Abstract
Antibody responses to influenza viruses are critical for protection, but the ways in which repeated viral exposures shape antibody evolution and effectiveness over time remain controversial. Early observations demonstrated that the history of viral exposures has a profound effect on the specificity and magnitude of antibody responses to a new viral strain, a phenomenon called “original antigenic sin.” Although “sin” might suppress some aspects of the immune response, so far there is little indication that hosts with pre-existing immunity are more susceptible to viral infections compared to naïve hosts. However, the tendency of the immune response to focus on previously recognized conserved epitopes when encountering new viral strains can create an opportunity cost when mutations arise in these conserved epitopes. Hosts with different exposure histories may continue to experience distinct patterns of infection over time, which may influence influenza viruses’ continued antigenic evolution. Understanding the dynamics of B cell competition that underlie the development of antibody responses might help explain the low effectiveness of current influenza vaccines and lead to better vaccination strategies.
Introduction
Antibodies impose strong selection on influenza viruses and largely determine susceptibility to infection. Frequent mutations in viral surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) allow influenza viruses to continuously evade antibodies and infect human hosts repeatedly during their lifetime. Despite nearly seventy years of research, a coherent picture of the induction of human antibody responses and how these antibodies shape viral evolution and vaccine effectiveness is still emerging.
In this review, we propose that immunological and epidemiological evidence is remarkably consistent with one of the oldest and most notorious theories in influenza virus literature. In a series of studies in the 1940s and 1950s, Thomas Francis and colleagues demonstrated that humans have high antibody titers to influenza virus strains that they likely encountered early in life and that subsequent exposures with antigenically drifted viral strains boost antibody responses initiated by early childhood infections [1,2]. They also found that compared to primary exposures, antibodies generated during subsequent infections were more likely to cross-react with previous strains. Francis coined the phrase “original antigenic sin” to describe the preferential boosting of antibody responses to viral strains encountered early in life. Here, we review studies that led to the concept of original antigenic sin, and we describe more generally how prior viral exposures can have positive and negative effects on the generation of antibody responses. We present a working model of how prior exposures influence susceptibility to new influenza virus strains, which has important implications for viral evolution and vaccination strategies.
A short history of original antigenic sin
In 1947, a new antigenic variant of H1N1 influenza A viruses caused a severe epidemic. College students who had been vaccinated a few months earlier with the previously circulating viral strain (PR8) and naturally infected with the new viral strain developed higher acute antibody titers to PR8 upon infection than did unvaccinated students [3]. Infected students from both groups had higher acute and convalescent antibody titers to PR8 than to the new viral strain, and antibody titers to the new strain did not differ between the two groups. A preliminary explanation for these phenomena would take several years to unfold.
Davenport et al. [4] soon found that humans of all ages have higher antibody titers to strains they likely encountered in childhood. Sera from 1,250 Michigan residents showed that children possessed a narrower range of antibodies specific to recent strains of influenza A and B viruses, whereas older cohorts had higher antibody titers to older strains and more cross-reactive responses against recent strains. A cross-sectional study in Sheffield, England, revealed similar trends [5]. For each age cohort, antibody titers were usually highest against viral strains circulating in childhood and declined steadily against more recent viral strains [6,7]. Nearly sixty years later, studies of H3N2 antibody responses also found higher titers to older viral strains, although titers were not necessarily highest to strains from childhood [8,9].
As early as 1953, it was suspected that preexisting antibody responses were boosted when new strains shared cross-reactive antigens [4], but the first confirmation appeared when Jensen et al. analyzed the composition of sera from immunized humans and sequentially infected ferrets [10]. Sera from secondary exposures contained a high fraction of antibodies that cross-reacted with early viral strains and relatively few antibodies specific to later viral strains. Ten years later, de St. Groth and Webster showed that the secondary response, in contrast to the primary, was highly cross-reactive and surprisingly uniform in its affinity [11]. These results provided preliminary support for Francis’s claim that the response to the “first dominant antigen” would be repeatedly stimulated over a person’s lifetime, even as the original antigen became a “secondary or lesser component” of subsequent strains [2,12].
Is original antigenic sin detrimental?
