Abstract
Pancreatic cancer is characterized by a dense desmoplastic reaction in which extracellular matrix proteins accumulate and surround tumor cells. Integrins and their related signaling molecules are associated with progression of pancreatic cancer. In the present study, the association between the metastasis of pancreatic cancer and the expression of hnRNP E1 and integrin β1 was evaluated. In vitro and in vivo experiments were designed to study the mechanism underlying the regulation of integrin β1 splicing by the Fyn/hnRNP E1 spliceosome. Expression of hnRNP E1 and integrin β1A were associated with metastasis of pancreatic cancer. Inhibition of Fyn activity upregulated the expression of P21-activated kinase 1 and promoted the phosphorylation and nuclear localization of hnRNP E1, leading to the construction of a spliceosome complex that affected the alterative splicing of integrin β1. In the hnRNP E1 spliceosome complex, hnRNP A1 and serine/arginine-rich splicing factor 1 were responsible for binding to the pre-mRNA of integrin β1. Suppression of Fyn activity and/or overexpression of hnRNP E1 decreased the metastasis of pancreatic cancer cells. In pancreatic cancer, the present study demonstrated a novel mechanism by which Fyn/hnRNP E1 signaling regulates pancreatic cancer metastasis by affecting the alternative splicing of integrin β1. hnRNP E1 and integrin β1A are associated with the metastasis of pancreatic cancer and may be novel molecular targets for pancreatic cancer treatment.
Keywords: pancreatic cancer, Fyn, integrin, alternative splicing, hnRNPs
Introduction
Pancreatic cancer is characterized by a dense desmoplastic reaction in which extracellular matrix (ECM) proteins accumulate and surround tumor cells. These ECM proteins not only provide physical support but also affect the biological properties of pancreatic cancer cells (1–3). Recent studies have demonstrated that the integrin family of heterodimeric transmembrane glycoproteins, each of which are composed of an α- and a β-subunit, and their related signaling molecules are associated with the progression of pancreatic cancer (4–7). Among these signaling molecules, the Src family kinases (SFKs), a group of non-receptor tyrosine kinases, are overexpressed in 70% of human pancreatic cancer cases and their activities are critical for tumor progression (8–11).
Fyn, a key member of the SFKs, mediates 'outside-in' signaling from the ECM-integrin interaction to affect cellular processes, including T-cell receptor differentiation, cell adhesion, migration and apoptosis regulation (12–15). Our previous study showed that the activation of Fyn promotes cell proliferation and metastasis in pancreatic cancer (16). However, the role of Fyn in 'inside-out' signaling to regulate pancreatic cancer metastasis is not completely understood.
Integrins are heterodimers of two type-I membrane glycoproteins, α and β, which bind to each other non-covalently. Currently, seventeen α subunits and eight β subunits have been identified in vertebrates, and different components of integrin signaling have been shown to be vital for cell survival and cancer metastasis (17–19). It is well established that alternatively spliced variants of the α- and β-subunits contribute to the variety of biological functions of the integrin receptors (20). In endothelial cells, alternative splicing of integrin αv have been analyzed by Gauck et al (21) who found Cdc2-like kinases as well as DNA topoisomerase I to modulate the differential isoform expression of Cyr61 and its receptor integrin αv, the protein expression and secretion in resting as well as in TNF-α-stimulated human microvascular endothelial cells. Moreover, these processes affected the endothelial cell proliferation and pro-angiogenic tube formation by HMEC-1. In pancreatic cancer, the integrin β1-ECM interaction has been associated with metastasis (4). Four splice variants (A, B, C and D), which differ only in the amino acid sequence of the cytoplasmic domain, have been identified in the integrin β1 family, and the β1A isoform is widely expressed among all mammalian species assessed so far. The expression of splice variants B, C and D, which can inhibit β1A-mediated focal adhesion formation, cell spreading and motility, has been found to be downregulated or even lost in various tumor tissues, and may be involved in tumor progression (22–24). However, the formation of splice variants of integrin β1 is not yet fully understood.
Splicing of individual precursor messenger ribonucleic acid (pre-mRNA) is determined by spliceosome proteins including serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) (25). The SR proteins and hnRNPs function as trans-acting factors by binding to exonic splicing enhancers (ESEs) or exonic splicing silencers (ESSs) to regulate alternative splicing (26–29). Recent reports have shown that hnRNP E1 binds to the CD44 pre-mRNA, which affects its alternative splicing (30). However, the participation of spliceosome proteins in integrin β1 splicing regulation and the resultant effect on cancer metastasis are unknown.
As a group of RNA-binding proteins, hnRNPs regulate the splicing and transportation of mRNA and participate in growth regulation and carcinogenesis (31–33). Expression of hnRNP E1 has been detected in several cancer types (34,35). However, the mechanism by which hnRNP E1 regulates tumor metastasis is not clear. Our preliminary study indicated that inhibition of the activity of Fyn significantly increased hnRNP E1 expression in human pancreatic cancer cells (36). To determine whether Fyn modulates pancreatic cancer metastasis via hnRNP E1, the present study evaluated the association between the metastasis of pancreatic cancer and the expression of hnRNP E1 and integrin β1 in human pancreatic cancer tissues. Subsequently, the mechanism of integrin β1 splicing regulation by Fyn/hnRNP E1 signaling in pancreatic cancer cells was investigated using multiple experimental approaches.
