Abstract
The objective of this study was to determine whether the Food and Drug Administration’s Adverse Event Reporting System (FAERS) data set could serve as the basis of automated electronic health record (EHR) monitoring for the adverse drug reaction (ADR) subset of adverse drug events. We retrospectively collected EHR entries for 71 909 pediatric inpatient visits at Cincinnati Children’s Hospital Medical Center. Natural language processing (NLP) techniques were used to identify positive diseases/disorders and signs/symptoms (DDSSs) from the patients’ clinical narratives. We downloaded all FAERS reports submitted by medical providers and extracted the reported drug-DDSS pairs. For each patient, we aligned the drug-DDSS pairs extracted from their clinical notes with the corresponding drug-DDSS pairs from the FAERS data set to identify Drug-Reaction Pair Sentences (DRPSs). The DRPSs were processed by NLP techniques to identify ADR-related DRPSs. We used clinician annotated, real-world EHR data as reference standard to evaluate the proposed algorithm. During evaluation, the algorithm achieved promising performance and showed great potential in identifying ADRs accurately for pediatric patients.
Keywords: Adverse drug reaction, clinical notes, electronic health records, natural language processing
Introduction
We focused on the adverse drug reaction (ADR) subset of adverse drug events (ADEs). We defined ADE and ADR according to the Nebecker definitions.1 An ADE is “an injury resulting from the use of a drug.” Under this definition, the term ADE includes harm caused by the drug (ADEs and over-doses) and harm from the use of the drug (including dose reductions and discontinuations of drug therapy). An ADR is defined as “response to a drug which is noxious and unintended and which occurs at doses normally used in man for prophylaxis, diagnosis, or therapy of disease or for the modification of physiologic function.”1 “In sum, an ADR is harm directly caused by the drug at normal doses, during normal use.”2
It remains challenging to incorporate clinical evidence in electronic ADE alerting systems, especially in pediatric settings.3,4 Adverse drug reactions can happen in all health care settings including inpatient, outpatient, and nursing homes, causing more than 700 000 injuries in the United States annually.5–7 Many ADRs could be detected and mitigated if we could identify patients’ clinical status changes promptly. However, it is a big challenge to detect such changes and ultimately ADRs from complex clinical documents. An automated or even semiautomated system could be an advancement if the positive predictive value (PPV) achieved is higher than provided by current techniques.8,9
Several studies have been conducted in identifying ADRs. In the past, researchers manually reviewed clinical documents to identify ADRs.10 With the rapid growth of information technology, the researchers started leveraging the information from electronic health record (EHR) and newly emerging techniques to identify ADRs. The EHR data are a rich resource for data mining and knowledge discovery on ADRs.11 It stores both structured and unstructured information related to a patient over time, including demographics, diagnoses and medical histories, medications taken, and clinical notes. The ADR-related information is often contained in clinical narratives. It remains challenging to identify clinical evidence from the EHR sources to ascertain ADR events.
The Food and Drug Administration’s Adverse Event Reporting System (FAERS) collects ADEs from health professionals and patients.12 Both structured reports and unstructured plain text are provided to describe ADR-related information on the Food and Drug Administration (FDA) Web site. In this study, we focused on using the FAERS information to help identify reactions that might be caused by a specific medication. We then identified ADR-related Drug-Reaction Pair Sentences (DRPSs) from clinical notes in the EHRs with natural language processing (NLP) technologies.
By leveraging the information from the FAERS reports, our objective was to build an NLP-based, sentence-level pipeline to detect potential ADRs from the EHR data. The null hypothesis (H0) of the study stated that leveraging drug-reaction pairs from the FAERS reports would not improve the performance of EHR-based ADR detection. As a proof-of-concept study, we focused on detecting ADRs on patients treated at an urban tertiary care pediatric hospital.
Usually, NLP studies report both recall and PPV in their evaluation sections. However, in studies similar to ours, calculating recall requires to accurately identify all the sentences with true-positive ADR mentions, at least in a randomly selected sample of sentences in the EHR. This poses a serious challenge because the true-positive ADR mentions are extremely rare compared with the number of sentences in the EHR. Consequently, calculating the recall statistics in those studies would require such a large physician work effort that is usually not feasible. Our evaluation was modeled on the published studies that faced similar challenges,13–20 where we calculated PPV and approximated recall using the relative recall (RR) measure.21
The following sections describe our work in detail. First, we introduce the related work for the study; second, we describe the data sets used to detect ADRs and the implementation processes of the ADR detector; then, we present the results and findings, evaluate, and specify our limitations. Finally, we conclude the work and propose potential improvements for future studies.
Related work
Rule-based ADR detection has been widely used in recent works. Chazard et al22 generated potential rules for ADE detection using decision trees and association rules based on structured and unstructured data from the EHR. The rules were then filtered, validated, and reorganized by domain experts, resulting in 236 validated ADE detection rules. In their experiments, the rules were applied to automatically detect 27 types of ADEs. One limitation for Chazard method was that generating ADE detection rules with data mining was complex. By applying the 236 validated ADE detection rules and statistical analysis, Chazard et al23 concluded that contextualization was necessary for ADE detection and prevention. Instead of extracting context, our study presented a new technique to detect ADRs by leveraging additional knowledge sources.
Sohn et al8 developed a system to extract physician-asserted drug side effect sentences from clinical notes for psychiatry and psychology patients. They examined key words and expression patterns of side effects manually and developed matching rules to discover a side effect and its causative drug relationship. They then used decision trees (C4.5) to extract sentences that contain side effect and causative drug pairs. The approach achieved a 64% PPV. Nevertheless, there were 2 limitations for the study by Sohn et al. First, they focused on a small data set from a particular specialty (psychiatry). The generalizability of the rules for identifying side effects on other specialties was unknown. Second, the approach only identified well-defined side effects and causative drugs with limited indication words and patterns, whereas the complicated or indirect description of side effect might be missed.
Our study was inspired by the work by Sohn et al. However, instead of developing a limited set of drug-side effect relationships manually, we attempted to leverage the drug-reaction pairs from the FAERS reports, allowing us to obtain more comprehensive drug-reaction relationships in a cost-effective manner. In addition, the FAERS database is continuously updated, and potential drug-reaction relationships are continuously entered. Consequently, it provides a mean for new drug-reaction relationships to be discovered. The second difference from the work by Sohn et al is that we applied named entity recognition to identify complicated or indirect DDSSs from clinical notes. Our hypothesis was that the NLP technology allows us to rule out non–ADR-related sentences that also contain drugs and reactions, hence increasing the PPV of ADR detection.
Natural language processing has been used in several studies in the area of ADR detection.24 Aramaki et al studied patterns of ADEs from clinical records, and the results showed that 7.7% of the clinical records include adverse event information, in which 59% (4.5% in total) can be extracted automatically by NLP techniques. The finding was encouraging due to the massive amounts of clinical records.25 Wang et al26 performed an NLP-based method to detect ADEs in clinical discharge summaries and applied contextual filtering to improve performance. Their evaluation suggested that applying filters improved recall (from 43% to 75%) and PPV (from 16% to 31%) of ADE detection. One limitation for the work by Wang et al was that they only studied discharge summaries.
Park et al27 proposed an algorithm for detecting laboratory abnormalities after treatment with medications. The PPV of the algorithm was between 22% and 75%. Epstein et al28 developed a high-performance NLP algorithm in Transact-SQL to identify medication and food allergies and sensitivities from unstructured allergy registries in EHRs. The algorithm was trained on a set of 9445 records and tested on a separate set of 9430 records. Positive predictive value for medication allergy matches was 99.9% on both the training and the test sets. One limitation of the study by Epstein et al was that the lookup tables developed might not be fully portable to other institutions.
Pathak et al applied linked data principles and technologies to represent patient EHR data as resource description frame-work. They then identified potential drug-drug interactions for cardiovascular and gastroenterology drugs.29 The limitation of the study was that it only used drug prescription data. Because a patient could have a prescription without actually taking the medication, we used both drug prescription and medication administration information in our study. Time window (time course, censoring periods) was used to exclude false-positive ADRs in the studies.30,31 The limitations for the study by Sauzet et al31 included that all patients who were prescribed were considered at risk, even if they did not take the drug. Other works focused on a specific or a specific type of drug or reaction.32,33 As such, the algorithms and the conclusions were not generalizable to a larger scope of medications and reactions. Machine learning methods have also been applied to detect ADRs from EHR data. Ji et al34 introduced causal association rules to represent potential causal relationships between a drug and International Classification of Diseases, Ninth Revision (ICD-9)–coded signs or symptoms representing potential ADRs. In total, 8 out of 50 top drug-symptom pairs were validated to represent true causal associations. The PPV for the study by Ji et al was 16%.
