Abstract
Metabolically engineered Escherichia coli JM109 harboring plasmid pBPP1 and expressing the nonnatural BPEC pathway for synthesis of thermoplastic polyhydroxyalkanoates (PHA) and novel polythioesters (PTE) to provide suitable substrates of PHA synthase was investigated with respect to biotechnological production of poly(3-mercaptopropionate) [poly(3MP)]. Fed-batch fermentation processes were established at the 30- and 500-liter scales in stirred tank bioreactors to produce kilogram amounts of poly(3MP). Cultivation was done in a modified M9 mineral salts medium containing glucose or glycerol as the carbon and energy source and with 3-mercaptopropionic acid (3MP) as the precursor substrate for poly(3MP) biosynthesis provided from the late exponential growth phase. Approximately 23 g of cell dry matter (CDM) per liter and poly(3MP) cell contents of up to 45% (wt/wt) were the highest cell densities and polymer contents obtained, respectively. At best, 69.1% (wt/wt) of 3MP was converted into poly(3MP), indicating that 3MP was mostly used for poly(3MP) biosynthesis. Furthermore, a novel in situ process for rapid and convenient isolation of poly(3MP) from the cells in the bioreactor was developed. This was achieved by addition of sodium dodecyl sulfate to the cultivation broth immediately after the fermentation, heating to 90°C for 20 min with intensive stirring, and subsequent washing steps. The purity of such in situ isolated poly(3MP) was more than 98%, as revealed by gas chromatographic and elemental sulfur analyses of the material isolated.
Polyhydroxyalkanoates (PHAs) are intracellular storage polymers that are accumulated by a wide variety of bacteria under nutrient limitation and carbon excess conditions via different pathways (11, 25, 30, 31). Naturally, polyoxoesters are synthesized from coenzyme A (CoA) thioesters of the respective hydroxyalkanoates, which are synthesized from acetyl-CoA or are derived from metabolites of fatty acid metabolism (11, 30, 31, 33, 34). Generally, PHAs are water insoluble, thermoplastic and/or elastomeric, enantiomerically pure, nontoxic, biocompatible, and, in particular, biodegradable (29). More than 140 hydroxyalkanoic acids have been identified as constituents of PHAs, representing a versatile class of microbial polymers (32).
Intensive research has been carried out on the physiology, biochemistry, and molecular genetics of the metabolism of PHAs in various naturally PHA-producing bacteria during the last two decades (26, 33). High-cell-density culture techniques for culturing recombinant strains of Escherichia coli were developed for production of various PHAs to reduce the production cost and to increase productivity (9, 10, 11, 16, 27). Cell densities of up to 174 g (dry cell weight)/liter have been achieved in a dialysis reactor (9). However, high-cell-density techniques have several drawbacks, including substrate inhibition, limited oxygen transfer capacity, and formation of growth-inhibiting by-products, like acetate (9, 10, 14, 22, 27).
Recently, novel biopolymers with thioester linkages in the polymer backbone were isolated from PHA-accumulating Ralstonia eutropha. These biopolymers contained 3-mercaptoalkanoates as constituents in addition to 3-hydroxybutyrate and were referred to as polythioesters (PTEs) (17, 19). PTEs isolated from R. eutropha always contained more than 25 mol% 3-hydroxybutyrate (18), because the poly(3-hydroxybutyrate) biosynthesis pathway could not be suppressed in this bacterium. Therefore, a recombinant strain of E. coli that expressed an alternative nonnatural biosynthesis pathway for production of PTE homopolymers was used (15). This pathway was constructed by cloning three genes from two different bacteria, resulting in plasmid pBPP1, and it was heterologously expressed in E. coli JM109. The biosynthesis pathway included a butyrate kinase (Buk) and a phosphotransbutyrylase (Ptb) from Clostridium acetobutylicum and a PHA synthase (PhaEC) from Thiococcus pfennigii to synthesize polyoxoesters and polythioesters independently from the central metabolism of the host cells (12, 13). The respective hydroxyalkanoates or 3-mercaptoalkanoates were phosphorylated by Buk after uptake, subsequently converted to the corresponding hydroxyalkanoate-CoA or 3-mercaptoalkanoate-CoA by Ptb, and finally polymerized by PhaEC. By using this nonnatural BPEC pathway, different PHA and PTE homopolymers were synthesized depending on the precursor substrates provided (12, 13, 15, 16).
So far, the novel PTEs have been available only in milligram or gram quantities. This is the first report of biotechnological production of poly(3-mercaptopropionate) [poly(3MP)]. Fermentations were done in 30- and 500-liter pilot scale plants by employing the BPEC pathway. In this paper, developments in improving the biotechnological process for poly(3MP) production, including improvements in medium composition, feeding regimen, etc., are described. Since the previously described enzymatic method for isolation and purification of poly(3MP) (15) was not applicable to this large scale, a novel in situ isolation method was developed for downstream processing and for purifying poly(3MP) at a technical scale.
MATERIALS AND METHODS
Bacterial strain and cultivation media.
E. coli JM109 harboring plasmid pBPP1 (12, 13) was used in all experiments. The basic medium used in this study was the M9 mineral salts medium (M9-MSM) described by Sambrook et al. (28), which was supplemented with yeast extract and the SL6 trace elements solution (23), as shown in Table 1, and with 75 μg of ampicillin per ml as indicated below. This medium was modified in different cultivation experiments as described below. To inoculate 30- and 500-liter cultures, starter cultures grown in 3 and 27 liters of M9-MSM, respectively, were used.
