Abstract
The difficulty in isolating and propagating functional primary cholangiocytes is a major limitation in studying biliary disorders and testing novel therapeutic agents. To overcome this problem, we have developed a platform for the differentiation of human Pluripotent Stem Cells (hPSCs) into functional cholangiocyte-like cells (CLCs). We have previously reported that our 26-day protocol closely recapitulates key stages of biliary development starting with the differentiation of hPSCs into endoderm and subsequently foregut progenitor cells, followed by the generation of hepatoblasts, cholangiocyte progenitors expressing early biliary markers and mature CLCs displaying cholangiocyte functionality. Compared to alternative protocols for biliary differentiation of hPSCs, our system does not require co-culture with other cell types and relies on chemically defined conditions up to and including the generation of cholangiocyte progenitors. A complex extracellular matrix is used for the maturation of CLCs, therefore experience in hPSC culture and 3D organoid systems may be necessary for optimal results. Finally, the capacity of our platform for generating large amounts of disease-specific functional cholangiocytes will have broad applications for cholangiopathies, in disease modeling and for screening of therapeutic compounds.
Introduction
Adult bile ducts consist of highly functional biliary epithelial cells1 which regulate bile homeostasis and modulate inflammatory responses. These cells are also known as cholangiocytes and represent the main cell type affected in cholangiopathies2,3; a diverse group of liver disorders including diseases such as Primary Biliary Cirrhosis and Primary Sclerosing Cholangitis. Despite the growing importance of these diseases, research in biliary physiology and the development of new therapeutics have been hampered by the lack of robust platforms for disease modeling and high-throughput drug screening3,4. Although animal models exist, their capacity for fully reproducing human pathophysiology is limited5,6; while access to primary biliary tissue remains problematic prohibiting large scale experiments. Here, we describe a protocol for generating large quantities of CLCs from human hPSCs, which can be applied to model cholangiopathies in vitro and validate the effects of therapeutic compounds6.
Development of the protocol
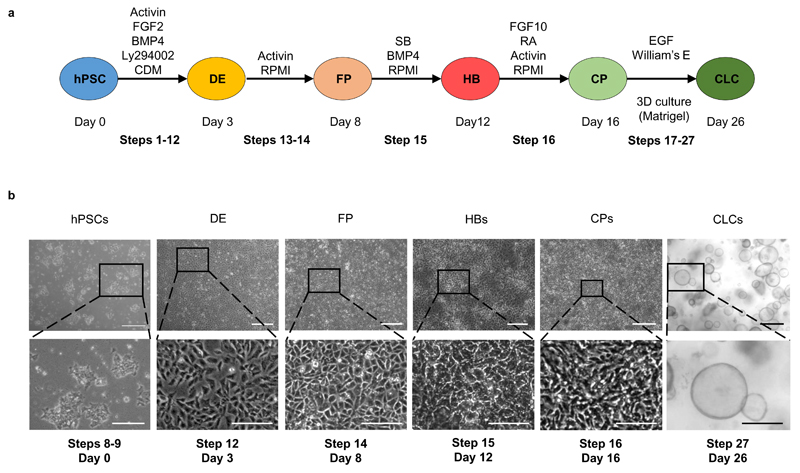
The protocol for the generation of cholangiocyte-like cells7 was developed by recapitulating key stages of native bile duct development (Figure 1a). Cholangiocytes originate from hepatoblasts (HBs), a bipotent population of embryonic liver progenitor cells8, which can also differentiate into hepatocytes. Hepatoblasts surrounding the portal vein give rise to a monolayer of immature cholangiocyte progenitor cells (the ductal plate)8, which undergoes a process of 3D remodeling and maturation resulting in functional bile ducts.
Figure 1.
Generation of Cholangiocyte-like Cells (CLCs) from human Pluripotent Stem Cells (hPSCs). (a) Schematic representation of the protocol for the generation of hPSC-derived CLCs. DE: Definitive Endoderm, FP: Foregut progenitors, HB: Hepatoblasts, CP: Cholangiocyte Progenitors; BMP, bone morphogenetic protein; Ly294002 is a phosphatidylinositol-3-OH kinase inhibitor; CDM, chemically defined medium; RPMI, Roswell Park Memorial Institute medium; SB, SB-431542; HGF, hepatocyte growth factor; RA, retinoic acid; EGF, epidermal growth factor; FGF, fibroblast growth factor. Schematic modified from 7. The procedure steps corresponding to each stage are noted for reference. (b) Light microscopy images of cells at key stages of CLC differentiation. Scale bars for hPSCs, DE, FPs, CPs: 500 μm; HBs: 100 μm; zoomed in images: 50μm. The procedure steps and day numbers corresponding to each image are noted for reference.
The generation of bipotent HBs was based on our established methodology for producing hPSC-derived hepatocyte-like cells9. To achieve biliary commitment of HBs, we used physiological cues reported to control biliary specification such as Activin-A (a member of the TGFbeta superfamily)8,10 and Fibroblast Growth Factor (FGF) 1011. Screening a variety of growth factors, we also identified a requirement for Retinoic Acid7. The combined activation of these signaling pathways was sufficient to promote differentiation of HBs to cholangiocyte progenitors expressing early biliary markers including KRT19 and SOX97.
Maturation of native cholangiocytes happens in synchrony with 3D rearrangement of the ductal plate into tubular structures8. Most of the functional properties of the biliary epithelium are associated with absorption and secretion processes, which require a polarized epithelium forming a lumen and therefore cannot be accurately reproduced by cells organized in monolayer12,13. Consequently, for the final stage of our protocol promoting CP maturation to CLCs, we developed a 3D culture system, based on previous studies using matrigel and Epidermal Growth Factor (EGF)14,15 which promote spontaneous differentiation of hepatoblasts into cystic structures expressing early biliary markers, such as KRT1914,15. Prolonged culture of CPs under these conditions resulted in CLC organoids with a central lumen demonstrating characteristic functional properties, such as GGT activity7
Applications
The mechanisms controlling development of the human biliary tree remain poorly understood. Indeed, developmental studies in humans is limited by minimal access to fetal tissue, while animal models fail to fully recapitulate the development of the human biliary tree or the phenotype of developmental disorders6. Our in vitro system could address some of these challenges, as it relies on a step-wise differentiation protocol which closely mimics embryonic bile duct development. Therefore, significant numbers of cells corresponding to different embryological stages can be easily generated, enabling mechanistic large scale studies in biliary specification or developmental disorders. Accordingly, we applied this methodology to interrogate the role of TGFβ and Notch signaling in biliary tubulogenesis and reproduce the phenotype of Alagille Syndrome in vitro7. The same principle could be used in future studies to explore a broad spectrum of pathways which could be involved in bile duct development and pathogenesis.
CLCs also recapitulate many physiological functions of cholangiocytes in vitro as well as their defects in the context of disease when using hPSCs derived from patients with cholangiopathies7. Consequently, CLCs could present an optimal platform for modeling biliary disease, validating therapeutic compounds and screening for novel treatment agents. We have already demonstrated proof-of-principle for the feasibility of this application by reproducing the effects of the drugs verapamil and octreotide in our culture system7 and using patient specific hiPSCs to identify a new application for the experimental compound VX809 in the management of Cystic Fibrosis Cholangiopathy7. Importantly, the capacity of our system for generating significant numbers of CLCs7 combined with its compatibility with large scale experiment formats (24 and 48 well plates)7 could set the foundation for the development of high-throughput drug screening platforms for cholangiopathies in the future using patient-derived CLCs.
Comparison with other methods
Primary cholangiocyte isolation has been reported16–18. However, these methodologies are technically challenging, only support short term growth with limited expansion and generate limited numbers of cells, all of which are not compatible with large scale experiments16–18. Furthermore, primary cholangiocytes cultured in monolayer systems have not been shown to maintain their functional properties16–18.
Two other protocols have been described for generating biliary epithelium from hPSCs19,20. The method by Dianat et al. results in cells with a transcriptional signature7,20 compatible with a sub-population of cholangiocytes located in the canals of Hering known as small cholangiocytes21. Therefore, this approach is optimized for studies on small cholangiocytes and complements our protocol which is aimed towards the production of large cholangiocytes. The method by Ogawa et al. generates cholangiocyte organoids expressing mature markers and demonstrating biliary functionality; however, biliary specification is based in a co-culture system with mouse OP9 cells13. Although a mixed culture system may recapitulate more closely the native niche of hepatoblasts/cholangiocytes, it is technically more challenging and presents several limitations. Indeed, OP9 cells are derived from bone marrow and are known to promote hematopoietic differentiation of ESCs by secreting factors such as M-CSF. This poses significant limitations for mechanistic studies in biliary specification and early biliary development since unknown secreted factors could interfere with experimental outcomes. Furthermore, the heterogeneity of the cell population in a co-culture system renders –omic studies, such as genome wide analyses more challenging. Consequently, the platform by Ogawa et al may be better suited for studies where accurate reproduction of a complex cellular niche is crucial, while our system is more optimized for mechanistic studies in biliary development and therapeutics.
