Abstract
The effects of the inorganic medium components, the initial pH, the incubation temperature, the oxygen supply, the carbon-to-nitrogen ratio, and chloramphenicol on the synthesis of cyanophycin (CGP) by Acinetobacter calcoaceticus strain ADP1 were studied in a mineral salts medium containing sodium glutamate and ammonium sulfate as carbon and nitrogen sources, respectively. Variation of all these factors resulted in maximum CGP contents of only about 3.5% (wt/wt) of the cell dry matter (CDM), and phosphate depletion triggered CGP accumulation most substantially. However, addition of arginine to the medium as the sole carbon source for growth promoted CGP accumulation most strikingly. This effect was systematically studied, and an optimized phosphate-limited medium containing 75 mM arginine and 10 mM ammonium sulfate yielded a CGP content of 41.4% (wt/wt) of the CDM at 30°C. The CGP content of the cells was further increased to 46.0% (wt/wt) of the CDM by adding 2.5 μg of chloramphenicol per ml of medium in the accumulation phase. These contents are by far the highest CGP contents of bacterial cells ever reported. CGP was easily isolated from the cells by using an acid extraction method, and this CGP contained about equimolar amounts of aspartic acid and arginine and no detectable lysine; the molecular masses ranged from 21 to 29 kDa, and the average molecular mass was about 25 kDa. Transmission electron micrographs of thin sections of cells revealed large CGP granules that frequently had an irregular shape with protuberances at the surface and often severely deformed the cells. A cphI::ΩKm mutant of strain ADP1 with a disrupted putative cyanophycinase gene accumulated significantly less CGP than the wild type accumulated, although the cells expressed cyanophycin synthetase at about the same high level. It is possible that the intact CphI protein is involved in the release of CGP primer molecules from initially synthesized CGP. The resulting lower concentration of primer molecules could explain the observed low rate of accumulation at similar specific activities.
Cyanophycin, which is also referred to as cyanophycin granule polypeptide (CGP), is a branched nonribosomally synthesized polypeptide consisting of about equimolar amounts of aspartic acid and arginine. This copolymer consists of a polyaspartate backbone, and arginine residues are linked to the β-carboxyl group of each aspartic acid by their α-amino groups (44). Purified CGP can be chemically converted to a derivative with a reduced arginine content or even to poly(aspartic acid) (19). The latter is produced by the chemical industry as a biodegradable substitute for nonbiodegradable polyacrylic acid (5, 39), which is used for many technical and medical applications (19, 35, 39). CGP occurs as insoluble inclusions in the cytoplasm and serves as a storage compound for carbon, nitrogen, and energy (24, 25). CGP is insoluble in water at physiological pH and is soluble under acidic (pH <2) or alkaline (pH >9) conditions (23). The molecular mass of CGP ranges from 25 to 100 kDa, as estimated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. It is not affected by proteases, such as pronase, pepsin, chymotrypsin, α- or β-carboxypeptidase, leucin aminopeptidase, and clostripeptidase B (34, 44, 45). However, CGP is susceptible to hydrolysis to Asp-Arg dimers by cyanophycinases. Intracellular cyanophycinases (CphB) are involved in mobilization of this storage compound (24, 31, 34), whereas extracellular cyanophycinases (CphE) degrade CGP released upon lysis from other cells (27, 28).
The key enzyme of CGP biosynthesis in cyanobacteria is cyanophycin synthetase (CphA). This enzyme covalently links the aspartate and arginine residues in a consecutive step-by-step reaction and is strictly dependent upon ATP, Mg2+, and a CGP primer (8). The corresponding cyanophycin synthetase structural genes (cphA) have been cloned from several cyanobacteria, such as Anabaena variabilis ATCC 29413, Anabaena sp. strain PCC7120, Synechocystis sp. strain PCC6803, Synechocystis sp. strain PCC6308, Synechococcus elongatus, and Synechococcus sp. strain MA19 (1, 8, 16, 29, 48). Cyanophycin synthetases have also been heterologously expressed in several heterotrophic bacteria, such as Escherichia coli, Ralstonia eutropha, Corynebacterium glutamicum, and Pseudomonas putida (3). This enabled production of CGP at a technical scale and allowed isolation of large amounts of the polyamide so that physical and material properties of this interesting biopolymer could be revealed (14). However, high-performance liquid chromatography (HPLC) analysis revealed that the hydrolyzed CGP isolated from recombinant bacteria contained, in addition to aspartate and arginine, approximately 10% lysine, which partially replaced arginine (2, 29, 48).
CGP was first discovered by Borzi (10) in cyanobacteria, and microbiology textbooks mention it as a polymer that exclusively occurs in cyanobacteria (37). However, recently, the occurrence of genes encoding proteins homologous to cyanobacterial cyanophycin synthetases has been described for other eubacteria not belonging to the cyanobacterial group, such as Acinetobacter sp., Bordetella bronchiseptica, Bordetella pertussis, Bordetella parapertussis, Clostridium botulinum, Desulfitobacterium hafniense, and Nitrosomonas europaea (20, 49). It was also shown that the cphA genes of Acinetobacter calcoaceticus strain ADP1 and D. hafniense encode functionally active enzymes and that cells of Acinetobacter accumulated CGP when they were cultivated under phosphate-limiting conditions, albeit at low levels (up to 1.4% [wt/wt] of the cell protein content) (20). The functionality of D. hafniense CphA was studied only in recombinant E. coli (49). When the Acinetobacter cphA gene was cloned and expressed in E. coli, a cyanophycin synthetase activity of 1.2 U per mg of protein was measured, and CGP accounted for up to 7.5% of the cell dry matter (CDM) (20). Interestingly, the CGP produced in recombinant E. coli or obtained by in vitro synthesis by using the purified enzyme of A. calcoaceticus did not contain any detectable lysine, indicating that the A. calcoaceticus cyanophycin synthetase exhibits much higher substrate specificity than other cyanophycin synthetases exhibit (21). It is also worth mentioning that downstream of cphA a gene that putatively encodes a cyanophycinase (cphI) was detected in the Acinetobacter genome (20). The genes for CphA and CphB are also clustered in many cyanobacteria, but with cphB preceding cphA. This study was aimed at evaluating the limits of cyanophycin synthesis and accumulation in A. calcoaceticus strain ADP1.
MATERIALS AND METHODS
Bacterial strains, plasmids, DNA fragments, and cultivation conditions.
All bacterial strains, plasmids, and DNA fragments used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Acinetobacter calcoaceticus strain ADP1 | Wild type | ATCC 33305a |
| A. calcoaceticus mutant ΔcphI | Cyanophycinase-negative mutant of ADP1, Kmr | This study |
| Escherichia coli Top10 | F−araD139 Δ(ara, leu)7697 ΔlacX74 galU galK rpsL deoR Φ80dlacZΔM15 endA1 nupG recA1 mdrA Δ(mrr hsdRMS mcrBC) | Invitrogen (San Diego, Calif.) |
| pBluescript SK(−) | Apr, lacPOZ′ | Stratagene (San Diego, Calif.) |
| pKOS3 | Apr, cphI from A. calcoaceticus ADP1 (DSM 587) colinear with lacPO | This study |
ATCC, American Type Culture Collection.