While it is clear that antibody responses against childhood viral strains are efficiently boosted by antigenically novel strains, early reports conflicted about whether boosting comes at the expense of generating strong antibody responses against the new strain. The original study by Francis in 1947 found no difference in post-infection antibody titers to the new viral strain between recent recipients of the mismatched vaccine strain, whose titers were boosted, and non-recipients [3]. Similar results were found in animals sequentially infected with different influenza viruses [11]. The magnitude of the responses elicited by an antigenically distinct influenza virus in these studies was the same in animals with and without prior influenza exposure.
Other studies have suggested that prior exposures actively suppress the magnitude or quality of antibody responses to new viral strains. For example, Davenport & Hennessy [6] noted a “suppressive effect” on the antibody response to some viral strains in children, depending on the order in which they received monovalent vaccinations, and cited similar patterns of apparent suppression in other immunization studies [4,13]. Antibody responses tend to decline during repeated vaccinations [14]. de St. Groth & Webster [11] described the secondary response in immunized rabbits as “inadequate” because antibodies in the secondary response reacted better with the first antigen than the second. However, most studies that report inhibitory effects of prior exposures rely on the hemagglutination-inhibition assay, which only measures antibodies that block viral attachment to sialic acid. It is possible that sequential vaccinations in these studies elicit cross-reactive antibodies against other epitopes (such as the HA stalk) that are not detected in classical hemagglutination-inhibition assays. Thus, these studies might indicate that prior exposures affect the specificity of antibody responses, but this change in specificity might not affect overall protection.
There is currently minimal evidence that hosts with preexisting, cross-reactive immunity to influenza viruses experience greater susceptibility or more severe infections compared to naïve hosts. Cross-reactive antibody responses to influenza viruses appear generally beneficial. Early studies speculated that antibodies elicited against older viral strains were partially protective and that these cross-reactive antibodies reduced susceptibility and the opportunity to develop immunity to new strains [4,5,13]. A robust relationship between pre-existing antibody titers and reduced susceptibility has been repeatedly observed [15–17]. Cross-reactive antibodies elicited by initial infections limit virus replication during secondary viral exposures and reduce disease in experimental infections [18,19]. However, as discussed below, the direct benefits of preexisting responses against influenza viruses may be inevitably associated with opportunity costs. These costs can make some types of pre-existing antibody responses appear less beneficial than others, but they do not demonstrate that original antigenic sin has a net cost.
A contemporary synthesis
Nearly seventy years of accumulated evidence suggests how pre-existing responses, coupled with repeated exposures to antigenically evolving influenza viruses, might generate the immunological and epidemiological patterns associated with original antigenic sin. A central element is the competitive dominance of memory versus naïve B cells for antigen. The anamnestic basis of secondary responses to influenza viruses has been demonstrated by Jensen et al. [10], de St. Groth and Webster [11], and others [20–23]. Memory B cells targeting epitopes shared with the original strain are reactivated, and these cells dominate secondary immune responses because they presumably outcompete naïve B cells, which have a higher threshold of activation [24,25]. The recall of memory B cells can be advantageous because these cells can acquire additional somatic mutations that increase affinity to new viral strains [22]. The level of activation of naïve B cells in secondary immune responses is likely partially dependent on antigen dose. For example, naïve B cells can be activated and the antibody response broadened if high doses of secondary antigen are administered [11], the antigen is given with adjuvants [26], or repeated doses of antigen are given [7].
From these immune dynamics, complex patterns of serology and infection can arise as a function of hosts’ exposure histories. Due to influenza viruses’ rapid spread and fast antigenic evolution, these differences are partly recognizable as contrasting patterns by birth year (Figure 1A). Hosts infected for the first time develop antibodies that target multiple epitopes on the surface of influenza viruses’ HA, although antibodies with particular specificities may dominate due to differences in epitopes’ immunogenicity, chance, or host-specific factors [27]. Hosts remain protected as long as circulating viral strains conserve at least one epitope to which hosts have high concentrations of neutralizing antibodies. If exposure induces mild infection (many influenza virus infections are mild [28,29]), then these responses are boosted, and additional cross-reactive antibodies may continue to evolve. This model is consistent with the gradual increase with age in concentrations of cross-reactive anti-HA stalk antibodies [30], which are normally subdominant [21]. It also shares features with other models of immune dynamics that allow preexisting responses to outcompete new responses via resource limitation or suppression [31–33].
