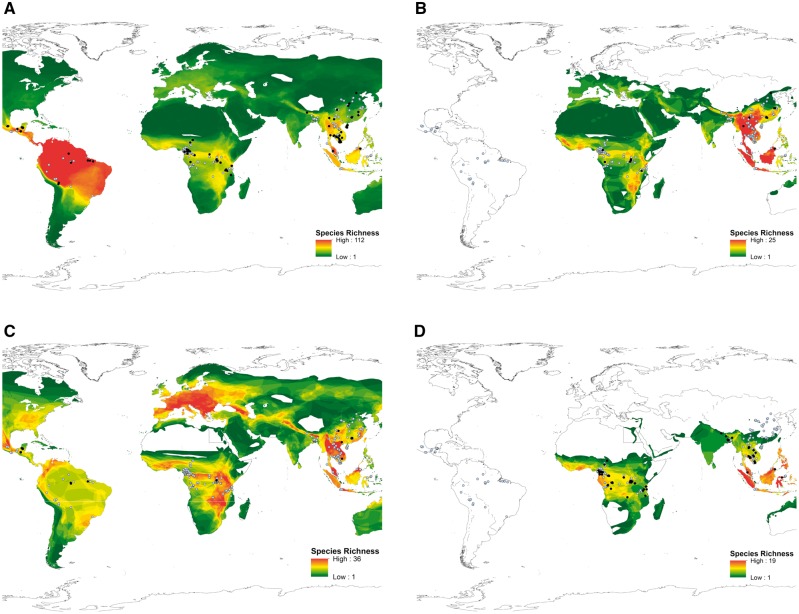
Figure 7.
Viral diversity ‘hotspot’ maps. Panel A shows the potential hotspots for CoVs based on the distribution of bats worldwide. Location of alpha CoV sequences from this study are shown in black and beta CoV sequences in blue, indicating there is no geographical bias based on viral genus (i.e. alpha and beta CoVs are equally likely to be found in all regions). Some viral sub-clades were associated with particular bat families, and the spatial distribution data of all species belonging to these families were plotted to indicate the potential hotspots of viral diversity (richness) for these sub-clades. Panel B indicates the potential distribution of 2b CoVs based on the distribution of rhinolphus and hipposideros bats. Locations of 2b-positive animals identified in this study are indicated in black, and correlate with areas of high species richness (for these families). CoV-positive animals for other sub-clades shown in light blue. Panel C indicates the potential distribution of 2c CoVs based on the distribution of vespertilionid bats. Locations of 2c-positive animals identified in this study are indicated in black. This map suggests there are hotspots of 2c diversity in regions not covered in this study (e.g. Europe). Panel D indicates the potential distribution of 2d viruses based on the distribution of pteropid bats. The map suggests these viruses may have a more limited distribution, compared with viruses of other sub-clades. Locations of 2d-positive animals identified in this study are shown in black.
4. Discussion
The emergence of SARS and MERS has driven a need to understand more about the diversity, ecology and evolution of coronaviruses, particularly at so-called ‘hotspots’ of zoonotic emergence (Jones et al. 2008; Drexler et al. 2014). To this end, we surveyed the diversity of CoVs from twenty countries in Latin America, Africa, and Asia to identify global factors driving viral diversity and to look for regional differences in factors that contribute to the risk of emergence, such as host switching. In total, we identified sequences from 100 discrete phylogenetic clusters, ninety-one of which were found in bats.
Our data suggest that the diversity of bat CoVs has been driven primarily by host ecology. First, viral richness was strongly correlated with bat richness, suggesting that most CoVs will be found in regions where bat diversity is highest. Second, we showed that CoV diversity separates into three distinct communities by region, echoing the distribution of bats and suggesting an ecological dependence on their hosts. And third, we identified particular associations between viral sub-clade and bat family, indicating that CoVs have evolved with (or adapted to) preferred families. Collectively, these data show that the global diversity and distribution of CoVs in bats is non-random and is driven by variation in the biogeography of bats.
Regional variation was also observed in the proportion of host switching events, relative to other evolutionary mechanisms such as co-speciation, duplication, or sharing (see Methods for definitions). Overall, host switching was the dominant mechanism, supporting the general trend observed for CoVs (Vijaykrishna et al. 2007; Woo et al. 2009; Lau et al. 2012) as well as similar findings for other viral families, including paramxyoviruses (Melade et al. 2016), hantaviruses (Ramsden et al. 2009), and arenaviruses (Coulibaly-N'Golo et al. 2011; Irwin et al. 2012). However, when analyzed by region, there were proportionally fewer events in Latin America compared with Africa or Asia. Given that host switching is the first critical step in zoonotic emergence (Li et al. 2005;Pfefferle et al. 2009; Woo et al. 2012; Azhar et al. 2014), we suggest this finding could reflect regional differences in the risk of disease emergence.