A. calcoaceticus strain ADP1 was cultivated at 30°C in complex Luria-Bertani (LB) medium (36) or in a Tris-HCl-buffered mineral salts medium (MSM) as described by Krehenbrink et al. (20). The basic MSM contained 50 mM Tris-HCl (pH 7.3), 107 mM sodium glutamate, 30 mM (NH4)2SO4, 20 mM KCl, 0.8 mM MgSO4 · 7H2O, 6 mg of FeCl3 · 6H2O per liter, 6 mg of MnCl2 · 4H2O per liter, 6 mg of ZnCl2 per liter, 6 mg of CuSO4 · 5H2O per liter, 6 mg of CaCl2 · 2H2O per liter, and 6 mg of NaCl per liter. The composition of this medium was changed as described below to study the effects of phosphate, ammonium, sulfate, carbon source, nitrogen source, carbon-to-nitrogen ratio, or effectors on cyanophycin accumulation. Cultivation was done in Erlenmeyer flasks, which were agitated on shakers at 130 rpm with a horizontal amplitude of about 5 cm.
E. coli was grown at 37°C in LB medium.
Growth of all bacterial strains was monitored by measuring the turbidity at 600 nm.
Determination of cell dry matter.
For determination of bacterial CDM, a known volume of a culture was centrifuged with a bench centrifuge at 3,500 × g and 4°C. The supernatant was carefully discarded, and the cell pellet was washed after suspension in a saline solution (0.9% [wt/vol] NaCl) by centrifugation. The cell pellet was then lyophilized, and the mass was gravimetrically determined.
Purification and analysis of CGP.
CGP was isolated from cells of A. calcoaceticus strain ADP1 by the procedure described by Simon (43). The amino acid constituents of the isolated material were determined by HPLC as described by Frey et al. (14).
A novel method was developed for routine analysis and fast determination of the CGP content of small fresh or lyophilized samples of A. calcoaceticus strain ADP1. An appropriate amount (1 to 5 mg) of cells was suspended in acetone and vigorously agitated in order to dissolve the cell wall lipids and to increase cell wall and membrane permeability. The cells were then centrifuged and washed two times with 50 mM Tris-HCl buffer (pH 7.5) to remove all soluble proteins and other compounds. To dissolve CGP, the washed cells were then suspended in 0.52 ml of 0.1 M HCl, shaken for 30 min at room temperature, and centrifuged for 10 min at 14,000 × g. Then 500 μl of the supernatant was transferred to a clean plastic tube to which 500 μl of precipitation buffer (0.1 M Tris-HCl [pH 7.5] brought to pH 12 by addition of 0.1 M NaOH) was added to precipitate the CGP from the supernatant at neutral pH. The mixture was incubated for 10 min on ice to complete CGP precipitation and was then centrifuged at 14,000 × g. The supernatant was discarded, and 500 μl of 0.1 M HCl was added to the CGP-containing pellet to dissolve the CGP again before it was centrifuged for 1 min at 14,000 × g to remove insoluble proteins. The concentration of CGP was measured spectrophotometrically by the method described by Bradford (11), as modified by Obst et al. (28).
Preparation of cell extract and assay of cyanophycin synthetase activity.
The enzyme activity of cyanophycin synthetase in the soluble cell fraction was assayed by the radiometric method described previously (1).
Electrophoresis and determination of protein concentrations.
SDS-polyacrylamide gel electrophoresis was performed in 11.5% (wt/vol) polyacrylamide gels by using the standard method (22). Proteins and cyanophycin were stained with Serva Blue R (47). The concentrations of soluble protein were determined as described by Bradford (11), while the total protein contents were determined by the method of Schmidt et al. (38).
Isolation, manipulation, and transfer of DNA.
Genomic DNA was isolated from A. calcoaceticus strain ADP1 by the procedure of Rao et al. (33). Plasmid DNA was isolated from E. coli by alkaline lysis (9). Restriction enzymes and other DNA-manipulating enzymes were used as described by the manufacturers.
For transformation of E. coli Top10, competent cells were prepared by the CaCl2 method of Hanahan (17). Transformation of A. calcoaceticus strain ADP1 was done essentially as described by Palmen et al. (30). Cells were grown in 100 ml of LB medium at 30°C for 12 h, and 1 ml of this culture was then used to inoculate 24 ml of prewarmed LB medium and incubated for 2 h at 30°C with shaking. For transformation, 500 μl of this suspension was incubated with 8 μg of linear DNA for 2 h at 30°C, and the cells were then spread on LB agar plates containing 50 μg of kanamycin ml−1.
Generation of a cphI mutant.
A 2,768-bp DNA fragment containing the cphI gene was amplified by PCR from genomic DNA of A. calcoaceticus strain ADP1 by using primers P1 (ATTTTAGATTACGGACACAATGAAG) and P2 (CGACCTTATTAAATCATGATAATGG) and Platinum Pfx DNA polymerase (Invitrogen). The PCR product was directly ligated into Eco32I-digested pBluescript SK(−) DNA (Stratagene). A recombinant plasmid (pKOS3) containing the cphI fragment in a colinear orientation with respect to the lacZ promoter was chosen and digested with Eco32I, which resulted in the loss of a 1,440-bp fragment. The digested vector was then ligated with a blunt-end kanamycin resistance cassette. By using primers P1 and P2, a fragment flanked by cphI sequences was amplified from the ligation reaction mixture and directly used to transform competent cells of A. calcoaceticus strain ADP1. PCR with primers P1 and P2 was employed to confirm the presence of a 2,308-bp fragment (corresponding to replacement of the 1,440-bp Eco32I fragment with the 980-bp kanamycin resistance cassette) and the absence of a full-length 2,768-bp cphI fragment in kanamycin-resistant clones.
Transmission electron microscopic studies.
Cells were fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate-buffered saline (PBS) (pH 7.3) for 45 min by the method of Sørensen (7). After three washes with PBS, each for 20 min, cells were postfixed in 1% (wt/vol) osmium tetroxide in 0.1 M PBS (pH 7.3) and washed once with PBS for 20 min. Then the water was removed by using a graded water-ethanol series (30, 50, 70, 90, and 96% and absolute ethanol); each step lasted about 15 min. To obtain thin sections, the samples were embedded in Spurr resin without propylene oxide (46). Sections that were 70 to 80 nm thick were cut with an ultramicrotome (Leica Mikroskopie und Systeme GmbH, Wetzlar, Germany) by using a diamond knife and were placed on a 200-mesh copper grid. Imaging was performed with an H-500 transmission electron microscope (Hitachi, Tokyo, Japan) in the bright-field mode at an acceleration voltage of 80 kV at room temperature.
RESULTS
Effects of phosphate, sulfate, or ammonium limitation and variation of the initial pH.