Harpaz et al35 also applied association rule mining on the FAERS reports to identify multidrug ADR associations. The algorithm identified 2603 multidrug ADR associations, and the PPV was 67%. However, the study focused on the FAERS reports, most of which were potential ADR events. Lingren36 first identified sentences containing DDSSs from clinical notes, after which he applied support vector machines to classify the sentences into adverse and non–ADE-related sentences. In his work, the PPV for identifying ADEs was 80.4%. However, the work was performed on the data from only 10 patients. Other data mining methods have also been used to detect ADRs.37–40 Several studies worked on creating annotated corpus of ADR events to facilitate ADR detection and evaluation.41–43
Electronic health record data are complex, and a substantial portion of the meaningful information (eg, symptoms, treatments, and ADRs) is represented only in narrative text. As such, NLP plays an important role in extracting clinical information from the EHR data. Natural language processing–based methods have been applied to a broad range of clinical informatics topics, including patient protected health information (PHI) identification, medication detection, and clinical trial eligibility screening.44–46
Based on existing open-source technologies of the Unstructured Information Management Architecture frame-work and OpenNLP toolkit, Savova et al47 developed an open-source NLP system, named the clinical Text Analysis and Knowledge Extraction System (cTAKES), for information extraction from unstructured clinical notes. The cTAKES was specifically trained for clinical text, enabling it to automatically extract rich linguistic and semantic annotations that can be used in clinical research. In our study, cTAKES was used to process clinical notes and identify clinical named entities such as DDSSs.
Identifying medical conditions from clinical notes is important but not sufficient for identifying ADRs. The context information around a medical condition is also critical to determine an ADR. For instance, approximately half of the medical conditions described in clinical notes are negated, which could cause false positives in ADR detection.48 Therefore, the contextual properties for a medical condition including negation should also be analyzed. Methods for identifying negation expressions have been developed in recent years.49–53 Chapman et al50,54 developed a regular expression-based algorithm, NegEx, to search specific expressions (eg, not, without, and absence of) around a phrase to identify negated phrases. By evaluating different methods for negation detection, Goryachev et al55 concluded that the accuracy of regular expressions methods (91.9%–92.3%) was higher than that of machine learning–based methods (83.5%–89.9%). Based on this conclusion, we used the regular expression-based algorithm NegEx to detect negation in the study.
Methods
This study focused on pediatric patients. The institutional review board of Cincinnati Children’s Hospital Medical Center (CCHMC) approved the study and a waiver of consent was authorized (study ID: 2010-3031).
We retrospectively extracted EHR data for 71 909 inpatient and emergency department (ED) visits (referred to as “encounters”) of 42 995 pediatric patients treated at CCHMC between January 1, 2010 and August 31, 2012. The collected data included structured demographics and medication administration records, and unstructured clinical notes created during the patients’ hospitalization. The medication administration records were used to obtain information of medications given to a patient during the hospitalization. The free text clinical notes were used to retrieve information about medical conditions (eg, DDSSs) for the patient.
The FAERS is a database that collects information of adverse events and medication error reports submitted to the FDA.12 Some reports were received from health care professionals (eg, physicians, pharmacists, and nurses), whereas others from consumers (eg, patients, family members, and lawyers) and manufacturers. We downloaded 10 years (2004–2013) of ADE reports from the FAERS. Each report included information such as demographics, reported drugs, reactions, patient outcomes, report sources, drug therapies, and indications for use (diagnoses).
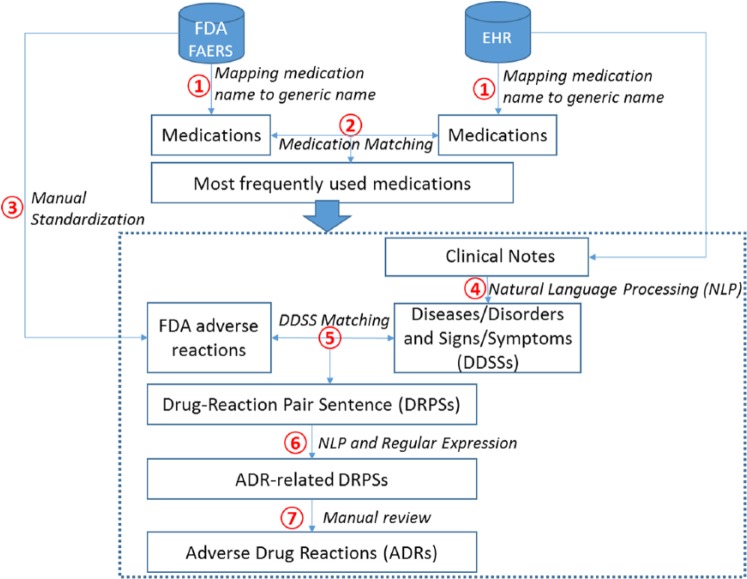
Figure 1 illustrates the overview of the study. Our objective was to detect ADRs in the EHR data with the aid of information from the FAERS reports. We applied the following 7 steps (steps 1–7 in Figure 1) to achieve the goal. (1) We first preprocessed medications and mapped brand names to generic names for both the EHR and the FAERS reports. (2) We then identified the most frequent medications from both the EHR and the FAERS reports based on frequencies of the medications’ generic names. Because our study was data-driven, we excluded low-frequency medications. The medications extracted from the EHR data and the FAERS reports were then matched using their generic names. (3) To match FDA reactions with DDSSs in the EHR data, we manually standardized the reaction terms from the FAERS reports. (4) Natural language processing techniques were leveraged to identify DDSSs from clinical notes. (5) Given a medication taken by the patient, the corresponding FDA reactions were considered potential ADR candidates and matched with the DDSSs identified from the patient’s clinical notes. A sentence containing any of these reactions was called a DRPS for the medication. (6) Given all DRPSs, we performed NLP and regular expression techniques (section analysis, sentence-based postprocessing) to rule out non-ADR DRPSs. The remaining sentences were considered ADR-related DRPSs. (7) Two independent reviewers manually reviewed the ADR-related DRPSs generated by the proposed algorithm and identified true ADRs. One reviewer had a medical degree. The results of the independent reviews were consolidated and the disagreements resolved. The algorithm performance was then calculated based on the adjudicated reference standard. Supplementary Appendix Table 3 clarifies that each step was completed manually or automatically.
Figure 1.
Overview of the study. EHR indicates electronic health record; FDA, Food and Drug Administration.
To clarify steps 1–6, we present an example for identifying ADR-related events for oxycodone: from the FAERS reports, our algorithm automatically extracted a drug name and potentially associated adverse event of “Roxicodone caused scratch.” The medication name Roxicodone was then manually converted to its generic name oxycodone (step 1) using a commercial generics-brand drug names database. We also manually determined the concept unique identifier (CUI) for “scratch” in the Unified Medical Language System (UMLS) browser and obtained C1384489 for “scratch” (step 3). Assuming that our algorithm identified a sentence of “Oxycodone use also makes him itch/scratch” from an EHR clinical note that contained the targeted medication oxycodone, automated NLP techniques were used to identify and convert the DDSS “itch/scratch” to the CUI C1384489 (step 4). Because the sentence contained the drug-reaction pair (oxycodone-C1384489) and the reaction was not negated, the algorithm considered it an ADR-related DRPS (steps 5 and 6).