TABLE 1.
Medium components and yields of cell mass and poly(3MP) obtained during different fed batch fermentations of E. coli JM109(pBPP1)
| Parameter | Fed batch fermentation
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Vol at beginning (liters)a | 27 | 27 | 400 | 400 | 400 | 22 | 25 | 15 | 18 |
| Vol at end (liters)b | 30 | 30 | 470 | 470 | 500 | 25 | 30 | 25 | 28 |
| Medium components added (total amt)c | |||||||||
| Glucose (g) | 480 | 600 | 10,500 | 12,000 | 23,500 | ||||
| Glycerol (g) | 1,638 | 4,183 | 3,250 | 3,780 | |||||
| Na2HPO4 · 12H2O (g) | 162 | 162 | 2,400 | 2,400 | 2,400 | 150 | 360 | 405 | 60 |
| KH2PO4 (g) | 81 | 81 | 1,200 | 1,200 | 1,200 | 75 | 180 | 202.5 | 195 |
| NH4Cl (g) | 67 | 81 | 900 | 700 | 2,400 | 110 | 190 | 165 | 180 |
| NaCl (g) | 13.5 | 13.5 | 207 | 207 | 200 | 12.5 | 12.5 | 11.25 | 7.5 |
| MgSO4 · 7H2O (g) | 40.5 | 40.5 | 616 | 616 | 600 | 37.5 | 90 | 135 | 240 |
| CaCl2 · 2H2O (g) | 5.4 | 5.4 | 37 | 37 | 80 | 5 | 12 | 2.25 | |
| Yeast extract (g) | 18.9 | 18.9 | 267 | 267 | 280 | 25 | 30 | 18 | 30 |
| SL6 solution (ml) | 1.1 | 3 | 3 | 3 | |||||
| Fe(III) NH4+ citrate (ml) | 22 | 30 | 30 | 30 | |||||
| 3MP (ml) | 140 | 140 | 2,500 | 2,500 | 3,000 | 150 | 200 | 450 | 400 |
| Amt of CDM (g/liter) | 3.3 | 3.7 | 2.6 | 2.5 | 7 | 7.3 | 17 | 22 | 23 |
| Poly(3MP) content (%, wt/wt) | 14 | 41 | 38 | 45 | 40 | 37 | 27 | 42 | 30 |
| Glycogen content of CDM (%, wt/wt) | 3 | 18 | 19 | NDe | ND | ND | ND | ND | |
| Acetic acid concn (g/liter) | 8 | 8 | |||||||
| Total amt of poly(3MP) isolated (g)d | 380 | 570 | 1,050 | 50 | 140 | 147 | 180 | ||
| Poly(3MP) concn (g/liter)d | 0.8 | 1.2 | 2.1 | 2 | 4.7 | 5.9 | 6.4 | ||
| Conversion of 3MP to poly(3MP) (%, wt/wt) | 9 | 29.7 | 18.6 | 21.2 | 46.9 | 45.2 | 69.1 | 51.5 | 48.5 |
| Glycogen content of CDM (%, wt/wt) | 3 | 18 | 19 | ND | ND | ND | ND | ND | |
Volume at the beginning of the fermentation after all of the medium components were added and before the medium was inoculated with the preculture.
Volume at the end of the fermentation before the beginning of downstream processing. Differences from the data for the volume at the beginning resulted from addition of nutrients and HCl or NaOH during the fermentation and the withdrawal of samples for analysis.
Total amounts of individual medium components that were added to the medium.
Amount of poly(3MP) isilated by the in situ isolation procedure described in Materials and Methods.
ND, not detected.
Cultivation in Erlenmeyer flasks.
Cultivation was done at 37°C in 250-ml and 2-liter Erlenmeyer flasks equipped with baffles and containing 50 and 600 ml of medium, respectively. The flasks were incubated on a Pilotshake RC-4/6-W horizontal shaker (Kuehner, Birsfelden, Switzerland) at 200 rpm and at an amplitude of 5 cm.
Cultivation at the 30-liter scale.
Cultivation at the 30-liter scale was done in a Biostat DL30 stainless steel reactor (B. Braun Biotech International, Melsungen, Germany), which had a total volume of 42 liters (inside diameter, 28 cm; height, 71 cm) and d/D value calculated from the quotient of the stirrer diameter (10.5 cm) to the vessel diameter (28 cm) of 0.375. The bioreactor was equipped with three stirrers, each containing six paddles, and a Funda-Foam mechanical foam destroyer (B. Braun Biotech International). In addition, ports for sterilizable probes to measure the dissolved oxygen (pO2) concentration (model 25; Mettler Toledo, Steinbach, Switzerland), pH (model Pa/25; Mettler Toledo), foam (model L300/Rd.28; B. Braun Biotech International), temperature (pt 100 electrode; M. K. Juchheim GmbH, Fulda, Germany), and optical density (model CT66 operating at 890 nm; Sentex/Monitek Technology, Hayward, Calif.) were available. The operations were controlled and recorded with a DCU-3 digital control unit in combination with the MFCS/win software package (B. Braun Biotech International). Carbon dioxide and oxygen concentrations in the spent gas leaving the bioreactor were measured with a URAS 10 P NDIR spectrophotometer (Mannesmann, Hartmann and Braun, Frankfurt, Germany) and a Mangos 6 G oxygen analyzer (Mannesmann, Hartmann and Braun), respectively. Cultivation was done at 37°C unless specified otherwise and at a pO2 range from 0 to 100% saturation in the medium, which was controlled by agitation rates between 100 and 700 rpm and aeration rates of 0.5 to 1.0 volume per volume per min (vvm). The pH of the medium was kept at 7.0 automatically by controlled addition of 4 N HCl or NaOH. Foam was controlled by a mechanical foam destroyer; if this was not sufficient, the antifoam agent Silikon Antischaum emulsion SLE (Wacker, Darwin Vertriebs GmbH, Ottobrunn, Germany) was added. Small samples withdrawn from the culture fluid for analytical purposes were separated into a cell pellet and a cell-free supernatant by 15 min of centrifugation at 3,500 × g.