It is important to note that we distinguish ‘host switching’ (inter-genus or family switching) from ‘sharing’ (intra-genus switching) in our analysis and that while the proportion of host switches was lower in Latin America, the proportion of sharing events was concomitantly higher. This indicates that CoVs in this region still switch hosts, just like they do in Africa and Asia, but that they preferentially move between closely related species. While we do not yet understand the factors driving this difference, if we assume that the risk of spillover to humans reflects the taxonomic distances that CoVs are able to jump (Kreuder Johnson et al. 2015), our results would indicate a higher risk for zoonotic emergence in Africa and Asia [acknowledging that host switching is not the only important factor driving zoonotic emergence (Jones et al. 2008; Keesing et al. 2010; Karesh et al. 2012)]. While evidence of regional variation in host switching has not been reported previously for viruses (to our knowledge), similar patterns have been observed in hemosporidian parasites (Ellis et al. 2015).
In total, we estimated that there are at least 3,204 CoVs (based on the cluster definition used in this study) in bats. This suggests that although there is still a large number of coronaviruses that remain to be discovered, their detection remains an achievable goal, and we advocate for their discovery given the potential for even greater insights into the global ecology and evolution of these viruses (e.g. further investigation into regional differences in host switching), and the opportunity to elucidate their individual zoonotic potential (Ge et al. 2013; Wang et al. 2014; Yang et al. 2014; Anthony et al. 2017). Building on our study, we suggest that future surveillance efforts should consider the number of samples carefully and include up to 400 individuals (of either sex) per species in order to maximize the chance of detecting all CoVs, and that including less than 154 individuals per species might reduce the chance of finding even one virus and would likely produce a poor return on investment. We also recommend that feces (or rectal swabs) followed by saliva (or oral swabs) should be prioritized if resources are limited and all sample types cannot be processed, and that sampling should be biased towards immature animals and in the dry season.
To predict where this unknown diversity is likely to be, we plotted the known distribution of all bat families associated with each CoV sub-clade. The 2b SARS clade was associated with rhinolophid and hipposiderid bats, so we plotted the distribution of all bat species belonging to these families to generate a map of viral diversity (richness). Based on this map, we would expect the greatest diversity of unknown 2b viruses to be found in South East Asia, which is consistent with previous studies describing 2b viruses in bats, but also in isolated pockets throughout Africa. In contrast, the 2c MERS clade was associated with vespertilionid bats, predicting that 2c diversity will be highest in Mexico, Europe, Central and South East Africa, and parts of South East Asia. Again, this is consistent with the distribution of 2c viruses reported here, and previously in the literature (Woo et al. 2007; Reusken et al. 2010; Anthony et al. 2012; Lelli et al. 2013; Corman et al. 2014a,b; Anthony et al. 2017 ).
Certain limitations should be considered in the interpretation of our data. First, the diversity of CoV sequences detected is almost certainly biased by our sampling effort. A small number of species were sampled abundantly, thus maximizing our chances of finding a CoV (27/282 species had >110 individuals); however, most were under-sampled (232/282 with ≤50 individuals). While this biases the probability of finding a CoV towards the more abundantly sampled species, we stress that it also reflects the uneven distribution of naturally occurring communities (Magurran 2011), and that targeting the very rare species would be prohibitively expensive. It is also unclear if population size has an effect on the number of viruses present in a species—e.g. do larger populations accommodate more viral diversity? Second our analysis only represents the ‘last’ evolutionary event identified. It does not represent, or account for, the complete evolutionary history of these lineages. Therefore, while a single event has been ascribed to each association (see ‘Methods’), it does not preclude other events having an equal or greater impact in the past. We are limited to using this most recent event type in order to assign a single evolutionary event to each unique association. It should also be noted that events are dependent on the relationships observed in the reconstructed trees and could change as more viruses are added or longer sequences are used. Third, we have only generated partial sequences for analysis, which limits our ability to comment on the specific zoonotic potential of each virus or provide a comprehensive analysis of their genetic histories and taxonomy, for example, estimating the frequency or impact of recombination, which can be an important driver of host switching and disease emergence (Anthony et al. 2017). We certainly support full-genome sequencing wherever possible [and have such efforts underway (Anthony et al. 2017)], however difficulties in virus isolation or sequencing directly from swabs (where material and viral load can be low, yielding variable results) can often preclude extending sequences much beyond the short fragments amplified by consensus PCR (Drexler et al. 2014). Equally, logistic and permitting issues in moving biological samples across borders, and the need to first develop in-country capacity to fully sequence these viruses, have all limited our ability to characterize (most of) these viruses further.
