Abstract
Background
The effects of PPI are variable owing to the CYP2C19 polymorphisms. However, whether the polymorphisms could affect the Hp eradication efficacy of triple therapy is still not clear. The present study aimed to assess the effects of CYP2C19 gene polymorphisms on proton pump inhibitor (PPI), amoxicillin, and levofloxacin triple therapy for Helicobacter pylori (Hp) eradication.
Material/Methods
We randomly assigned 160 Hp-positive patients with chronic gastritis to 2 groups to receive either 20 mg bid omeprazole (OAL group, n=80) or 10 mg bid rabeprazole (RAL group, n=80), combined with 1000 mg bid amoxicillin and 500 mg qd levofloxacin. The 2 groups were treated for 10 days. The CYP2C19 genotypes included wild-type, M1 mutant gene (*2, the mutation of exon 5), and M2 mutant gene (*3, the mutation of exon 4) identified by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFIP). According to CYP2C19 genotype combinations, the patients were divided into extensive metabolizer (EM), intermediate metabolizer (IM), and poor metabolizer (PM) subgroups. The eradication efficacy of Hp was evaluated by 14C-UBT at 28 days after treatment.
Results
The trial was completed by 155 patients. Hp eradication rates in OAL and RAL groups were 78.2% and 88.3%, respectively, on per-protocol (PP) analysis, indicating no significant difference (P>0.05). Regarding CYP2C19 genotypes, eradication rates of 60.7%, 84.2%, and 100% were obtained for EM, IM, and PM subgroups, respectively, of the OAL group. EM group eradication rates were significantly lower than IM and PM group values (P<0.05). In the RAL group, no such difference was observed (P>0.05). Hp eradication rates were significantly lower in the EM subgroup of the OAL group compared with that of the RAL group.
Conclusions
Hp eradication rates were higher in the RAL group than in OAL-treated patients. Interestingly, omeprazole-based therapy was significantly affected by the CYP2C19 genotype, unlike the rabeprazole-based therapy.
MeSH Keywords: Amoxicillin, Helicobacter pylori, Levofloxacin, Proton Pump Inhibitors
Background
Helicobacter pylori (Hp) infection is an important risk factor for digestive ulcer, gastric mucosa-associated lymphoid tissue lymphoma (MALT), chronic gastritis with indigestion, gastric mucosa atrophy or erosion, gastric polyps, malignant tumors, and other diseases of the digestive system [1]. Long-term follow-up showed that Hp eradication has an important role in peptic ulcer treatment and Hp-related chronic gastritis prognosis, as well as in reversing gastric mucosa-associated lymphoid tissue lymphoma progression. Hp eradication has a preventive effect by eliminating the precancerous lesions of gastric cancer [2].
Proton pump inhibitors (PPIs) increase gastric pH by inhibiting acid secretions, and enhance the activity of oral antibiotics while inhibiting Hp; therefore, they are widely used in the clinical treatment of acid-related diseases. Currently, triple therapy based on PPI combination with 2 antibiotics is commonly used in Hp eradication treatment [3]. In the traditional standard triple therapy, the most commonly used antibiotics are amoxicillin, metronidazole, and clarithromycin. Recently, a Chinese Hp resistance survey indicated that the Hp resistance rates towards metronidazole and clarithromycin were 75.6% and 27.6%, respectively, in many areas of China [4].
Therefore, Hp eradication with standard triple therapy is becoming less common and cannot satisfy the target demand. In recent years, a number of studies suggested that triple therapy containing levofloxacin has better Hp eradication rates [5,6]. The latest Maastricht IV/Florence Consensus Report and Fourth Chinese National Consensus Report on the management of Helicobacter pylori infection both recommend triple therapy containing levofloxacin in Hp eradication strategies, especially amoxicillin combined with levofloxacin based on PPI as the initial treatment scheme [6].