FDA sources
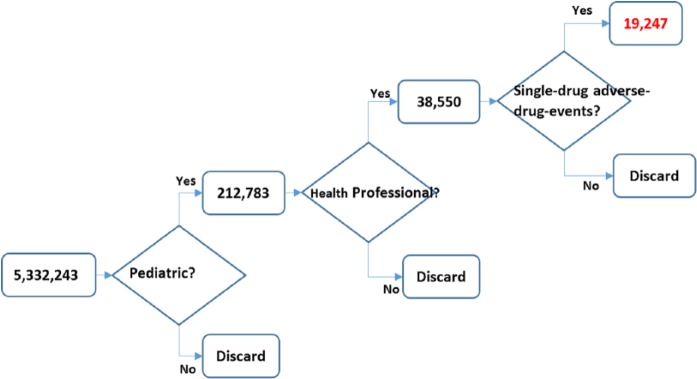
Because the study focused on pediatric patients, we used the FAERS reports that were associated with patients between 0 and 18 years old. We only used reports submitted by health professionals to assure the reliability of the sources. In addition, we focused on single-drug ADE (ADE caused by only 1 drug) to avoid complex drug interactions. Figure 2 illustrates the data preprocessing of the FDA reports. Among the 5 332 243 downloaded unique FDA-reported events, 4.0% (212 783 events) were recorded as pediatric events. About 18.1% (38 550 events) of these pediatric events were reported by a health professional, among which, 49.9% were single-drug ADEs. Consequently, a total of 19 247 events were single-drug ADEs on pediatric patients and were reported by health professionals. These events were used as a reference standard of drug-reaction relations in the automated algorithm.
Figure 2.
Data preprocessing on the Food and Drug Administration reports.
Identifying most frequently used medications
In the FAERS reports and EHR data, the medications were recorded using either generic name or brand name. To merge brand and generic names (eg, “Tylenol” and “acetaminophen”), we mapped each medication to its generic name using the Lexicomp Online Database56 (step 1 in Figure 1). The Lexicomp Online Database is a commercial database that provides search for medication information.
Based on their generic names, the top 50 most frequently used medications were identified (step 2 in Figure 1). We excluded 9 medications from the list due to no FAERS reports on the medications. The exclusion resulted in 41 target medications for the study. We then focused on detecting ADRs for the 63 043 encounters (covering 93% of all encounters) in which the patients (93.7% of all patients) were given these 41 medications.
Standardizing DDSSs and reactions in FAERS reports
The FAERS reports and the EHR data might have used different descriptions for the same DDSS and reaction. For instance, the DDSS of “headache” in the EHR did not match the FDA reaction “pain in head,” although they were the same symptom and had the same CUI (C0018681).57 To match FDA reactions with DDSSs in the EHR data, we converted the textual terms from both sources into CUIs corresponding to the UMLS58 (step 3 in Figure 1).
For the 41 most frequently used medications, we first excluded non–ADR-related reactions from their FAERS reports (Supplementary Appendix Table 1). For instance, overdose of any medications was excluded because overdose is not an ADR.1,59 In addition, some drug-reaction pairs in the FAERS reports (eg, levetiracetam—seizures) were excluded because they indicated that the medication was used to treat rather than cause the reaction. Finally, we manually excluded the following types of reactions (Supplementary Appendix Table 2): overdose, indications, accidents, reactions mostly relevant for adults, known wrong medication or medication errors, and irrelevant or vague description of the reactions. A pediatrician reviewed and confirmed the list of excluded reactions. After the exclusions, a total of 2646 unique FDA drug-reaction pairs was used in the study. To correctly identify the corresponding CUIs, we manually searched the reactions in the UMLS terminology browser60 and extracted the returned CUIs. A reaction-CUI mapping dictionary was then created for all reaction terms.
Detecting ADRs in clinical notes
In current EHR systems, a substantial portion of the meaningful information (eg, medical conditions) is represented only in clinical notes. As such, clinical notes are valuable resource for detecting ADRs. In this study, we collected all 2 647 746 clinical notes for the 71 909 encounters. The note data covered 54 documentation types such as History and Physical (H&P) Notes, Discharge Summaries, ED Notes, and Progress Notes. On average, there were 37 notes per encounter and the average length of a note was 309 words. Every note had a time stamp in structure format that recorded its time of filing.
Based on the guideline created by our physician champion, we focused on processing 4 types of clinical notes for the targeted encounters: H&P notes, discharge summaries, ED notes, and progress notes. We excluded notes from the encounters that did not use the targeted medications. In addition, patients with more than 1000 notes during their hospitalization tended to have diseases that required a significantly larger number of medications and had higher possibility to have ADRs caused by drug-drug interaction. Our proof-of-concept study did not investigate drug-drug interaction and hence we excluded these encounters. Ten (0.02%) of the encounters were excluded under this selection criterion. After this processing, 1 168 397 (44.1%) of the clinical notes were left for investigation.
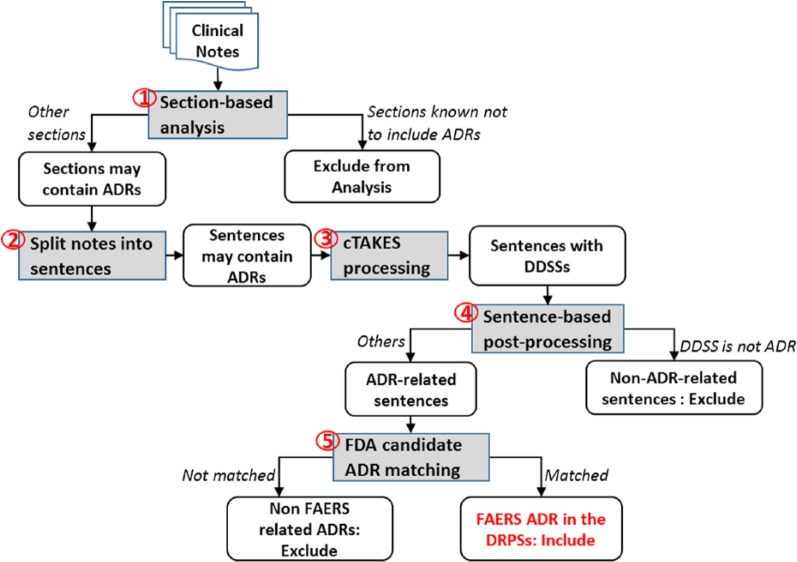
We developed an NLP pipeline to process clinical notes and identify potential ADR events (steps 4–6 in Figure 1). Figure 3 illustrates the NLP processes performed. We first applied section-based analysis (process 1 in Figure 3) to exclude sections that never mentioned ADRs (eg, plan of care sections). For the remaining sections, the content was split into sentences (process 2), and the mentions of DDSSs were identified from each sentence and standardized into CUIs using cTAKES47 (process 3). The semantic modifiers (eg, negation, hypothetical events, and therapies) on the reactions were also detected by NLP postprocessing (process 4), and the reactions were converted to the corresponding CUIs. Given each drug-reaction pair from the FAERS reports, we searched both the medication names and the corresponding CUIs in each output sentence (process 5). If the medication and the reaction were found in a sentence, the sentence was classified as a DRPS. Finally, using the time stamps documented in the medication administration records, we filtered out DRPSs written before a medication was given. The remaining DRPSs were then considered ADR-related DRPSs.
Figure 3.
Overview of the natural language processing processes. ADRs indicate adverse drug reactions; cTAKES, clinical Text Analysis and Knowledge Extraction System; DDSSs, diseases/disorders and signs/symptoms; DRPSs, Drug-Reaction Pair Sentences; FAERS, Food and Drug Administration’s Adverse Event Reporting System; FDA, Food and Drug Administration.
Section-based analysis
Some sections in a clinical note specified future plans and prescription information (eg, plan of care, medications, and instruction). Sentences in these sections would never describe an ADR event even if they contained the drug-reaction pairs from the FAERS reports. To reduce false-positive matches, we excluded these sections from the ADR detection. In the NLP pipeline, we developed a list of regular expressions on section headers to rule out the sections.
NLP of clinical notes
We split the remaining content into sentences using rules and applied cTAKES to identify and convert DDSSs to CUIs. Unlike the reactions extracted from the FAERS reports, the DDSSs documented in clinical notes could be modified by semantic modifiers. For instance, a sentence could contain negation that represented the absence of the DDSS (eg, “no overnight events-pain well controlled with ibuprofen with no nausea/vomiting”). It could also indicate a hypothetical event that commonly existed in discharge instruction and plan of care (eg, “I will increase dose of ibuprofen if pain continues”). In addition, some sentences describing therapies indicated that the medications were used to treat rather than caused the reactions (eg, “pain was well controlled by oxycodone”).