Our ‘state of the art’ is in the unique scope of our study, focusing on a single viral family at a global scale. To achieve this, the USAID PREDICT project first had to establish a strategy for viral discovery in twenty different countries, many of which had no prior capacity to do this work at all. For this reason, we used a simple yet highly implementable approach based on cPCR. Initially, this approach might not appear to be as powerful as high throughput sequencing (HTS) techniques; however, this study has demonstrated the particular and considerable strengths of cPCR as an affordable screening and discovery tool in resource-poor settings, where HTS is still largely unsupported.
Studying viral diversity on a global scale is just one component of a larger strategy to understand the factors driving viral diversity. Ecological and evolutionary mechanisms can contribute differently across scales, and we therefore advocate complimenting the global perspective with studies that focus on entire viral communities (rather than only one viral family) within single individuals or host species (rather than only on a global scale) (Anthony et al. 2015). We propose that it is critical to understand both the component parts of the ‘zoonotic pool’ (Morse et al. 2012) and the nature of their interactions at different scales if we are to move towards better predictive models, and ultimately to reducing zoonotic emergence.
Finally, we offer a specific comment regarding the public health threat posed by bats. While it is tempting to conclude that bats harbor a large number of potentially zoonotic CoVs, most of the putative viruses detected in this study are unlikely to pose any threat to humans—either because they lack the biological pre-requisites to infect human cells or because the ecology of their hosts limits the opportunity for spillover. Studies such as this are intended to advance our understanding of the fundamental biology of viruses, not to create alarm or incite the retaliatory culling of bats. Indeed, such actions often have unanticipated consequences; and can even enhance disease transmission, as seen with rabies in vampire bats (Streicker et al. 2012; Blackwood et al. 2013). Bats are important insectivores, pollinators and seed dispersers and we underscore both their vital ecosystem role and the need to consider any public health interventions carefully.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
Supplementary Material
Acknowledgements
This study was made possible by the generous support of the American people through the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT project (cooperative agreement number GHN-A-OO-09-00010-00). We thank the governments of Cameroon, Gabon, Democratic Republic of Congo, Republic of Congo, Rwanda, Tanzania, Uganda, Peru, Bolivia, Brazil, Mexico, Bangladesh, Cambodia, China, Indonesia, Laos, Malaysia, Nepal, Thailand, and Viet Nam for permission to conduct this study, and the field teams and collaborating laboratories that performed sample collection and testing.
References
- Anthony S. J. et al. (2012) ‘Coronaviruses in bats from Mexico’, Journal of General Virology, 94: 1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S. J., et al. (2015) ‘Non-random patterns in viral diversity’, Nature Communications, 6: 8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S. J., et al. (2017) ‘Further evidence for bats as the evolutionary source of MERS coronavirus’, mBio, 8/2. pii: e00373-17. doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E. I. et al. (2014) ‘Evidence for camel-to-human transmission of MERS coronavirus’, The New England Journal of Medicine, 370: 2499–505. [DOI] [PubMed] [Google Scholar]
- Blackwood J. C. et al. (2013) ‘Resolving the roles of immunity, pathogenesis, and immigration for rabies persistence in vampire bats’, Proceedings of the National Academy of Sciences of the United States of America, 110: 20837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham K. P., Anderson D. R. (2002) Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. New York: Springer. [Google Scholar]
- Charleston M. A. (1998) ‘Jungles: a new solution to the host/parasite phylogeny reconciliation problem’, Mathematical Biosciences, 149: 191–223. [DOI] [PubMed] [Google Scholar]
- Conow C. et al. (2010) ‘Jane: a new tool for the cophylogeny reconstruction problem’, Algorithms for Molecular Biology: AMB, 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M. et al. (2014a) ‘Rooting the phylogenetic tree of middle east respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat’, Journal of Virology, 88: 11297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M. et al. (2014b) ‘Characterization of a novel betacoronavirus related to middle East respiratory syndrome coronavirus in European hedgehogs’, Journal of Virology, 88: 717–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly-N'Golo D. et al. (2011) ‘Novel arenavirus sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from Cote d'Ivoire: implications for evolution of arenaviruses in Africa’, PLoS One, 6: e20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D. et al. (2012) ‘jModelTest 2: more models, new heuristics and parallel computing’, Nature Methods, 9: 772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R. J. et al. (2009) ‘Virus taxonomy: classification and nomenclature of viruses’, in King A. M. Q., Adams M. J., Carstens E. B., Lefkkowitch J. (eds) Ninth Report of the International Committee for the Taxonomy of Viruses. Amsterdam: Elsevier. [Google Scholar]