Proton pump inhibitor (PPI) is an important basic part in Hp eradication schemes [7]. Omeprazole, as the first-generation representative PPI drug, is one of the most commonly used proton pump inhibitors and has been widely studied for its pharmacogenetics properties; it is mainly involved in CYP2C19 and CYP3A4 metabolic pathways [8]. The affinity of omeprazole for CYP3A4 is 10 times lower than for CYP2C19, and its metabolism is mainly through CYP2C19 in vivo [9]; therefore, omeprazole efficacy is significantly affected by CYP2C19 gene polymorphisms. Rabeprazole is a new-generation PPI developed in recent years, with unique pharmacokinetic characteristics; its dependence on liver drug metabolizing enzymes is reduced, but it is transformed into sulfide rabeprazole mainly through the non-liver enzyme pathway. Therefore, CYP2C19 gene polymorphisms minimally affect rabeprazole metabolism. After oral rabeprazole intake, AUC values for the PM group are only 1.2 times higher than those of the EM group; anti-acid effects for both groups do not reach ideal levels, suggesting that the effects on intragastric pH do not depend on the CYP2C19 genotype [10].
For the past few years, studies have demonstrated great diversity in pharmacokinetic and pharmacodynamic aspects (e.g., bioavailability, metabolism, drug interactions, and biological specificity) among different PPIs [11–13]. The therapeutic effects observed in different populations using the same PPI are variable, mainly due to the interactions between the drugs and genetic factors, including Cytochrome P450 2C19 (CYP2C19) polymorphisms [14,15]. CYP2C19 (NCBI gene ID: 1557), also known as S-mephenytoin hydroxylase, belongs to the protein family of the cytochrome P450 mixed-function oxidase system, and is involved in the metabolism of xenobiotics such as PPIs and anti-epileptics [16]. Its gene defects directly affect enzyme activity and, consequently, drug efficacy. CYP2C19 has been shown to display at least 30 alleles, including 23 encoding a protein (CYP2C19*1 is the gene yielding the normal enzyme activity) [17,18]. De Morais et al. found that CYP2C19 mainly shows 2 mutant alleles [19], CYP2C19*2 and CYP2C19*3, which were previously known as CYP2C19 M1 and CYP2C19 M2; both of them encode nonfunctional proteins due to single-nucleotide mutations. CYP2C19*2 is the main CYP2C19 mutation in Chinese individuals [18]. According to CYP2C19 genotype differences, 3 metabolic types can be distinguished: no mutation, strong metabolism type (EM); one-locus mutated, intermediate metabolizer (IM); and 2 loci mutated, weak metabotropic (PM).
The present study aimed to explore the effects of CYP2C19 gene polymorphisms on efficacy of triple therapy with amoxicillin combined with levofloxacin based on proton pump inhibitor (PPI) for the eradication treatment of Hp-positive chronic gastritis, and provide new insights to guide individual treatments and reasonable adjustment of drug dosage to ensure drug efficacy.
Material and Methods
Patients
This study enrolled 160 patients with dyspeptic symptoms, including 77 males and 83 females, diagnosed with Hp-positive chronic gastritis by endoscopy in Guangzhou First Municipal People’s Hospital from May to December 2014. The subjects were all of Han ethnicity, with an average age of 45.2±12.6 years (18 to 70 years).
Inclusion criteria were: (1) diagnosis of Hp-positive chronic gastritis by gastroscopy in parallel to rapid urease test, (2) aged from 18 to 70 years, (3) no use of antimicrobial drugs within 2 weeks of enrolment, and (4) willingness to undergo Hp eradication therapy.
Exclusion criteria were: (1) comorbidity with peptic ulcer, Zollinger Ellison syndrome, reflux esophagitis, esophageal varices, gallstones; a history of stomach surgery and other digestive diseases; (2) coexistence of serious underlying diseases such as severe liver disease, pulmonary heart disease, kidney disease, malignant tumors; (3) treatment with antibiotics and bismuth, H2 receptor antagonists, and proton pump inhibitors within 2 weeks of enrolment; (4) treatment with nonsteroidal anti-inflammatory drugs, antiplatelet drugs, anticoagulant drugs for gastric mucosal damage, or phenytoin, diazepam and other drugs that affect CYP2C19 within 3 months of enrolment; (5) allergies to study drugs; and (6) pregnant or lactating women.
Exit criteria included: (1) condition aggravation or occurrence of other serious complications; (2) inability to tolerate adverse drug reactions; (3) pregnancy or other symptoms interfering with the study arising during treatment; and (4) loss to follow-up and voluntary withdrawal.