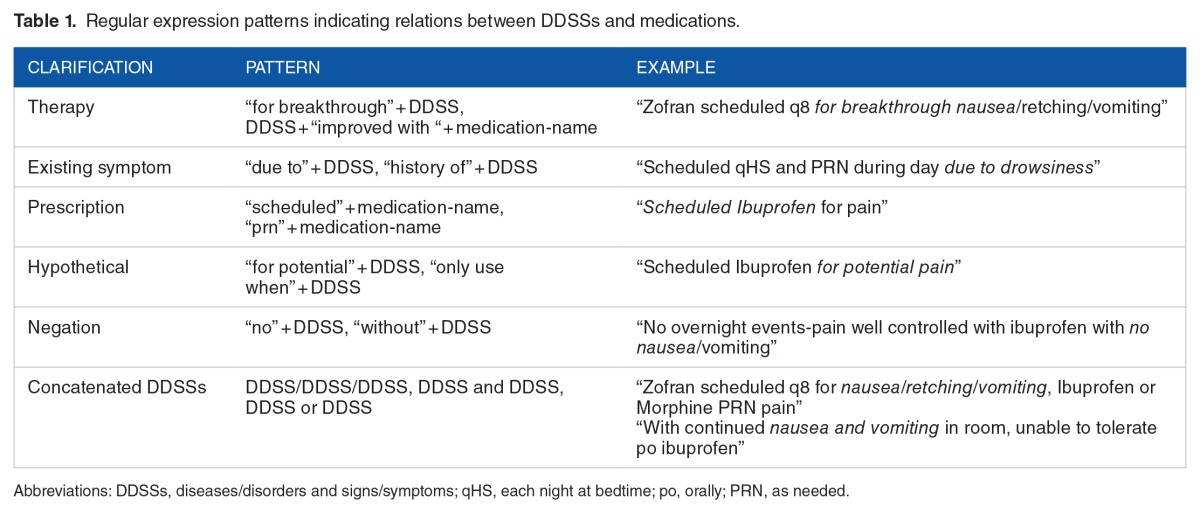
We used 3 methods to address these issues and ultimately reduce false-positive matches. First, we customized the Java implementation (GeneralNegEx)61 of the NegEx algorithm54 to identify and rule out negated DDSSs. To exclude hypothetical DDSSs and therapies that often occurred in the instruction and plan of care sections, we developed an additional list of regular expressions: (1) to exclude sentences with key words that indicated plans, hypothetical events, and instructions, etc and (2) to identify patterns indicating relations between DDSSs and medications and exclude sentences with patterns not related to ADRs. Table 1 shows the complete list of regular expression patterns in addition to NegEx. After the NLP post-processing, we searched the drug-reaction pairs in the output sentences to identify DPRSs.
Table 1.
Regular expression patterns indicating relations between DDSSs and medications.
Time-window filtering
Time windows are important because an ADR usually happens within a specific time period after the medication is given. Using all notes created during an encounter to detect ADRs could potentially cause false-positive alerts (eg, capturing sentences containing drug-reaction pairs before a medication was given). Although different ADRs could have different time windows even for the same medication, as a proof of concept, we used a flexible time window for all medications. That is, we only analyzed the clinical notes that were written after a medication was given and before a patient’s discharge. After the filtering, we considered the remaining DRPSs as ADR-related DRPSs.
Baseline system for identifying ADR-related DRPSs
For performance comparison, we generated a set of baseline ADR-related DRPSs without using the FAERS reports. In this baseline, we used a list of gold standard drugs and DDSSs annotations developed in our earlier study as candidate drug-reaction pairs.62 The drug-reaction set contained 4345 CUI represented DDSSs for 29 medications. The combination of 29 medications and 4345 DDSSs yielded a total number of 126 005 baseline candidate drug-reaction pairs. The workflow of generating ADR-related DRPSs was similar to the work-flow described in previous sections, except that we used the derived drug-DDSS set as candidate drug-reaction pairs instead of using drug-reaction pairs from the FAERS reports.
Results
Evaluation metrics
Given the ADR-related DRPSs predicted by the algorithms, we assessed the numbers of true-positive and false-positive predictions via manual chart review. A clinical informatics expert with a medical degree and a graduate student independently reviewed the targeted sentences to make the decisions. The F measure of inter-reviewer agreement was 81.5%. Differences were resolved and an adjudicated reference standard was created. We used PPV = true positive/(true positive + false positive) as the primary evaluation measure. To approximate recall measure, we calculated the RR between the proposed algorithm and the baseline by the following formula:
 |
Performance of ADR detection with FAERS reports
The algorithm identified 2475 ADR-related DRPSs from 1055 encounters. By removing repetitive sentences, there were a total of 1492 unique ADR-related DRPSs. We performed a stratified random sampling based on numbers of DRPSs for each medication and selected 528 (35.4%) unique ADR-related DRPSs for manual review (step 7 in Figure 1). Twelve medications did not have ADR-related DRPSs and were excluded from the evaluation. For the remaining 29 medications, Table 2 shows the true positives and PPVs achieved for each medication. In total, 224 DRPSs were marked as sentences associated with real ADRs and the overall PPV was 42.4% (95% confidence interval [CI]: [38.2%–46.6%]). The algorithm achieved PPVs of greater than or equal to 70% on 2 medications (morphine sulfate and lorazepam), PPVs of greater than or equal to 60% on 8 medications, and PPVs of greater than or equal to 40% on 11 medications. The algorithm had PPVs of 0% on 8 medications. However, the number of reviewed DRPSs on those medications was less than 10 (<1.8%). The PPVs for the other 11 medications were between 3.6% and 37.5%. Table 3 shows the true positives and PPVs on note types. The algorithm achieved the highest PPV on detecting ADRs from the ED notes (80%), followed by progress notes (45.8%), discharge summaries (38.1%), and H&P notes (25.9%).
Table 2.
Algorithm performance for each medication.
Table 3.
Algorithm performance for each note type.