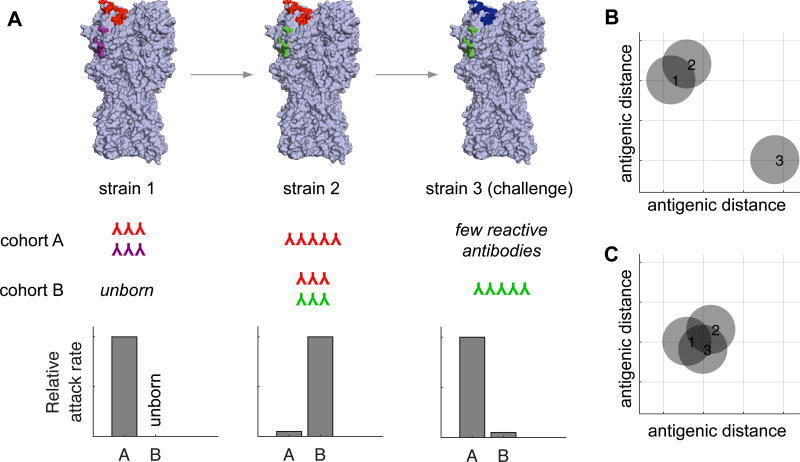
Figure 1. The development of antibody response to drifted influenza virus strains.
(A) The population begins completely susceptible to strain 1, which has two epitopes in this example. Upon infection, individuals in cohort A generate antibodies against the red and purple epitopes. The strain then acquires a mutation in the purple epitope (and becomes strain 2). New susceptible hosts (cohort B) that become infected with strain 2 develop antibodies to the red and green epitopes. In contrast, older individuals (cohort A) that are exposed to strain 2 develop an antibody response focused primarily on the red epitope that was conserved in strain 1. Older individuals likely experience mild infections with strain 2 since these individuals possess cross-reactive antibodies against the red epitope. Eventually, immune pressure at the red epitope selects for a virus (strain 3) that possesses a red→blue mutation. Older individuals (cohort A) regain susceptibility since they have an antibody response focused on the former red epitope, and younger individuals (cohort B) are protected against this strain because they possess antibodies against the green epitope conserved in strain 2. (B–C) After exposure to strain 2, antigenic cartography based on the sera from cohort A (B) and cohort B (C) reveals different patterns. Antibodies from individuals in cohort B recognize the red and green epitopes and perceive all strains as identical, whereas antibodies from individuals in cohort A recognize the red epitope and perceive strain 3 to be distinct from strains 1 and 2.
The focusing of antibody responses to epitopes conserved in older influenza virus strains can have dangerous consequences when viruses acquire mutations in these epitopes. An example of this was seen following the 2009 H1N1 pandemic. Most individuals infected with the 2009 pandemic H1N1 virus mounted antibody responses against epitopes that were conserved in older seasonal H1N1 viruses [20,23,34,35]. Following exposure with the 2009 H1N1 virus, humans produced antibodies of different specificities depending on the specific seasonal H1N1 virus that they were exposed to in childhood [20,35,36]. In some individuals, this led to a very focused antibody response. This focus became a problem during the 2013–2014 influenza season when pandemic H1N1 viruses acquired a new mutation in an exposed region of HA that was targeted by antibodies present in many middle-aged individuals [20]. This region of HA was conserved between seasonal H1N1 viruses from the late 1970s and the 2009 pandemic H1N1 virus. Since antibodies failed to bind to the 2013–2014 H1N1 strain that possessed a mutation in this epitope, middle-aged individuals were disproportionately affected by the new drifted H1N1 strain [37]. This season revealed the opportunity cost of preexisting immunity: because middle-aged humans were more protected than younger cohorts to the original 2009 strain, they missed opportunities to develop antibodies to other epitopes that would have protected them in 2013–2014.