Materials and methods
Antibodies
Rabbit monoclonal antibodies against SRC family-phospho Y418 (reactive to Fyn-pY419, ab40660; 1:1,000 for western blot analysis), integrin β1 (reactive to the extracellular domain, ab179471, 1:1,000 for western blot analysis) and p21-activated kinase 1 (ab40852, 1:1,000 for western blot analysis), rabbit polyclonal antibodies against serine/arginine-rich splicing factor 1 (ab38017, 1:1,000 for western blot analysis) and SRp20 (ab73891, 1:1,000 for western blot analysis) and a mouse monoclonal antibody against hnRNP A1 (ab5832, 1:1000 for western blot analysis) were purchased from Abcam (Cambridge, UK). Rabbit polyclonal antibodies against hnRNP E1 (sc-28725, 1:500 for western blot analysis) and β-actin (sc-130657, 1:5,000 for western blot analysis) were purchased from Santa Cruz (Santa Cruz Biotechnology, Dallas, TX, USA). Mouse monoclonal antibodies against Fyn (P2992, 1:1000 for western blot analysis) and green fluorescent protein (GFP) (SAB5300167, 1:5000 for western blot analysis) were purchased from Sigma-Aldrich (St. Louis, MQ, USA). A rabbit polyclonal antibody against phosphothreonine (71-8200, 1:500 for western blot analysis) was purchased from Invitrogen/Thermo Fisher Scientific (Waltham, MA, USA). Goat anti-rabbit IgG rhodamine-conjugated (31670) and goat anti-rabbit (31460) or anti-mouse (31430) IgG (1:5,000 for western blot analysis) peroxidase-conjugated antibodies were purchased from Pierce Biotechnology (Rockford, IL, USA).
Adenoviral expression of kinase-dead Fyn (KdFyn) and hnRNP E1-GFP (E1-GFP)
The KdFyn recombinant adenovirus was previously prepared in our laboratory (16). The full-length hnRNP E1 coding sequence was obtained from HEK293 RNA using a PrimeScript™ RT-PCR kit (RR014A; Takara Bio, Dalian, China) according to the manufacturer's protocol. The polymerase chain reaction (PCR) products were identified by sequencing and cloned into the multiple cloning site of the pEGFP-N2 plasmid (6081-1 BD; Biosciences, Bedford, MA, USA) to construct an hnRNP E1-GFP fusion protein. The hnRNP E1-GFP recombinant adenovirus was generated using the AdEasy Vector System (240009; Stratagene, La Jolla, CA, USA) according to the manufacturer's protocol. The GFP recombinant adenovirus was used as a control vector in the cell study. Sequences of the hnRNP E1-GFP fusion protein PCR primers are listed in Table I.
Table I.
Sequences of the integrin β1, β1A, β1C, hnRNP E1-GFP and β-actin PCR primers.
| Forward primer | Reverse primer | Size (bp) | EFF% | |
|---|---|---|---|---|
| Integrin β1A | AGAATCCAGAGTGTCCCACTGG | TTTCCCTCATACTTCGGATTG | 238 | 92.4 |
| Integrin β1C | TCTGTCGCCCAGCCTGGAGTG | TTTCCCTCATACTTCGGATTG | 172 | 96.3 |
| integrin β1 | CCTCATAACAGTCCTGTGCCTAGAAT | CCAGCCAATGTGGTGAAACCC | 270 | 91.2 |
| hnRNP E1-GFP | GC(AGATCT)CTCGCCATGGATGCCGGTGT | CA(GAATTC)GCCCTTCTCAGAGGAAAGCCTGG | 1078 | 93.5 |
| β-actin | CGGGAAATCGTGCGTGAC | TGGAAGGTGGACAGCGAGG | 443 | 90.7 |
EFF%, amplification efficiency.
Cell lines and tissue samples
The human pancreatic cancer cell lines AsPC1, BxPC3, CFPAC1 and Panc1, and the HEK293 cell line, were cultured in RPMI-1640 medium (11875093; Thermo Fisher Scientific, Waltham, MA, USA), Iscove's modified Dulbecco's medium (12440053; Thermo Fisher Scientific) or Dulbecco's modified Eagle's medium (41965062; Thermo Fisher Scientific), each supplemented with 10% fetal bovine serum (FBS, 12657; Thermo Fisher Scientific) and antibiotic-antimycotic (15240-062; Thermo Fisher Scientific). All cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) characterized according to the cell line authentication testing and used within 6 months after resuscitation.
A total of 152 pancreatic cancer specimens from our institution were used in the present study, including 76 specimens that had been cryopreserved in liquid nitrogen, which were used for RNA analysis, and 76 paraffin-embedded specimens (8330; Thermo Fisher Scientific), which were used for histological analysis. Each pancreatic cancer specimen was reviewed by two pathologists.
All human studies were reviewed and approved by the Ethics Committee of Southwest Hospital and were therefore performed in accordance with the ethical standards described in the 1975 Declaration of Helsinki, as revised in 1983. Informed consent was obtained from all individual participants that were included in the study.
Cell staining for confocal microscopy
Cells were cultured to 50–70% confluence on glass-bottom tissue culture dishes (150680; Thermo Fisher Scientific) and then washed with phosphate-buffered solution (PBS, 10010023; Thermo Fisher Scientific). The cells were fixed with 2% paraformaldehyde (FB002; Thermo Fisher Scientific), followed by the addition of glycine buffer (0.1 mM glycine) (G7126; Sigma-Aldrich) for paraformaldehyde quenching and blocked in 1% bovine serum albumin (BSA, TS-38839; Thermo Fisher Scientific) in PBS for 1 h. The hnRNP E1 rabbit polyclonal antibody (sc-28725; Santa Cruz Biotechnology) was diluted in PBS (1:200) with 1% BSA and incubated with the cells at 4°C overnight. The cells were then washed with cold PBS three times prior to incubation with rhodamine-conjugated goat anti-rabbit IgG (1:500; Pierce Biotechnology) for 45 min at room temperature. The labeled cells were washed with cold PBS and imaged using a confocal microscope (Carl Zeiss LSM780; Carl Zeiss Microscopy GmbH, Jena, Germany). The images were analyzed using ZEN 2012 software (Carl Zeiss Microscopy GmbH).
Cell invasion assay
Cell invasion assays were performed using 24-well Transwell inserts with 8 µm pore size (PI8P01250; Merck Millipore, Billerica, MA, USA) coated with a thin layer of matrigel (356234; BD Biosciences, Bedford, MA, USA) (1.5 µg/mm2). Cells (1×105 cells in 300 µl serum-free medium) were seeded into the upper chamber, and the lower compartment was filled with 500 µl of complete medium. After 24 h at 37°C, non-invading cells were removed by wiping the upper side of the membrane. Invading cells were fixed, stained and counted.