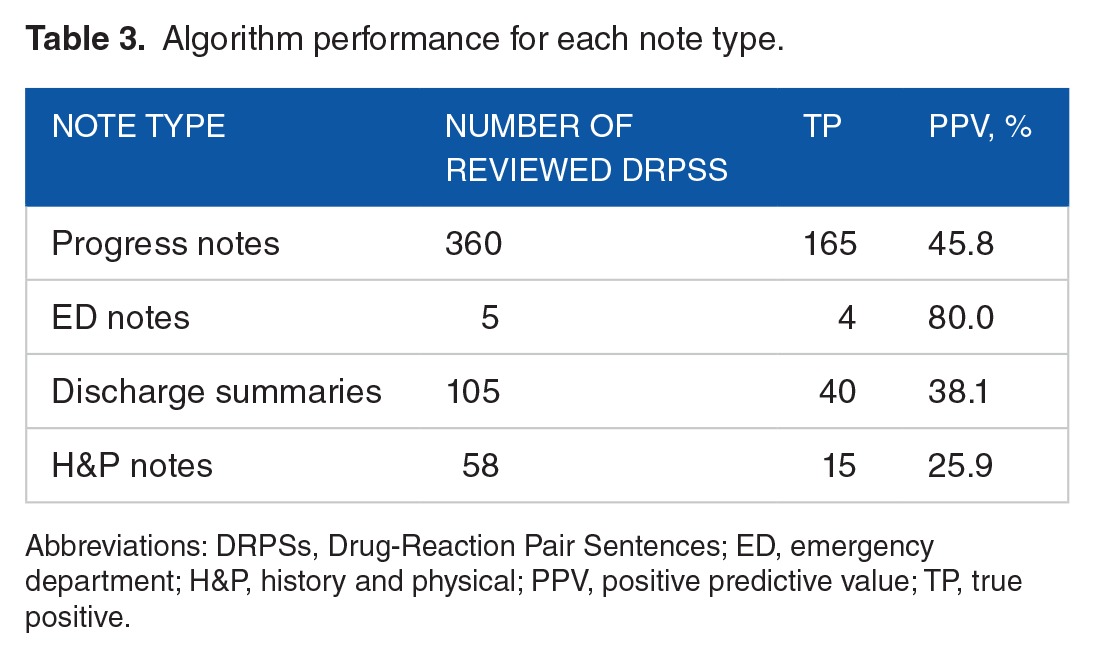
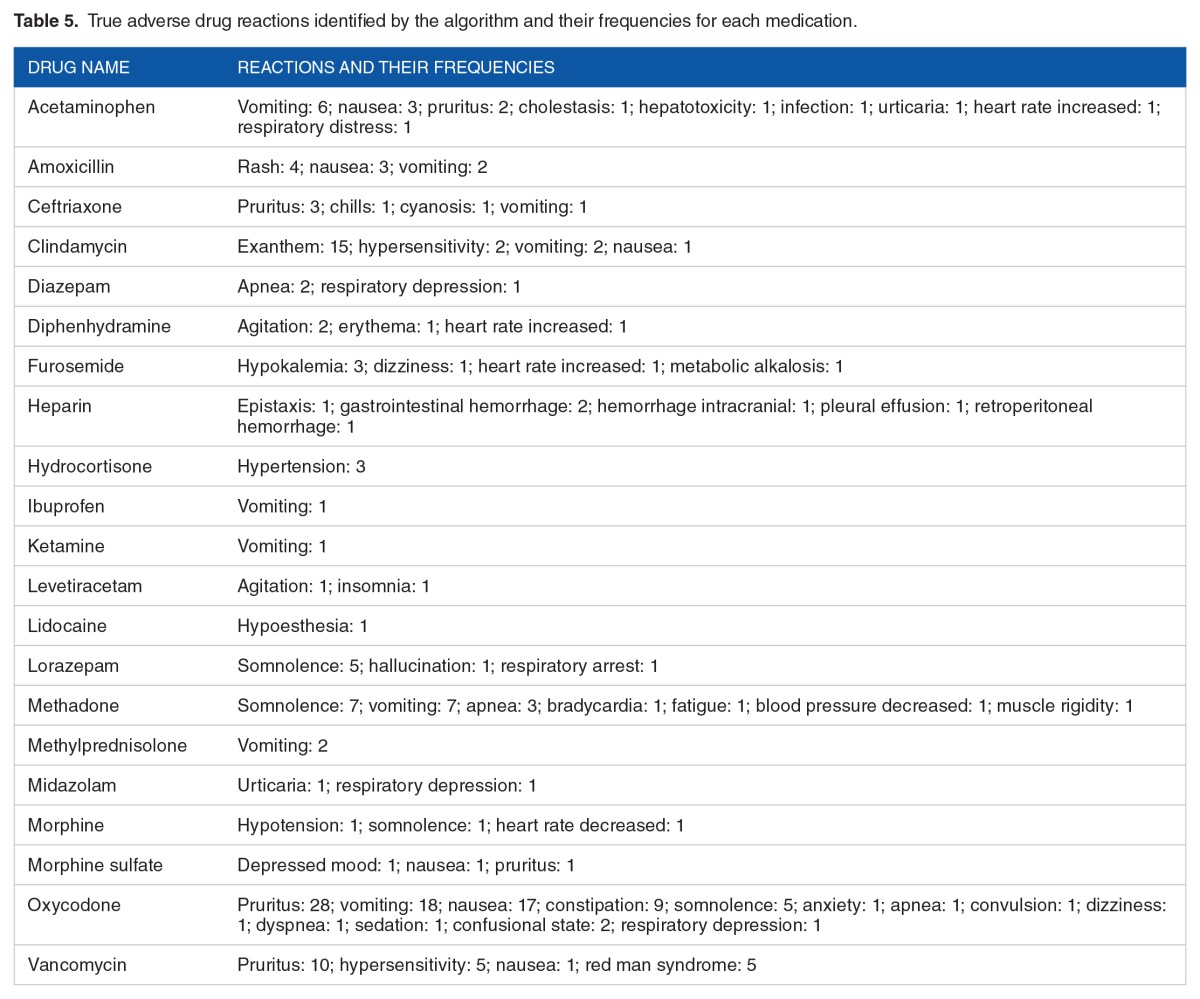
Table 4 shows the algorithm performance on each medication-note–type pair. There were 68 medication-note–type pairs in the evaluation set. Among them, the algorithm achieved 100% PPV on 9 medication-note–type pairs: acetaminophen, midazolam, vancomycin, and lidocaine in the ED notes; heparin, diphenhydramine, furosemide, and morphine in the discharge summaries; and methadone in the H&P notes. Table 5 lists the true ADRs identified by the algorithm for each medication and the frequencies of each reaction.
Table 4.
Algorithm performance for each medication in each note type.
Table 5.
True adverse drug reactions identified by the algorithm and their frequencies for each medication.
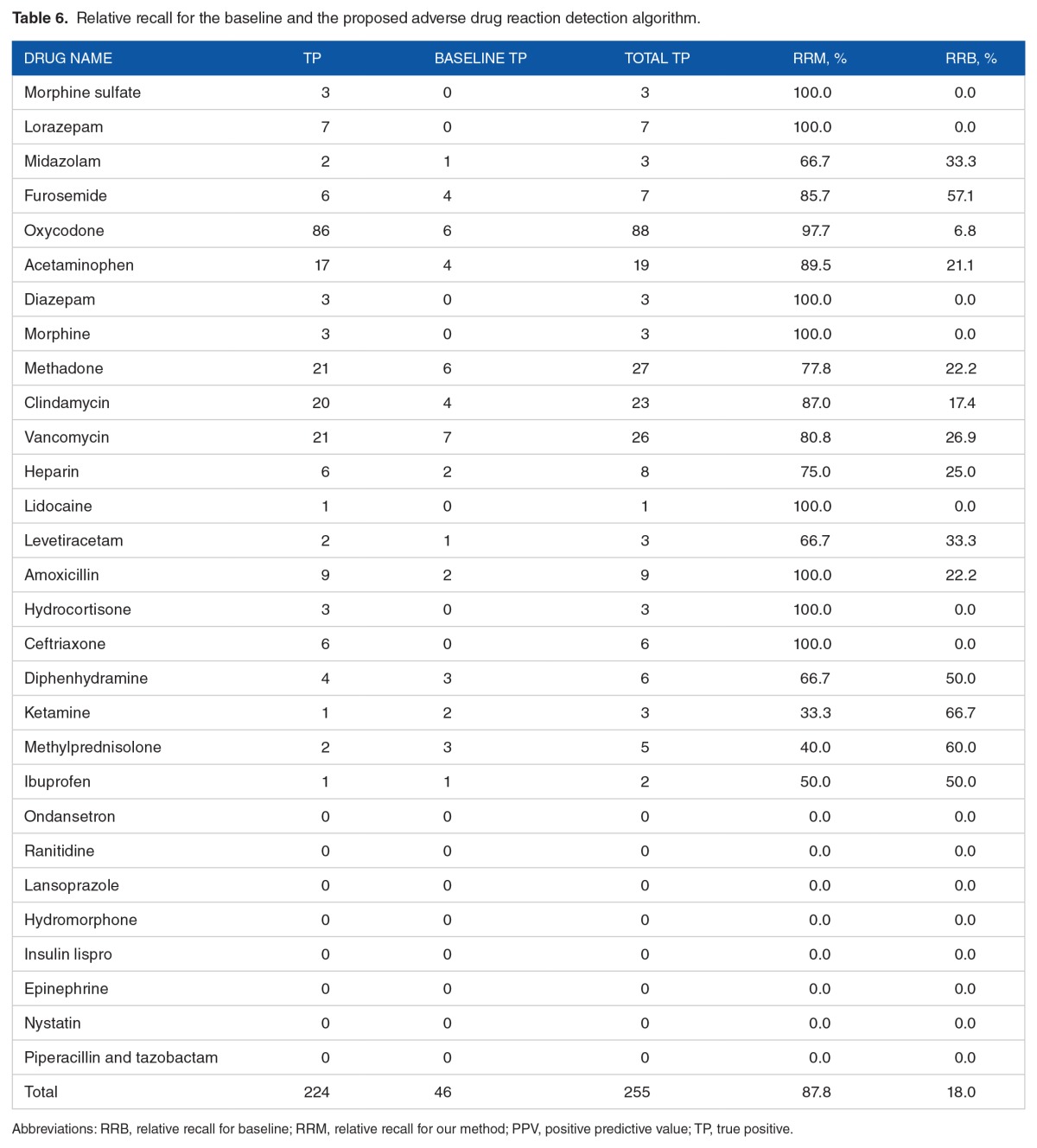
Finally, Table 6 presents the true-positive ADRs identified by the proposed algorithm and the baseline, and the derived RRs. The algorithm achieved an overall RR of 83% (95% CI: [79.8%–86.2%]), and the RR was more than 75% for 14 of the 29 medications.