Immune history may shape patterns of infection not only with different strains within the same subtype but also with different subtypes. Several lines of evidence suggest that birth year, a proxy for early exposure to particular subtypes, affects susceptibility to others. In 1953, Francis speculated that the peculiarly high incidence in young adults of a pandemic influenza-like illness in 1782 resulted from preexisting immunity [1]. Francis and others proposed that in 1918, primary exposures to previously circulating H1 subtypes lowered susceptibility in children and older adults [1,38]: although young adults had probably already been infected with other H1 viruses, their first exposure (presumably to an H3 virus that emerged in 1889–1890) may have precluded the development of a robust response to H1. Other evidence suggesting that the subtype of first exposure affects immunity to other subtypes in an original antigenic sin-like way comes from age-specific mortality patterns in 2009 [39] and the age distribution of H5N1 and H7N9 cases [40]. Neutralizing cross-reactive heterosubtypic antibodies appear uncommon [21], and thus a reduction in heterosubtypic antigen availability mediated by other cross-reactive immune responses, such as memory T cells, might explain this sin-like phenomenon. Repeated exposures may gradually erode this effect [41,42]. This erosion is consistent with the observation that subtype-specific stalk antibodies accumulate in proportion to total exposure to each subtype [30].
Implications for viral evolution and vaccination
Influenza virus populations evolve through competition for susceptible hosts, and the existence of host subpopulations targeting different epitopes suggests a mechanism for influenza viruses’ regular antigenic evolution. In theory, assuming mutations that change the antigenic phenotype do not otherwise affect viral fitness, mutated strains that have the most susceptible hosts should spread fastest. If antigenic mutations occur slowly relative to the timescale of transmission, then influenza viruses could evolve to escape immunity in one subpopulation after another [43,44].
The effects of immune history on susceptibility to influenza viruses have several consequences for current vaccination strategies. Antigenic distances are typically measured using sera isolated from ferrets recovering from influenza virus infections [45]. With epitope-specific immunity, the antigenic distance between two strains can differ among hosts with different immune histories (Figure 1B,C) [46,47]. Thus, strains that appear antigenically similar according to antibodies raised in ferrets (i.e., in animals without prior influenza virus exposures) might be distinct from antibodies in adults [36]. The World Health Organization has recognized this problem and recently updated the 2017 Southern Hemisphere H1N1 vaccine strain based on human serology (http://www.who.int/influenza/vaccines/virus/recommendations/2017_south/en/). Antibodies elicited in ferrets do not antigenically distinguish the old and new H1N1 vaccine strains, but antibodies elicited in a subset of humans do differentially recognize these H1N1 strains. Due to the primacy of antibody titers in determining susceptibility and strain fitness, cross-sectional serologic testing could be useful not only for identifying the need for vaccine updates but also as a complementary—or even alternative—method to predict the evolution of seasonal viruses [48–52].
Immune history also matters for the development of new vaccination strategies. In 1957, Davenport and colleagues immunized differently aged individuals with a polyvalent vaccine containing four antigenic variants of H1N1 [7]. Broad antibody titers arose after repeated immunizations, and more recent studies confirm that multiple immunizations can elicit antibodies that target conserved epitopes [23,53]. An important consideration is whether such responses are protective, because it is possible that sequential exposures might elicit antibody responses toward conserved epitopes that are non-neutralizing. It will also be important to determine how to maintain levels of specific types of antibodies via vaccination. For example, in 2009 many individuals mounted antibodies that recognize the conserved HA stalk of the pandemic H1N1 virus [23]. However, these HA stalk-specific antibody responses dissipated after repeated vaccinations [21]. Understanding interactions between preexisting and new responses might also illuminate seemingly low vaccine effectiveness among repeat vaccinees [54–58].
Future directions
The majority of this manuscript has focused on neutralizing HA antibodies, but it is clear that other types of antibodies can limit influenza virus replication and spread. For example, some anti-HA antibodies limit virus replication in vivo through mechanisms involving ADCC [59,60]. These antibodies are not accounted for in most influenza virus serological assays. Similarly, NA antibodies can limit influenza virus spread and disease severity [61], and there is evidence that NAs of human influenza viruses undergo antigenic drift [62]. It will be important to continue to identify new correlates of protection against influenza virus infection and determine how prior exposures influence these processes.