This study was approved by the Ethics Committee of Guangzhou First Municipal People’s Hospital, and each patient provided signed informed consent.
Hp testing standards
Each piece of mucosa from the gastric antrum to body of stomach was examined by gastroscopy, and a rapid urease test was carried out. The effect of HP eradication therapy was detected with 14C urea breath test at 4 weeks after therapy end and withdrawal of acid-suppressive drugs.
Rapid urease test
Phenol reagent and urea were mixed evenly to pale yellow according at the proportion of 10 ml: 0.2 g as a pH indicator. The gastric mucosal tissue was immediately placed in the mixture and color change was observed after 30 min; Hp presence was reflected by the color changing from pale yellow to red.
14C urea breath assay
Urea [14C] breath test kits were purchased from Haidewei Biotechnology Co., Ltd. (China), and used according to the manufacturer’s instructions. Fasting subjects took a capsule containing urea [14C] with 20 ml of cold water, and sat still for 25 min before blowing through a catheter to the CO2 collecting agent until it became colorless. Then, 4.5 ml diluted scintillation fluid was added to the sample bottle and stamped with a seal, shaken up before assessing radioactivity of 14C samples for 2 min on a liquid flash instrument. Hp was considered to be present when 14C-UBT was greater than or equal to 100 dpm/mmol CO2.
Grouping and treatment scheme
Patients were randomly divided into 2 groups, and received levofloxacin triple therapy for Hp eradication therapy. The omeprazole group (OAL group, 80 cases) was treated with 20 mg omeprazole (Losec, AstraZeneca Pharmaceutical Co., Ltd) twice daily, in combination with 1000 mg amoxicillin (AMO Xian, Zhuhai federal Pharmaceutical Co. Ltd.) twice a day and 500 mg levofloxacin (Cravit, Daiichi Sankyo Co. Ltd.) once daily, for 10 days. The rabeprazole group (RAL group, 80 cases) received 10 mg rabeprazole (Pollitt Lei Bei, Eisai Co. Ltd.) per os twice a day, in combination with amoxicillin and levofloxacin at the above regimens, also for 10 days.
CYP 2C19 genotype assessment
Genomic DNA was extracted from 2 ml EDTA anticoagulated peripheral venous blood obtained from fasting patients, using the Blood DNA TIANamp Kit (Tiangen Biotech Co., Ltd., Beijing, China). CYP2C19 (NCBI gene ID: 1557) genotype analysis was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technology on a Bio-Rad S1000 PCR instrument, and the primer sequences are shown in Table 1. Three main alleles were found: wild-type (CYP2C19*1) and 2 mutants (mutations in exon 5 for CYP2C19*2 and in exon 4 for CYP2C19*3) (Supplementary Figure 1).
Table 1.
PCR primers.
| Genotype | Primer sequence |
|---|---|
| CYP2C19*2 | Forward 5′-AATTACAACCAGAGCTTGGC-3′ |
| Reverse 5′-TATCACTTTCCATAAAAGCAAG-3′ | |
| CYP2C19*3 | Forward 5′-TATTATTATCTGTTAACTAATATGA-3′ |
| Reverse 5′-ACTTCAGGGCTTGGTCAATA-3′ |
Genomic DNA characterization
DNA samples were examined for concentration and purity on an ultraviolet spectrophotometer and agarose gel electrophoresis, and qualified specimens were selected: concentration >50 ng/μL, OD260/OD280 ratio between 1.5 and 2.0, gel electrophoresis bands clear, and integrity >20 kb, ensuring DNA integrity and purity compatible with the PCR method.
PCR amplification
Genomic DNA from patient blood samples was used as template with the specific primers described in Table 2. PCR was carried out with the Taq PCR Master Mix kit (TianGen Biotech Co., Ltd.) according to the manufacturer’s instructions. Briefly, a total reaction volume of 25 μl contained DNA template (5 μl), upstream and downstream primers (10 μM, 1 μl each), 2×Taq PCR Master Mix (12.5 μl), and ddH2O to 25 μl. The reaction conditions were: 94°C for 3 min (initial denaturation); 30 cycles of 94°C for 30 s (denaturation), 56 °C (exon 5) or 53 °C (exon 4) for 30 s (annealing), and 72°C for 1 min (extension); and final extension at 72°C for 5 min. PCR products of exon 5 carrying CYP2C19*2 mutation sites and exon 4 with CYP2C19*3 mutation sites were assessed by 2% agarose gel electrophoresis.