- Donaldson E. F. et al. (2010) ‘Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat’, Journal of Virology, 84: 13004–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J. F., Corman V. M., Drosten C. (2014) ‘Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS’, Antiviral Research, 101: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C. et al. (2003) ‘Identification of a novel coronavirus in patients with severe acute respiratory syndrome’, The New England Journal of Medicine, 348: 1967–76. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: multiple sequence alignment with high accuracy and high throughput’, Nucleic Acids Research, 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis V. A. et al. (2015) ‘Local host specialization, host-switching, and dispersal shape the regional distributions of avian haemosporidian parasites’, Proceedings of the National Academy of Sciences of the United States of America, 112: 11294–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESRI. (2011) ArcGIS Desktop: Release 10. Redlands, CA: Environmental Systems Research Institute.
- Ge X. Y. et al. (2013) ‘Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor’, Nature, 503: 535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans R. J., Williams E., Vennes C. (2015) Geosphere: Spherical Trigonometry v. R Package version 1.5. [Google Scholar]
- Hagberg A. A., Schult D. A., Swart P. J. (2008) ‘Exploring network structure, dynamics, and function using NetworkX’, in Varoquaux G., Vaught T., Millman J. (eds.) Proceedings of 7th Python in Science Conference (SciPy2008), pp. 11–15. Pasadena, CA. [Google Scholar]
- Hu B. et al. (2015) ‘Bat origin of human coronaviruses’, Virology Journal, 12: 221.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. W. et al. (2013) ‘Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States’, mBio, 4: e00737–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J. et al. (2012) ‘Evidence supporting a zoonotic origin of human coronavirus strain NL63’, Journal of Virology, 86: 12816–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin N. R. et al. (2012) ‘Complex patterns of host switching in New World arenaviruses’, Molecular Ecology, 21: 4137–50. [DOI] [PubMed] [Google Scholar]
- Jacomy M. et al. (2014) ‘ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software’, PLoS One, 9: e98679.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. E. et al. (2008) ‘Global trends in emerging infectious diseases’, Nature, 451: 990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L., Chao A., Chazdon R. L. (2011) ‘Compositional similarity and beta diversity’ in Magurran A. E., McGill B. J. (eds.) Biological Diversity: Frontiers in Measurement and Assessment. Oxford: Oxford University Press. [Google Scholar]
- Karesh W. B. et al. (2012) ‘Ecology of zoonoses: natural and unnatural histories’, Lancet, 380: 1936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F. et al. (2010) ‘Impacts of biodiversity on the emergence and transmission of infectious diseases’, Nature, 468: 647–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M. Q. et al. (2012) Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press. [Google Scholar]
- Kreuder Johnson C. et al. (2015) ‘Spillover and pandemic properties of zoonotic viruses with high host plasticity’, Scientific Reports, 5: 14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T. G. et al. (2003) ‘A novel coronavirus associated with severe acute respiratory syndrome’, The New England Journal of Medicine, 348: 1953–66. [DOI] [PubMed] [Google Scholar]
- Lau S. K. et al. (2005) ‘Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats’, Proceedings of the National Academy of Sciences of the United States of America, 102: 14040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K., et al. (2012) ‘Recent transmission of a novel alphacoronavirus, bat coronavirus HKU10, from Leschenault's rousettes to pomona leaf-nosed bats: first evidence of interspecies transmission of coronavirus between bats of different suborders’, Journal of Virology, 86: 11906–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K., et al. (2015) ‘Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A’, Journal of Virology, 89: 3076–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli D. et al. (2013) ‘Detection of coronaviruses in bats of various species in Italy’, Viruses, 5: 2679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. (2005) ‘Bats are natural reservoirs of SARS-like coronaviruses’, Science, 310: 676–9. [DOI] [PubMed] [Google Scholar]
- Liang K., Zeger S. L. (1986) ‘Longitudinal data analysis using generalized linear models’, Biometrika, 73: 13–22. [Google Scholar]