Flow cytometry
BxPC3 pancreatic cancer cells were cultured in RPMI-1640 medium with 10% FBS and antibiotic-anti-mycotic, prior to harvesteding by trypsinization (25200056; Thermo Fisher Scientific). Subsequently, the cells were washed with PBS containing 1% normal goat serum (01-6201; Thermo Fisher Scientific) and incubated with integrin β1 rabbit monoclonal antibody (ab179471, 1:500; Abcam) at 4°C for 1 h, followed by rhodamine-conjugated goat anti-rabbit IgG (31670, 1:500; Pierce Biotechnology) for 30 min. The stained cells were resuspended in 100 µl PBS and analyzed with a Becton Dickinson FACSort flow cytometer.
Immunoprecipitation and RNA-protein immunoprecipitation
Immunoprecipitation was performed by incubating 1 µg hnRNP E1 rabbit polyclonal antibody (sc-28725; Santa Cruz Biotechnology) or Fyn mouse monoclonal antibody (P2992; Sigma-Aldrich) with 500 µg extracted proteins overnight at 4°C with constant rotation. The immunocomplexes were captured by adding protein A agarose (15918014; Invitrogen/Thermo Fisher Scientific) for 1 h at 4°C with constant rotation. The immunoprecipitates were analyzed by western blotting.
For RNA-protein immunoprecipitation, 1 mg extracted proteins were pre-cleared with 20 µl protein A/G plus agarose (20423; Invitrogen/Thermo Fisher Scientific) and then incubated with 5 µg of hnRNP E1 rabbit polyclonal antibody (sc-28725; Santa Cruz Biotechnology), SF2/ASF rabbit polyclonal antibody (ab38017; Abcam) or hnRNP A1 mouse monoclonal antibody (ab5832; Abcam) overnight at 4°C with constant rotation. The co-precipitated RNA was then extracted by phenol (AM9712; Thermo Fisher Scientific)/chloroform (1.02444; Sigma-Aldrich) and used for further analysis.
Immunohistochemical staining
Specimens were fixed in formalin (9990916; Thermo Fisher Scientific), embedded in paraffin and cut into 3-mm sections. Sections were deparaffinized in xylene (9990501; Thermo Fisher Scientific), rehydrated in a graded series of ethanol (E7023; Sigma-Aldrich) solutions and incubated in 3.0% hydrogen peroxide (TA-060-HP; Thermo Fisher Scientific) in methanol (960055; Thermo Fisher Scientific) for 30 min to block endogenous peroxidase activity. Slides were heated at 120°C in an autoclave in 10 mM sodium citrate (pH 6.0) (71497; Sigma-Aldrich) for 130 sec and then cooled to room temperature. After blocking with 10% goat serum for 30 min, the sections were incubated overnight at 4°C with the hnRNP E1 rabbit polyclonal antibody (sc-28725, 1:200; Santa Cruz Biotechnology). Negative controls were obtained by omitting the primary antibody. The sections were incubated with peroxidase-conjugated anti-mouse/rabbit immunoglobulins (K5007; Dako EnVision™ System; Dako, Copenhagen, Denmark) according to the manufacturer's protocol for 60 min at 37°C. The peroxidase reaction was developed with 3,3′-diaminobenzidine as the chromogen, followed by counterstaining with hematoxylin.
Mass spectrometry
Briefly, 5 mg extracted proteins were pre-cleared with 50 µl protein A/G Plus agarose and then incubated with 10 µg hnRNP E1 rabbit polyclonal antibody (sc-28725; Santa Cruz Biotechnology) overnight at 4°C with constant rotation. The immunocomplexes were captured by adding protein A/G Plus agarose for 1 h at 4°C. The immunoprecipitates were sent to the BGI (Shenzhen, China) for mass spectrometry analysis.
Nude mouse xenograft model
Male athymic nu/nu mice, 4–6 weeks of age, were obtained from the Beijing Animal Facility and provided with food and water ad libitum. Tumor cells (2×105 tumor cells in 100 µl of sterile PBS) were transplanted into each mouse via subcutaneous or intrasplenic injection. Animals were euthanized after 5–8 weeks, and the subcutaneous tumors and liver were resected. Primary tumor mass was determined by measuring the wet weight of the resected tumors and metastasis was determined as the incidence of tumor nodes present in the liver.
Procedures involving animals and their care were conducted in conformity with NIH guidelines (NIH Pub. No. 85-23, revised 1996) and were approved by the Institutional Animal Care and Use Committee of the Third Military Medical University.
RNA extraction and quantitative real-time-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues or cultured cells using RNAiso Plus reagent (9108; Takara) according to the manufacturer's protocol. The RNA was stored at −80°C, and reverse transcription of the extracted RNA was performed using the PrimeScript™ RT reagent kit with genomic DNA Eraser (RR047A; Takara) according to the manufacturer's protocol. The cDNA was stored at −20°C. qRT-PCR assays were performed to detect integrin β1 expression using the One Step SYBR® PrimeScript™ RT-PCR kit (RR086A; Takara) according to the manufacturer's protocol. The results were normalized to the expression of β-actin. For quantitative measurement of integrin β1 expression, qRT-PCR was performed using a CFX96 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) with the following cycling conditions: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. The qRT-PCR results were analyzed and expressed relative to a pancreatic cancer sample and converted to fold-change value. The PCR products were separated on a 2% agarose gel, stained with ethidium bromide and imaged with a video camera (Vilber Lourmat, Marne-la-Vallée, France). The primers used and the sizes of all the PCR products for integrin β1 and β-actin are presented in Table I.