Table 6.
Relative recall for the baseline and the proposed adverse drug reaction detection algorithm.
Performance of ADR detection using the baseline
The baseline algorithm identified 93 598 ADR-related DRPSs, in which there were a total of 32 778 unique DRPSs. Again, we performed a stratified random sampling to obtain 528 ADR-related DRPSs output by the baseline algorithm. For each medication, the number of review cases sampled for the baseline was identical to the number of reviewed cases randomly sampled for the algorithm leveraging FAERS reports. Based on our review, 46 out of 528 ADR-related DRPSs identified by the baseline algorithm were true ADRs. The PPV for the baseline was 8.7% (95% CI: [6.3%–11.1%]). Table 5 presents the RRs achieved by the baseline. The overall RR was 17.0% (95% CI: [13.8%–20.2%]), and the RR was less than 50% for most of the medications.
Discussion
By leveraging the FAERS reports, we developed an NLP-based algorithm to semiautomatically detect ADRs retrospectively in the EHR data. Our objective was to reduce the cost of manual development of drug-ADR pairs and mitigate the need to chart review clinical notes in the EHR. The use of FAERS reports for detecting ADRs in the EHR data is novel and the results are promising. Among a total of 60 085 encounters that contained 1 168 397 clinical notes and 161 817 736 sentences, our algorithm identified a total of 2475 sentences (from 2089 notes, 1055 encounters) as positive ADR cases. Leveraging information from the FAERS reports, the performance of ADR detection (PPV = 42.4%, RR = 83.0%) was statistically significantly better than that of the baseline (PPV = 8.7%, RR = 17.0%; P value < .001). In particular, the proposed algorithm yielded both better PPV and RR than the baseline for most of the medications.
The best PPV is 75% for morphine sulfate and the overall PPV is 42.4%. As a like-for-like comparison, the NLP-based algorithm developed by Wang et al26 achieved PPV of 31%. The NLP algorithm developed by Park et al27 for detecting laboratory abnormalities after treatment with medications has PPVs between 22% and 75%. The machine learning methods built by Ji et al34 and Lingren reached 16% and 80.4% PPV36 for identifying potential ADRs from EHRs, respectively. Most of the existing algorithms for ADR detection focused on certain medications. In contrast, our method provided a more generalizable solution for ADR detection that was not limited to certain medications or reactions. Ours is also more future proof approach because the FAERS database is constantly updated and provides the possibility of semiautomation to incorporate new drugs as they are introduced to the consumer market. It only requires an automated download of the newly reported FAERS drug-reaction pairs and a manual-based or script-based conversion of the brand-generic drug names. This is either a one-time or minimal continuous investment of funds and human resources. Compared with the machine learning algorithms that require manual annotation, our approach was almost fully automated and hence more cost-effective.
The algorithm performed better on the ED notes and the progress notes (Table 3), suggesting that the medication-reaction pairs found in these notes are more likely to indicate ADR events. This is because the ADR events occurred during patients’ hospitalization were usually documented in these notes other than H&P notes and discharge summaries. In addition, the algorithm achieved 100% PPV on 9 medication-note–type pairs (Table 4). The promising results show the great potential for integrating the algorithm into clinical practice to detect ADR events for these medications. Patient-specific correct ADRs listed in a table similar to Table 5, if shown in the EHR, could potentially benefit clinicians. It shows which reaction(s) could have been caused by a medication as well as the frequency/probability of the reactions on a medication and patient-specific basis. This is particularly important as health care moves toward personalized medicine.
Error analysis
We performed error analysis for the proposed algorithm by reviewing all 304 false-positive ADR predictions. Approximately 74% of the errors were due to lack of semantic analysis. For instance, the algorithm triggered a false positive (vancomycin-nausea) on “his nausea was worse on flagyl, so he was switched back to oral vancomycin” due to the miss of clause boundaries. In addition, the algorithm enumerated all medication-reaction pairs from a DRPS without analyzing the syntax of the sentence. For instance, the algorithm would detect 4 ADR-related events from the sentence “patient had vomiting last night, now having itching related to oxycodone use, discontinued oxycodone and started ibuprofen”: oxycodone-vomiting, oxycodone-itching, ibuprofen-vomiting, and ibuprofen-itching. Although it captured the correct ADR (oxycodone-itching), it also caused 3 false positives due to the sentence structure. To alleviate this problem in future works, we will implement advanced semantic parsing algorithms to analyze semantic and temporal relations in sentences.63–65 The omission of negation, temporal, and hypothetical expression detection on DRPSs also triggered a notable amount (22.69%) of false positives. For example, the algorithm identified an ADR-related event ibuprofen-allergy from the sentence “Patient also gives ibuprofen after Okay’ed by mom (has used in the past with no allergy)” due to the miss of the negated expression. Finally, the omission of excluding patient allergy section that never mentioned ADRs also contributed to 3.31% of the errors. In the future, we will add additional regular expression rules to these NLP components to improve their functionalities.
Limitations
The utilization of FAERS reports limits the use cases of the proposed ADR detection system. Because the FAERS only collects reported ADRs, the system could not detect events with unknown ADRs. In addition, the FAERS is not a gold standard collection of drug-reaction relations. Although we restricted the spectrum of FAERS drug-reaction pairs to physician-submitted reports only, some of these drug-reaction pairs might still be unrelated from a drug-reaction perspective. We will leverage sources additional to FAERS reports to improve the spectrum and accuracy of ADR detection in future studies.
Another limitation of our study is that the severity of the reactions was not analyzed. Severe reactions (eg, “severe pain”) should be treated immediately, other reactions (eg, “vomiting”) need attention and followed up by clinicians, and mild reactions (eg, “a little sleepiness”) might be assigned a low severity score and given low priority to be reviewed and clinically followed. Future work is in progress to identify severity and attribution of the ADRs to the medications. In addition, the study did not consider drug-drug interactions or dosage information. The patients were likely to take multiple drugs, and some drugs might interact with others and cause ADRs. Although drug dosage was collected in the study, we did not leverage it due to the complexity and computational power requirements. We will study drug-drug interaction and incorporate dosage information in future works. Furthermore, we used a fixed time window to detect ADR events in the study. In reality, different ADRs could have different reaction times even for the same medication. The PPV would be improved if we would develop specific and precise time windows for each drug-reaction pair.
Finally, the study has limitations with respect to algorithm evaluation. Selection bias may exist because we only used the EHR data from 1 pediatric institution. In addition, we could not fully evaluate algorithm recall results because no gold standard ADR reports existed for the data set. In future studies, we will involve more clinicians and pharmacists to generate gold standard sets and perform a more comprehensive evaluation including recall across health care institutions. The PPV-only evaluation statistics is a limitation but not unprecedented,13,66–69 and our study with additional RR statistics would contribute to the body of literature on EHR-based ADR detection.
Conclusions
By leveraging drug-reaction pairs from the FDA ADE reports, we developed an NLP-based, semiautomated algorithm to identify ADRs in the EHR data for the most frequently used medications. Using a clinician-generated, reference standard–based evaluation of real-world clinical data, the proposed algorithm achieved promising performance and showed great potential in identifying ADRs accurately. The experimental results suggested that leveraging drug-reaction pairs from the FAERS improves the performance of EHR-based ADR detection. The traditional methods to identify ADR in the EHR are manual, which are not scalable to handle a set of 161 817 736 sentences to find approximately 1000 ADRs. However, the proposed algorithm, when implemented in the clinical environments, could result in a substantial workload reduction for clinicians looking for ADRs in patients’ clinical notes.
Although the study focused on the most frequently used medications in an urban tertiary care pediatric institution, the algorithm is generalizable to all medications and a wide range of institutions with EHRs and clinical notes. Our study will pave the way to automate ADR detection in a more cost-effective manner.