It was recently shown that A. calcoaceticus strain DSM 587 possesses a gene that codes for a functional active cyanophycin synthetase (CphAAc), as revealed by detection of enzyme activity and CGP in the cells of A. calcoaceticus (20). However, only small amounts of CGP were detected in the cells (the maximum amount was 1.4% [wt/wt] of the total protein), and these small amounts of CGP occurred only if there was phosphate limitation (20).
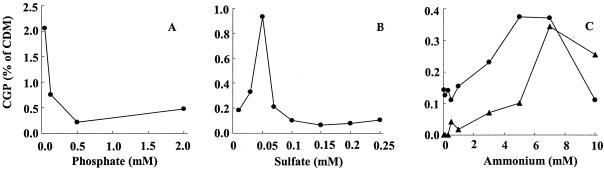
This physiological study was aimed at identification of cultivation conditions under which cells of A. calcoaceticus accumulate the maximum possible amounts of CGP. Cultivation experiments were done in Erlenmeyer flasks and by using mineral salts medium in which the concentration of one of the nutrients (PO43−, NH4+, SO42−) was varied, while all other constituents of the medium (potassium, magnesium, iron, calcium, trace elements) and of the carbon source (sodium glutamate or sodium gluconate) were kept constant. By varying the concentrations of phosphate (from 42 μM to 2.0 mM), sulfate (from 0.01 to 0.25 mM), and ammonium (from 0.1 to 10 mM), maximum amounts of CGP of only 2.2, 0.9, and 0.4% (wt/wt) of the CDM were obtained, respectively (Fig. 1). Although the amounts of CGP were small, phosphate limitation was obviously most suitable for promoting CGP biosynthesis and accumulation. Ammonium limitation gave the lowest CGP contents, as expected, since nitrogen is a major constituent of the CGP molecule. These small amounts of CGP occurred irrespective of whether gluconate or glutamate was used as the carbon source. Therefore, all further cultivation experiments were done under phosphate limitation conditions.
FIG. 1.
Effects of phosphate, sulfate, and ammonium limitation on cyanophycin accumulation in A. calcoaceticus. Cells were cultivated for 44 h at 30°C in 250-ml Erlenmeyer flasks without baffles containing 100 ml of Tris-HCl-buffered MSM with 107 mM sodium glutamate (•) or 107 mM sodium gluconate (▴) as the sole carbon source. The basic medium was prepared as described in Materials and Methods except for the nutrient investigated. At the end of the experiment, the cells were harvested by centrifugation, and the cyanophycin content was analyzed. (A) The phosphate concentration was varied between 42 μM and 2.0 mM. (B) The sulfate concentration was varied between 0.01 and 0.25 mM by varying the amount of (NH4)2SO4 added to the medium. The concentration of ammonium was kept constant at 30 mM by adding the appropriate amount of NH4Cl to the medium. (C) The ammonium concentration was varied between 0.1 and 10 mM by varying the amount of (NH4)2SO4 added to the medium. The concentration of sulfate ions was kept constant at 15 mM by adding the appropriate amount of MgSO4 · 7H2O to the medium.
Variation of the initial pH of the basic medium (containing 107 mM sodium glutamate and 42 μM Na2HPO4 for phosphate limitation) did not enhance the accumulation of CGP significantly. However, there was a tendency toward increasing CGP contents in the cells if an alkaline initial pH (pH 8 or greater) was used (data not shown).
Effect of temperature.
In addition, we varied the incubation temperature between 25 and 40°C. The optimum temperature for growth and CGP accumulation was 30°C. Therefore, all further experiments were done at this temperature.
Effect of oxygen supply and chloramphenicol.
To investigate the effect of oxygen supply, cultivation experiments were done in parallel in Erlenmeyer flasks with and without baffles. After 20 and 40 h of cultivation, the CGP contents of cells cultivated in Erlenmeyer flasks equipped with baffles were significantly higher and reached values of 1.6 and 3.4% (wt/wt) of the CDM, respectively, whereas the CGP contents were 25 and 50% lower, respectively, in cells cultivated in flasks without baffles (Table 2). Addition of chloramphenicol, an inhibitor of ribosomal protein biosynthesis known to stimulate CGP synthesis (13, 40, 42), had no significant effect on the CGP content of the cells after 20 h of cultivation. Only in cells cultivated for 40 h in Erlenmeyer flasks without baffles was a markedly higher CGP content measured in the presence of chloramphenicol (Table 2).
TABLE 2.
Effects of aeration and addition of chloramphenicol on cyanophycin synthesisa
| Incubation period (h) | Type of flask | CGP content (% of CDM)
|
|
|---|---|---|---|
| With chloramphenicol (2.5 μg/ml) | Without chloramphenicol | ||
| 20 | No baffles | 1.0 ± 0.025 | 1.2 ± 0.04 |
| With baffles | 1.8 ± 0.15 | 1.7 ± 0.03 | |
| 40 | No baffles | 3.0 ± 0.07 | 1.7 ± 0.03 |
| With baffles | 3.1 ± 0.01 | 3.4 ± 0.3 | |
Cells of A. calcoaceticus strain ADP1 were grown in 250-ml Erlenmeyer flasks with and without baffles which contained 100 ml of MSM supplemented with sodium glutamate as the sole source of carbon and were incubated at 30°C for 40 h. Chloramphenicol was added after 16 h of growth. The data are means and standard deviations of three independent experiments.
Variation of the nitrogen-to-carbon ratio.
Varying the nitrogen-to-carbon ratio in MSM by using sodium glutamate and ammonium sulfate as the carbon and nitrogen sources, respectively, and varying the absolute amounts of these two nutrients in the medium had no significant effect on the amounts of CGP accumulated by cells of A. calcoaceticus (Table 3). The amounts of CGP in the cells varied only slightly between about 1.8 and 2.8% (wt/wt) of the CDM and remained at a low level. Using other carbon sources, such as glucose, sucrose, fructose, sodium acetate, sodium citrate, and ethanol, instead of glutamate led to even lower CGP contents (between 0.2 and 0.8% [wt/wt] of the CDM). If other nitrogen sources, such as potassium nitrate (0.5%, wt/vol), whey (1.0%, wt/vol), yeast extract (0.5% wt/vol), or tryptone (0.5%, wt/vol), were used instead of ammonium sulfate, the CGP contents of the cells were even lower (between 0 and 0.4% [wt/wt] of the CDM).
TABLE 3.
Effect of variation of the carbon-to-nitrogen ratio on accumulation of cyanophycina
| Sodium glutamate/ammonium sulfate ratio (g/g) | CGP content (% of CDM) |
|---|---|
| 0.5/0.1 | 2.8 ± 0.01 |
| 1.0/0.1 | 2.3 ± 0.2 |
| 1.5/0.1 | 1.7 ± 0.5 |
| 0.5/0.2 | 2.3 ± 0.1 |
| 1.0/0.2 | 2.2 ± 0.04 |
| 1.5/0.2 | 1.8 ± 0.04 |
| 2.0/0.2 | 2.4 ± 0.06 |
| 1.0/0.4 | 1.7 ± 0.05 |
| 1.5/0.4 | 1.9 ± 0.1 |
| 2.0/0.4 | 2.1 ± 0.04 |
Cells of A. calcoaceticus strain ADP1 were cultivated for 44 h at 30°C in 250-ml Erlenmeyer flasks without baffles containing 100 ml of Tris-HCl-buffered MSM with different amounts of sodium glutamate and ammonium sulfate. The basic medium was prepared as described in Materials and Methods except for the concentration of the carbon and nitrogen source. At the end of the experiment the cells were harvested by centrifugation, and the cyanophycin content was analyzed. The data are means and standard deviations of three independent experiments.