Small interfering RNA (siRNA) transfection
Target-specific siRNAs for hnRNP E1 (sc-43843), hnRNP A1 (sc-270345), SF2/ASF (sc-38319) and SRp20 (sc-38338) were purchased from Santa Cruz Biotechnology and transfected into pancreatic cancer cells according to the manufacturer's protocol. In a 6-well tissue culture plate (PIDL06P05; Merck Millipore), 2×105 cells were seeded per well in 2 ml antibiotic-free normal growth medium supplemented with FBS. The cells were incubated at 37°C in a CO2 incubator until the cells were 60–80% confluent. For each transfection, 2–8 µl of siRNA duplexes were diluted into 100 µl siRNA transfection medium (31985070; Thermo Fisher Scientific), and 2–8 µl siRNA transfection reagent (11668019; Thermo Fisher Scientific) was diluted into 100 µl siRNA transfection medium. The siRNA duplex was added directly to the diluted transfection reagent using a pipette and mixed gently by pipetting the solution up and down, followed by incubation for 45 min at room temperature. The cells were washed once with 2 ml of siRNA transfection medium, and 0.8 ml of siRNA transfection medium was added to each tube containing the siRNA transfection reagent mixture. The solutions were mixed gently and overlaid onto the washed cells. The cells were incubated for 5–7 h at 37°C in a CO2 incubator. Subsequently, 1 ml of normal growth medium containing two times the normal serum and antibiotic concentration (2X normal growth medium) was added without removing the transfection mixture. The cells were incubated for an additional 18–24 h. The medium was aspirated and replaced with fresh 1X normal growth medium. The cells were assayed using the appropriate protocol at 72 h after the addition of normal growth medium (2X normal growth medium) in the step above.
Scratch wound assay
Cells were seeded in 6-well plates (PIDL06P05; Merck Millipore) at a density of >80% confluence. Cells were transfected with KdFyn, E1-GFP or hnRNP E1 siRNA or the control vector 24 h after seeding. Wounds were made in confluent monolayers 48 h after transfection by using a pipette tip under vacuum suction. A straight line was drawn across the bottom of each well, and then pictures were captured with the line at the bottom of the viewing field. Wounds were measured from the exact vertical middle of each image such that the initial measurement and the measurement 24 h later were taken from the same vertical spot in each well. Representative images are shown for each group, and results are reported as the percentage of the wound closed.
Western blot analysis
Whole cell protein was extracted with lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol), and 30 µg protein, determined using a BCA protein assay kit (23227; Pierce Biotechnology), was resolved on a 12% acrylamide gel and then transferred onto nylon membranes (LC2003; Thermo Fisher Scientific). The blots were incubated overnight at 4°C with 1% blocking solution in TBS buffer (50 mM Tris-HCl and 150 mM NaCl) and 0.1% Tween-20. The membranes were then incubated with the primary antibodies. After 2 h at room temperature, the membranes were washed twice with TBS plus 0.1% Tween-20 and 0.5% blocking buffer and incubated for 1 h with horseradish peroxidase-conjugated secondary antibody at room temperature. Following incubation with the appropriate secondary antibodies, the membranes were washed and the signals were visualized with Amersham ECL plus Western blotting detection reagents (28-9829-42 AB; GE Healthcare Bio-Sciences AB, Stockholm, Sweden). The specific bands on the autoradiograms were quantitated by densitometry.
Statistical analysis
All analyses were performed using SPSS 19.0 software. The correlation between categorical variables was evaluated using the χ2 test. Normally distributed data were analyzed using a Student's t-test; otherwise, the non-parametric Mann-Whitney test was applied. P<0.05 was considered to indicate a statistically significant difference.
Results
Activity of Fyn affects alterative splicing of integrin β1
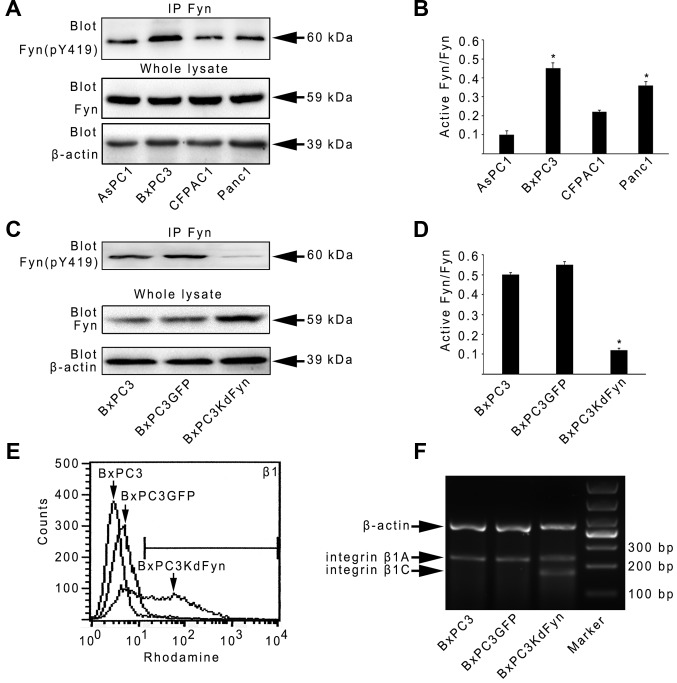
To determine whether the activity of Fyn affects the expression of integrin β1 in pancreatic cancer cells, the expression of active Fyn was analyzed in four pancreatic cancer cell lines, and BxPC3 pancreatic cancer cells were selected for the investigation of integrin β1 expression when Fyn activity was suppressed. As shown in Fig. 1A–E, BxPC3 cells expressed higher levels of active Fyn than the other three pancreatic cancer cells. In the KdFyn-transfected BxPC3 cells (BxPC3KdFyn), the expression of integrin β1 did not decrease; however, the level of a certain subtype of integrin β1 was increased, suggesting that suppressing Fyn activity may result in a change in integrin β1 splicing. Therefore, the splicing of integrin β1 in BxPC3 cells with different activity levels of Fyn was further analyzed. Integrin β1B and D were not expressed in these pancreatic cancer cells, and decreasing the activity of Fyn significantly promoted the expression of integrin β1C (Fig. 1F). These results suggested that the activity of Fyn affected the alterative splicing of integrin β1.
Figure 1.