Footnotes
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 3606 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was partially supported by 1R01LM012230-01 and 1U01HG008666-01 from the National Institutes of Health. H.T., I.S., E.K., H.Z., T.L., and Y.N. were also supported by internal funds from CCHMC.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
IS, YN, HT, EK, and JM conceived and designed the experiments; jointly developed the structure and arguments for the paper; and made critical revisions and approved final version. HT, YN, IS, HZ, TL analyzed the data. HT wrote the first draft of the manuscript. YN, IS, and HT contributed to the writing of the manuscript. HT, IS, EK, HZ, TL, JM, and YN agree with manuscript results and conclusions. All authors reviewed and approved the final manuscript.
Availability of Data and Materials
The FAERS reports analyzed are available at the FDA Adverse Event Reporting System, http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm083765.htm. The EHR data collected in this study are not publicly available because the data contain patient protected health information that cannot be shared outside the institution.
Consent for Publication
The study was approved by the institutional review board of CCHMC and a waiver of consent was authorized (study ID: 2010-3031).
Disclosures and Ethics
As a requirement of publication, author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality, and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Ethics Approval and Consent to Participate
The CCHMC institutional review board approved the study and a waived consent (study ID: 2012-2769).
REFERENCES
- 1.Nebeker JR, Barach P, Samore MH. Clarifying adverse drug events: a clinician’s guide to terminology, documentation, and reporting. Ann Intern Med. 2004;140:795–801. doi: 10.7326/0003-4819-140-10-200405180-00009. [DOI] [PubMed] [Google Scholar]
- 2.VA Center for Medication Safety Adverse Drug Events, Adverse Drug Reactions and Medication Errors: FAQ. 2006. pp. 1–4. https://www.va.gov/MS/Professionals/medications/Adverse_Drug_Reaction_FAQ.pdf.
- 3.Carspecken CW, Sharek PJ, Longhurst C, Pageler NM. A clinical case of electronic health record drug alert fatigue: consequences for patient outcome. Pediatrics. 2013;131:e1970–e1973. doi: 10.1542/peds.2012-3252. [DOI] [PubMed] [Google Scholar]
- 4.Honig PK. Advancing the science of pharmacovigilance. Clin Pharmacol Ther. 2013;93:474–475. doi: 10.1038/clpt.2013.60. [DOI] [PubMed] [Google Scholar]
- 5.Cullen DJ, Sweitzer BJ, Bates DW, Burdick E, Edmondson A, Leape LL. Preventable adverse drug events in hospitalized patients: a comparative study of intensive care and general care units. Crit Care Med. 1997;25:1289–1297. doi: 10.1097/00003246-199708000-00014. [DOI] [PubMed] [Google Scholar]
- 6.Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 277:301–306. [PubMed] [Google Scholar]
- 7.HHS Reducing and Preventing Adverse Drug Events to Decrease Hospital Costs. [Accessed January 1, 2015]. http://archive.ahrq.gov/research/findings/factsheets/errors-safety/aderia/ade.html. Published 2015.
- 8.Sohn S, Kocher J-P, Chute CG, Savova GK. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J Am Med Inform Assoc. 2011;18:i144–i149. doi: 10.1136/amiajnl-2011-000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagne J, Wang S, Rassen J, Schneeweiss S. A modular, prospective, semi-automated drug safety monitoring system for use in a distributed data environment. Pharmacoepidemiol Drug Saf. 2015;23:619–627. doi: 10.1002/pds.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates DW, Leape LL, Petrycki S. Incidence and preventability of adverse drug events in hospitalized adults. J Gen Intern Med. 1993;8:289–294. doi: 10.1007/BF02600138. [DOI] [PubMed] [Google Scholar]
- 11.Roque FS, Jensen PB, Schmock H, et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput Biol. 2011;7:e1002141. doi: 10.1371/journal.pcbi.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FDA The FDA Adverse Event Reporting System (FAERS) http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/. Published 2015.
- 13.Haerian K, Salmasian H, Friedman C. Methods for identifying suicide or suicidal ideation in EHRs. AMIA Annu Symp Proc. 2012;2012:1244–1253. [PMC free article] [PubMed] [Google Scholar]
- 14.Field TS, Gurwitz JH, Harrold LR, et al. Strategies for detecting adverse drug events among older persons in the ambulatory setting. J Am Med Inform Assoc. 2004;11:492–498. doi: 10.1197/jamia.M1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koppel R, Leonard CE, Localio AR, Cohen A, Auten R, Strom BL. Identifying and quantifying medication errors: evaluation of rapidly discontinued medication orders submitted to a computerized physician order entry system. J Am Med Inform Assoc. 2008;15:461–465. doi: 10.1197/jamia.M2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boockvar KS, Liu S, Goldstein N, Nebeker J, Siu A, Fried T. Prescribing discrepancies likely to cause adverse drug events after patient transfer. Qual Saf Health Care. 2009;18:32–36. doi: 10.1136/qshc.2007.025957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nwulu U, Nirantharakumar K, Odesanya R, McDowell SE, Coleman JJ. Improvement in the detection of adverse drug events by the use of electronic health and prescription records: an evaluation of two trigger tools. Eur J Clin Pharmacol. 2013;69:255–259. doi: 10.1007/s00228-012-1327-1. [DOI] [PubMed] [Google Scholar]
- 18.Carnevali L, Krug B, Amant F, et al. Performance of the adverse drug event trigger tool and the global trigger tool for identifying adverse drug events: experience in a Belgian hospital. Ann Pharmacother. 2013;47:1414–1419. doi: 10.1177/1060028013500939. [DOI] [PubMed] [Google Scholar]
- 19.Hébert G, Netzer F, Ferrua M, Ducreux M, Lemare F, Minvielle E. Evaluating iatrogenic prescribing: development of an oncology-focused trigger tool. Eur J Cancer. 2015;51:427–435. doi: 10.1016/j.ejca.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 20.DiPoto JP, Buckley MS, Kane-Gill SL. Evaluation of an automated surveillance system using trigger alerts to prevent adverse drug events in the intensive care unit and general ward. Drug Saf. 2015;38:311–317. doi: 10.1007/s40264-015-0272-1. [DOI] [PubMed] [Google Scholar]
- 21.Sampson M, Zhang L, Morrison A, et al. An alternative to the hand searching gold standard: validating methodological search filters using relative recall. BMC Med Res Methodol. 2006;6:33. doi: 10.1186/1471-2288-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chazard E, Ficheur G, Bernonville S, Luyckx M, Beuscart R. Data mining to generate adverse drug events detection rules. IEEE Trans Inf Technol Biomed. 2011;15:823–830. doi: 10.1109/TITB.2011.2165727. [DOI] [PubMed] [Google Scholar]
- 23.Chazard E, Bernonville S, Ficheur G, Beuscart R. A statistics-based approach of contextualization for adverse drug events detection and prevention. Stud Health Technol Inform. 2012;180:766–770. [PubMed] [Google Scholar]
- 24.Sarker A, Gonzalez G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J Biomed Inf. 2015;53:196–207. doi: 10.1016/j.jbi.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aramaki E, Miura Y, Tonoike M, et al. Extraction of adverse drug effects from clinical records. Stud Health Technol Inform. 2010;160:739–743. [PubMed] [Google Scholar]
- 26.Wang X, Chase H, Markatou M, Hripcsak G, Friedman C. Selecting information in electronic health records for knowledge acquisition. J Biomed Inform. 2010;43:595–601. doi: 10.1016/j.jbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park MY, Yoon D, Lee K, et al. A novel algorithm for detection of adverse drug reaction signals using a hospital electronic medical record database. Pharmacoepidemiol Drug Saf. 2011;20:598–607. doi: 10.1002/pds.2139. [DOI] [PubMed] [Google Scholar]
- 28.Epstein RH, St Jacques P, Stockin M, Rothman B, Ehrenfeld JM, Denny JC. Automated identification of drug and food allergies entered using non-standard terminology. J Am Med Inform Assoc. 2013;20:962–968. doi: 10.1136/amiajnl-2013-001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pathak J, Kiefer RC, Chute CG. Using linked data for mining drug-drug interactions in electronic health records. Stud Health Technol Inform. 2013;192:682–686. [PMC free article] [PubMed] [Google Scholar]