We propose that quantitative, predictive models that relate previous exposures to susceptibility to different strains are within reach. The main hurdle is to understand fundamental dynamics of the immune system. There is evidence that “antigenic sin” occurs in humans, but the mechanisms involved in this process remain underdeveloped. What determines which viral epitopes are targeted by antibodies in primary infections, what determines variation between individuals, and how do immune repertoires evolve over time? New sequencing methods to examine B cell receptors, combined with animal experiments and longitudinal studies in humans, have the potential to provide fine-scale observations of the development of immunity to influenza viruses and related pathogens [21,63–65]. These large data sets can be used to evaluate mathematical models that capture the complex immune dynamics involved in secondary responses to influenza viruses [31,32,66]. Understanding the interactions that shape immunity over time will aid in our understanding of the selective pressures that shape the fitness of circulating influenza virus strains, and could potentially reveal strategies to increase vaccine effectiveness.
Highlights.
Early viral infections shape B cell response recalled against future viral strains
Competition between memory and naïve B cells occurs in secondary viral exposures
Antibodies become focused on epitopes conserved in past influenza virus strains
Focused antibody responses fail to protect against mutated viral strains
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (1R01AI113047, SEH; 1R01AI108686, SEH; DP2AI117921, SC; CEIRS HHSN272201400005C, SEH and SC). Scott E. Hensley, Ph.D. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
*special interest, **outstanding interest
- 1.Francis T., Jr Influenza: the new acquayantance. Ann Intern Med. 1953;39:203–221. doi: 10.7326/0003-4819-39-2-203. [DOI] [PubMed] [Google Scholar]
- 2.Francis T., Jr The current status of the control of influenza. Ann Intern Med. 1955;43:534–538. doi: 10.7326/0003-4819-43-3-534. [DOI] [PubMed] [Google Scholar]
- 3.Francis T, Salk JE, Quilligan JJ. Experience with Vaccination Against Influenza in the Spring of 1947: A Preliminary Report. Am J Public Health Nations Health. 1947;37:1013–1016. doi: 10.2105/ajph.37.8.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davenport FM, Hennessy AV, Francis T., Jr Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J Exp Med. 1953;98:641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport FM, Hennessy AV, Stuart-Harris CH, Francis T., Jr Epidemiology of influenza; comparative serological observations in England and the United States. Lancet. 1955;269:469–474. doi: 10.1016/s0140-6736(55)93328-6. [DOI] [PubMed] [Google Scholar]
- 6.Davenport FM, Hennessy AV. A serologic recapitulation of past experiences with influenza A; antibody response to monovalent vaccine. J Exp Med. 1956;104:85–97. doi: 10.1084/jem.104.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davenport FM, Hennessy AV. Predetermination by infection and by vaccination of antibody response to influenza virus vaccines. J Exp Med. 1957;106:835–850. doi: 10.1084/jem.106.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessler J, Riley S, Read JM, Wang S, Zhu H, Smith GJ, Guan Y, Jiang CQ, Cummings DA. Evidence for antigenic seniority in influenza A (H3N2) antibody responses in southern China. PLoS Pathog. 2012;8:e1002802. doi: 10.1371/journal.ppat.1002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, Xue L, Jones TC, Le NM, Pham QT, et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346:996–1000. doi: 10.1126/science.1256427.* This longitudinal analysis of human sera demonstrates that exposures to new viral strains generally boost responses to old viral strains, consistent with many studies from the 1950s. In contrast to observations of H1N1 and Lessler et al. (2012), however, there is a suggestion that this boosting may be focused on more recent viral strains.