Limitations
There are two main limitations to our platform. Our system relies on a complex extracellular matrix (Matrigel). The composition of Matrigel is not fully defined while variation in the growth factor and protein contents of each batch could affect the efficiency of the final stage of our protocol. Furthermore, the use of matrigel could render the translation of our platform to Good Manufacturing Practice (GMP) conditions challenging and prevent in vivo applications towards cell based therapy and regenerative medicine. Another important consideration is the maturity of the generated cells. CLC organoids express both early and mature biliary markers and maintain some fetal characteristics corresponding more accurately to a stage between fetal and fully mature bile ducts. Consequently, prior to modeling adult biliary disorders CLCs should be tested for the presence of the relevant mature markers and functionality.
Experimental Design
Our method describes the generation of hPSC-derived CLC organoids over a period of 26 days. Biliary differentiation is achieved through 5 key stages of recapitulating bile duct development (Figure 1). Our protocol starts with the plating of hPSCs on day 0 (d0), while we refer to the first day of differentiation as day 1 (d1). The first stage (d1-3) results in the generation of definitive endoderm (DE) cells. These cells correspond to the common progenitor from which the liver, lung, pancreas, alimentary tract and thyroid arise. Subsequently, DE cells are differentiated into Foregut Progenitor (FP) cells (stage 2, d4-8), which correspond to precursors of the liver, pancreas, lung and thyroid lineages found in the anterior portion of the embryonic alimentary canal. In the third stage (d9-12) FP cells are differentiated to hepatoblasts (HBs), bipotent progenitors of hepatocytes and cholangiocytes, which can give rise to both. The fourth stage (d13-16) results in biliary commitment of HBs and the generation of cholangiocyte progenitors (CPs), which represent early cholangiocytes forming the ductal plate in vivo. In the final stage of our method (d17-26) CPs form functional CLC organoids in 3D culture conditions.
Starting population considerations
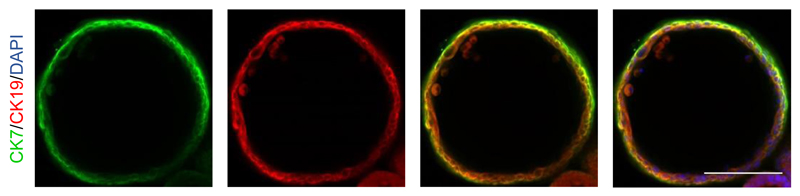
We have demonstrated that this protocol is reproducible with 4 different hPSC lines 7 and embryonic stem (ES) cells22 (Figure 2). Variability in differentiation capacity is a common issue with hPSC lines, which may reflect on the efficiency and timing of our protocol. Therefore, some minor optimization steps may be required for each hPSC-line as described in the following sections.
Figure 2.
Derivation of CLCs from embryonic stem (ES) cells. IF analyses demonstrating the expression of key biliary markers (CK7, CK19) in a CLC organoid generated from ES cells (H9). Scale bars: 100 μm. See table 2 for a detailed list of the antibodies and concentrations used.
Preparation of hPSCs
To achieve high differentiation efficiency the generation of a near homogeneous DE population is crucial. For that hPSCs have to exhibit optimal morphology and minimal background differentiation. They should first be allowed to grow to near confluence (70-80%), then they are broken into small clumps and plated at high density as described in the sections below (Figure 1b; Steps 1-9). Clump size and density plays a critical role in this step. Very small clumps or single cells are not viable after the first day of differentiation, while large clumps differentiate only partially, maintaining the expression of pluripotency markers at their center. Low densities prevent the cells from reaching near confluence by the end of the first stage. This can have a negative impact on paracrine signaling, cell migration and cell to cell contact, which are crucial factors for efficient formation of the foregut epithelium. A minimum of 24 hours is allowed for the hPSCs to adhere to the plate before starting differentiation; however this period can be extended to a maximum of 48 hours if the clump size is thought to be too small.
Generation of Definitive Endoderm and Foregut Progenitors
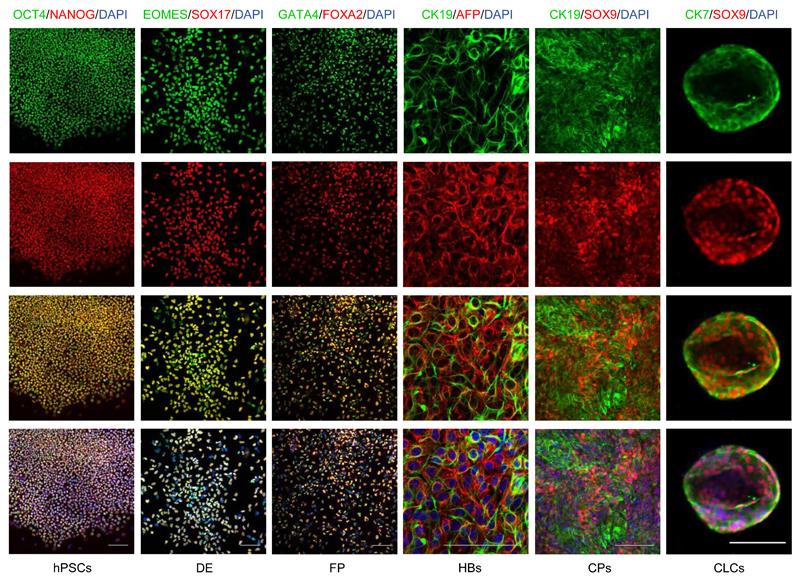
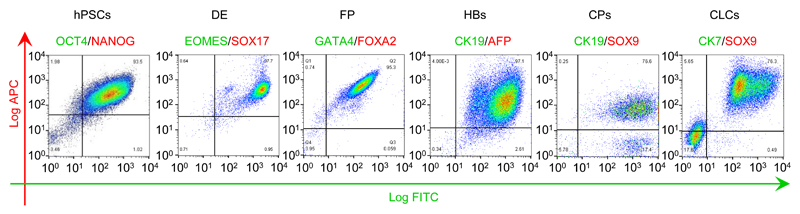
Definitive endoderm differentiation is characterized by morphological changes; epithelial-mesenchymal transition (EMT; Figure 1b); significant proliferation of the cells and increased death of cells that fail to differentiate. By the end of day 3 the cells should be approaching confluence (Figure 1b) and express Sox17 and EOMES homogeneously (>90%, Figure 3-4). Cell proliferation continues during the FP stage and by the end of day 8 the cells should be forming a confluent epithelium with cells exhibiting a characteristic rhomboidal morphology (Figure 1b). The generation of a near homogeneous population of FP cells is crucial for the efficiency of later stages. Therefore, we recommend that differentiations are optimized to generate cell populations with >95% purity for endoderm and foregut markers (Figure 3-4), such as GATA4 and FOXA2. In particular, for resistant hPSC lines with significant contamination from partially differentiated cells, we recommend splitting the cells at the foregut stage (d6). For lines with lower proliferation rates and good differentiation efficiency this step is optional. If the cells are split at this stage it is very important that they are dissociated to single cells and re-plated at a density allowing the formation of a fully confluent epithelium by d8.
Figure 3.
Immunofluorescence analyses demonstrating the expression of characteristic markers at key stages of CLC differentiation. Scale bars: 100 μm. See table 2 for a detailed list of the antibodies and concentrations used The method for staining CLC organoids is described in procedure steps 28-39.
Figure 4.
Flow cytometry analyses demonstrating the expression of characteristic markers at key stages of CLC differentiation. CLC organoids were harvested as described in procedure steps 40-47. Cells were dissociated into single cells following incubation with TrypLE for 5 minutes at 37°C and fixed with 4% PFA for 20 minutes at 4°C. The cells were stained for IF as previously described7, using the antibodies provided in table 2. A standard gating strategy was used25 demonstrated in Supplementary Fig 2. A minimum of 2x104 gated events were used for analysis. Post sort fractions are indicated in the quadrants of each graph. The average differentiation efficiency from hPSCs to CLCs across three lines (CK7+/Sox9+ organoids) was 77% (s.d. = 6.5%)7.