Cultivation at the 500-liter scale.
Cultivation at the 500-liter scale was performed with a Biostat D650 stainless steel bioreactor (B. Braun Biotech International), which had a total volume of 650 liters (inside diameter, 64 cm; height, 198 cm) and a d/D value calculated from the quotient of the stirrer diameter (24 cm) to the vessel diameter (64 cm) of 0.375. For online optical density measurements a model CT6 probe (Sentex/Monitek Technology) operating at 850 nm was available. All other equipment is described above; however, the corresponding tip speed of the stirrer blades was between 1.3 and 5.0 m s−1 with stirrer speeds between 100 and 400 rpm. The aeration rate was adjusted to between 0.3 and 0.5 vvm depending on the oxygen requirement of the culture.
Cell and poly(3MP) harvesting from 30- and 500-liter cultures.
Cells or poly(3MP) was harvested by centrifugation with a CEPA type Z41 or type Z61 continuous centrifuge (Carl Padberg Zentrifugenbau GmbH, Lahr, Germany).
Isolation and purification of poly(3MP).
A novel in situ isolation procedure was developed to isolate poly(3MP) from the cells inside the bioreactor. This method involved the following steps. Sodium dodecyl sulfate (SDS) was added to the cultivation broth immediately after fermentation. The amount of SDS used was based on the amount of cellular dry matter (CDM), and the optimum SDS-to-CDM ratio was 0.5. After the suspension was stirred for 2 h, the bioreactor was heated to 90°C for 20 min with intensive stirring and finally cooled to 37°C. Poly(3MP) was collected by continuous centrifugation and was further purified by three washes with distilled water. This crude poly(3MP) preparation was then treated with 3 N HCl to remove the remaining contaminants, such as proteins or glycogen, and again repeatedly washed with distilled water and ethanol. The polymer was then subjected to extraction with a mixture of acetone and diethyl ether (2:1, vol/vol) at 60°C in a soxhlet apparatus for 6 h to remove any remaining lipids and to obtain highly purified poly(3MP).
Analysis of ammonium, acetate, glucose, and glycerol.
The concentrations of ammonium in cell-free supernatants were determined by employing a gas-sensitive type 152303000 ammonium electrode (Mettler Toledo GmbH, Greifensee, Switzerland). The concentrations of glycerol and acetic acid in cell-free supernatants were determined enzymatically by using corresponding test kits (Roche Diagnostics GmbH, Mannheim, Germany), whereas the concentration of glucose was determined by using glucose test bars (Roche Diagnostics GmbH) according to the instructions provided by the manufacturer.
Analysis of 3MP.
The concentrations of 3-mercaptopropionic acid (3MP) in the medium were determined with cell-free supernatants by high-performance liquid chromatography (HPLC) (Kontron Instruments SpA, Milan, Italy) by employing the KromaSystem 2000 software package and a Nucleosil RP column (length, 250 mm; particle size, 5 μm; Macherey-Nagel, Düren, Germany). The HPLC apparatus was equipped with a diode array detector operating at 230 nm. Demineralized water and acetonitrile were acidified by addition of 0.35% (vol/vol) phosphoric acid and used for elution at a flow rate of 1 ml/min with 12% (vol/vol) acetonitrile at the beginning and 35% (vol/vol) acetonitrile at the end after 25 min.
Analysis of poly(3MP).
Poly(3MP) contents of cells were determined by gas chromatography after methanolysis of freeze-dried cells (2, 19). The purity of poly(3MP) was controlled by gas chromatography and also by elemental sulfur analysis at the Mikroanalytisches Labor Beller (Göttingen, Germany) performed by the method of Grote and Kerkeler (DIN 51768). For determination of protein contamination in poly(3MP) samples, 10 mg of polymer was denatured, SDS-polyacrylamide gel electrophoresis was performed (8), and the gel was stained with silver (6).
Determination of glycogen content.
The glycogen content of bacterial cells was determined by the modified method described by Good et al. (4). Bacterial cells were digested with 1 M KOH for 20 min at 100°C. After cooling, the glycogen was precipitated with ethanol (60%, vol/vol). The pellet was collected by centrifugation and then resolubilized in 1.0 ml of distilled water. Aliquots of this solution were diluted 1:10 with an iodine reagent containing 0.1% (wt/vol) iodine, 100 mM KI, and 50 mM CaCl2. After 5 min of incubation at room temperature, the glycogen-iodine complex formed was measured spectrophotometrically at 460 nm and compared with standard glycogen (Sigma, St. Louis, Mo.) concentrations.