Activity of Fyn affects alterative splicing of integrin β1. (A–D) Active Fyn was analyzed by detecting the expression of tyrosine 419-phosphorylated Fyn in different pancreatic cancer cell lines. (E) Expression of integrin β1 was analyzed by fluorescence-activated cell sorting using an antibody specific for the extracellular amino acid sequence of integrin β1. (F) Splicing of integrin β1 was analyzed in BxPC3, BxPC3GFP and BxPC3KdFyn cells. *P<0.05. Data are presented as the mean ± standard deviation from ≥3 independent experiments. Sequences of the PCR primers for integrin β1A and C (Fig. 4A) and β-actin are listed in Table I.
Fyn regulates phosphorylation and nuclear localization of hnRNP E1 via P21-activated kinase 1 (PAK1), thus affecting the alterative splicing of integrin β1
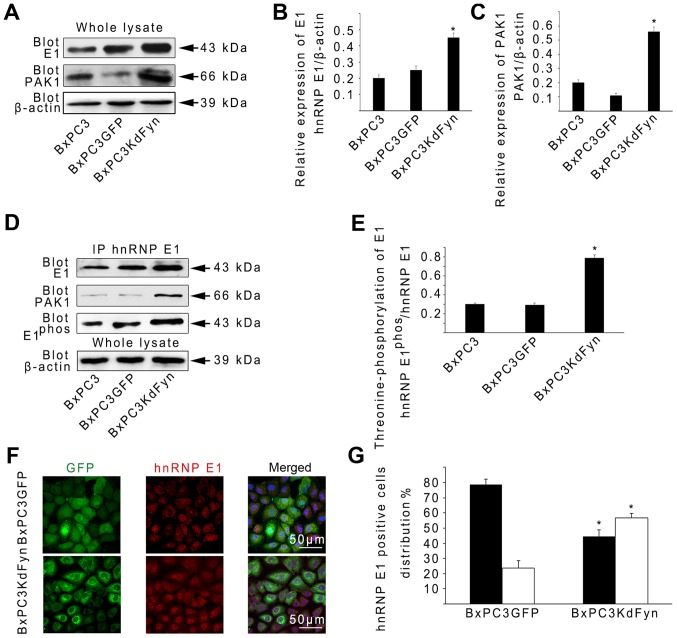
To determine whether Fyn regulates the biological function of hnRNP E1, the protein levels of hnRNP E1 in BxPC3 cancer cells with different Fyn activities were evaluated. The expression of hnRNP E1 was revealed to be increased ~2-fold in BxPC3KdFyn cells compared with BxPC3GFP or BxPC3 cells (Fig. 2A and B). It has been shown that PAK1, one of the evolutionarily conserved families of serine/threonine protein kinases, contributes to the threonine-phosphorylation of hnRNP E1 thus activating hnRNP E1 for RNA processing (37). To further explore whether the inhibition of Fyn activity affects PAK1 expression, thus promoting hnRNP E1 phosphorylation, PAK1 expression was analyzed in BxPC3 cells with different Fyn activity levels. PAK1 expression was found to be increased significantly in BxPC3KdFyn cells compared with the control cells (Fig. 2A and C). Subsequently, co-immunoprecipitation was used to analyze the threonine-phosphorylation of hnRNP E1 and the interaction between hnRNP E1 and PAK1 in BxPC3 cells with different Fyn activity levels. As shown in Fig. 2D and E, threonine-phosphorylation of hnRNP E1 was increased ~2.5-fold in BxPC3KdFyn cells compared with BxPC3 or BxPC3GFP cells. In addition, inhibition of Fyn activity promoted the interaction of hnRNP E1 with PAK1 in BxPC3KdFyn cells. Phosphorylated hnRNP E1 is involved in nuclear localization and mediates a variety of functions associated with mRNA splicing and stability (37,38). Therefore, the effect of Fyn activity on hnRNP E1 nuclear localization was next investigated using confocal immunofluorescence, revealing that the nuclear distribution of hnRNP E1 was increased ~2-fold in BxPC3KdFyn cells compared with BxPC3GFP cells (Fig. 2F and G). These results suggested that Fyn regulated the phosphorylation and nuclear localization of hnRNP E1 via PAK1 in pancreatic cancer cells.
Figure 2.
Activity of Fyn regulates the phosphorylation and nuclear localization of hnRNP E1 via PAK1. (A–C) Western blot analysis revealed the relative expression levels of hnRNP E1 and PAK1 protein among BxPC3, BxPC3GFP and BxPC3KdFyn cells. β-actin blotting was used as a loading control. (D and E) Co-immunoprecipitation analysis showed the threonine-phosphorylation of hnRNP E1 and the interaction of hnRNP E1 with PAK1 among BxPC3, BxPC3GFP and BxPC3KdFyn cells. (F and G) Confocal microscopy analysis showed that the nuclear distribution of hnRNP E1 was increased in BxPC3KdFyn cells compared with BxPC3GFP cells. *P<0.05. Data are presented as the mean ± standard deviation from ≥3 independent experiments.