Generation of bipotent hepatoblasts
Cell proliferation begins to reduce at this stage, although cells should continue to proliferate at a lower rate. We have noticed variability in proliferation rate between different hPSC lines. Differentiation of FP to HBs should result in a near homogeneous population (>95%) expressing hepatoblast markers (CK19, AFP) (Figure 3-4), which is important for the efficiency of subsequent steps. High cell density of FPs forming a monolayer of relatively small cells is crucial for the success of this stage. For resistant hPSC lines this stage could be prolonged by 24hrs to improve differentiation efficiency. However, significant prolongation of HB differentiation carries the risk of committing a significant proportion of cells to the hepatic lineage which results in increased hepatoblast/hepatocyte contamination in the next stage and reduced biliary lineage commitment.
Generation of cholangiocyte progenitors
Cell proliferation should increase compared to the previous stage and by the fourth day of cholangiocyte progenitors differentiation significant overgrowth should be seen (100% confluence and/or areas of cells forming multiple layers). Differentiation of HBs to CPs is heterogeneous, resulting in a mixed population of CPs (75%), HBs (15%) and cells at intermediate stages (Figure 4). Consequently, hepatic markers, such as AFP can still be detected, but early biliary markers such as CK19 and SOX9 should also be expressed almost homogeneously (Figure 2, 3). Poor efficiency at this stage (<75% SOX9 positive cells) could result in incomplete maturation of CLC organoids in the next step. The quality and duration of the HB stage is critical to limit HB contamination and ensure biliary commitment.
Therefore, for resistant hPSC lines we recommend optimizing the duration and efficiency of HB differentiation as described in the previous section (Generation of bipotent hepatoblasts) and in the troubleshooting section (Table 1).
Table 1. TROUBLESHOOTING.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 10 | Poor attachment of hPSCs | Longer attachment time required | Repeat step 9 incubating the cells for 1 more day before proceeding to step 10 |
| Colony size too small | Break colonies into slightly bigger clumps which gravitate to the bottom of the plate more easily, facilitating attachment | ||
| Variability between hPSC lines | Add Rho kinase inhibitor Y-27632 in the medium during passaging | ||
| Poor FBS batch | If this problem occurs with more than one line change FBS batch. Screening FBS batches may be required as outlined in the Reagents section | ||
| 12 | Poor endoderm differentiation efficiency | Suboptimal plating of hPSCs for differentiation | Decrease clump size and increase plating density |
| Variability between lines with different sensitivity to activin or Wnt signaling | For particularly resistant lines optimize the dose of activin A in steps 10-12 and CHIR in step 10, by monitoring the impact of increased doses in the efficiency of endoderm differentiation | ||
| 14A, 14B | Poor FP differentiation efficiency | Variability between lines | For persistent contamination with poorly differentiated cells split the cells using step 14B |
| Suboptimal plating of cells following split in step 14B | For poor differentiation efficiency following a split optimize cell density ensuring the cells are confluent by the following day | ||
| Reduced activity of growth factors | Check Activin-A activity | ||
| Use growth factors that are within 5 freeze-thaw cycles | |||
| Poor B27 batch | For particularly resistant lines change B27 batch. Screening B27 batches may be required as outlined in the Reagents section | ||
| 14B | Poor attachment following split | Suboptimal plating of cells following split in step 14B | Increase cell density |
| Use a viability dye such as trypan blue when counting the cells to ensure the appropriate number of live cells are plated. | |||
| Poor FBS batch | If this problem occurs with more than one lines change FBS batch. Screening FBS batches may be required as outlined in the Reagents section | ||
| 15 | Poor hepatoblast differentiation | Suboptimal previous steps | Check and optimize the differentiation efficiency to Foregut Progenitors |
| Increase plating density in step 14B to ensure the cells are confluent by step 15 | |||
| Reduced activity of growth factors | Check SB431542 activity | ||
| Use growth factors that are within 5 freeze-thaw cycles | |||
| Variability between lines with different sensitivity to activin signaling | For particularly resistant lines the dose of SB431542 can be increased | ||
| Poor B27 batch | For particularly resistant lines change B27 batch. Screening B27 batches may be required as outlined in the Reagents section | ||
| 16 | Poor cholangiocyte progenitor differentiation | Variability between lines with increased sensitivity to the previous differentiation stage resulting in hepatic commitment of the generated hepatoblasts | Minimize hepatoblast contamination by optimizing the duration of the previous stage to avoid hepatic commitment of the cells. Aim for the minimum duration that allows upregulation of hepatoblast markers |
| Reduced activity of growth factors | Check activin-A and retinoic acid activity | ||
| Increase the dose of Activin-A | |||
| Use growth factors that are within 5 freeze-thaw cycles | |||
| Variability between lines | The differentiation efficiency of hPSC-derived hepatoblasts into cholangiocyte progenitors is dependent on the culture media. Optimization may be required for particularly resistant lines. Advanced DMEM/F12 can replace RPMI/B27 for selected lines. | ||
| Poor B27 batch | For particularly resistant lines change B27 batch. Screening B27 batches may be required as outlined in the Reagents section | ||
| 19 | Clump size too small/single cells following incubation with cell dissociation buffer | Variability between lines | Reduce the cell dissociation buffer incubation time |
| For sensitive lines omit step 18 (incubation with cell dissociation buffer) and proceed to mechanical dissociation only in step 19 | |||
| If even mechanical dissociation alone results in single cells/small clumps check cell density and plate the cells at higher density in step 14B | |||
| 26 | Poor cell survival following transfer to 3D culture | Variability between lines | Optimize clump size. More sensitive lines may require larger clump sizes |
| Optimize plating density. Low plating densities are associated with poor survival | |||
| Reduced activity of reagents | Check Y-27632 activity and ensure it is freshly prepared | ||
| Increased cell death secondary to vigorous mechanical dissociation | Avoid vigorous mechanical dissociation of the cells during passaging | ||
| 27 | Poor organoid formation and/or poor CLC differentiation and/or poor CLC function | Suboptimal previous steps | Check the differentiation efficiency of cholangiocyte progenitors |
| Variability between lines | Optimize clump size and plating density‥ Aim for smaller clumps and lower density if there is significant attachment of cells to the bottom of the plate | ||
| For resistant lines consider adding forskolin to the culture medium | |||
| Poor matrigel batch | For particularly resistant lines change matrigel batch. Screening matrigel batches may be required as outlined in the Reagents section | ||
| 39 | Unsuccessful staining | Poor antibody penetration | Decrease the matrigel dilution to 40% |
| Place the plate on a lab rocker during the staining and washing steps | |||
| Difficulties acquiring optimal images in 3D while CLC organoids are embedded in matrigel | Grow CLC organoids on chamber slides and use alternative staining method; remove chambers and snap freeze or repeat steps 27-37 and use a cover slip to flatten organoids | ||
| Inadequate optimization for antibodies | Optimize antibody concentration, duration of washing and staining steps |
Generation of CLC organoids
For the final stage of our protocol CPs are dissociated into small clumps and transferred to 3D culture conditions. Density and clump size are the most critical factors for the success of this step. Very high densities do not allow adequate space for the cells to expand and re-organize into organoids with a central lumen. Instead, proliferating clumps of cells merge together into large aggregates. Single cells or very small clumps may not be viable, while large clumps gradually migrate and attach to the bottom of the plate forming a monolayer. Consequently, the efficiency of this phase depends on careful manipulation of the quantity of cells, matrigel and media. Minor adjustments to density and clump size may be required for different hPSC lines (see troubleshooting and procedures sections, steps 17-26). For resistant lines such as H9, we recommend adding forskolin (optional step), which promotes intraluminal fluid secretion and facilitates the formation of organoids with a lumen.
Of note this step starts with a heterogeneous population of cells including HBs and CPs and thus some hepatic contamination is expected. However, cells expressing hepatic markers, such as AFP fail to form organoids and usually gravitate to the bottom of the plate. On the contrary, by the end of this stage CLC organoids should express biliary (CK19, CK7, SOX9) (Figure 3-4) but not hepatic markers (AFP) and demonstrate functional properties characteristic of biliary epithelium, such as GGT and ALP activity (Figure 5). Importantly, we have noticed differences in differentiation efficiency with different batches of Matrigel. For resistant hPSC lines Matrigel should be screened for batches which support organoid formation and cholangiocyte functionality.
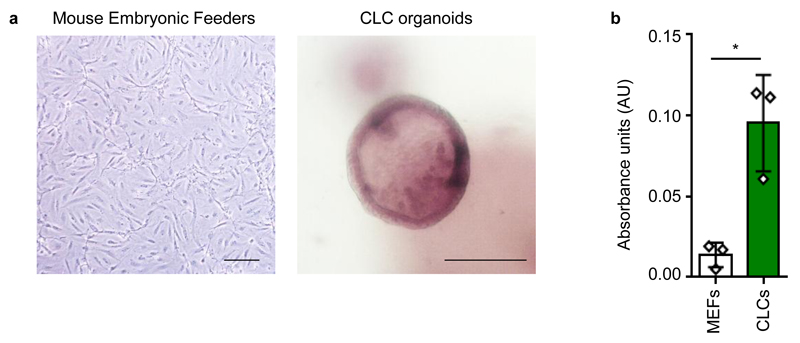
Figure 5.