Effects of aspartate and arginine.
Since CGP consists of about equimolar amounts of aspartate and arginine, the effects of these two amino acids on CGP accumulation were also studied. We also studied the effect of glutamate as an alternative carbon and nitrogen source and as a potential alternative constituent of CGP, as reported for a CGP-like polymer isolated from Synechocystis sp. strain PCC 6308 (26) under phosphate-limiting conditions. Aspartate (10 mM) and arginine (10 mM) were added either alone or together or in combination with 10 mM glutamate, ammonium sulfate was omitted, and phosphate was used at a concentration of 42 μM. Addition of arginine had a substantial positive effect on accumulation of CGP. Cells which were cultivated in the presence of 10 mM arginine as the sole nitrogen source accumulated CGP at levels up to 15% (wt/wt) of the CDM (Table 4). In contrast, aspartate had almost no effect. Moreover, aspartate and particularly glutamate prevented the positive effect of arginine almost completely (Table 4). The inhibitory effect of aspartate could not be overcome by using Erlenmeyer flasks equipped with baffles to provide a better oxygen supply.
TABLE 4.
Effects of aspartate, arginine, and glutamate on accumulation of cyanophycin by A. calcoaceticusa
| Substrate(s)b | CGP content (% of CDM) |
|---|---|
| Aspartate | 2.3 ± 0.04 |
| Arginine | 14.9 ± 0.16 |
| Aspartate + arginine | 4.0 ± 0.18 |
| Arginine + glutamate | 2.6 ± 0.10 |
| Aspartate + glutamate | 1.2 ± 0.08 |
| Aspartate + argininec | 4.5 ± 0.08 |
Cells of A. calcoaceticus strain ADP1 were cultivated for 44 h at 30°C in 250-ml Erlenmeyer flasks without baffles containing 100 ml of unbuffered MSM with an initial pH of 7.3 as described for the basic medium in Materials and Methods except that sodium glutamate and ammonium sulfate were omitted as the carbon and nitrogen sources respectively. At the end of the experiment, the cells were harvested by centrifugation, and the cyanophycin content was analyzed. The data are the means and standard deviations of three independent experiments.
Sodium aspartate (10 mM), arginine (10 mM), and sodium glutamate (10 mM) were added alone or in combination to the medium as carbon and nitrogen sources.
This test was performed in flasks equipped with baffles.
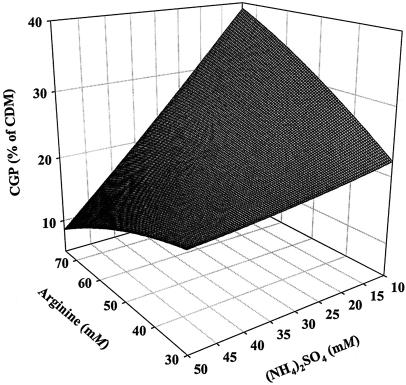
Subsequently, the positive effect of arginine on CGP accumulation was studied in more detail by increasing the concentration of arginine and varying the concentration of aspartate (Table 5). With 15 and 30 mM arginine added and in the absence of aspartate, the cells accumulated CGP at levels that were about 23 or 28% of the CDM, respectively. Furthermore, the strong inhibitory effect of aspartate on CGP accumulation occurred at either of the concentrations used (10 and 20 mM). The effect of arginine on CGP accumulation was then also studied in dependency on the concentration of ammonium sulfate by varying the concentrations of arginine and ammonium sulfate in the medium from 30 to 70 mM and from 10 to 50 mM, respectively (Fig. 2). As shown by a three-dimensional graph (Fig. 2), a high ratio of arginine to ammonium sulfate resulted in a high CGP content, and with 75 mM arginine and 10 mM ammonium sulfate in the medium, a CGP content as high as 41% (wt/wt) of the CDM was obtained.
TABLE 5.
Effects of arginine and aspartate on biosynthesis and accumulation of CGPa
| Arginine concn (mM) | Aspartate concn (mM) | CGP content (% of CDM) |
|---|---|---|
| 0 | 10 | 1.6 ± 0.16 |
| 0 | 20 | 1.0 ± 0.05 |
| 15 | 0 | 22.9 ± 0.13 |
| 15 | 10 | 3.9 ± 0.09 |
| 15 | 20 | 3.0 ± 0.11 |
| 30 | 0 | 27.8 ± 0.16 |
| 30 | 10 | 6.8 ± 0.02 |
| 30 | 20 | 3.0 ± 0.54 |
Cells of A. calcoaceticus strain ADP1 were cultivated at 30°C in 250-ml Erlenmeyer flasks without baffles containing 100 ml of unbuffered MSM without sodium glutamate and different concentrations of arginine and aspartic acid; the initial pH was 7.3. At the end of the experiment the cells were harvested by centrifugation, and the cyanophycin content was analyzed. The data are means and standard deviations of three independent experiments.
FIG. 2.
Three-dimensional graph illustrating the effects of arginine and ammonium sulfate on cyanophycin accumulation. Cells of A. calcoaceticus strain ADP1 were cultivated for 44 h at 30°C in 250-ml Erlenmeyer flasks without baffles containing 100 ml of unbuffered MSM containing no sodium glutamate but different concentrations of ammonium sulfate (10 to 50 mM) as the nitrogen source. In addition, arginine was added at concentrations of 30 to 75 mM. All other medium constituents used were as described for the basic medium in Materials and Method. At the end of the experiment the cells were harvested by centrifugation, and the cyanophycin content was analyzed.
Construction of a mutant defective in a putative cyanophycinase gene.
As in cyanobacteria, the cyanophycin synthetase gene (cphA) of A. calcoaceticus strain ADP1 is clustered with a putative intracellular cyanophycinase gene (cphI). In cyanobacteria, the cyanophycinases are responsible for mobilization of the storage polymer (24, 31, 34). However, in contrast to cyanobacteria, cphI is located downstream and not upstream of cphA. Another peculiarity of cphI is that its putative translational product resembles a fusion protein consisting of two cyanobacterial cyanophycinases fused with a hypothetical Ser-Glu-His catalytic triad in the N-terminal region. The putative protein is therefore much larger (80 kDa) than cyanobacterial cyanophycinases (about 27 to 30 kDa). cphI is the only gene in the genome of A. calcoaceticus strain ADP1 encoding a protein homologous to cyanophycinase. It was therefore expected that this protein is the only enzyme capable of catalyzing the degradation of CGP in this bacterium and that disruption of the gene could further improve CGP accumulation. We therefore generated a cphI mutant by insertion of a kanamycin resistance cassette as described in Materials and Methods. Disruption of cphI by this cassette, resulting in cphI::ΩKm, was confirmed by PCR as described in Materials and Methods. This mutant was therefore included in the following physiological experiments aimed at increasing the CGP content further.