Table 2.
Baseline patient data.
| OAL group (n= 78) | RAL group (n=77) | P value | |
|---|---|---|---|
| Male (%) | 37 (47.4%) | 39 (50.6%) | 0.160 |
| Age (year) | 44.62±12.23 | 45±12.83 | 0.848 |
| Smoking (%) | 22 (28.2%) | 24 (31.2%) | 0.163 |
| Alcohol taking (%) | 9 (11.5%) | 10 (13.0%) | 0.076 |
| CYP2C19 genotype | |||
| EM (%) | 28 (35.9%) | 26 (33.8%) | 0.078 |
| IM (%) | 38 (48.7%) | 40 (51.9%) | 0.162 |
| PM (%) | 12 (15.4%) | 11 (14.3%) | 0.37 |
Alcohol taking: Drink at least 4 times a week for the past 30 days.
Enzyme digestion
Each PCR product (exon 5 and exon 4) was cut by restriction endonucleases SmaI and BamHI (NEB Co., Ltd.), respectively. Exons 5 and 4 were digested (37°C) for 15 min according to the manufacturer’s instructions.
Agarose gel electrophoresis
Electrophoresis of enzyme digestion products was carried out on 2% agarose gel followed by band observation under ultraviolet light; the genotypes were assessed by standard DNA molecular techniques. Figure 1 shows polymorphism bands of CYP2C19 *2 and CYP2C19*3.
Figure 1.
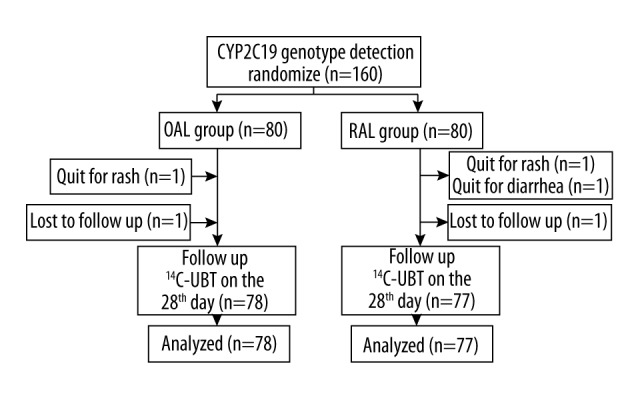
Flow chart of patient enrolment.
Statistical analysis
General patient information was compared between the 2 groups using the t test and χ2 test, as appropriate; HP eradication rate differences were analyzed by the χ2 test. Intention to treat (ITT) and per-protocol (PP) analyses were performed in comparing total HP eradication rates of the 2 treatment groups. In ITT analysis, a sample was retained in the original group for analysis regardless of whether treatment was completed, but PP analysis only included samples with completed treatment in the final analysis. The statistical software SPSS 18 was employed in this study; P<0.05 was considered statistically significant.
Results
General patient data
A total of 160 subjects were enrolled in this study, with 155 completing the treatment, including 76 males and 79 females, aged between 18 and 70 years (average 44.8±12.5 years). There were 46/155 smokers (29.6%) and 19/155 drinkers (12.3%). In the OAL group, 78 cases completed the treatment: 1 case withdrew due to a rash that healed without special treatment, while another was lost to follow-up. The RAL group included 77 cases who completed the treatment: 2 cases dropped out due to adverse reactions (1 case each of skin rash and diarrhea, who were healed without special treatment) and a third patient was lost to follow-up (Figure 1). There were no significant differences in age, sex, genotype, smoking, drinking, and compliance between the 2 treatment groups (Table 2, P>0.05). Of the 155 patients, a total of 54 (34.8%) were of EM type (genotype, *1/*1), including 28 and 26 from OAL and RAL groups, respectively. There were 78 cases (50.3%) of IM type (genotype, *1/*3 or *1/*2), with 38 and 40 from the OAL and RAL groups, respectively. Finally, a total of 23 cases (14.8%) were of PM type (genotype, *2/*3, *1/*2 or *3/*3), with 12 OAL group cases and 11 RAL group individuals. There were no significant differences between the 2 groups (Table 2, P>0.05).