- 10.Jensen KE, Davenport FM, Hennessy AV, Francis T., Jr Characterization of influenza antibodies by serum absorption. J Exp Med. 1956;104:199–209. doi: 10.1084/jem.104.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fazekas de St G, Webster RG. Disquisitions on Original Antigenic Sin. II. Proof in lower creatures. J Exp Med. 1966;124:347–361. doi: 10.1084/jem.124.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis T. On the doctrine of original antigenic sin. Proceedings of the American Philosophical Society. 1960;104:572–578. [Google Scholar]
- 13.Hennessy AV, Davenport FM, Francis T., Jr Studies of antibodies to strains of influenza virus in persons of different ages in sera collected in a postepidemic period. J Immunol. 1955;75:401–409. [PubMed] [Google Scholar]
- 14.Thompson MG, Naleway A, Fry AM, Ball S, Spencer SM, Reynolds S, Bozeman S, Levine M, Katz JM, Gaglani M. Effects of Repeated Annual Inactivated Influenza Vaccination among Healthcare Personnel on Serum Hemagglutinin Inhibition Antibody Response to A/Perth/16/2009 (H3N2)-like virus during 2010–11. Vaccine. 2016;34:981–988. doi: 10.1016/j.vaccine.2015.10.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wikramaratna PS, Rambaut A. Relationship between haemagglutination inhibition titre and immunity to influenza in ferrets. Vaccine. 2015;33:5380–5385. doi: 10.1016/j.vaccine.2015.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coudeville L, Bailleux F, Riche B, Megas F, Andre P, Ecochard R. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: development and application of a bayesian random-effects model. BMC Med Res Methodol. 2010;10:18. doi: 10.1186/1471-2288-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox A, Mai le Q, Thanh le T, Wolbers M, Le Khanh Hang N, Thai PQ, Thi Thu Yen N, Minh Hoa le N, Bryant JE, Duong TN, et al. Hemagglutination inhibiting antibodies and protection against seasonal and pandemic influenza infection. J Infect. 2015;70:187–196. doi: 10.1016/j.jinf.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JH, Skountzou I, Compans R, Jacob J. Original antigenic sin responses to influenza viruses. J Immunol. 2009;183:3294–3301. doi: 10.4049/jimmunol.0900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, Liepkalns J, Reber AJ, Lu X, Music N, Jacob J, Sambhara S. Prior infection with influenza virus but not vaccination leaves a long-term immunological imprint that intensifies the protective efficacy of antigenically drifted vaccine strains. Vaccine. 2016;34:495–502. doi: 10.1016/j.vaccine.2015.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, Ellebedy AH, Carter DM, Andrews SF, Zheng NY, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc Natl Acad Sci U S A. 2014;111:15798–15803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linderman SL, Hensley SE. Antibodies with 'Original Antigenic Sin' Properties Are Valuable Components of Secondary Immune Responses to Influenza Viruses. PLoS Pathog. 2016;12:e1005806. doi: 10.1371/journal.ppat.1005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tangye SG, Avery DT, Deenick EK, Hodgkin PD. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J Immunol. 2003;170:686–694. doi: 10.4049/jimmunol.170.2.686. [DOI] [PubMed] [Google Scholar]
- 25.Tomayko MM, Anderson SM, Brayton CE, Sadanand S, Steinel NC, Behrens TW, Shlomchik MJ. Systematic comparison of gene expression between murine memory and naive B cells demonstrates that memory B cells have unique signaling capabilities. J Immunol. 2008;181:27–38. doi: 10.4049/jimmunol.181.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JH, Davis WG, Sambhara S, Jacob J. Strategies to alleviate original antigenic sin responses to influenza viruses. Proc Natl Acad Sci U S A. 2012;109:13751–13756. doi: 10.1073/pnas.0912458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cobey S, Wilson P, Matsen FAt. The evolution within us. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeb M, Singh PK, Fox J, Russell ML, Pabbaraju K, Zarra D, Wong S, Neupane B, Singh P, Webby R, et al. Longitudinal study of influenza molecular viral shedding in Hutterite communities. J Infect Dis. 2012;206:1078–1084. doi: 10.1093/infdis/jis450. [DOI] [PubMed] [Google Scholar]
- 29.Carrat F, Vergu E, Ferguson NM, Lemaitre M, Cauchemez S, Leach S, Valleron AJ. Time lines of infection and disease in human influenza: A review of volunteer challenge studies. American Journal of Epidemiology. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 30.Miller MS, Gardner TJ, Krammer F, Aguado LC, Tortorella D, Basler CF, Palese P. Neutralizing antibodies against previously encountered influenza virus strains increase over time: a longitudinal analysis. Sci Transl Med. 2013;5:198ra107. doi: 10.1126/scitranslmed.3006637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zarnitsyna VI, Lavine J, Ellebedy A, Ahmed R, Antia R. Multi-epitope Models Explain How Pre-existing Antibodies Affect the Generation of Broadly Protective Responses to Influenza. PLoS Pathog. 2016;12:e1005692. doi: 10.1371/journal.ppat.1005692.* Using antibody titers before and after vaccination, the study evaluates several mechanisms by which antibodies to the HA head outcompete antibodies to the HA stalk. They find the most support for negative density-dependent feedback on HA head antibodies due to epitope masking.