Effect of chloramphenicol and comparison of the wild type with the cphI mutant with regard to CGP accumulation.
During cultivation in optimized MSM (with 75 mM arginine as the carbon source and 10 mM ammonium sulfate as the nitrogen source), the cphI mutant reproducibly produced significantly less CGP than the wild type produced during 48 h of cultivation (Table 6). Whereas CGP in the wild type accounted for about 38% of the CDM, in the mutant CGP accounted for only about 14% of the CDM (i.e., the CGP content was only about 37% of the CGP content of the wild type). In parallel, chloramphenicol was applied under the optimized conditions. Its addition at a concentration of 2.5 mg/liter increased the CGP content in wild-type cells by about 18%, resulting in a CGP content of about 46% (wt/wt) of the CDM (Table 6). The CGP content of the cphI mutant was also slightly increased to 15.5% (wt/wt) of the CDM. However, in comparison to the wild type, the CGP content remained rather low and was only about 35% of the CGP content of the wild type (Table 6). The CGP productivities of the wild type and the mutant (expressed in grams per liter) were also compared. Cells of the wild type produced about 8 and 28 times more CGP than the mutant produced in the absence and in the presence of chloramphenicol, respectively (Table 6).
TABLE 6.
Effects of cphI gene and chloramphenicol on cyanophycin accumulation in A. calcoaceticus strain ADP1a
| Strain | Chloramphenicolb | CGP content (% of CDM) | CGP concn (g/liter) |
|---|---|---|---|
| A. calcoaceticus ADP1 | − | 38.0 ± 0.5 | 0.23 ± 9 × 10−3 |
| + | 46.0 ± 0.6 | 0.28 ± 1 × 10−2 | |
| A. calcoaceticus ΔcphIc | − | 14.3 ± 0.4 | 0.03 ± 16 × 10−4 |
| + | 15.5 ± 0.5 | 0.01 ± 4 × 10−4 |
Cells of A. calcoaceticus strain ADP1 and of the ΔcphI mutant were cultivated at 30°C in 250-ml Erlenmeyer flasks without baffles containing 100 ml of unbuffered MSM containing 10 mM ammonium sulfate as the nitrogen source; the initial pH was 7.3. In addition, the medium contained 75 mM arginine. At the end of the experiment the cells were harvested by centrifugation, and the cyanophycin content was analyzed. The data are means and standard deviations of three independent experiments.
Chloramphenicol (2.5 μg/ml) was added after 24 h of cultivation, and cultivation was then continued for a further 24 h.
For the cphI mutant, the medium also contained 50 μg of kanamycin per ml.
To further reveal the phenotype of the cphI mutant, cells of the wild type and of the mutant were cultivated for 4 days under the conditions optimized for CGP accumulation, and the growth and CGP contents of the cells were monitored. The CGP content of cells of the wild type increased over 2 days of incubation, when it accounted for 40% (wt/wt) of the CDM, equivalent to 0.25 g liter of culture−1. Thereafter, the CGP content of the culture decreased steadily until the fourth day (Table 7), clearly indicating that degradation of CGP occurred. The cphI mutant, in contrast, continued to accumulate CGP at a low rate during the entire 4-day period. On the fourth day, the mutant cells and the wild-type cells had about the same CGP content.
TABLE 7.
Time course of CGP accumulation in A. calcoaceticus strain ADP1 and the cphI mutanta
| Incubation period (days) | CGP concn (g/liter)
|
|
|---|---|---|
| A. calcoaceticus ADP1 | A. calcoaceticus ΔcphI | |
| 1 | 0.15 ± 2 × 10−3 | 0.001 ± 8 × 10−4 |
| 2 | 0.24 ± 6 × 10−3 | 0.006 ± 9 × 10−4 |
| 3 | 0.13 ± 4 × 10−3 | 0.042 ± 15 × 10−4 |
| 4 | 0.06 ± 2 × 10−3 | 0.061 ± 18 × 10−4 |
Cells of A. calcoaceticus strain ADP1 and of the ΔcphI mutant were cultivated for 96 h at 30°C in 250-ml Erlenmeyer flasks without baffles containing 100 ml of unbuffered MSM with 10 mM ammonium sulfate as the nitrogen source. In addition, 75 mM arginine was added. The initial pH of the medium was 7.3. After 24, 48, 72, and 96 h of cultivation aliquots of the cells were harvested by centrifugation, and the cyanophycin content was analyzed by determining the amount of cyanophycin isolated by the acid extraction method. The data are means and standard deviations of three independent experiments.
Although it was rather unlikely that insertion of the kanamycin resistance cassette had a polar effect on expression of cphA due to the position of cphI in relation to cphA, the activities of the cyanophycin synthetase were measured in the wild type and in the mutant. Cells of both strains exhibited very high specific activities of this enzyme. The specific activity of cyanophycin synthetase was only slightly less in the mutant (18.8 U per mg of protein), in which CGP accounted for 14.3% (wt/wt) of the CDM, than in the wild type (23.3 U per mg of protein), in which the polymer accounted for 38.0% (wt/wt) of the CDM, when the organisms were cultivated in MSM containing arginine.
Analysis of CGP.
The CGP synthesized by A. calcoaceticus strain ADP1 grown for 44 h under optimized conditions (75 mM arginine and 10 mM ammonium sulfate) was isolated from cells that contained CGP at a level of 40% (wt/wt) of the CDM, and its composition and molecular weight were analyzed. HPLC analysis revealed almost equimolar amounts of aspartate and arginine; lysine or other amino acids could not be detected. SDS-polyacrylamide gel electrophoresis showed that the apparent molecular masses of CGP molecules were in the range from about 21 to 29 kDa.
Electron microscopic studies of CGP-rich cells.