RESULTS
Only a few properties of the novel PTEs are currently known. To determine additional properties and in particular the material properties of PTEs and also to investigate putative applications, it is necessary to obtain these polymers in kilogram quantities. Since biosynthesis of PTEs depends on the use of precursor substrates, PTEs cannot be produced from a simple carbon source and sulfate. 3MP is the only precursor substrate for PTE biosynthesis which is available at a reasonably low cost because it is produced on a large scale by the chemical industry for various applications (3) and for various organic syntheses (5).
Production of poly(3MP) at the 30-liter scale by using glucose and 3MP.
Various fed batch cultivation experiments with E. coli JM109 harboring plasmid pBPP1 were done in M9-MSM with glucose as the carbon source. The medium composition and yield based on the poly(3MP) content, the rate of conversion of 3MP to poly(3MP), and other data for two fermentations (fermentations 1 and 2) are summarized in Table 1. 3MP (24 g) was provided as a precursor substrate for poly(3MP) biosynthesis in the late exponential growth phase. The cultivation conditions were optimized for high poly(3MP) contents of the cells. This was achieved by feeding glucose along with 3MP from the late exponential growth phase, which resulted in poly(3MP) contents of up to 41% (wt/wt) of the CDM. In addition, the E. coli cells accumulated small amounts of glycogen (for example 3% [wt/wt] of the CDM in fermentation 1).
An increase in the dissolved oxygen concentration in the medium and a decrease in the carbon dioxide concentration in the spent gas indicated exhaustion of glucose; the concentration of glucose in the medium was also measured. Additional glucose was fed accordingly in all experiments before depletion. In addition, approximately 10-g portions of ammonium chloride were fed if the ammonium concentration in the medium fell below 0.3 g/liter. The amount of 3MP consumed was revealed by HPLC analysis of cell-free supernatants of the medium.
Since E. coli cannot use 3MP as a sole carbon source for growth and since there was also no evidence that other products were formed from 3MP, the cells obviously used 3MP only as a substrate for poly(3MP) synthesis. Therefore, E. coli should theoretically convert 3MP into poly(3MP) according to the following equation: n3MP → (3MP)n + (n − 1)H2O. According to this equation and in terms of mass, about 100 g of 3MP is converted into 83 g of poly(3MP). However, the conversion rates obtained in fed batch fermentations 1 and 2 were much lower (9 and 29.7% [wt/wt], respectively) and deviated significantly from the theoretical values.
Production of poly(3MP) at the 500-liter scale by using glucose and 3MP.
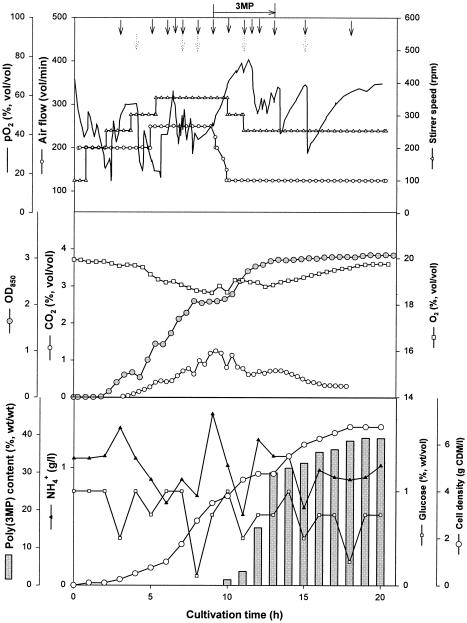
The BPEC process was also used for poly(3MP) production at the kilogram scale, and three cultivation experiments (fermentations 3, 4, and 5) with glucose as the carbon source and 3MP as the precursor for poly(3MP) were carried out at the 500-liter scale in a pilot plant. Table 1 and Fig. 1 show the details of these fermentations and the time course of fermentation 5. Approximately 200-g portions of ammonium chloride were fed when the ammonium chloride concentration in the medium fell below 0.3 g/liter in fermentations 3 and 4, whereas in fermentation 5 approximately 400 g was fed when the concentration in the medium fell below 1.0 g/liter. Approximately 375-g portions of 3MP were fed in fermentations 3 and 4, whereas in fermentation 5 approximately 514 g of 3MP was fed. Fermentations 3 and 4 yielded cell densities of 2.6 and 2.5 g of CDM/liter, and the cells contained 38 and 45% (wt/wt) poly(3MP), respectively. According to the equation shown above, these values corresponded to 18.6 and 21.2% (wt/wt) conversion of the 3MP in fermentations 3 and 4, respectively, and the CDM contained 18 and 19% glycogen (wt/wt), respectively. From the cells obtained in these fed batch cultivations, approximately 380 and 570 g of poly(3MP), respectively, were purified by a new rapid in situ isolation procedure (see below).
FIG. 1.
Time course of fed batch fermentation 5 of E. coli JM109(pBPP1) expressing the BPEC pathway for production of poly(3MP) at the 500-liter scale. Cultivation was done in a 650-liter stirred tank reactor containing 400 liters of modified M9-MSM with 0.6% (wt/vol) glucose and 2 g of NH4Cl per liter at the beginning of the fermentation. The temperature (37°C) and pH (pH 7.0) were kept constant. The aeration (0.3 to 0.5 vvm) and the stirrer speed (100 to 400 rpm) were adjusted according to the oxygen demand of the culture. During fermentation glucose (solid arrows) and NH4Cl (dotted arrows) were repeatedly fed before they became limiting at concentrations of 2 to 3 and 0.5 g/liter, respectively. The availability of the carbon source was estimated by monitoring the carbon dioxide concentration in the spent gas and the dissolved oxygen level in the medium. 3MP (514 g) was fed several times in the late exponential phase (horizontal arrow). Table 1 shows the total amounts of medium components used in this fermentation. OD850, optical density at 850 nm.