Hp eradication rates
Hp eradication rates by PP and ITT analyses of the OAL group were 78.2% and 76.3%, respectively; in the RAL group, eradication rates of 88.3% and 85%, respectively, were obtained. The differences in HP eradication rates between the 2 treatment groups were not statistically significant by any analysis method (Table 3, P>0.05).
Table 3.
Hp eradication rates in different treatment groups.
| Analyze method | OAL group | RAL group | P value |
|---|---|---|---|
| PP analysis | 61/78 (78.2%) | 68/77 (88.3%) | 0.092 |
| ITT analysis | 61/80 (76.3%) | 68/80 (85%) | 0.161 |
Genotype distribution and HP eradication rate
Hp eradication rates of different genotypes were compared between the 2 treatment groups (Table 4). In the OAL group, Hp eradication rates of EM, IM, and PM subgroups were 60.7%, 84.2%, and 100%, respectively, indicating statistically significant differences among the 3 genotypes. Indeed, the EM type showed significantly lower eradication rates compared with the IM and PM subgroups (P<0.05); however, the differences between IM and PM showed no statistical significance (P>0.05). Hp eradication rates of EM, IM, and PM subgroups were 84.6%, 86%, and 100%, respectively, in the RAL group; there was no statistically significant difference among these 3 genotypes (P>0.05). A comparison of Hp eradication rates of the same genotype between both groups suggested that the EM type of the OAL group had marginally lower values than that of the RAL group (P=0.05) (Table 5), while HP eradication rates of IM and PM genotypes did not differ significantly between treatment groups (P>0.05).
Table 4.
Hp eradication rates of patients in the same treatment based on genotypes.
| Treatment groups | Genotype | Hp eradication rate | P value | |
|---|---|---|---|---|
| Multi comparison | Comparison between two groups | |||
| OAL group | EM | 60.7% (17/28) | 0.010 | 0.031* |
| IM | 84.2% (32/38) | 0.338** | ||
| PM | 100% (12/12) | 0.030*** | ||
| RAL group | EM | 84.6% (22/26) | 0.401 | 1.000* |
| IM | 86.0% (35/40) | 0.508** | ||
| PM | 100% (11/11) | 0.296*** | ||
EM vs. IM;
IM vs. PM;
EM vs. PM.
Table 5.
Hp eradication rate of patients with the same genotypes based on treatment groups.
| Genotype | Treatment group | Hp eradication rate | P value |
|---|---|---|---|
| EM | OAL | 60.7% (17/28) | 0.050 |
| RAL | 84.6% (22/26) | ||
| IM | OAL | 84.2% (32/38) | 0.677 |
| RAL | 86.0% (35/40) | ||
| PM | OAL | 100% (12/12) | 1.000 |
| RAL | 100% (11/11) |
Discussion
CYP2C19 metabolic polymorphisms show individual and population differences. For instance, multiple studies showed that PM mutation rates in whites and Asians are 3–5% and 13–23%, respectively, with blacks showing intermediate rates [20]. Wang et al. found that CYP2C19 mainly shows 2 point mutations (M1 and M2) in 155 unrelated healthy Han volunteers, with incidence rates for EM, IM, and PM of 36.8% (57/155), 47.7% (74/155), and 15.5% (24/155), respectively [21]. The present study showed that the genotype distribution was 34.8% EM (54/155), 50.3% IM (78/155), and 14.9% PM (23/155), corroborating the above findings.
This study showed that HP eradication by triple therapy containing levofloxacin and based on the first generation of PPI omeprazole is markedly affected by CYP2C19 gene polymorphism, while a similar triple therapy based on rabeprazole is more stable and less affected by CYP2C19 gene polymorphism. These findings indicate that the impact of CYP2C19 gene polymorphism on different PPIs is similar to that observed in standard triple therapy, with treatment containing levofloxacin and based on the new generation PPI rabeprazole having higher Hp eradication ratio and smaller individual differences.