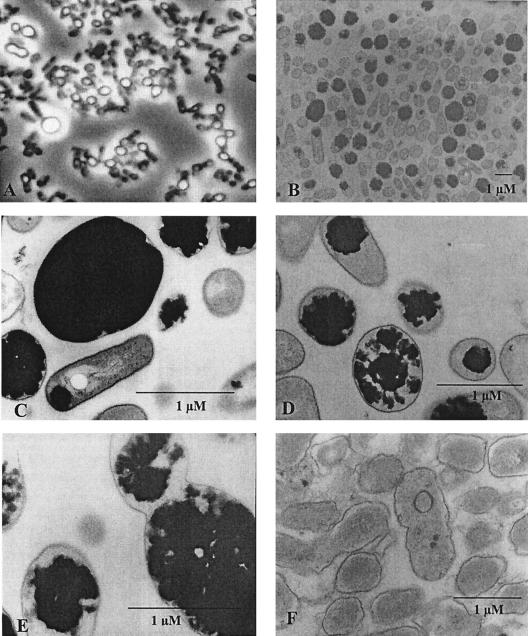
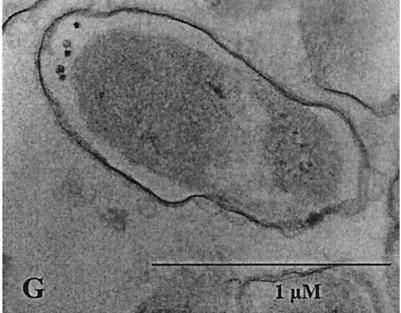
Cells of A. calcoaceticus strain ADP1 grown under optimized conditions (CGP content, 40% [wt/wt] of the CDM) were investigated by phase-contrast light microscopy (Fig. 3A). Most cells contained large refractile granules, and some cells were strongly deformed by the large granules in the cells. Transmission electron micrographs of thin sections of the cells (Fig. 3B to E) gave a more detailed view of the cells and the granules. These micrographs revealed that in many cells there were only single large CGP granules, some of which occupied the entire cytoplasm (Fig. 3B and C). Interestingly, some of the granules that had not yet occupied the entire cytoplasm had an irregular surface (Fig. 3C to E). Other cells contained very small granules near the poles of the cells, and very few cells showed no sign of CGP accumulation.
FIG. 3.
Microscopic inspection of cells of A. calcoaceticus. Cells of A. calcoaceticus were cultivated for 44 h at 30°C in MSM containing 75 mM arginine and 10 mM ammonium sulfate. After this, the cells were harvested and examined by phase-contrast light microscopy (A) or were fixed and examined by electron microscopy as described in Materials and Methods (B to E). Two samples were subjected to acid extraction before they were fixed and examined by electron microscopy (F and G). Bars = 1 μM.
An acid extraction method recently developed to isolate CGP from cells of E. coli without mechanical disruption (14) showed high efficiency with regard to CGP extraction when it was applied to cells of A. calcoaceticus. About 99% of the CGP could be extracted in a single step, as revealed by gravimetric and chemical analysis of the extracted polymer. Nearly complete extraction of the polymer with apparently intact cells remaining was also illustrated by electron micrographs of thin sections of these cells (Fig. 3F and G).
DISCUSSION
It was only recently reported that noncyanobacterial genome sequences harbor genes encoding proteins homologous to cyanophycin synthetases and cyanophycinases (20, 49). A. calcoaceticus was one of these bacteria and also was the first bacterium not belonging to the cyanobacteria in which the presence of CGP could be demonstrated (20). In this study, we investigated the ability of A. calcoaceticus strain ADP1 to accumulate larger amounts of CGP. Our analysis included varying the cultivation conditions and targeted mutagenesis. We demonstrated that A. calcoaceticus strain ADP1 is capable of accumulating CGP at levels up to 46% of the cell dry matter. This was more than the level in any cyanobacterium or recombinant strain harboring cphA. In recombinant P. putida KT2440, C. glutamicum, and R. eutropha HF39 harboring cphA from Synechocystis sp. strain PCC6803 CGP contents of 11, 3.6, and 7.0% of the CDM, respectively, were measured (3). Furthermore, a costly medium was used to produce CGP at a level of 24% of the CDM by using E. coli DH1 expressing cphA from Synechocystis sp. strain PCC 6803 (14). With the highest cellular CGP content ever reported, A. calcoaceticus strain ADP1 is a good candidate for production of CGP at a large scale in industry. Another peculiarity of A. calcoaceticus with regard to CGP biosynthesis is the restricted substrate specificity of its cyanophycin synthetase, which allows production of CGP consisting of only aspartic acid and arginine and not containing lysine, thus confirming in vitro studies recently performed with the purified enzyme (21). In contrast, cyanobacterial cyanophycin synthetases generally seem to incorporate significant amounts of lysine in place of arginine in vitro and in vivo when they are expressed heterologously. In recombinant strains a significant fraction of arginine is replaced by lysine (up to 10 mol%), yielding a CGP-like biopolymer consisting of aspartic acid, arginine, and lysine (2, 14, 20, 29, 48).
So far, one disadvantage of A. calcoaceticus strain ADP1 is its inability to synthesize these large amounts of CGP without supplementation of the medium with arginine. All initial experiments to increase the CGP contents of A. calcoaceticus cells by varying the cultivation conditions, such as temperature and oxygen supply, by varying the concentrations of medium components, such as phosphate, ammonium, or sulfate, by varying the pH, or by adding chloramphenicol as an inhibitor of ribosomal protein biosynthesis failed to result in CGP contents greater than 3% (wt/wt) of the CDM. While the relatively small changes in CGP contents in A. calcoaceticus cells obtained in this study by the variations mentioned above may not seem interesting from a biotechnological perspective, they revealed aspects of the regulation of CphA in A. calcoaceticus ADP1 that could be employed to enhance productivity. The results clearly indicated the need to uncouple cphA expression from nutrient starvation to achieve high cyanophycin synthetase activity at high cell densities. Physiological studies on the occurrence and regulation of cyanophycin synthesis and accumulation were performed in much more detail with cyanobacteria, particularly nitrogen-fixing species (12, 15, 24, 32, 40, 45). However, in these bacteria the situation is very different, since CGP serves primarily as a temporary storage molecule for newly fixed nitrogen (12, 18, 25). In contrast, A. calcoaceticus does not fix nitrogen; therefore, depletion of a source of bound nitrogen could not trigger CGP biosynthesis as it does in cyanobacteria (4, 6, 20) because then the nitrogen for CGP would also be missing.
The only conditions under which significantly larger amounts of CGP were accumulated was addition of arginine as a carbon source at relatively high concentrations (75 mM). Addition of arginine and aspartic acid to mineral salts media or the use of complex media, such as Luria-Bertani broth or nutrient broth, also increased the CGP contents of recombinant strains of E. coli (14), P. putida KT2440 (3), and R. eutropha (3) harboring one of the various cloned cyanobacterial cyanophycin synthetase genes; however, the maximum CGP contents of the cells were much lower (11, 7, and 24%, respectively) than the maximum CGP content obtained with A. calcoaceticus strain ADP1 in this study (46%). In summary, the optimal conditions for CGP accumulation in A. calcoaceticus strain ADP1 were as follows: 75 mM arginine as a carbon source, 10 mM ammonium sulfate as a nitrogen source, phosphate limitation of growth, and addition of chloramphenicol. Free arginine must be supplied in the medium. Addition of complex nitrogen sources did not significantly promote CGP accumulation. One reason for this may be that this strain is not capable of utilizing the complex nitrogen sources for CGP biosynthesis; another reason may be the insufficiently high arginine contents of the complex nitrogen sources, considering that arginine had to be supplied at a concentration of 75 mM for optimal CGP production. One further explanation may be the presence of aspartic and glutamic acids in complex nitrogen sources, since both of these acids have a severe negative effect on CGP accumulation in A. calcoaceticus. The suppression of CGP synthesis by aspartic acid or glutamic acid was profound and cannot be explained so far.