In the third fed batch experiment done at this scale (fermentation 5), the amounts of glucose, ammonium, and 3MP provided to the cells were increased by 96, 240, and 20% (wt/wt), respectively, compared to fermentation 4. Consequently, this fermentation yielded a cell density of 7.0 g of CDM/liter and a poly(3MP) content of 40% (wt/wt) of the CDM (Fig. 1). According to the equation shown above, this corresponded to 46.9% (wt/wt) conversion of the 3MP provided to the cells. Cells from fermentation 5 did not accumulate glycogen. From this batch approximately 1,050 g of poly(3MP) was purified by the in situ isolation procedure, corresponding to recovery of 75% (wt/wt) of the calculated polymer present in the cells. These data demonstrated that there was successful optimization of the BPEC process, including the poly(3MP) isolation method.
Production of poly(3MP) at the 30-liter scale by using glycerol and 3MP.
Although acetate formation was determined for only two fed batch experiments (fermentations 8 and 9), the lower rate of glycerol transport into the cells compared to the rate of glucose transport led to a reduction in the flux of carbon through glycolysis, and therefore, the formation of acetate was greatly reduced (9). Glycerol is also a cheap carbon source because it is a residual compound obtained during production of biodiesel from rape seed (fatty acid methyl or ethyl esters) (20). Therefore, glycerol is often used for cultivation of E. coli and other bacteria in biotechnological processes (7, 9); hence, it was used in this study as a carbon source for growth instead of glucose.
The same strain of E. coli (JM109 harboring plasmid pBPP1) was cultivated in modified M9-MSM at the 30-liter scale with glycerol as the carbon source for growth, and 3MP was provided as the precursor substrate for poly(3MP) biosynthesis in the late exponential growth phase. Since no online method for glycerol analysis was available, the concentrations of glycerol in the cell-free supernatant were determined at the end of the fermentation with samples withdrawn at regular intervals. In fermentations 6, 7, 8, and 9, 36-, 48-, 67-, and 53-g portions of 3MP, respectively, were fed. The concentrations of 3MP in the medium were determined by HPLC. In addition, approximately 15-g portions of ammonium chloride were fed if the ammonium concentration in the medium fell below 1.0 g/liter.
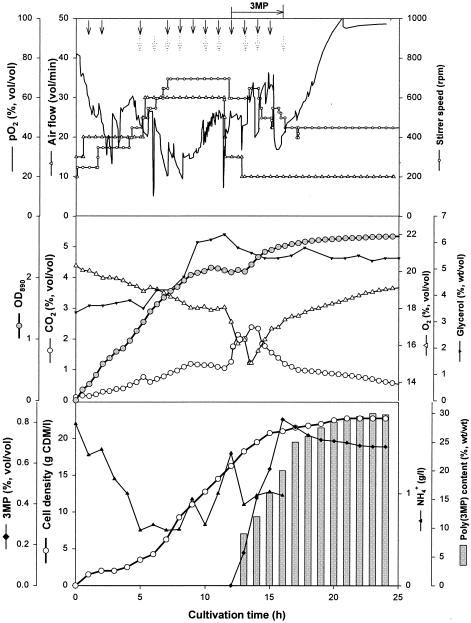
The medium compositions and yields based on the poly(3MP) content, the amounts of poly(3MP) isolated, the amounts of poly(3MP) per liter of medium, and other data for fermentations 6, 7, 8, and 9 are summarized in Table 1. The experiments yielded cell densities of 7.3, 17, 22, and 23 g of CDM/liter and cells containing 37, 27, 42 and 30% (wt/wt) poly(3MP) in fermentations 6, 7, 8, and 9, respectively. Glycogen could not be detected in any of the cells. According to the equation shown above, this corresponded to 45.2, 69.1, 51.5, and 48.5% (wt/wt) conversion of the 3MP provided to the cells in fermentations 6, 7, 8, and 9, respectively. From these fed batch fermentations, approximately 50, 140, 147, and 180 g of poly(3MP), respectively, were purified by the in situ isolation procedure. In fed batch fermentation 7, the concentrations of Na2HPO4 · 12H2O, KH2PO4, MgSO4 · 7H2O, and CaCl2 · 2H2O were increased by 100% (wt/wt) compared to the concentrations in fermentation 6 to get higher cell densities. In fermentation 8 the concentrations of Na2HPO4 · 12H2O and KH2PO4 were increased by 35% (wt/wt) compared to the concentrations in fermentation 7. The concentration of MgSO4 · 7H2O was increased by 80% and the concentration of CaCl2 · 2H2O was reduced by 79% compared to the concentrations in fermentation 7. Figure 2 shows the results for fed batch fermentation 9, which yielded the highest cell density (23 g of CDM/liter), with poly(3MP) contributing 30% (wt/wt) of the CDM. In this cultivation experiment the concentrations of Na2HPO4 · 12H2O and KH2PO4 were decreased by 87 and 13.5% (wt/wt), respectively, compared to the concentrations in fermentation 8. CaCl2 · 2H2O was completely removed from the medium components to avoid precipitation, whereas the concentration of MgSO4 · 7H2O was increased by 18.5% (wt/wt) compared to the concentration in fermentation 8. In fed batch fermentations 8 and 9 E. coli produced up to 8 g of acetic acid/liter at the end of fermentation.