Functional properties of CLC organoids. (a) CLC organoids demonstrating characteristic ALP staining. Mouse embryonic feeders are used as a negative control. Scale bars: 100 μm. (b) GGT activity of CLC organoids measured in absorbance units (AU); n=3; Mouse Embryonic Feeders (MEFs) are used as a negative control. Error bars, standard deviation; individual data points are demonstrated; *P<0.05, two tailed student’s t-test; F-test used to compare variances, P=0.1218 (no significant difference in variance). GGT and ALP activity were assessed using commercially available kits (MaxDiscovery™ gamma-Glutamyl Transferase (GGT) Enzymatic Assay Kit and BCIP/NBT Color Development Substrate respectively) according to the manufacturer’s instructions.
Controls
Intrahepatic cholangiocytes are not commercially available. Therefore, we recommend the use of fresh bile duct tissue obtained from liver donors, or frozen isolated common bile duct cholangiocytes commercially available (Celprogen) as a positive control for the expression of biliary markers.
Materials
Reagents
CRITICAL All the reagents listed are reconstituted and stored as per the manufacturer’s instructions unless specifically stated
hPSCs. All hPSC lines were derived by the Cambridge Biomedical Research Campus (BRC) hIPSC core facility (ethics reference no. 08/H0311/201 for Hertfordshire Regional Ethics Committee (REC) and 09/H0304/77 for National Research Ethics Service (NRES) Committee East of England, Cambridge East) CAUTION HPSC derivation should always occur in compliance with appropriate national laws and institutional regulations. Informed consent must be obtained from human subjects. CAUTION The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Gelatin (Sigma, cat. no. G1890)
Water for embryo transfer (Sigma, cat. no. W1503)
Advanced DMEM F12 (Life Technologies, cat. no. 12634028)
Penicillin-streptomycin (Life Technologies, cat. no. 15140122)
L-Glutamine (Life Technologies, cat. no. 25030024)
β-Mercaptoethanol (Sigma, cat. no. M6250) CAUTION β-Mercaptoethanol is toxic if ingested, inhaled, or following prolonged skin exposure. Wear protective clothing and use a fume hood.
FBS (Life Technologies, cat. no. 10500064) CRITICAL Due to batch-to-batch variability in serum, serum batches should be screened for their capacity to maintain pluripotency for a minimum of 2 passages. Key features of pluripotent cell growth include characteristic colony morphology, differentiation potential to all 3 germ layers and expression of pluripotency markers such as NANOG, POU5F1 and SOX2.
Ham's F-12 Nutrient Mix, GlutaMAX™ Supplement (Life Technologies, cat. no. 31765068)
Iscove's Modified Dulbecco's Medium (IMDM) (Life Technologies, cat. no. 21980065)
Chemically defined lipid concentrate (Life Technologies, cat. no. 11905031)
Monothioglycerol (Sigma, cat. no. M6145)
Transferrin (30 mg/ml, Roche, cat. no. 652202)
Insulin, 10 mg/ml (Roche, cat. no. 1376497)
Poly(vinyl alcohol) (PVA) 87-90% hydrolyzed (Sigma, cat. no. P8136)
KnockOut serum replacement (KOSR; Life Technologies, cat. no. 10828028)
Collagenase IV (Life Technologies, cat. no. 17104019)
Dispase (Invitrogen, cat. no. 17105041)
DMEM F-12 (Life Technologies, cat. no. 11330032)
RPMI 1640 + GlutaMAX (Gibco, cat. no. 61870)
B-27 supplement containing insulin (Gibco, cat. no. 17504-044) CRITICAL Due to batch-to-batch variability in B27, B27 batches should be screened for their capacity to support HB and CP differentiation in a minimum of 2 different differentiation experiments. HB and CP differentiation should be assessed based on appropriate markers on flow cytometry analyses. These include >95% expression of CK19 and AFP for HBs and >75% expression of Sox9 and CK19 for CPs (Figure 4).
MEM non-essential amino acids (MEM-NEAA; Gibco, cat. no. 1140)
Dulbecco’s PBS (DPBS; Life Technologies, cat. no. 14190)
Cell Dissociation Buffer, enzyme-free, PBS (Gibco, cat. no. 13151014)
William's E Medium, no phenol Red (Invitrogen, cat. no. A12176-01)
Dexamethasone (R&D systems, cat. no. 1126/100)
DMSO (Sigma, cat. no. D2650)
ITS+ Universal Cell Culture Supplement Premix, 20 ml, 2 L equivalent (Corning, cat. no. 354352)
Nicotinamide (Sigma, cat. no. N0636)
D-Glucose (Invitrogen, cat. no. 15023021)
Sodium bicarbonate powder (Sigma, cat. no. S5761)
2-Phospho-L-Ascorbic Acid Trisodium Salt (Sigma, cat. no. 49752)
HEPES Solution (Sigma, cat. no. H0887-20ML)
Sodium Pyruvate (Invitrogen, cat. no. 11360-070)
Recombinant human Activin A (R&D Systems, cat. no. 338-AC)
Recombinant human BMP4 (R&D Systems, cat. no. 314-BP)
Recombinant human FGF basic, 146 aa (R&D Systems, cat. no. 233-FB)
LY294002 (Promega, cat. no. V1201)
CHIR99021 (Tocris, cat. no. 4423)
SB431542 (Tocris bioscience, cat. no. 1614)
Recombinant Human Keratinocyte Growth Factor-2 (FGF10) (Source Bioscience, cat. no. ABC144)
Retinoic acid (Sigma, cat. no. R2625)
Y27632 (ROCK Inhibitor) (Selleck, cat. no. S1049)
Matrigel (BD Biosciences, cat. no. 356237) CRITICAL Due to batch-to-batch variability in matrigel, matrigel batches should be screened for their capacity to support organoid formation and maturation in a minimum of 2 different differentiation experiments. Organoids should be clearly identified following 5 days of culture in matrigel, while small ring structures can be seen as early as 48-72 hours. CLC maturation should be assessed based on appropriate marker expression on flow cytometry analyses and functional assays. These include >75% expression of Sox9 and CK7, ALP and GGT activity (Figure 3-5).
Recombinant Human EGF Protein (R&D Systems, cat. no. 236-EG)
Cell recovery solution (SLS, cat. no. 354253)
Donkey serum (abd serotec, cat. no. c06sb)
Triton-X100 solution (Sigma, cat. no. X100-500ML)
Parafolmadehyde 16% w/v (PFA; Alfa Aesar, cat. no. 30525-89-4)
GenElute™ Mammalian Total RNA Miniprep Kit (Sigma, cat. no. RTN-350)
TrypLE™ Express Enzyme (1X), no phenol red (Gibco, cat. no. 12604021)
Cytokeratin 7 antibody (RCK105) (Abcam, cat. no. ab9021; Table 2)
Table 2. ANTIBODY LIST.