One of the most striking results of this study besides the accumulation of CGP at levels up to 46% of the CDM in A. calcoaceticus strain ADP1 was the significantly lower rate of CGP accumulation in the cphI mutant (Table 7). Since cyanophycin synthetase exhibited similar high specific activities in the wild type and in the mutant, the possibility of a polar effect caused by insertion of the kanamycin resistance gene cassette into cphI on expression of cphA can be excluded. It was demonstrated that mobilization of CGP occurs in the wild type but not in the mutant (Table 7). Therefore, and since no other gene coding for a putative cyanophycinase gene could be identified in the genome of this bacterium, it is most likely that the cphI translational product is responsible for intracellular CGP degradation. Therefore, it may be speculated that this cyanophycinase has a second function besides the mobilization of CGP. This could be the generation of primers for CGP synthesis (24). If CGP is partially hydrolyzed during the initial phase of CGP accumulation, then the number of CGP primers should increase. Several studies have shown that the activities of all cyanophycin synthetases investigated, including the enzyme of A. calcoaceticus, strictly depend on the presence of a CGP primer (2, 8, 20, 24, 43, 48). If this hypothesis for a dual function of CphI is true, disruption of the cphI gene should result in a lower CGP synthesis rate, and it would also explain the lower CGP content of the mutant cells than of the wild-type cells.
The molecular mass of the CGP produced by A. calcoaceticus ADP1 under optimized conditions was in the same range (21 to 29 kDa, with an average molecular mass of about 25 kDa) as the molecular mass obtained previously without optimization (18). Biosynthesis of such a low-molecular-mass CGP by a natural strain is another peculiarity of A. calcoaceticus, because the molecular mass range for CGP accumulated by cyanobacteria generally is 30 to 100 kDa and the average molecular mass is about 60 kDa (41). Low-molecular-mass CGP (range, 25 to 35 kDa) is otherwise synthesized only by recombinant bacteria expressing cyanobacterial cyanophycin synthetases, but the average molecular mass is slightly higher, about 30 kDa. A water-soluble CGP-like polymer produced by a recombinant strain of E. coli expressing the cyanophycin synthetase of D. hafniense (49) showed a heterogeneous distribution of molecular masses; SDS-polyacrylamide gel electrophoresis revealed two bands representing CGP molecules, one at molecular masses ranging from 22 to 26 kDa and a second at molecular masses ranging from 30 to 35 kDa.
In conclusion, we found that A. calcoaceticus has a high capacity for CGP synthesis and accumulation. Provision of arginine to the cyanophycin synthetase is the most limiting bottleneck for CGP production if this amino acid must be synthesized de novo and is not provided externally. Therefore, for further optimization of CGP synthesis by A. calcoaceticus workers must focus on overcoming this bottleneck, preferably by metabolic engineering of the arginine biosynthesis pathway and its regulation.
Acknowledgments
This study was supported by grants from the Bundesminsterium für Verbraucherschutz, Ernährung und Landwirtschaft (grant OONR125) and BAYER AG (Leverkusen) and by a fellowship to Yasser Elbahloul provided by the Deutsche Akademischer Austauschdienst and the government of the Arabic Republic of Egypt.
REFERENCES
- 1.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2000. Molecular characterization of the cyanophycin synthetase from Synechocystis sp. strain PCC6308. Arch. Microbiol. 174:297-306. [DOI] [PubMed] [Google Scholar]
- 2.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Purification of Synechocystis sp. strain PCC 6308 cyanophycin synthetase and its characterization with respect to substrate and primer specificity. Appl. Environ. Microbiol. 67:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aboulmagd, E., I. Voss, F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Heterologous expression of cyanophycin synthetase and cyanophycin synthesis in the industrial relevant bacteria Corynebacterium glutamicum and Ralstonia eutropha and in Pseudomonas putida. Biomacromolecules 2:1338-1342. [DOI] [PubMed] [Google Scholar]
- 4.Aldor, I., and J. D. Keasling. 2001. Metabolic engineering of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) composition in recombinant Salmonella enterica serovar Typhimurium. Biotechnol. Bioeng. 76:108-114. [DOI] [PubMed] [Google Scholar]
- 5.Alford, D. D., A. P. Wheeler, and C. A. Pettigrew. 1994. Biodegradation of thermally synthesized polyaspartate. J. Environ. Polym. Degrad. 2:225-236. [Google Scholar]
- 6.Alvarez, H. M., O. H. Pucci, and A. Steinbüchel. 1997. Lipid storage compounds in marine bacteria. Appl. Microbiol. Biotechnol. 47:132-139. [Google Scholar]
- 7.Arnold, M. 1968. Histochemie. Einführung in die Grundlagen und Prinzipien der Methoden. Springer-Verlag, Berlin, Germany.
- 8.Berg, H., K. Ziegler, K. Piotukh, K. Baier, W. Lockau, and R. Volkmer-Engert. 2000. Biosynthesis of the cyanobacterial reserve polymer multi-l-arginylpoly-l-aspartic acid (cyanophycin). Mechanism of the cyanophycin synthetase reaction studied with synthetic primers. Eur. J. Biochem. 267:5561-5570. [DOI] [PubMed] [Google Scholar]
- 9.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borzi, A. 1887. Le comunicazioni intracellulari delle Nostochinee. Malpighia 1:28-74. [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Carr, N. G. 1988. Nitrogen reserves and dynamics reservoirs in cyanobacteria, p. 13-21. In L. J. Rogers and J. R. Gallon (ed.), Biochemistry of the algae and cyanobacteria. Annual Proceedings of the Phytochemical Society of Europe. Clarendon, Oxford, United Kingdom.
- 13.Colón-López, M. S., D. M. Sherman, and L. A. Sherman. 1997. Transcriptional and translational regulation of nitrogenase in light-dark- and continuous-light-grown cultures of the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 179:4319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey, K. M., F. B. Oppermann-Sanio, H. Schmidt. and A. Steinbüchel. 2002. Technical scale production of cyanophycin with recombinant strains of Escherichia coli. Appl. Environ. Microbiol. 68:3377-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta, M., and N. G. Carr. 1981. Enzyme activities related to cyanophycin metabolism in heterocysts and vegetative cells of Anabaena sp. J. Gen. Microbiol. 125:17-23. [Google Scholar]
- 16.Hai, T., F. B. Oppermann-Sanio, and A. Steinbüchel. 1999. Purification and characterization of cyanophycin and cyanophycin synthetase from the thermophilic Synechococcus sp. MA19. FEMS Microbiol. Lett. 181:229-236. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on the transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Howarth, R. W., and J. J. Cole. 1985. Molybdenum availability, nitrogen limitation, and phytoplankton growth in natural waters. Science 229:635-655. [DOI] [PubMed] [Google Scholar]
- 19.Joentgen, W., T. Groth, A. Steinbüchel, T. Hai, and F. B. Oppermann. 1998. Polyaspartic acid homopolymers and copolymers, biotechnological production and use thereof. International patent application WO 98/39090.