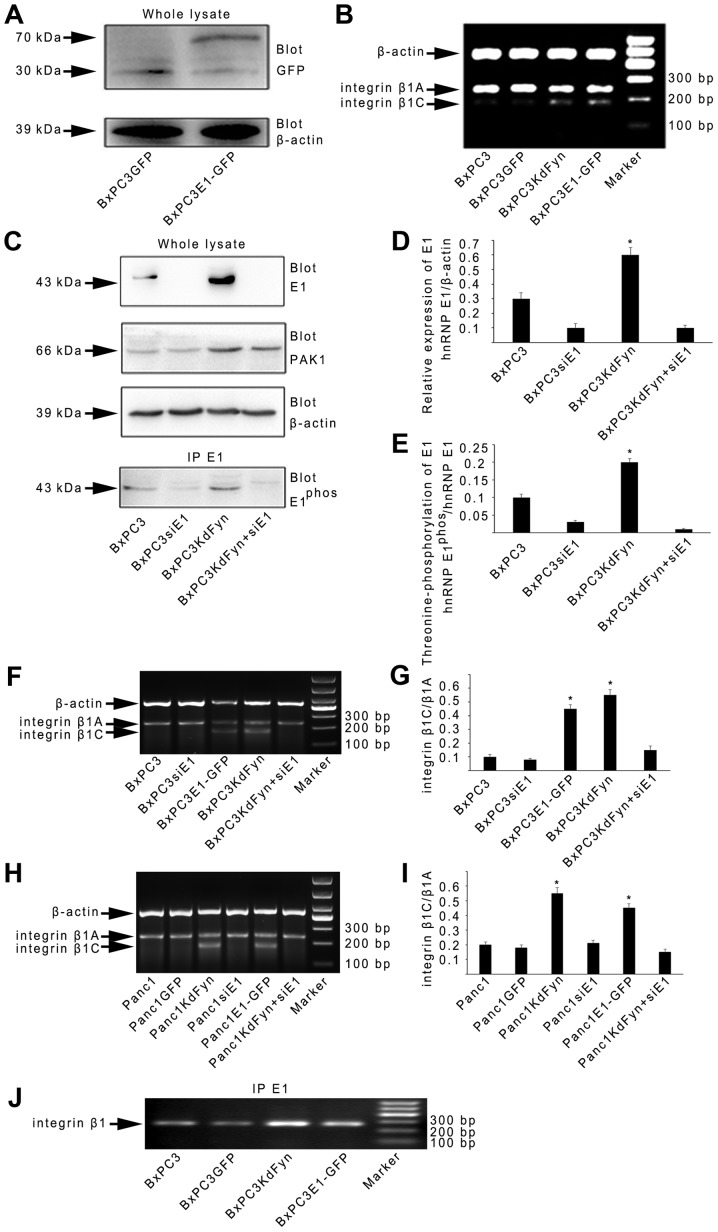
However, to date, the cellular function of hnRNP E1 in integrin β1 splicing has not been reported. Therefore, whether changes in hnRNP E1 expression could modulate the splicing of integrin β1 was next investigated. A GFP-hnRNP E1 fusion protein was expressed from an adenoviral system in BxPC3 cells (BxPC3E1-GFP) (Fig. 3A). The expression of integrin β1C was markedly increased in BxPC3E1-GFP cells (Fig. 3B). Furthermore, knockdown of the expression of hnRNP E1 by transfection specific hnRNP E1 siRNA in the BxPC3KdFyn cells (BxPC3KdFyn+siE1) decreased the expression of integrin β1C (Fig. 3C–G). It was also noted that inhibition of the expression of hnRNP E1 did not affect PAK1 expression, but significantly decreased the phosphorylation of hnRNP E1 (Fig. 3E). In addition, inhibition of the activity of Fyn or overexpression of hnRNP E1 in the Panc1 pancreatic cancer cells which expressed active Fyn (Fig. 1A) also promoted the splicing of integrin β1C (Fig. 3H and I). These results suggested that Fyn affected the splicing of integrin β1 by regulating the phosphorylation and nuclear localization of hnRNP E1.
Figure 3.
Fyn regulates the alternative splicing of integrin β1 via hnRNP E1. (A) Co-immunoprecipitation analysis showed the expression of GFP-hnRNP E1 fusion protein in BxPC3 cells. (B, F and G) Splicing of integrin β1 was analyzed under different Fyn activity levels or hnRNP E1 expression levels in BxPC3 cells. (C–E) Knockdown of hnRNP E1 expression decreased the phosphorylation of hnRNP E1 in BxPC3 cells. (H and I) Splicing of integrin β1 was analyzed under different Fyn activity levels or hnRNP E1 expression levels in Panc1 cells. (J) RNA immunoprecipitation showed the interaction between hnRNP E1 and the pre-mRNA of integrin β1. *P<0.05. Data are presented as the mean ± standard deviation from ≥3 independent experiments. Sequences of the PCR primers for integrin β1 pre-mRNA, which cover the exon and intron sequences of integrin β1C (Fig. 4A) are listed in Table I.
hnRNP E1 spliceosome regulates integrin β1 splicing by using hnRNP A1 and SF2/ASF to recognize the pre-mRNA of integrin β1
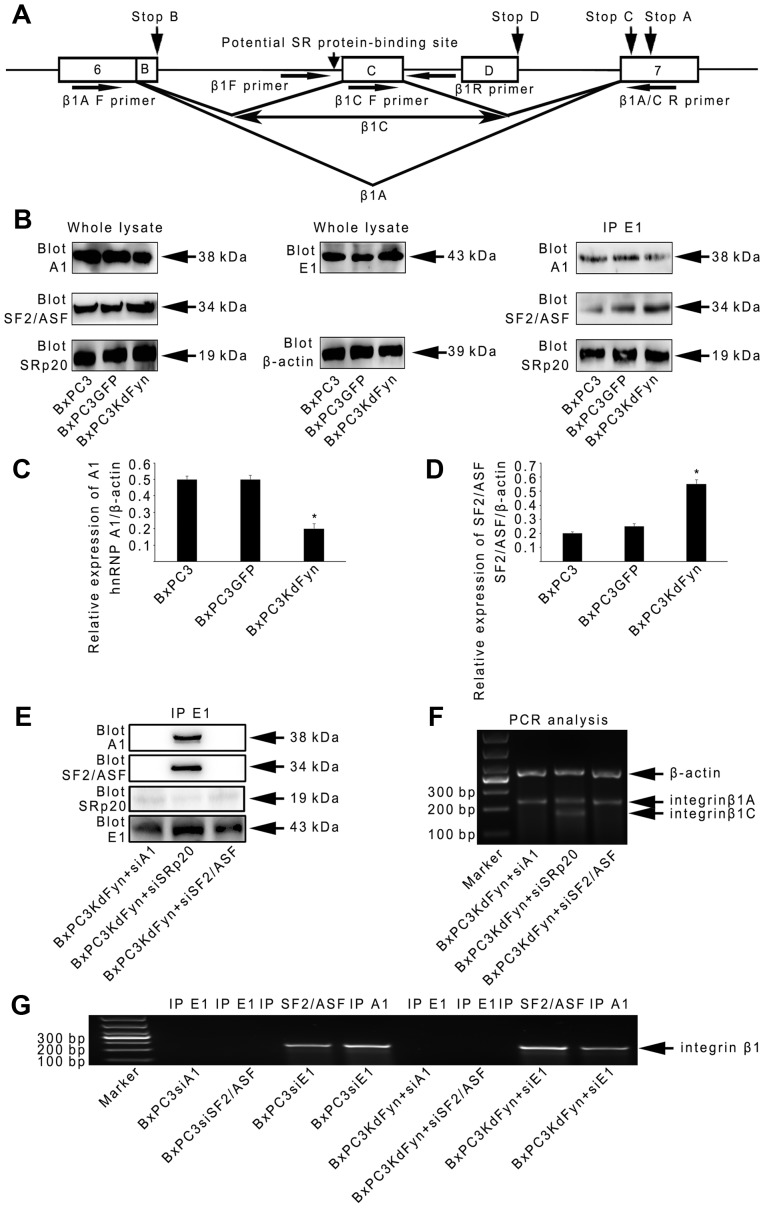
It is established that splicing is performed by the spliceosome, which is composed of five small ribonucleoproteins (snRNPs) and hundreds of additional proteins, including the SR proteins and the hnRNPs. The SR proteins and hnRNPs function as trans-acting factors by binding to the ESEs or ESSs of target pre-mRNAs to regulate alternative splicing (39–43). As the results of the present study identified that hnRNP E1 participated in the regulation of integrin β1 splicing, whether hnRNP E1 binds to the pre-mRNA of integrin β1 was further analyzed by RNA immunoprecipitation. As shown in Fig. 3J, the binding of hnRNP E1 to integrin β1 pre-mRNA in BxPC3 pancreatic cancer cells with different Fyn activity or hnRNP E1 protein levels suggested that hnRNP E1 may be a component of the spliceosome complex that regulates integrin β1 splicing. Therefore, the components of the hnRNP E1 spliceosome complex extracted from BxPC3KdFyn cells were next analyzed by mass spectrometry. This revealed two SR proteins (SRSF1 and SRSF3, also known as SF2/ASF and SRp20) and six other hnRNPs (hnRNP A1, A3, C1, K, M and U) in the hnRNP E1 spliceosome complex (Table II). Previous studies showed a potential SR protein-binding site in the pre-mRNA of integrin β1 for the splicing of β1C (Fig. 4A (44). Therefore, whether these two SR proteins and three other hnRNPs (hnRNP A1, C1 and M, which have been demonstrated to participate in splicing regulation) affect integrin β1 splicing was examined in this study.