- 32.Ndifon W. A simple mechanistic explanation for original antigenic sin and its alleviation by adjuvants. J R Soc Interface. 2015;12 doi: 10.1098/rsif.2015.0627.* This paper uses experimental observations of Kim et al. (2009, 2012) to propose a simple mathematical model of original antigenic sin. T regulatory cells reduce available antigen in secondary infections, promoting the reactivation and expansion of memory B cells at the expense of naïve B cells.
- 33.Smith DJ, Forrest S, Ackley DH, Perelson AS. Variable efficacy of repeated annual influenza vaccination. Proc Natl Acad Sci U S A. 1999;96:14001–14006. doi: 10.1073/pnas.96.24.14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang KY, Rijal P, Schimanski L, Powell TJ, Lin TY, McCauley JW, Daniels RS, Townsend AR. Focused antibody response to influenza linked to antigenic drift. J Clin Invest. 2015;125:2631–2645. doi: 10.1172/JCI81104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Myers JL, Bostick DL, Sullivan CB, Madara J, Linderman SL, Liu Q, Carter DM, Wrammert J, Esposito S, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med. 2013;210:1493–1500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hensley SE. Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr Opin Virol. 2014;8C:85–89. doi: 10.1016/j.coviro.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrie JG, Parkhouse K, Ohmit SE, Malosh RE, Monto AS, Hensley SE. Antibodies against the current influenza A H1N1 vaccine strain do not protect some individuals from infection with contemporary circulating H1N1 viral strains. J Infect Dis. 2016 doi: 10.1093/infdis/jiw479. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Worobey M, Han GZ, Rambaut A. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci U S A. 2014;111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs JH, Archer BN, Baker MG, Cowling BJ, Heffernan RT, Mercer G, Uez O, Hanshaoworakul W, Viboud C, Schwartz J, et al. Searching for sharp drops in the incidence of pandemic A/H1N1 influenza by single year of age. PLoS One. 2012;7:e42328. doi: 10.1371/journal.pone.0042328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. Potent Protection against H5N1 and H7N9 Influenza via Childhood Hemagglutinin Imprinting. Science. doi: 10.1126/science.aag1322. in press.** The authors demonstrate that birth year, a proxy for primary exposures to different subtypes, correlates well with the risk of severe infection or death with avian influenza subtypes H5N1 and H7N9. They propose that primary exposure to group 1 HA (H1 or H2) induces strong immunity to the HA that provides some protection against other HAs from this group, such as H5. The complement is true for group 2 (H3 and H7).
- 41.Gagnon A, Miller MS, Hallman SA, Bourbeau R, Herring DA, Earn DJ, Madrenas J. Age-specific mortality during the 1918 influenza pandemic: unravelling the mystery of high young adult mortality. PLoS One. 2013;8:e69586. doi: 10.1371/journal.pone.0069586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gagnon A, Acosta JE, Madrenas J, Miller MS. Is antigenic sin always "original?" Re-examining the evidence regarding circulation of a human H1 influenza virus immediately prior to the 1918 Spanish flu. PLoS Pathog. 2015;11:e1004615. doi: 10.1371/journal.ppat.1004615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato K, Morishita T, Nobusawa E, Tonegawa K, Sakae K, Nakajima S, Nakajima K. Amino-acid change on the antigenic region B1 of H3 haemagglutinin may be a trigger for the emergence of drift strain of influenza A virus. Epidemiology and Infection. 2004;132:399–406. doi: 10.1017/s0950268803001821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang ML, Skehel JJ, Wiley DC. Comparative analyses of the specificities of anti-influenza hemagglutinin antibodies in human sera. J Virol. 1986;57:124–128. doi: 10.1128/jvi.57.1.124-128.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 46.Cobey S, Pascual M. Consequences of host heterogeneity, epitope immunodominance, and immune breadth for strain competition. J Theor Biol. 2011;270:80–87. doi: 10.1016/j.jtbi.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cobey S. Pathogen evolution and the immunological niche. Ann N Y Acad Sci. 2014;1320:1–15. doi: 10.1111/nyas.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luksza M, Lassig M. A predictive fitness model for influenza. Nature. 2014;507:57–61. doi: 10.1038/nature13087.* This manuscript proposes and evaluates models to predict the evolution of influenza virus strains. The models are based entirely on sequence analyses. This and similar models might be even more accurate after taking into account the specificity of antibodies prevelant in different human cohorts.