- Maes P. et al. (2009) ‘A proposal for new criteria for the classification of hantaviruses, based on S and M segment protein sequences’, Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 9: 813–20. [DOI] [PubMed] [Google Scholar]
- Magurran A. E. (2011) Biological Diversity: Frontiers in Measurement and Assessment. Oxford: Oxford University Press. [Google Scholar]
- Melade J. et al. (2016) ‘An eco-epidemiological study of Morbilli-related paramyxovirus infection in Madagascar bats reveals host-switching as the dominant macro-evolutionary mechanism’, Scientific Reports, 6: 23752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle D., Middendorf M., Wieseke N. (2010) ‘A parameter-adaptive dynamic programming approach for inferring cophylogenies’, BMC Bioinformatics, 11 Suppl 1: S60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S. S. et al. (2012) ‘Prediction and prevention of the next pandemic zoonosis’, The Lancet, 380: 1956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S. et al. (2009) ‘Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana’, Emerging Infectious Diseases, 15: 1377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PREDICT_Consortium. (2014) Reducing Pandemic Risk. Promoting Global Health. One Health Institute, University of California Davis, Davis, CA. [Google Scholar]
- Quan P. L. et al. (2010) ‘Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria’, mBio, 1: pii: e00208-10. doi:10.1128/mBio.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Foundation for Statistical Computing. (2008). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ramsden C., Holmes E. C., Charleston M. A. (2009) ‘Hantavirus evolution in relation to its rodent and insectivore hosts: no evidence for codivergence’, Molecular Biology and Evolution, 26: 143–53. [DOI] [PubMed] [Google Scholar]
- Reusken C. B. et al. (2010) ‘Circulation of group 2 coronaviruses in a bat species common to urban areas in Western Europe’, Vector Borne Zoonotic Dis, 10: 785–91. [DOI] [PubMed] [Google Scholar]
- Reusken C. B., et al. (2013) ‘Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study’, The Lancet. Infectious Diseases, 13: 859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D. B. (1987) Multiple Imputation for Nonresponse in Surveys. New York: John Wiley and Sons. [Google Scholar]
- Sabir J. S. et al. (2016) ‘Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia’, Science, 351: 81–4. [DOI] [PubMed] [Google Scholar]
- Streicker D. G. et al. (2012) ‘Ecological and anthropogenic drivers of rabies exposure in vampire bats: implications for transmission and control’, Proceedings. Biological Sciences/the Royal Society, 279: 3384–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. (2013) ‘MEGA6: molecular evolutionary genetics analysis version 6.0’, Molecular Biology and Evolution, 30: 2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X. C. et al. (2006) ‘Prevalence and genetic diversity of coronaviruses in bats from China’, Journal of Virology, 80: 7481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoleridis T. et al. (2016) ‘Discovery of novel alphacoronaviruses in European rodents and shrews’, Viruses, 8: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D. et al. (2007) ‘Evolutionary insights into the ecology of coronaviruses’, Journal of Virology, 81: 4012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L. et al. (2005) ‘Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event’, Journal of Virology, 79: 1595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S. et al. (2015) ‘Diversity of coronavirus in bats from Eastern Thailand’, Virology Journal, 12: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. et al. (2014) ‘Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26’, Cell Host & Microbe, 16: 328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. (2015) ‘Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China’, Virology, 474: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S. et al. (2010) ‘Bat Coronaviruses and experimental infection of bats, the Philippines’, Emerging Infectious Diseases, 16: 1217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. et al. (2006) ‘Molecular diversity of coronaviruses in bats’, Virology, 351: 180–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C., et al. (2007) ‘Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features’, Journal of Virology, 81: 1574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C., et al. (2009) ‘Coronavirus diversity, phylogeny and interspecies jumping’, Experimental Biology and Medicine, 234: 1117–27. [DOI] [PubMed] [Google Scholar]
- Woo P. C., et al. (2012) ‘Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5’, Emerging Microbes and Infection, 1: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. (2014) ‘Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus’, Proceedings of the National Academy of Sciences of the United States of America, 111: 12516–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A. M. et al. (2012) ‘Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia’, The New England Journal of Medicine, 367: 1814–20 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.