FIG. 2.
Time course of fed batch fermentation 9 of E. coli JM109(pBPP1) expressing the BPEC pathway for production of poly(3MP) at the 30-liter scale. Cultivation was done in a 42-liter stirred-tank reactor containing 18 liters of modified M9-MSM with 3% (wt/vol) glycerol and 2 g of NH4Cl per liter at the beginning of the fermentation. The temperature (35°C) and pH (pH 7.0) were kept constant. The aeration (0.5 to 1.0 vvm) and agitation (100 to 700 rpm) were adjusted according to the oxygen demand of the culture. Glycerol was used as the sole carbon source. Glycerol (5 to 10 g/liter) and MgSO4 · 7H2O (0.9 g/liter) were fed together (solid arrows) and NH4Cl (0.5 g/liter) (dotted arrows) was fed repeatedly during the fermentation before these compounds became limiting. The availability of the carbon source was estimated by monitoring the carbon dioxide concentration in the spent gas and the dissolved oxygen level in the medium. 3MP (53 g) was fed several times in the late exponential phase (horizontal arrow). Table 1 shows the total amounts of medium components used in this fermentation. OD850, optical density at 850 nm.
Isolation of poly(3MP) by a novel rapid in situ isolation procedure.
The protocol of the enzymatic method for isolation and purification of poly(3MP) described previously (16) was very expensive and time-consuming and was applicable only at the small laboratory scale.
In this study, a novel in situ method for rapid and convenient isolation of poly(3MP), which was applicable to a large scale, was developed. This method allowed cell lysis in a bioreactor without previous cell harvesting. Before this method was applied to the reactor scale, the optimum concentration of SDS with regard to cell density was determined in small-scale experiments. To do this, 5-g portions of dry cells of recombinant E. coli containing poly(3MP), which were obtained from fed batch fermentation 1 (Table 1), were incubated in 200-ml (final volume) portions of distilled water containing various amounts of SDS in 1-liter screw-cap glass bottles. Table 2 summarizes the results for determining the optimum SDS-to-CDM ratio for complete cell lysis and removal of contaminants, as indicated by the poly(3MP) purity. The optimum ratio of SDS to CDM was 0.5 g/g, which yielded almost 100% pure poly(3MP) after the final washing step, as revealed by gas chromatography analysis. At this ratio, the efficiency of this procedure after the recovery of poly(3MP) in the isolated material compared to the poly(3MP) content of the cells was 99%. Protein contaminants could not be detected in the final purified poly(3MP) samples (data not shown), as revealed by an SDS-polyacrylamide gel stained with silver. The 3MP contents calculated from the data from elemental sulfur analyses revealed that poly(3MP) accounted for at least 98% (wt/wt) of the polymer isolated. This isolation procedure was routinely and successfully used to isolate poly(3MP) in quantities of several hundred grams to even 1 kg (Table 1).
TABLE 2.
Optimization of SDS/CDM ratio for in situ isolation of poly(3MP)a
| SDS/CDM ratio (g/g) | Purity of poly(3MP) (%, wt/wt) |
|---|---|
| 0.05 | 42 |
| 0.1 | 59 |
| 0.2 | 80 |
| 0.3 | 92 |
| 0.4 | 97 |
| 0.5 | 100 |
Each 5 g of cell dry matter obtained from fed batch fermentation 1 was incubated for 2 h at room temperature in 200 ml (final volume) of water containing 0.25, 0.5, 1.0, 1.5, 2.0, or 2.5 g of SDS in a 1-liter screw-cap glass bottle. After this, the suspensions were incubated for 20 min at 90°C and then cooled to room temperature. Insoluble material was collected by centrifugation and was washed three times with distilled water. The washed insoluble material was treated with 3 M HCl for 15 min at 90°C. The poly(3MP) obtained was then washed with distilled water and ethanol. After these steps the purity of each poly(3MP) sample was analyzed by gas chromatography.
DISCUSSION
In this study an efficient biotechnological process based on the nonnatural BPEC pathway (16) for production of poly(3MP) homopolymer in a recombinant strain of E. coli at a technical scale was established. This process consists of two parts: (i) production of poly(3MP) in the cells by using glucose or glycerol as the carbon source and 3MP as the precursor substrate; and (ii) application of a novel SDS-based in situ process for isolation of poly(3MP). Despite the limited available technical equipment for the 30- and 500-liter stirred tank bioreactors used, this BPEC process provided sufficient amounts of poly(3MP) for further investigation. In addition, the novel in situ isolation process provided a rapid, cheap, and convenient method for isolation of poly(3MP) and was applicable to the pilot scale. This allowed production of kilogram quantities of this novel, recently detected biopolymer (19), which previously could be obtained only at the laboratory scale for analytical purposes. It should now be possible to study the material properties and biodegradability of the novel biopolymer and also to establish putative applications.