As an important clinical treatment tool, CYP2C19 genotype detection method has been widely applied because it is rapid, simple, affordable, and suitable for use in all levels of hospital. Patients should be submitted to CYP2C19 genotyping before treatment of acid-related diseases and eradication of Hp. This would contribute to providing individualized PPI therapy for different patients, adjusting drug use based on gene polymorphism, and developing the best solutions for further treatment optimization. Selecting drug and administration regimen according to the genetic polymorphism of different individuals will provide a strong scientific basis for rational drug use and better efficacy, with fewer adverse reactions and lower costs.
Conclusion
Eradication rates of Hp were higher for levofloxacin-containing triple therapy in the RAL group than in OAL-treated patients. Interestingly, Hp eradication rates with omeprazole-based levofloxacin-containing triple therapy were significantly affected by the CYP2C19 genotype. The efficacy of rabeprazole-based levofloxacin-containing triple therapy was relatively stable and less affected by the CYP2C19 genotype. These findings provide additional insights that can help develop individualized therapies for Hp eradiation, with rational drug use, better efficacy, fewer adverse reactions, and lower costs.
Supplementary materials
PCR-RFLP electrophoretogram OF CYP2C19 genotypes. (A) M, 50 bp Marker; 1, PCR product; 2, *1/*1; 3, *1/*2; 4, *2/*2. (B) M, 50 bp Marker; 1, PCR product; 2, *1/*1; 3, *1/*3; 4, *3/*3.
Footnotes
Source of support: Departmental sources
Conflicts of interest
All authors declare that they have no conflict of interest and no funding support.
References
- 1.Kim SS, Ruiz VE, Carroll JD, Moss SF. Helicobacter pylori in the pathogenesis of gastric cancer and gastric lymphoma. Cancer Lett. 2011;305:228–38. doi: 10.1016/j.canlet.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrero R, Park JY, Forman D. The fight against gastric cancer – the IARC Working Group report. Best Pract Res Clin Gastroenterol. 2014;28:1107–14. doi: 10.1016/j.bpg.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Vallve M, Vergara M, Gisbert JP, Calvet X. Single vs. double dose of a proton pump inhibitor in triple therapy for Helicobacter pylori eradication: A meta-analysis. Aliment Pharmacol Ther. 2002;16:1149–56. doi: 10.1046/j.1365-2036.2002.01270.x. [DOI] [PubMed] [Google Scholar]
- 4.Group CHP, Hospital PUF, Beijing. Prevalence of Helicobacter pylori Resistance to Antibiotics and its Influence on the Treatment Outcome in China: A Multicenter Clinical Study. Chinese Journal of Gastroenterology. 2007;12:525–30. [in Chinese] [Google Scholar]
- 5.Gou QY, Shi RH, Yu RB. Gastroenterology DO. Levofloxacin containing triple therapy vs. standard triple therapy for eradication of Helicobacter pylori: A meta analysis. World Chinese Journal of Digestology. 2014;22 [in Chinese] [Google Scholar]
- 6.Cheng H, Hu FL, Zhang GX, et al. Levofloxacin-based triple therapy for first-line Helicobacter pylori eradication treatment: A multi-central, randomized, controlled clinical study. Zhonghua Yi Xue Za Zhi. 2010;90:79–82. [PubMed] [Google Scholar]
- 7.Kuo CH, Lu CY, Shih HY, et al. CYP2C19 polymorphism influences Helicobacter pylori eradication. World J Gastroenterol. 2014;20:16029–36. doi: 10.3748/wjg.v20.i43.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez HM, Romero EM, Peregrina AA, et al. CYP2C19- and CYP3A4-dependent omeprazole metabolism in West Mexicans. J Clin Pharmacol. 2003;43:1211–15. doi: 10.1177/0091270003258170. [DOI] [PubMed] [Google Scholar]