Table II.
Proteins identified by mass spectrometry in the hnRNP E1 spliceosome of BxPC3KdFyn cells.
| Group ID | Protein ID | Protein score | Protein mass | Coverage | No. of unique peptide | No. of unique spectrum | Isoelectric point | Description |
|---|---|---|---|---|---|---|---|---|
| 1 | IPI00003865 | 972.09 | 71082.31 | 0.243034056 | 11 | 24 | 5.16 | HSPA8 isoform 1 of heat shock cognate 71 kDa protein |
| 2 | IPI00216049 | 704.64 | 51229.51 | 0.265658747 | 9 | 17 | 5.18 | HNRNPK, isoform 1 of heterogeneous nuclear ribonucleoprotein K |
| 3 | IPI00017617 | 526.07 | 69617.93 | 0.140065147 | 7 | 9 | 9.21 | DDX5 probable ATP-dependent RNA helicase DDX5 |
| 4 | IPI00911039 | 440.23 | 64169.98 | 0.015358362 | 1 | 1 | 5.19 | HSPA1B, HSPA1A cDNA FLJ54408, highly similar to heat shock 70 kDa protein 1 |
| 5 | IPI00304925 | 440.12 | 70294.14 | 0.014040562 | 1 | 2 | 5.31 | HSPA1B, HSPA1A heat shock 70 kDa protein 1A/1B |
| 6 | IPI00479217 | 335.99 | 89665.43 | 0.079404467 | 5 | 8 | 5.51 | HNRNPU, isoform short of heterogeneous nuclear ribonucleoprotein U |
| 7 | IPI00171903 | 323.1 | 77749.42 | 0.115068493 | 6 | 9 | 9.1 | HNRNPM, isoform 1 of heteronuclear ribonucleoprotein M |
| 8 | IPI00215965 | 316.85 | 38837.09 | 0.112903226 | 3 | 6 | 9.46 | HNRNPA1, isoform A1-B of heterogeneous nuclear ribonucleoprotein A1 |
| 9 | IPI00420014 | 295.88 | 246006.24 | 0.033707865 | 5 | 5 | 5.95 | SNRNP200 isoform 1 of U5 small nuclear ribonucleoprotein 200 kDa helicase |
| 10 | IPI00216592 | 292.33 | 32374.89 | 0.160409556 | 4 | 10 | 4.69 | HNRNPC, isoform C1 of heterogeneous nuclear ribonucleoproteins C1/C2 |
| 11 | IPI00419373 | 278.6 | 39798.67 | 0.137566138 | 4 | 5 | 9.31 | HNRNPA3, isoform 1 of heterogeneous nuclear ribonucleoprotein A3 |
| 12 | IPI00300371 | 236.25 | 136575.15 | 0.045193098 | 4 | 4 | 4.91 | SF3B3 isoform 1 of splicing factor factor 3B subunit 3 |
| 13 | IPI00552938 | 208.36 | 22271.46 | 0.178571429 | 3 | 6 | 4.95 | RBMX heterogeneous nuclear ribonucleoprotein G isoform 2 |
| 14 | IPI00215884 | 208.03 | 27841.86 | 0.14516129 | 3 | 5 | 10.77 | SRSF1, isoform ASF-1 of serine/arginine-rich splicing factor 1 |
| 15 | IPI00641829 | 206.99 | 51103.09 | 0.0248307 | 1 | 1 | 5.67 | SNORD84, DDX39B isoform 2 of spliceosome RNA helicase DDX39B |
| 16 | IPI00328840 | 178.33 | 27540.9 | 0.189393939 | 3 | 5 | 11.6 | THOC4 THO complex subunit 4 |
| 17 | IPI00007423 | 173.48 | 28941.37 | 0.079681275 | 2 | 2 | 3.67 | ANP32B isoform 1 of acidic leucine-rich nuclear phosphoprotein 32 family member B |
| 18 | IPI00017964 | 167.06 | 14021.35 | 0.317460317 | 3 | 4 | 10.98 | SNRPD3 small nuclear ribonucleoprotein Sm D3 |
| 19 | IPI00010204 | 141.72 | 19545.99 | 0.140243902 | 2 | 4 | 12.15 | SRSF3, Serine/arginine-rich splicing factor 3 |
| 20 | IPI00031556 | 104.61 | 53809.32 | 0.044210526 | 2 | 2 | 9.49 | U2AF2 isoform 1 of splicing factor U2AF 65 kDa subunit |
| 21 | IPI00396435 | 91.92 | 91673.47 | 0.031446541 | 2 | 3 | 7.48 | DHX15 putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 |
| 22 | IPI00009328 | 86.29 | 47126.29 | 0.03406326 | 1 | 2 | 6.69 | EIF4A3 eukaryotic initiation factor 4A-III |
| 23 | IPI00001757 | 80.34 | 19933.75 | 0.097701149 | 1 | 2 | 5.43 | RBM8A isoform 1 of RNA-binding protein 8A |
| 24 | IPI00007928 | 79.34 | 274738.05 | 0.003426124 | 1 | 1 | 9.14 | PRPF8 Pre-mRNA-processing- splicing factor 8 |