- 20.Krehenbrink, M., F. B. Oppermann-Sanio, and A. Steinbüchel. 2002. Evaluation of non-cyanobacterial genome sequences for occurrence of genes encoding proteins homologous to cyanophycin synthetase and cloning of an active cyanophycin synthetase from Acinetobacter sp. strain DSM 587. Arch. Microbiol. 177:371-380. [DOI] [PubMed] [Google Scholar]
- 21.Krehenbrink, M., and A. Steinbüchel. 2004. Partial purification and characterization of a non-cyanobacterial cyanophycin synthetase from Acinetobacter calcoaceticus strain ADP1 with regard to substrate specificity, substrate affinity and binding to cyanophycin. Microbiology 150:2599-2608. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lang, N. J., R. D. Simon, and C. P. Wolk. 1972. Correspondence of cyanophycin granules with structured granules in Anabaena cylindrica. Arch. Microbiol. 83:313-320. [Google Scholar]
- 24.Li, H., D. M. Sherman, S. Bao, and L. A. Sherman. 2001. Pattern of cyanophycin accumulation in nitrogen-fixing and non-nitrogen-fixing cyanobacteria. Arch. Microbiol. 176:9-18. [DOI] [PubMed] [Google Scholar]
- 25.Mackerras, A. H., N. M. de Chazal, and G. D. Smith. 1990. Transient accumulation of cyanophycin in Anabaena cylindrical and Synechocystis 6308. J. Gen. Microbiol. 136:2057-2065. [Google Scholar]
- 26.Merritt, M. V., S. S. Sid, L. Mesh, and M. M. Allen. 1994. Variations in the amino acid composition of cyanophycin in the cyanobacterium Synechocystis sp. PCC6308 as a function of growth conditions. Arch. Microbiol. 162:158-166. [DOI] [PubMed] [Google Scholar]
- 27.Obst, M., F. B. Oppermann-Sanio, and A. Steinbüchel. 2002. Isolation of cyanophycin degrading bacteria, cloning and characterization of an extracellular cyanophycinase gene (cphE) from Pseudomonas anguilliseptica strain BI—the cphE gene from P. anguilliseptica BI encodes a cyanophycin-hydrolyzing enzyme. J. Biol. Chem. 277:25096-25105. [DOI] [PubMed] [Google Scholar]
- 28.Obst, M., A. Sallam, H. Luftmann, and A. Steinbüchel. 2003. Isolation and characterization of Gram-positive cyanophycin-degrading bacteria—kinetic studies on cyanophycin depolymerase activity in aerobic bacteria. Biomacromolecules 5:153-161. [DOI] [PubMed] [Google Scholar]
- 29.Oppermann-Sanio, F. B., T. Hai, E. Aboulmagd, F. F. Hezayen, S. Jossek, and A. Steinbüchel. 1999. Biochemistry of microbial polyamide metabolism, p. 185-193. In A. Steinbüchel (ed.), Biochemical principles and mechanisms of biosynthesis and biodegradation of polymers. Wiley-VCH, Weinheim, Germany.
- 30.Palmen, R., B. Vosman, P. Buijsman, C. K. Breek, and K. J. Hellingwerf. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295-305. [DOI] [PubMed] [Google Scholar]
- 31.Picossi, S., A. Valladares, E. Flores, and A. Herrero. 2004. Nitrogen-regulated genes for the metabolism of cyanophycin, a bacterial nitrogen reserve polymer. J. Biol. Chem. 279:11582-11592. [DOI] [PubMed] [Google Scholar]
- 32.Quintero, M. J., A. M. Muro-Pastor, A. Herrero, and E. Flores. 2000. Arginine catabolism in the cyanobacterium Synechocystis sp. strain PCC 6803 involves the urea cycle and argininase pathway. J. Bacteriol. 182:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao, N. R., M. A. Richardson, and S. Kuhstoss. 1987. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 153:166-198. [DOI] [PubMed] [Google Scholar]
- 34.Richter, R., M. Hejazi, R. Kraft, K. Ziegler, and W. Lockau. 1999. Cyanophycinase, a peptidase degrading the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartic acid (cyanophycin). Molecular cloning of the gene of Synechocystis sp. PCC6803, expression in Escherichia coli, and biochemical characterization of the purified enzyme. Eur. J. Biochem. 263:163-169. [DOI] [PubMed] [Google Scholar]
- 35.Roweton, S., S. J. Huang, and G. Swift. 1997. Poly(aspartic acid): synthesis, biodegradation, and current applications. J. Environ. Polym. Degrad. 5:175-181. [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Schlegel, H. G. 1992. Allgemeine Mikrobiologie, 2nd ed. Thieme Verlag, Stuttgart, Germany.
- 38.Schmidt, K., S. Liaanen-Jensen, and H. G. Schlegel. 1963. Die Carotinoide der Thiorhodaceae. Arch. Microbiol. 46:117-126. [PubMed] [Google Scholar]
- 39.Schwamborn, M. 1998. Chemical synthesis of polyaspartates: a biodegradable alternative to currently used polycarboxylate homo- and copolymers. Polym. Degrad. Stabil. 59:39-45. [Google Scholar]
- 40.Sherman, D. M., D. Tucker, and L. A. Sherman. 2000. Heterocyst development and localization of cyanophycin in N2-fixing cultures of Anabaena sp. PCC 7120 (Cyanobacteria). J. Phycol. 36:932-941. [Google Scholar]
- 41.Simon, R. D. 1971. Cyanophycin granules from the blue-green alga Anabaena cylindrica: a reserve material consisting of copolymer of aspartic acid and arginine. Proc. Natl. Acad. Sci. USA 68:265-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, R. D. 1973. The effect of chloramphenicol on the production of cyanophycin granule polypeptide in the blue-green alga Anabaena cylindrica. Arch. Microbiol. 92:115-122. [DOI] [PubMed] [Google Scholar]
- 43.Simon, R. D. 1976. The biosynthesis of multi-l-arginyl-poly(l-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 422:407-418. [DOI] [PubMed] [Google Scholar]
- 44.Simon, R. D., and P. Weathers. 1976. Determination of the structure of the novel polypeptide containing aspartic acid and arginine which is found in cyanobacteria. Biochim. Biophys. Acta 420:165-176. [DOI] [PubMed] [Google Scholar]
- 45.Simon, R. D. 1987. Inclusion bodies in the cyanobacteria: cyanophycin, polyphosphate, polyhedral bodies, p. 199-225. In P. Fay and C. van Baalen (ed.), The cyanobacteria. Elsevier, Amsterdam, The Netherlands.
- 46.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 47.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determination by dodecylsulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 48.Ziegler, K., A. Diener, C. Herpin, R. Richter, R. Deutzmann, and W. Lockau. 1998. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartate (cyanophycin). Eur. J. Biochem. 254:154-159. [DOI] [PubMed] [Google Scholar]
- 49.Ziegler, K., R. Deutzmann, and W. Lockau. 2002. Cyanophycin synthetase-like enzymes of non-cyanobacterial eubacteria: characterization of the polymer produced by a recombinant synthetase of Desulfitobacterium hafniense. Z. Naturforsch. Sect. C 57:522-529. [DOI] [PubMed] [Google Scholar]