- 49.Neher RA, Bedford T, Daniels RS, Russell CA, Shraiman BI. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc Natl Acad Sci U S A. 2016;113:E1701–E1709. doi: 10.1073/pnas.1525578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He J, Deem MW. Low-dimensional clustering detects incipient dominant influenza strain clusters. Protein Eng Des Sel. 2010;23:935–946. doi: 10.1093/protein/gzq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia Z, Jin G, Zhu J, Zhou R. Using a mutual information-based site transition network to map the genetic evolution of influenza A/H3N2 virus. Bioinformatics. 2009;25:2309–2317. doi: 10.1093/bioinformatics/btp423. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki Y. Predictability of antigenic evolution for H3N2 human influenza A virus. Genes Genet Syst. 2013;88:225–232. doi: 10.1266/ggs.88.225. [DOI] [PubMed] [Google Scholar]
- 53.Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, Palese P. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean HQ, Thompson MG, Sundaram ME, Meece JK, McClure DL, Friedrich TC, Belongia EA. Impact of repeated vaccination on vaccine effectiveness against influenza A(H3N2) and B during 8 seasons. Clin Infect Dis. 2014;59:1375–1385. doi: 10.1093/cid/ciu680.* This is one of several recent studies demonstrating that influenza vaccine effectiveness appears to be reduced among frequent vaccine recipients. This analysis is particularly strong because the outcome measure is confirmed infection, and the prospective design allowed careful tracking of vaccination and infection status over five years.
- 55.Ohmit SE, Thompson MG, Petrie JG, Thaker SN, Jackson ML, Belongia EA, Zimmerman RK, Gaglani M, Lamerato L, Spencer SM, et al. Influenza vaccine effectiveness in the 2011–2012 season: protection against each circulating virus and the effect of prior vaccination on estimates. Clin Infect Dis. 2014;58:319–327. doi: 10.1093/cid/cit736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohmit SE, Petrie JG, Malosh RE, Fry AM, Thompson MG, Monto AS. Influenza vaccine effectiveness in households with children during the 2012–2013 season: assessments of prior vaccination and serologic susceptibility. J Infect Dis. 2015;211:1519–1528. doi: 10.1093/infdis/jiu650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skowronski DM, Chambers C, Sabaiduc S, De Serres G, Winter AL, Dickinson JA, Krajden M, Gubbay JB, Drews SJ, Martineau C, et al. A Perfect Storm: Impact of Genomic Variation and Serial Vaccination on Low Influenza Vaccine Effectiveness During the 2014–2015 Season. Clin Infect Dis. 2016;63:21–32. doi: 10.1093/cid/ciw176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skowronski DM, De Serres G, Crowcroft NS, Janjua NZ, Boulianne N, Hottes TS, Rosella LC, Dickinson JA, Gilca R, Sethi P, et al. Association between the 2008–09 seasonal influenza vaccine and pandemic H1N1 illness during Spring-Summer 2009: four observational studies from Canada. PLoS Med. 2010;7:e1000258. doi: 10.1371/journal.pmed.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol. 2013;190:1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- 61.Eichelberger MC, Wan H. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol. 2015;386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- 62.Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A. 2011;108:20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- 64.DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, Ellington AD, Ippolito GC, Gray JJ, et al. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc Natl Acad Sci U S A. 2016;113:E2636–E2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laserson U, Vigneault F, Gadala-Maria D, Yaari G, Uduman M, Vander Heiden JA, Kelton W, Taek Jung S, Liu Y, Laserson J, et al. High-resolution antibody dynamics of vaccine-induced immune responses. Proc Natl Acad Sci U S A. 2014;111:4928–4933. doi: 10.1073/pnas.1323862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Childs LM, Baskerville EB, Cobey S. Trade-offs in antibody repertoires to complex antigens. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]