| Target antigen | Supplier | Cat No. | Cell Type | Analyses | Fluorophore type | Clone | Buffer | Concentration | Dilution |
|---|---|---|---|---|---|---|---|---|---|
| SOX17 | R&D | AF1924 | Endoderm | IF | Unconjugated | Polyclonal | Donkey Serum (DS) | 200 µg/ml | 1:100 |
| GATA4 | Santa Cruz | sc-25310 | FP | FC | Unconjugated | G-4 | DS | 200 µg/ml | 1:100 |
| HNF4A | Santa Cruz | sc-8987 | HB | IF | Unconjugated | H-171 | DS | 200 µg/ml | 1:100 |
| AFP | Dako | A-008 | HB | IF/FC | Unconjugated | Polyclonal | DS | 344 000 IU/mL | 1:100 |
| TBX3 | Santa Cruz | sc-17871 | HB | IF | Unconjugated | A-20 | DS | 200 µg/ml | 1:100 |
| SOX9 | Santa Cruz | sc-20095 | CP/CLCs | IF/FC | Unconjugated | H-90 | DS | 200 µg/ml | 1:100 |
| CK7 | Abcam | ab68459 | CLC | IF/FC | Unconjugated | EPR1619Y | DS | 0.111 mg/ml | 1:100 |
| CK7 | Abcam | ab9021 | CLC | IF/FC | Unconjugated | RCK105 | DS | 1 mg/ml | 1:100 |
| NANOG | R&D | AF1997 | hPSCs | IF/FC | Unconjugated | Polyclonal | DS | 200 µg/ml | 1:100 |
| Oct3-4 | Santa Cruz | sc-9081 | hPSCs | IF/FC | Unconjugated | H-134 | DS | 200 µg/ml | 1:100 |
| CK19 | Abcam | ab7754 | CLC | IF/FC | Unconjugated | A53-B/A2 | DS | 1 mg/ml | 1:100 |
| TBR2/EOMES | Abcam | ab23345 | DE | IF/FC | Unconjugated | Polyclonal | DS | 200 µg/ml | 1:100 |
| FOXA2 | R&D | AF2400 | FP | FC | Unconjugated | Polyclonal | DS | 200 µg/ml | 1:100 |
Cytokeratin 7 antibody (Abcam, cat. no. ab68459; Table 2)
Cytokeratin 19 antibody (Abcam, cat. no. ab7754; Table 2)
SOX9 H-90 antibody (Santa Cruz, cat. no. sc-20095; Table 2)
TBX3 (A-20) antibody (Santa Cruz, cat. no. sc-17871; Table 2)
HNF4 (H-171) antibody (Santa Cruz, cat. no. sc-8987; Table 2)
Alpha fetoprotein (AFP) antibody (DAKO, cat. no. A0008; Table 2)
Sox17 antibody (R&D, cat. no. AF1924; Table 2)
TBR2 / Eomes antibody - (Abcam, cat. no. ab23345; Table 2)
GATA4 (G-4) antibody (Santa Cruz, cat. no. sc-25310; Table 2)
HNF3b/FoxA2 antibody (R&D, cat. no. AF2400; Table 2)
Oct-3/4 (H-134) antibody (Santa Cruz, cat. no. sc-9081; Table 2)
Anti-human NANOG antibody (R&D, cat. no. AF1997; Table 2)
MaxDiscovery™ gamma-Glutamyl Transferase (GGT) Enzymatic Assay Kit (Bioo Scientific, 5601-01)
BCIP/NBT Color Development Substrate (Promega, S3771)
Equipment
CO2 incubator (Sanyo, cat. no. MCO-18AC)
Centrifuge (Eppendorf, cat. no. 5804)
Counting chamber (Superior Marienfeld, cat. no. 0640410)
Disposable serological pipettes, 5 10 and 25 ml (Corning, cat. nos. 4487, 4488 and 4489)
Graduated filter tips, 1,000 μl, 200 μl, 20 μl, 10 μl (Starlab, cat. nos. S1122-1830, S1120-8810, S1120-1810, S1120-3810)
Centrifuge tubes, 15 ml and 50 ml (Corning, cat. no. 430791 and 430291)
500mL Vacuum Filter/Storage Bottle System, 0.22µm Pore (Corning, cat. no. 431097)
100mm TC-Treated Culture Dish (Corning, cat. no. 430167)
Costar® 12 Well Clear TC-Treated Multiple Well Plates (Corning, cat. no. 3513)
Costar® 24 Well Clear TC-Treated Multiple Well Plates (Corning, cat. no. 3526)
Plate heater (TAP Biosystem, cat. no. 016-0R10)
Inverted microscope (Olympus, cat. no. CKX41)
Reagent Setup
Gelatin for coating tissue culture plates (500 ml)
Dissolve 0.5g of gelatin into 500mls of water for embryo transfer. Heat at 56°C until the gelatin has fully dissolved (approximately 30 minutes). CRITICAL Sterilize gelatin solution using a vacuum filter/storage bottle system. Store at room temperature (18–25 °C) for up to 1 month
Serum-containing medium for coating tissue culture plates (500 ml)
Add 50 ml of FBS, 5 ml of glutamine, 5 ml of penicillin-streptomycin (pen/strep) and 3.5 μl of β-mercaptoethanol in 450 ml of Advanced DMEM/F12. CRITICAL Sterilize serum-containing medium using a vacuum filter/storage bottle system. Mix well before filtration. Store at 4 °C for up to 1 month.
Chemically Defined Medium – PVA (CDM-PVA) medium for maintenance of hPSCs
Combine 0.5 g of PVA, 250 ml of F12 + GlutaMAX, 250 ml of IMDM, 5 ml of concentrated lipids, 20 μl of thioglycerol, 350 μl of insulin, 250 μl of transferrin and 5 ml of pen/strep. Store at 4 °C for up to 1 month. Dissolve PVA in IMDM by adding 0.5 g of PVA to 50 ml of IMDM and mixing overnight at 4 °C (e.g. using a 50 ml falcon on a roller). CRITICAL Sterilize CDM-PVA medium using a vacuum filter/storage bottle system. Mix well before filtration. Warm to 37 °C before use.
Collagenase, 500 ml
Dissolve 500 mg of Collagenase IV into 400 ml of Advanced DMEM/F12 combined with 100 ml of Knockout Serum Replacer, 5 ml of L-Glutamine, 3.5 μl of β-Mercaptoethanol. CRITICAL Sterilize collagenase using a vacuum filter/storage bottle system. Mix well before filtration. Store at 4 °C for up to 1 month. Warm to 37 °C before use.
Dispase, 500 ml
Dissolve 500 mg of Dispase into 500 ml of DMEM F-12. CRITICAL Sterilize dispase using a vacuum filter/storage bottle system. Mix well before filtration. Store at 4 °C for up to 1 month. Warm to 37 °C before use.
1:1 Collagenase/Dispase solution for dissociation of hPSCs
Warm collagenase and dispase to 37 °C. Mix 1 volume of collagenase with 1 volume of dispase immediately before use. The volumes used are dependent on the number and type of plates used. For each 10 cm plate mix 3 ml of collagenase with 3 ml of dispase.
RPMI/B-27 differentiation medium for the differentiation of FP, HBs and CPs (500 ml)
Add 10 ml of B-27, 5 ml of NEAA and 5 ml of pen/strep into 500 ml of RPMI-1640. CRITICAL We have noticed variation between different batches of B-27. B-27 batches should be screened for the capacity to support FP, HB and CP differentiation. Appropriate markers for the efficiency of each stage are provided in the experimental design section. Store at 4 °C for a maximum of 3 weeks. Warm to 37 °C before use.
Nicotinamide 0.4M stock solution
Dissolve 24.4 g of nicotinamide powder in 500 ml of embryo transfer water. CRITICAL Sterilize nicotinamide stock solution using a vacuum filter/storage bottle system. Mix well before filtration. Store at 4 °C for up to 3 months.
Sodium Bicarbonate 1M stock solution preparation
Dissolve 42 g of sodium bicarbonate powder in 500 ml of embryo transfer water. CRITICAL Sterilize sodium bicarbonate stock solution using a vacuum filter/storage bottle system. Mix well before filtration. Store at 4°C for up to 3 months.
Ascorbic acid trisodium salt 100mM stock solution preparation
Dissolve 16.1 g of Ascorbic acid trisodium salt powder in 500 ml of embryo transfer water. CRITICAL Sterilize ascorbic acid trisodium salt stock solution using a vacuum filter/storage bottle system. Mix well before filtration. Store at 4 °C for up to 3 months. Protect from light.
D-Glucose 1M stock solution preparation
Dissolve 90.1 g of D-glucose powder in 500 ml of embryo transfer water. Warm to 50°C to facilitate dissolution CRITICAL Sterilize D-glucose stock solution using a vacuum filter/storage bottle system. Mix well before filtration. Store at 4 °C for up to 3 months.
Dexamethasone 10 mM stock solution
Dissolve 100 mg of Dexamethasone in 25.4797 ml of DMSO. Aliquot in 50-100 μl aliquots. Store in -80°C for up to 12 months.
Supplemented William’s E medium for the maturation of CPs to CLCs in 3D culture
Combine 443 ml of William’s E (WE) medium with 12.5 ml nicotinamide stock solution, 8.5 ml sodium bicarbonate stock solution, 1 ml ascorbic acid trisodium salt stock solution, 7 ml glucose stock solution, 3.15 ml sodium pyruvate, 10 ml HEPES solution, 5 ml ITS+ premix, 5 μl dexamethasone (R&D Systems), 5.3 ml Glutamine and 5 ml pen/strep. CRITICAL Sterilize supplemented WE medium using a vacuum filter/storage bottle system. Mix well before filtration. Store at 4°C for up to 1 month. Warm to 37 °C before use.
Matrigel preparation
10 ml matrigel vials should be thawed slowly in an icebox placed at 4°C overnight. Thawed matrigel should be mixed well and then aliquoted in 1 ml aliquots. Aliquoting of matrigel should always happen in a tissue culture hood to avoid bacterial contamination. Matrigel should be kept constantly on ice to avoid solidification. All equipment coming in contact with matrigel should be pre-cooled to 4°C. This includes pipette tips and media for diluting matrigel. Tubes for aliquoting should be kept on ice. Store matrigel aliquots at -20°C or -80°C for up to 3 months. CRITICAL Each aliquot should undergo a maximum of 2 freeze thaw cycles. This can be achieved by adjusting aliquot volumes accordingly.