For the first fed batch cultivation experiments standard M9-MSM (28) was used, which yielded cell densities of only 2 to 3 g of CDM per liter. To obtain considerably higher cell densities and also to increase the poly(3MP) contents of the cells, the composition of this medium was modified as described above. Since butyrate kinase, which is the first enzyme of the BPEC pathway, requires ATP for phosphorylation of 3MP (12, 13, 15), glucose or glycerol was added along with 3MP as an energy source for ATP generation and to enhance poly(3MP) biosynthesis in the accumulation phase. The concentrations of nutrients required to obtain the desired cell densities were calculated based on the elemental composition of standard E. coli K-12 cells (1); we made sure that no nutritional element was limiting in subsequent fed batch cultivations. In the first four fed batch experiments the cell densities did not exceed approximately 2.5 to 3.7 g of CDM/liter due to limitation of the nitrogen source. In addition, cells accumulated glycogen due to nitrogen source limitation and carbon source excess (21, 24). Therefore, in subsequent experiments the concentration of ammonium was maintained at about 1 g/liter throughout the course of fermentation. This resulted in higher cell densities in fed batch fermentations 5 and 6, while accumulation of glycogen was inhibited in all subsequent cultivation experiments. Further modification of the medium composition resulted in higher cell densities in fed batch fermentations 7, 8, and 9. In addition, the latter fermentations were done at 35°C to reduce growth and thus consumption of oxygen (9, 27), which always became limiting at the end of the experiments, as also indicated by the excretion of considerable amounts of acetate. Acetate is produced by E. coli not only under oxygen-limiting conditions but also under aerobic conditions in the presence of excess glucose or glycerol (9). Acetate concentrations of more than 5 g/liter at pH 7 retard the growth of the cells; hence, high cell densities cannot be achieved (9). However, high cell densities can be obtained by using more sophisticated and automatic feeding methods, like indirect feedback control schemes that couple feeding with measurement of, for example, dissolved oxygen and the carbon dioxide evolution rate. By doing this, nutrients can be fed continuously at the required amounts, the specific growth rate can be controlled, and a decrease in acetate formation can be achieved; the latter may be also controlled by supplying pure oxygen instead of air (9, 11).
Previous studies of high-cell-density cultivation of E. coli have described cell densities of more then 100 g of CDM/liter when the sophisticated system described above was used (9, 10, 11, 14, 27). However, the aim of this study was to establish an efficient biotechnological process based on the nonnatural BPEC pathway for production of poly(3MP) homopolymer in a recombinant strain of E. coli at a technical scale. We succeeded in performing cultivation at a 500-liter scale, which yielded batches containing more than 1 kg of the recently detected novel biopolymer. This study also demonstrated the applicability of the novel in situ isolation procedure on a large scale. Further characterization of the material obtained will probably reveal applications for polythioesters.
Acknowledgments
We thank Bruno Bock Thiochemicals (Niedermarschacht, Germany) for a gift of 3-mercaptopropionic acid. The technical support of Herbert Ahlers, Ingo Voß, and Simone Diniz is gratefully acknowledged.
REFERENCES
- 1.Battley, E. H. 1991. Calculation of the heat of growth of Escherichia coli K-12 on succinic acid. Biotechnol. Bioeng. 37:334-343. [DOI] [PubMed] [Google Scholar]
- 2.Brandl, H., R. A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(3-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer, F. 2002. 3-Mercaptopropionic acid (3MPA). Synlett 8:1368-1369. [Google Scholar]
- 4.Good, C. A., H. Kramer, and M. Somogyi. 1933. The determination of glycogen. J. Biol. Chem. 100:485-491. [Google Scholar]
- 5.Gresham, T. L., J. E. Jansen, F. W. Shaver., and J. T. Gregory. 1948. β-Propiolactone. II. Reactions with salts of inorganic acids. J. Am. Chem. Soc. 70:998-999. [Google Scholar]
- 6.Heukeshofen, J., and R. Dernick. 1985. A simplified method for silver staining of proteins in polyacrylamide gels and its use as an immunosorbant for isolation of immunoglobulins. FEMS Lett. 28:73-76. [Google Scholar]
- 7.Himmi, E. H., A. Bories, A. Boussaid, and L. Hassani. 2000. Propionic acid fermentation of glycerol and glucose by Propionibacterium acidipropionici and Propionibacterium freudenreichii ssp. shermanii. Appl. Microbiol. Biotechnol. 53:435-440. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 9.Lee, S. Y. 1996. High cell-density culture of Escherichia coli. Tibtech 14:98-105. [DOI] [PubMed] [Google Scholar]
- 10.Lee, S. Y. 1996. Plastic bacteria? Progress and prospects for polyhydroxyalkanoate production in bacteria. Tibtech 14:431-438. [Google Scholar]