- 9.Meyer UA. Metabolic interactions of the proton-pump inhibitors lansoprazole, omeprazole and pantoprazole with other drugs. Eur J Gastroenterol Hepatol. 1996;8(Suppl 1):S21–25. doi: 10.1097/00042737-199610001-00005. [DOI] [PubMed] [Google Scholar]
- 10.Roman M, Ochoa D, Sanchez-Rojas SD, et al. Evaluation of the relationship between polymorphisms in CYP2C19 and the pharmacokinetics of omeprazole, pantoprazole and rabeprazole. Pharmacogenomics. 2014;15:1893–901. doi: 10.2217/pgs.14.141. [DOI] [PubMed] [Google Scholar]
- 11.de Bortoli N, Martinucci I, Giacchino M, et al. The pharmacokinetics of ilaprazole for gastro-esophageal reflux treatment. Expert Opin Drug Metab Toxicol. 2013;9:1361–69. doi: 10.1517/17425255.2013.813018. [DOI] [PubMed] [Google Scholar]
- 12.Wedemeyer RS, Blume H. Pharmacokinetic drug interaction profiles of proton pump inhibitors: an update. Drug Saf. 2014;37:201–11. doi: 10.1007/s40264-014-0144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JC, Lin CJ. CYP2C19 genotypes in the pharmacokinetics/pharmacodynamics of proton pump inhibitor-based therapy of Helicobacter pylori infection. Expert Opin Drug Metab Toxicol. 2010;6:29–41. doi: 10.1517/17425250903386251. [DOI] [PubMed] [Google Scholar]
- 14.Jainan W, Vilaichone RK. Effects of the CYP2C19 genetic polymorphism on gastritis, peptic ulcer disease, peptic ulcer bleeding and gastric cancer. Asian Pac J Cancer Prev. 2014;15:10957–60. doi: 10.7314/apjcp.2014.15.24.10957. [DOI] [PubMed] [Google Scholar]
- 15.Sugimoto M, Furuta T, Shirai N, et al. Poor metabolizer genotype status of CYP2C19 is a risk factor for developing gastric cancer in Japanese patients with Helicobacter pylori infection. Aliment Pharmacol Ther. 2005;22:1033–40. doi: 10.1111/j.1365-2036.2005.02678.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Zhang Y, Wu X, et al. In vitro assessment of cytochrome P450 2C19 potential of naoxintong. Evid Based Complement Alternat Med. 2012;2012:430262. doi: 10.1155/2012/430262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finta C, Zaphiropoulos PG. The human CYP2C locus: A prototype for intergenic and exon repetition splicing events. Genomics. 2000;63:433–38. doi: 10.1006/geno.1999.6063. [DOI] [PubMed] [Google Scholar]
- 18.Xiao ZS, Goldstein JA, Xie HG, et al. Differences in the incidence of the CYP2C19 polymorphism affecting the S-mephenytoin phenotype in Chinese Han and Bai populations and identification of a new rare CYP2C19 mutant allele. J Pharmacol Exp Ther. 1997;281:604–9. [PubMed] [Google Scholar]
- 19.de Morais SM, Wilkinson GR, Blaisdell J, et al. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419–22. [PubMed] [Google Scholar]
- 20.Yamada S, Shiohira H, Yasui-Furukori N, et al. The (R)-omeprazole hydroxylation index reflects CYP2C19 activity in healthy Japanese volunteers. Eur J Clin Pharmacol. 2013;69:1423–28. doi: 10.1007/s00228-013-1480-1. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Nie YQ, Dai SJ, et al. [The effect of proton pump inhibitor on intragastric acidity and it relation to S-mephenytoin hydroxylase genetic polymorphism]. Zhonghua Nei Ke Za Zhi. 2003;42:777–80. [PubMed] [Google Scholar]
- 22.Lou HY, Chang CC, Sheu MT, et al. Optimal dose regimens of esomeprazole for gastric acid suppression with minimal influence of the CYP2C19 polymorphism. Eur J Clin Pharmacol. 2009;65:55–64. doi: 10.1007/s00228-008-0552-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR-RFLP electrophoretogram OF CYP2C19 genotypes. (A) M, 50 bp Marker; 1, PCR product; 2, *1/*1; 3, *1/*2; 4, *2/*2. (B) M, 50 bp Marker; 1, PCR product; 2, *1/*1; 3, *1/*3; 4, *3/*3.