50%(vol/vol) matrigel solution preparation
Add 1 volume of supplemented WE medium to 1 volume of matrigel and mix thoroughly. CRITICAL The supplemented WE medium should be pre-cooled to 4°C. CRITICAL Both matrigel and the supplemented WE medium should be kept on ice during and after the preparation of the 50% (vol/vol) solution to avoid solidification.
To calculate the volume of supplemented WE medium and matrigel that need to be mixed please use the following formula:
Volume of supplemented WE = [(number of 24-plate wells) x 50 μl] / 2
The number of wells is multiplied by 50 μl which corresponds to the volume of each dome and divided by 2 to reflect the matrigel-media ratio (50% or 1:1)
Equipment Setup
Gelatin/serum-coated tissue culture plates
Add enough gelatin solution to fully cover the surface of the plate. Indicative volumes are 6 ml for a 10 cm plate and 1 ml for each well of a 12-well plate. Coat for a minimum of 30 min at room temperature, then aspirate the gelatin and replace with enough volume of serum-containing medium to fully cover the surface of the plate. Indicative volumes are 6 ml for a 10 cm plate and 1 ml for each well of a 12-well plate. Store in an incubator at 37 °C for up to 1 week. CRITICAL Allow a minimum of 24 hours at 37 °C before using the plate.
Plate heater setup
Clean the plate heater with trigene and 70%(vol/vol) ethanol and place in a tissue culture hood. Set the temperature to 37 °C and place a 24 well plate on the heating surface CRITICAL Allow a minimum of 30 minutes for the plate to warm up, prior to plating matrigel with cells. If you are using multiple plates these can be pre-warmed in an incubator for a minimum of 30 minutes with each plate placed on the plate heater immediately before plating.
Procedure
Passaging of hPSCs TIMING 1 d
1 Ensure that hPSC colonies are growing and maintaining their characteristic morphology23. We recommend using lines which have been stable in culture for at least 10 passages. Change the medium daily using CDM-PVA supplemented with Activin (10 ng/ml) and bFGF (12 ng/ml). Proceed to the next step when the cells are 70-80% confluent.
2 Aspirate the medium and wash the plate with Ca2 + /Mg2 + -free PBS. The volume of PBS depends on the type of plate used. Indicative minimum volumes are 6 ml for a 10 cm plate, 1-2 ml for a well of a 6-well plate and 0.5 ml for a well of a 12-well plate.
3 Aspirate the PBS and add the appropriate volume of 1:1 collagenase/dispase solution. Refer to step 2 for indicative volumes. Incubate at 37 °C for 30-60 min until the majority of the colonies (>90%) have detached.
4 Tilt the plate and wait for the colonies to gravitate to its lowest part forming a loose pellet. Using a 1000 μl pipette harvest the cells and transfer to a 15 ml tube containing 6 ml of CDM-PVA.
5 Allow 1-2 minutes for the colonies to settle to a loose pellet. Aspirate the supernatant and add 6 ml of CDM-PVA. Repeat this step twice for a total of 2 washes with CDM-PVA.
6 Aspirate the supernatant and re-suspend the pellet in 1 ml of CDM-PVA supplemented with Activin (10 ng/ml) and bFGF (12 ng/ml). CRITICAL STEP Using a 1000 μl pipette gently break the colonies into small clumps. Clump size can effect differentiation efficiency. Aim for clumps of 50–100 cells (Figure 1b).
7 Prepare new plates by washing gelatin coated plate with PBS as described in step 2. Aspirate the PBS and add the appropriate volume of CDM-PVA supplemented with Activin (10 ng/ml) and bFGF (12 ng/ml) as described in step 2
8 Add 100 μl of the cell suspension (step 6) to each 10 cm dish. CRITICAL STEP hPSCs should be plated in a density that will allow them to reach 80% confluence in 6 – 8 days for maintenance plates and 3-6 days for differentiation (Figure 1b). This is usually achieved by using a 1:6 – 1:10 split ratio. Adjust the volume of cell suspension added to each plate based on your split ratio. Optimal split ratios vary and need to be adjusted for each individual hPSC-line depending on its growth parameters. A typical plating density for our lines is 200,000 cells per 10 cm plate for maintenance and 500,000 – 1,000,000 cells for differentiation
9 Incubate the cells at 37 °C overnight
Differentiation of hPSCs into definitive endoderm TIMING 3 d
10 Day 1 Ensure hPSCs have fully attached after plating. Aspirate the medium and add freshly prepared CDM-PVA supplemented with Activin A (100 ng/ml), bFGF (80 ng/ml), BMP-4 (10 ng/ml), LY294002 (10 μM) and CHIR99021 (3 μM). Incubate the cells at 37 °C overnight
? TROUBLESHOOTING
11 Day 2 Replace the medium with freshly prepared CDM-PVA supplemented with Activin A (100 ng/ml), bFGF (80 ng/ml), BMP-4 (10 ng/ml), LY294002 (10 μM). Incubate the cells at 37 °C overnight
12 Day 3 Replace the medium with freshly prepared RPMI/B27 medium supplemented with Activin A (100 ng/ml) and bFGF (80 ng/ml). Incubate the cells at 37 °C overnight. The typical morphology of the cells at the end of this stage is demonstrated in figure 1b. A proportion of the cells can be further characterized with flow cytometry and IF for the expression of endoderm markers such as Sox17, anticipating >90% positive cells (Figure 3-4).
? TROUBLESHOOTING
Differentiation of definitive endoderm into foregut progenitor cells TIMING 5 d
13 Day 4-6 Replace the medium daily with freshly prepared RPMI/B27 medium supplemented with Activin A (50 ng/ml).
14 Day 7 Assess the homogeneity and morphology of the cells. The typical morphology of the cells on d7 is demonstrated in figure 1b. If cells exhibit optimal morphology with minimal contamination from undifferentiated or partially differentiated cells, then complete FP differentiation without splitting the cells (option A). For populations with sub-optimal morphological characteristics and significant contamination with poorly differentiated cells or if the cells are overgrown proceed to split the cells (option B).
(A) Completion of FP differentiation without splitting TIMING 2d
(i) Day 7-8 Replace the medium daily with freshly prepared RPMI/B27 medium supplemented with Activin A (50 ng/ml)
? TROUBLESHOOTING
(B) Splitting cells and completion of FP differentiation TIMING 2d
(i) Day 7 Prepare new plates as described in step 7
(ii) Wash the cells once with PBS as described in step 2. Add the appropriate volume of cell dissociation buffer as described in step 2 and incubate at 37 °C for 20 min until the cells have detached. Tap the plate to facilitate detachment.
(iii) Transfer the cells in a 15 ml tube. Gently aspirate and re-suspend the cell solution using a 5 ml serological pipette, to facilitate dissociation to single cells.
(iv) Wash the plate that contained the cells with 1 volume of RPMI/B27 medium and transfer the wash to the 15 ml tube
(v) Centrifuge at 444 g for 3 minutes. Aspirate the supernatant and resuspend the cells in 6 ml of RPMI/B27 medium.
(vi) Use a counting chamber to count the number of cells in the suspension
(vii) Centrifuge at 444 g for 3 minutes. Aspirate the supernatant and resuspend the cells the appropriate volume of freshly prepared RPMI/B27 medium supplemented with Activin A (50 ng/ml) and Rho kinase inhibitor Y-27632 (10 μm) for a final concentration of 1x106 cells/ml.
CRITICAL STEP Y-27632 should always be freshly added and kept in the culture for a minimum of 24 hours to improve cell survival.
(viii) Add the appropriate volume of cell suspension to the new plates to provide coverage of 150,000 cells / cm2. Ensure this is more than the minimum volume indicated in step 2 and supplement with freshly prepared RPMI/B27 medium supplemented with Activin A (50 ng/ml) if required.
(ix) Incubate at 37 °C overnight
CRITICAL STEP The density of the cells following the split may affect the efficiency of the later steps of differentiation. It is crucial to plate the cells at an appropriately high density so that the cells are almost confluent (90%) following the split. In some cases not all the cells attach therefore it is crucial to look at the plates and if necessary, increase the cell number plated to achieve the right confluence Very high densities promoting growth of cells in overlapping layers also have a negative impact on differentiation efficiency and should be avoided.