- 11.Lee, S. Y., J. Choi, and H. H. Wong. 1999. Recent advances in polyhydroxyalkanoate production by bacterial fermentation: mini-review. Int. J. Biol. Macromol. 25:31-36. [DOI] [PubMed] [Google Scholar]
- 12.Liu, S. J., and A. Steinbüchel. 2000. A novel genetically engineered pathway for synthesis of poly(hydroxyalkanoic acid) in Escherichia coli. Appl. Environ. Microbiol. 66:739-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, S. J., and A. Steinbüchel. 2000. Exploitation of butyrate kinase and phosphotransbutyrylase from Clostridium acetobutylicum for the in vitro biosynthesis of poly(hydroxyalkanoic acid). Appl. Microbiol. Biotechnol. 53:545-552. [DOI] [PubMed] [Google Scholar]
- 14.Liu, Y. C., L. C. Liao, and W. T. Wu. 2000. Cultivation of recombinant Escherichia coli to achieve high cell density with a high level of penicillin G acylase activity. Proc. Natl. Sci. Counc. Repub. China Part B 24:156-160. [PubMed] [Google Scholar]
- 15.Lütke-Eversloh, T., A. Fischer, U. Remminghorst, J. Kawada, R. H. Marchessault, A. Bögershausen, M. Kalwei, H. Eckert, R. Reichelt, S. J. Liu, and A. Steinbüchel. 2002. Biosynthesis of novel thermoplastic polythioesters by engineered Escherichia coli. Nat. Mater. 1:236-240. [DOI] [PubMed] [Google Scholar]
- 16.Lütke-Eversloh, T., and A. Steinbüchel. 2004. Microbial polythioesters. Macromol. Biosci. 4:165-174. [DOI] [PubMed] [Google Scholar]
- 17.Lütke-Eversloh, T., K. Bergander, H. Luftmann, and A. Steinbüchel. 2001. Biosynthesis of poly(3-hydroxybutyrate-co-3-mercaptobutyrate) as a sulfur analogue to poly(3-hydroxybutyrate) (PHB). Biomacromolecules 2:1061-1065. [DOI] [PubMed] [Google Scholar]
- 18.Lütke-Eversloh, T., J. Kawada, R. H. Marchessault, and A. Steinbüchel. 2002. Characterization of biological polythioesters: physical properties of novel copolymers synthesized by Ralstonia eutropha. Biomacromolecules 3:159-166. [DOI] [PubMed] [Google Scholar]
- 19.Lütke-Eversloh, T., K. Bergander, H. Luftmann, and A. Steinbüchel. 2001. Identification of a new class of biopolymer: bacterial synthesis of sulfur containing polymer with thioester linkages. Microbiology 147:11-19. [DOI] [PubMed] [Google Scholar]
- 20.Muniyappa, P. R., S. C. Brammer, and H. Noureddini. 1996. Improved conversion of plant oils and animal fats into biodiesel, and co-product. Bioresour. Technol. 56:19-24. [Google Scholar]
- 21.Okita, T. W., R. L. Rochiguez, and J. Preiss. 1981. Biosynthesis of bacterial glycogen: cloning of the glycogen biosynthetic enzyme structural genes of Escherichia coli. J. Biol. Chem. 256:6944-6952. [PubMed] [Google Scholar]
- 22.Park, S. J., J. P. Park, and S. Y. Lee. 2002. Production of poly(3-hydroxyalkanoate) from whey by fed-batch culture of recombinant Escherichia coli in pilot-scale fermenter. Biotechnol. Lett. 24:185-189. [Google Scholar]
- 23.Pfennig, N. 1974. Rhodopseudomonas globiformis sp. n., a new species of Rhodospirillaceae. Arch. Microbiol. 100:197-206. [Google Scholar]
- 24.Preiss, J. L., J. S. Ozbun, E. Hawker, and C. Lammel. 1973. ADPG synthase and ADPG glucan 4-glycosyl transferase: enzymes involved in bacterial glycogen and plant starch synthesis. Ann. N. Y. Acad. Sci. 210:265-278. [DOI] [PubMed] [Google Scholar]
- 25.Rehm, B. H. A., and A. Steinbüchel. 1999. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int. J. Biol. Macromol. 25:3-19. [DOI] [PubMed] [Google Scholar]
- 26.Rehm, B. H. A., and A. Steinbüchel. 2002. PHA synthases: the key enzymes of PHA synthesis, p. 173-215. In Y. Doi and A. Steinbüchel (ed.), Biopolymers, vol. 3a. Wiley-VCH, Weinheim, Germany.
- 27.Riesenberg, D., V. Schulz, W. A. Knorre, H.-D. Pohl, D. Korz, E. A. Sanders, A. Roß, and W.-D. Deckwer. 1991. High cell density cultivation of Escherichia coli at controlled specific growth rate. J. Biotechnol. 20:17-28. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Steinbüchel, A. 1995. Mikrobielle und chemische Synthese von biologisch abbaubaren Polyestern. Chem. Unserer Zeit 29:260-271. [Google Scholar]
- 30.Steinbüchel, A. 2001. Perspectives for biotechnological production and utilization of biopolymers: metabolic engineering of polyhydroxyalkanoate biosynthesis pathway as a successful example. Macromol. Biosci. 1:1-24. [Google Scholar]
- 31.Steinbüchel, A., and B. Füchtenbusch. 1998. Bacterial and other biological systems for polyester production. TibTech 16:419-427. [DOI] [PubMed] [Google Scholar]
- 32.Steinbüchel, A., and H. E. Valentin. 1995. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219-228. [Google Scholar]
- 33.Steinbüchel, A., and S. Hein. 2001. Biochemical and molecular basis of microbial synthesis of polyhydroxyalkanoates in microorganisms. Adv. Biochem. Eng. Biotechnol. 71:81-123. [DOI] [PubMed] [Google Scholar]
- 34.Steinbüchel, A., and T. Lütke-Eversloh. 2003. Metabolic engineering and pathway construction for biotechnological production of relevant polyhydroxyalkanoates in microorganisms. Biochem. Eng. J. 16:81-96. [Google Scholar]