- 30.Sai K, Hanatani T, Azuma Y, et al. Development of a detection algorithm for statin-induced myopathy using electronic medical records. J Clin Pharm Ther. 2013;38:230–235. doi: 10.1111/jcpt.12063. [DOI] [PubMed] [Google Scholar]
- 31.Sauzet O, Carvajal A, Escudero A, Molokhia M, Cornelius VR. Illustration of the weibull shape parameter signal detection tool using electronic healthcare record data. Drug Saf. 2013;36:995–1006. doi: 10.1007/s40264-013-0061-7. [DOI] [PubMed] [Google Scholar]
- 32.Overby CL, Pathak J, Gottesman O, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc. 2013;20:e243–e252. doi: 10.1136/amiajnl-2013-001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirkendall ES, Spires WL, Mottes TA, Schaffzin JK, Barclay C, Goldstein SL. Development and performance of electronic acute kidney injury triggers to identify pediatric patients at risk for nephrotoxic medication-associated harm. Appl Clin Inform. 2014;5:313–333. doi: 10.4338/ACI-2013-12-RA-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji Y, Ying H, Dews P, et al. A potential causal association mining algorithm for screening adverse drug reactions in postmarketing surveillance. IEEE Trans Inf Technol Biomed. 2011;15:428–437. doi: 10.1109/TITB.2011.2131669. [DOI] [PubMed] [Google Scholar]
- 35.Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinformatics. 2010;11(Suppl 9):S7. doi: 10.1186/1471-2105-11-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lingren TG. Detecting Adverse Events in Clinical Trial Free Text [thesis] University of Washington; 2013. [Accessed May 30, 2017]. https://digital.lib.washington.edu/research-works/handle/1773/24070. [Google Scholar]
- 37.Li Q, Melton K, Lingren T, et al. Phenotyping for patient safety: algorithm development for electronic health record based automated adverse event and medical error detection in neonatal intensive care. J Am Med Inform Assoc. 2014;21:776–784. doi: 10.1136/amiajnl-2013-001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon D, Park MY, Choi NK, Park BJ, Kim JH, Park RW. Detection of adverse drug reaction signals using an electronic health records database: comparison of the Laboratory Extreme Abnormality Ratio (CLEAR) algorithm. Clin Pharmacol Ther. 2012;91:467–474. doi: 10.1038/clpt.2011.248. [DOI] [PubMed] [Google Scholar]
- 39.Liu M, McPeek Hinz ER, Matheny ME, et al. Comparative analysis of pharmacovigilance methods in the detection of adverse drug reactions using electronic medical records. J Am Med Inform Assoc. 2013;20:420–426. doi: 10.1136/amiajnl-2012-001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henao R, Murray J, Ginsburg G, Carin L, Lucas JE. Patient clustering with uncoded text in electronic medical records. AMIA Annu Symp Proc. 2013;2013:592–599. [PMC free article] [PubMed] [Google Scholar]
- 41.Van Mulligen EM, Fourrier-Reglat A, Gurwitz D, et al. The EU-ADR corpus: annotated drugs, diseases, targets, and their relationships. J Biomed Inform. 2012;45:879–884. doi: 10.1016/j.jbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Ginn R, Pimpalkhute P, Nikfarjam A, et al. Mining Twitter for adverse drug reaction mentions: a corpus and classification benchmark; Proceedings of Fourth Workshop on Building and Evaluating Resources for Health and Biomedical Text Processing; Harpa, Iseland: 2014. [Google Scholar]
- 43.Karimi S, Metke-Jimenez A, Kemp M, Wang C. Cadec: a corpus of adverse drug event annotations. J Biomed Inform. 2015;55:73–81. doi: 10.1016/j.jbi.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Deleger L, Lingren T, Ni Y, et al. Preparing an annotated gold standard corpus to share with extramural investigators for de-identification research. J Biomed Inform. 2014;50:173–183. doi: 10.1016/j.jbi.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Spooner SA, Kaiser M, et al. An end-to-end hybrid algorithm for automated medication discrepancy detection. BMC Med Inform Decis Mak. 2015;15:37. doi: 10.1186/s12911-015-0160-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni Y, Kennebeck S, Dexheimer JW, et al. Automated clinical trial eligibility prescreening: increasing the efficiency of patient identification for clinical trials in the emergency department. J Am Med Inform Assoc. 2014;22:166–178. doi: 10.1136/amiajnl-2014-002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. Evaluation of negation phrases in narrative clinical reports. Proc AMIA Symp. 2001;2001:105–109. [PMC free article] [PubMed] [Google Scholar]
- 49.Auerbuch M, Karson TH, Ben-Ami B, Maimon O, Rokach L. Context-sensitive medical information retrieval. Stud Health Technol Inform. 2004;107:282–286. [PubMed] [Google Scholar]
- 50.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 51.Elkin PL, Brown SH, Bauer BA, et al. A controlled trial of automated classification of negation from clinical notes. BMC Med Inform Decis Mak. 2005;5:13. doi: 10.1186/1472-6947-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents. J Am Med Inform Assoc. 2001;8:598–609. doi: 10.1136/jamia.2001.0080598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42:839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman WW, Hillert D, Velupillai S, et al. Extending the NegEx lexicon for multiple languages. Stud Health Technol Inform. 2013;192:677–681. [PMC free article] [PubMed] [Google Scholar]
- 55.Goryachev S, Sordo M, Zeng Q, et al. Implementation and Evaluation of Four Different Methods of Negation Detection. Boston, MA: DSG; 2006. https://www.i2b2.org/software/projects/hitex/negation.pdf. [Google Scholar]
- 56.Lexicomp Lexicomp Online Database. http://online.lexi.com/. Published 2015.
- 57.NIH Concept unique identifier. [Accessed January 1, 2015]. http://www.nlm.nih.gov/research/umls/new_users/online_learning/Meta_005.html. Published 2015.
- 58.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edwards I, Aronson J. Adverse drug reactions: definitions, diagnosis, and management. Lancet. 2000;356:1255–1259. doi: 10.1016/S0140-6736(00)02799-9. [DOI] [PubMed] [Google Scholar]
- 60.NIH UMLS terminology browser. [Accessed January 1, 2015]. https://uts.nlm.nih.gov/home.html. Published 2015.
- 61.Solti I, Kye J. General NegEx Java Implementation v.2.0. https://code.google.com/p/negex/downloads/detail?name=GeneralNegEx.Java.v.2.0_10272010.zip. Published 2010.
- 62.Deleger L, Li Q, Lingren T, et al. Building gold standard corpora for medical natural language processing tasks. AMIA Annu Symp Proc. 2012;2012:144–153. [PMC free article] [PubMed] [Google Scholar]
- 63.Bundschus M, Dejori M, Stetter M, Tresp V, Kriegel HP. Extraction of semantic biomedical relations from text using conditional random fields. BMC Bioinform. 2008;9:207. doi: 10.1186/1471-2105-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ben Abacha A, Zweigenbaum P. Automatic extraction of semantic relations between medical entities: a rule based approach. J Biomed Semantics. 2011;2:S4. doi: 10.1186/2041-1480-2-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mintz M, Bills S, Snow R, et al. Distant supervision for relation extraction without labeled data; Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP; Singapore: Suntec; Aug, 2009. pp. 1003–1011. [DOI] [Google Scholar]
- 66.Handler SM, Becich MJ. Detecting Adverse Drug Reactions in the Nursing Home Setting Using a Clinical Event Monitor [doctoral dissertation] Pittsburgh, PA: Biomedical Informatics, University of Pittsburgh; 2010. [Google Scholar]
- 67.Oberaigner W, Buchberger W, Frede T, et al. Introduction of organised mammography screening in Tyrol: results of a one-year pilot phase. BMC Public Health. 2011;11:91. doi: 10.1186/1471-2458-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kunisaki KM, Bohn OA, Wetherbee EE, Rector TS. High-resolution wrist-worn overnight oximetry has high positive predictive value for obstructive sleep apnea in a sleep study referral population. Sleep Breath. 2015;20:583–587. doi: 10.1007/s11325-015-1251-6. [DOI] [PubMed] [Google Scholar]
- 69.Pérez-Topete SE, Miranda-Aquino T, Hernández-Portales JA. Valor predictivo positivo de la prueba de inmunoanálisis para detección de toxina A y B de Clostridium difficile en un hospital privado. Rev Gastroenterol México. 2016;81:190–194. doi: 10.1016/j.rgmx.2016.04.002. [DOI] [PubMed] [Google Scholar]