(x) Day 8 Replace the medium with freshly prepared RPMI/B27 medium supplemented with Activin A (50 ng/ml). Incubate at 37 °C overnight. Further characterize a proportion of the cells with IF and flow cytometry analyses for the expression of foregut markers such as GATA4 (Figure 3-4), anticipating >90% positive cells.
CRITICAL STEP The typical morphology of the cells at the end of this stage can be seen in figure 1b.
? TROUBLESHOOTING
Differentiation of foregut progenitor cells into hepatoblasts TIMING 4 d
15 Day 9-12 Replace the medium daily with freshly prepared RPMI/B27 medium supplemented with SB-431542 (10 μM) and BMP-4 (50 ng/ml). Monitor hepatoblast differentiation through the expression of HNF4A, AFP and TBX3 by IF and flow cytometry analyses.
CRITICAL STEP The typical morphology of the cells is demonstrated in figure 1b. Optimal hepatoblast differentiation is necessary for efficient differentiation of later stages. AFP expression should be observed in >95% of the cells by day 12 (Figure 3-4)
? TROUBLESHOOTING
Differentiation of hepatoblasts into cholangiocyte progenitors TIMING 4d
16 Day 13-16 Replace the medium daily with freshly prepared RPMI/B27 medium supplemented with FGF10 (50 ng/ml), Activin-A (50 ng/ml) and Retinoic acid (3μM). Monitor CP differentiation through the expression of Sox9 which should be observed in >75% of the cells by day 16 (Figure 4).
CRITICAL STEP The typical morphology of the cells is demonstrated in figure 1b. Optimal CP differentiation is necessary for efficient differentiation of later stages. ? TROUBLESHOOTING
Passaging of cholangiocyte progenitors and transfer to 3D culture conditions TIMING 1-2h
CRITICAL Prior to starting this step ensure that the matrigel and related equipment is prepared as described in the reagent setup section and the plate heater and the required number of plates are prepared as described in the equipment setup section.
17 Day 17 Wash the cells once with PBS and add the appropriate volume of cell dissociation buffer as described in step 2. Incubate at 37 °C for 20 min.
18 Tap the plate to facilitate detachment. The cells should detach as a monolayer or large clumps. If no detachment can be identified after 20 min proceed to mechanical dissociation with a pipette using a combination of horizontal, perpendicular and circular movements. We used a 1000 μl pipette for harvesting cells from 1 well of a 12-well plate.
19 Transfer the cells in a 15 ml tube. Gently aspirate and re-suspend the cell solution 2-3 times, using a 1000 μl pipette, to facilitate dissociation to small clumps.
CRITICAL STEP Clump size is crucial for the efficiency of the following differentiation step and the formation of organoids. Aim for clumps of 10-50 cells. Very small clumps and single cells exhibit poor survival, while large clumps gravitate to the bottom of the plate and fail to form organoids. Optimization of clump size may be required between different lines
? TROUBLESHOOTING
20 Wash the plate that contained the cells with 1 volume of RPMI/B27 medium and transfer the wash to the 15 ml tube Centrifuge at 444 g for 3 minutes. Aspirate the supernatant and resuspend the cells in 6 mls of RPMI/B27 medium.
21 Centrifuge at 444 g for 3 minutes. Aspirate the supernatant.
22 Re-suspend the cells in the appropriate volume of freshly prepared 50% (vol/vol) matrigel supplemented with EGF (20 ng/ml) and Rho kinase inhibitor Y-27632 (10 μm). Mix thoroughly. CRITICAL STEP Cholangiocyte progenitors should be plated in a density that will allow the emerging CLC organoids to reach 80% confluence in 10 days. This is usually achieved by using a 1:6 – 1:10 split ratio (1 well of a 12-well plate split to 10 wells of a 24-well plate). Optimal split ratios vary and need to be adjusted for each individual hPSC-line depending on its growth parameters and differentiation efficiency. A typical plating density for our lines is 1 – 2 x105 cells
CRITICAL STEP The 50% (vol/vol) matrigel cell suspension should be kept on ice at all times to avoid solidification
23 Mix the 50% (vol/vol) matrigel cell suspension thoroughly while keeping on ice. CRITICAL STEP Ensure 24 well plates have been placed on a plate heater or an incubator at least 30 min prior to plating, as described in the equipment setup section. Plating of the 50% (vol/vol) matrigel cell suspension should happen with the plate on the plate heater.
24 To form a matrigel dome in one well of a 24 well plate hold the tip of the 1000 μl pipette close to the surface of a well and start pipetting 50 μl of the 50% (vol/vol) matrigel cell suspension until a small droplet forms. Lower the pipette tip so that the droplet touches the warm plate surface and gently pipette the remainder of 50 μl. CRITICAL STEP Ensure that the droplet does not touch the walls of the well, which could lead to collapse of the matrigel dome.
25 Allow 1-2 minutes for the 50% (vol/vol) matrigel cell suspension to solidify. This can be assessed by gently tilting the plate. Turn the plate upside down and incubate at 37 °C for 30 min.
26 Add enough supplemented WE with EGF (20 ng/ml) and Rho kinase inhibitor Y-27632 (10 μm) to cover the matrigel domes. For 1 well of a 24 well plate we use 1ml of media.
CRITICAL STEP Y-27632 should always be freshly added and kept in the culture for a minimum of 24 hours to improve cell survival.
? TROUBLESHOOTING
Differentiation of cholangiocyte progenitors into Cholangiocyte-like Cell (CLC) organoids TIMING 10d
27 Day 17-26 Replace the medium every 2 days daily with freshly prepared supplemented WE medium with EGF (20 ng/ml). Organoids should start forming following 2-4 days of culture. Monitor CLC differentiation can be through the expression of CK7 which should be observed in >75% of the cells by day 26 (Figure 4), positive ALP staining (Figure 5a) and GGT activity (Figure 5b) of CLC organoids.
CRITICAL STEP The typical morphology of the cells is demonstrated in figure 1b and Supplementary Fig. 1.
? TROUBLESHOOTING
Characterization of CLC organoids
Immunofluorescence TIMING 2d
CRITICAL A matrigel dilution of 50% (vol/vol) or less should be used for the generation of CLC organoids for staining to allow adequate antibody penetration
28 Day 1 Aspirate the culture medium
29 Add 1ml of 4% (wt/vol) PFA per well of a 24-well plate, for 20 minutes at room temperature to fix CLC organoids in matrigel
30 Aspirate the PFA
31 Wash twice with PBS (10 minutes/wash)
32 Permeabilize and block for 1 hour with a 10% (vol/vol) donkey serum and 0.1% (vol/vol) Triton-X100 solution in PBS at room temperature
33 Stain overnight at 4°C with primary antibody diluted in a solution of 1% (vol/vol) donkey serum and 0.1% TritonX-100 in PBS.
34 Day 2 Wash 3 times with PBS (45 mins/wash)
35 Stain the CLC organoids for 1 hour at room temperature with secondary antibody raised in donkey and diluted 1:1000 (vol/vol) in a solution of 1% (vol/vol) donkey serum and 0.1% TritonX-100 in PBS.
36 Aspirate the secondary antibody
37 Add a solution of Hoechst 33258 1:10000 (vol/vol) in PBS for 10 minutes at room temperature
38 Wash 3 times with PBS (45 mins/wash).
39 Image using a confocal microscope. All IF images (Figure 2-3) were acquired using a Zeiss LSM 700 confocal microscope. Imagej 1.48k software (Wayne Rasband, NIHR, USA, http://imagej.nih.gov/ij) was used for image processing such as merging of different channels.
? TROUBLESHOOTING
Extraction of CLCs from matrigel for further analyses TIMING 40min
40 Aspirate the medium
41 Add 500μl/well of a 24-well plate cell recovery solution
42 Mechanically dissociate the matrigel and CLC organoids by scrapping with the tip of a P1000 pipette and transfer to a 15 ml falcon tube.
43 Incubate the resulting suspension of fragments of matrigel/CLC organoids in cell recovery solution for 30 minutes at 4°C
44 Centrifuge at 444 g, for 4 minutes
45 Aspirate the supernatant
46 Wash twice with supplemented WE medium.
47 Harvest and lyse CLC organoids for RNA extraction using any commercially available kit (we used the GenElute™ Mammalian Total RNA Miniprep Kit) or dissociate into single cells for flow cytometry following incubation with TrypLE for 5 minutes at 37°C.
TIMING
| Steps | Description | Timing |
|---|---|---|
| 1-9 | Passaging of hPSCs | 1 day |
| 10-12 | Differentiation of hPSCs into definitive endoderm | 3 days |
| 13-14 | Differentiation of definitive endoderm into foregut progenitor cells | 5 days |
| 15 | Differentiation of foregut progenitor cells into hepatoblasts | 4 days |
| 16 | Differentiation of hepatoblasts into cholangiocyte progenitors | 4 days |
| 17-26 | Passaging of cholangiocyte progenitors and transfer to 3D culture conditions | 1-2 hrs |
| 27 | Differentiation of cholangiocyte progenitors into CLC organoids | 10 days |
| 28-39 | IF staining of CLC organoids | 2 days |
| 40-47 | Extraction of cells from matrigel for further analyses | 40mins |
See Table 1 for Troubleshoot guidance
Anticipated Results
We describe a methodology for the differentiation of hPSCs into functional CLC organoids in 26 days. The early stages of our protocol (DE, FP, HB) result in > 90% cells expressing endoderm and then FP markers (Figure 4). However, biliary specification of hepatoblasts results in 75% CK19+/SOX9+ CPs, which mature to a population of 75% CK7+ CLCs during the final step of our differentiation (Figure 4). The resulting CLC organoids should express biliary markers such as CK19 and CK7 in immunofluorescence (IF) analyses (Figure 2-3). Hepatic markers (AFP, Albumin) can still be detected in these stages due to the presence of a contaminating population of hepatic lineage cells, but these should be identified only in clumps of cells without a lumen or attached to the bottom of the plate. Furthermore CLC organoids could be validated further for additional cholangiocyte markers such as CFTR, AE2, Secretin receptor7,24 and should demonstrate functional properties such as luminal accumulation of Rhodamine-123, GGT and ALP activity (Figure 5). The methods used to characterize CLC organoids (flow cytometry, immunofluorescence, Rhodamine-123 accumulation, GGT activity and ALP staining) have been described elsewhere 7.
Our platform promotes significant cell expansion. Using 3 different hPSC lines, we observed an average yield of >50x106 CLCs per 1x106 hPSCs. Proliferation should be particularly evident during the generation of CLC organoids. 1x105 CPs should give rise to 50-100 CLC organoids with diameters ranging between 100-1000μm. However, variations in terms of the expansion potential and the differentiation efficiency of our protocol can occur. This can be attributed to inherent differences between hPSC lines and batch-to-batch variability for some of the reagents including matrigel. For reproducible results the use of fresh medium and well-preserved small-molecule, recombinant protein and matrigel stocks is essential.
Supplementary Material
Morphology of CLC organoids. CLC organoids exhibit a typical cystic or branching tubular morphology. The black arrow indicates a tubular organoid, while the white arrow indicates a branching point. Scale bars: 100μm
Gating strategy used for the flow cytometry analyses demonstrated in Figure 4. Viable cells were gated based on forward scatter and side scatter and single cells were then gated based of forward scatter and pulse width. 2ary only controls were used to set the threshold for the FITC and APC channels.
Acknowledgements
This work was funded by ERC starting grant Relieve IMDs (L.V., N.H.), the Cambridge Hospitals National Institute for Health Research Biomedical Research Center (L.V., N.H., F.S.), the Evelyn trust (N.H.) and the EU Fp7 grant TissuGEN (M.CDB.). FS has been supported by an Addenbrooke’s Charitable Trust Clinical Research Training Fellowship and a joint MRC-Sparks Clinical Research Training Fellowship.
The authors would like to thank the Cambridge BRC hIPSCs core facility for the derivation of the Cystic Fibrosis hIPSC line, Dr Paulina Materek for the provision of cells used as negative controls, Dr Daniel Ortmann for his input in the design of the figures and Dr Petroula-Anastasia Tsagkaraki for her help with the generation of the manuscript figures and statistical analyses.
Footnotes
Author contributions: FS: Design and concept of study, execution of experiments and data acquisition, development of protocols and validation, collection of data, production of figures, manuscript writing, editing and final approval of manuscript. MCDB, IG, AB: Execution of experiments, collection and provision of data. NRFH: Design and concept of study, editing and final approval of manuscript. LV: Design and concept of study, editing and final approval of manuscript.
Competing financial interests: LV is a founder and shareholder of DefiniGEN. The remaining authors have nothing to disclose.
References
- 1.O’Hara SP, Tabibian JH, Splinter PL, Larusso NF. The dynamic biliary epithelia: Molecules, pathways, and disease. Journal of Hepatology. 2013;58:575–582. doi: 10.1016/j.jhep.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park SM. The crucial role of cholangiocytes in cholangiopathies. Gut Liver. 2012;6:295–304. doi: 10.5009/gnl.2012.6.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazaridis KN. The Cholangiopathies Konstantinos. Mayo Clin Proc. 2015;90:791–800. doi: 10.1016/j.mayocp.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sampaziotis F, Segeritz C-P, Vallier L. Potential of human induced pluripotent stem cells in studies of liver disease. Hepatology. 2015;62:303–311. doi: 10.1002/hep.27651. [DOI] [PubMed] [Google Scholar]
- 5.Pollheimer MJ, Trauner M, Fickert P. Will we ever model PSC? - ‘It’s hard to be a PSC model!’. Clin Res Hepatol Gastroenterol. 2011;35:792–804. doi: 10.1016/j.clinre.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Lemaigre FP. Notch signaling in bile duct development: new insights raise new questions. Hepatology. 2008;48:358–360. doi: 10.1002/hep.22480. [DOI] [PubMed] [Google Scholar]
- 7.Sampaziotis F, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and Development of the Liver. Developmental Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Hannan NRF, Segeritz C-P, Touboul T, Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat Protoc. 2013;8:430–437. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clotman F, et al. Control of liver cell fate decision by a gradient of TGF?? signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanai M, et al. FGF signaling segregates biliary cell-lineage from chick hepatoblasts cooperatively with BMP4 and ECM components in vitro. Dev Dyn. 2008;237:1268–1283. doi: 10.1002/dvdy.21520. [DOI] [PubMed] [Google Scholar]
- 12.Tabibian JH, Masyuk AI, Masyuk TV, O’Hara SP, LaRusso NF. Physiology of cholangiocytes. Compr Physiol. 2013;3:541–565. doi: 10.1002/cphy.c120019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa M, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol. 2015;33:853–61. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 14.Tanimizu N, Miyajima A, Mostov KE. Liver Progenitor Cells Develop Cholangiocyte-Type Epithelial Polarity in Three-dimensional Culture. Mol Biol Cell. 2007;18:1472–1479. doi: 10.1091/mbc.E06-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao D, et al. Derivation and characterization of hepatic progenitor cells from human embryonic stem cells. PLoS One. 2009;4:e6468. doi: 10.1371/journal.pone.0006468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabibian JH, et al. Characterization of cultured cholangiocytes isolated from livers of patients with primary sclerosing cholangitis. Lab Invest. 2014;0:1–8. doi: 10.1038/labinvest.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant AG, Billing BH. The isolation and characterization of a bile ductule cell population from normal and bile-duct ligated rat livers. Br J Exp Pathol. 1977;58:301–10. [PMC free article] [PubMed] [Google Scholar]
- 18.Joplin R, Strain AJ, Neuberger JM. Immuno-isolation and culture of biliary epithelial cells from normal human liver. In Vitro Cell Dev Biol. 1989;25:1189–92. doi: 10.1007/BF02621273. [DOI] [PubMed] [Google Scholar]
- 19.Zaret KS, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Eur J Cell Biol. 2015;33:1–48. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 20.Dianat N, et al. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology. 2014;60:700–14. doi: 10.1002/hep.27165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser S, et al. Heterogeneity of the intrahepatic biliary epithelium. World Journal of Gastroenterology. 2006;12:3523–3536. doi: 10.3748/wjg.v12.i22.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertero A, et al. Optimized inducible shRNA and CRISPR/Cas9 platforms for in vitro studies of human development using hPSCs. Development. 2016;143 doi: 10.1242/dev.138081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent L. Culture and maintenance of human embryonic stem cells. J Vis Exp. 2009:2–5. doi: 10.3791/1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannan NRF, Sampaziotis F, Segeritz C-P, Hanley NA, Vallier L. Generation of Distal Airway Epithelium from Multipotent Human Foregut Stem Cells. Stem Cells Dev. 2015;24:1680–90. doi: 10.1089/scd.2014.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agudo J, et al. GFP-specific CD8 T cells enable targeted cell depletion and visualization of T-cell interactions. Nat Biotechnol. 2015;33:1287–1292. doi: 10.1038/nbt.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphology of CLC organoids. CLC organoids exhibit a typical cystic or branching tubular morphology. The black arrow indicates a tubular organoid, while the white arrow indicates a branching point. Scale bars: 100μm
Gating strategy used for the flow cytometry analyses demonstrated in Figure 4. Viable cells were gated based on forward scatter and side scatter and single cells were then gated based of forward scatter and pulse width. 2ary only controls were used to set the threshold for the FITC and APC channels.