| 25 | IPI00026089 | 70.91 | 146479.37 | 0.013803681 | 1 | 1 | 7.1 | SF3B1 splicing factor 3B subunit 1 |
| 26 | IPI00302850 | 64.13 | 13273.36 | 0.092436975 | 1 | 1 | 12.09 | SNRPD1 small nuclear ribonucleo-protein Sm D1 |
| 27 | IPI00016610 | 60.99 | 37987.14 | 0.04494382 | 1 | 1 | 7.11 | PCBP1, Poly(rC)-binding protein 1 (hnRNP E1) |
| 28 | IPI00003519 | 60.81 | 110335.65 | 0.012345679 | 1 | 1 | 4.6 | EFTUD2 116 kDa U5 small nuclear ribonucleoprotein component |
| 29 | IPI00027285 | 54.91 | 24764.75 | 0.033333333 | 1 | 1 | 11.75 | SNRPB isoform SM-B' of small nuclear ribonucleoprotein associated proteins B and B' |
| 30 | IPI00016572 | 54.82 | 8547.45 | 0.157894737 | 1 | 1 | 9.41 | SNRPG small nuclear ribonucleo-protein G |
| 31 | IPI00305068 | 43.69 | 107656.09 | 0.015940489 | 1 | 1 | 8.4 | PRPF6 Pre-mRNA-processing factor 6 |
| 32 | IPI00221106 | 41.48 | 100279.02 | 0.013407821 | 1 | 1 | 5.37 | SF3B2 splicing factor 3B subunit 2 |
| 33 | IPI00329791 | 39.34 | 117802.7 | 0.010669253 | 1 | 1 | 9.87 | DDX46 probable ATP-dependent RNA helicase DDX46 |
| 34 | IPI00019380 | 38.82 | 92863.93 | 0.013924051 | 1 | 1 | 6.4 | NCBP1 nuclear cap-binding protein subunit 1 |
| 35 | IPI00012442 | 38.16 | 52189.1 | 0.036480687 | 1 | 1 | 5.21 | G3BP1 Ras GTPase-activating protein-binding protein 1 |
| 36 | IPI00008943 | 37.53 | 54348.96 | 0.022964509 | 1 | 1 | 6.21 | DDX19B isoform 1 of ATP-dependent RNA helicase DDX19B |
| 37 | IPI00013180 | 36.66 | 17558.8 | 0.0625 | 1 | 1 | 9 | BUD31 protein BUD31 homolog |
| 38 | IPI00010158 | 34.07 | 14758.48 | 0.061068702 | 1 | 2 | 4.73 | CHRAC1 chromatin accessibility complex protein 1 |
| 39 | IPI00029266 | 32.15 | 10853.66 | 0.119565217 | 1 | 1 | 9.86 | SNRPE small nuclear ribonucleo-protein E |
Figure 4.
hnRNP E1 spliceosome regulates integrin β1 splicing by hnRNP A1 and SF2/ASF. (A) Schematic representation of the genomic sequence of integrin β1. Exon sequences are shown as boxes and introns as solid lines. The splicing pattern used to generate β1A and C transcripts are depicted, and the potential SR protein-binding site is indicated with a vertical arrow. The positions of the stop codons for the various splice variants are also indicated by arrows. The horizontal arrows indicate the primers used in PCR experiments (Table I). (B–D) Western blot analysis and co-immunoprecipitation were used to analyze the expression levels and abundance of hnRNP E1-specific spliceosome components among BxPC3, BxPC3GFP and BxPC3KdFyn cells. β-actin blotting was used as a loading control. (E and F) Co-immunoprecipitation and splicing analysis were used to explore the functions of hnRNP A1 and SF2/ASF on the construction of the hnRNP E1 spliceosome complex and the splicing of integrin β1. (G) RNA immunoprecipitation showed that hnRNP A1 and SF2/ASF, but not hnRNP E1, were directly bound to the pre-mRNA of integrin β1. *P<0.05. Data are presented as the mean ± standard deviation from ≥3 independent experiments.
Generally, the alternative splicing of genes is determined by SR proteins and hnRNPs in a concentration-dependent manner (45). Therefore, we analyzed the expression of these SR and hnRNP proteins and their abundance in the hnRNP E1 complex in pancreatic cancer cells. As shown in Fig. 4B–D, the expression of SF2/ASF was significantly increased in BxPC3KdFyn cells compared with control cells. However, the expression of SRp20 did not significantly differ among these groups of cells. With regard to the three hnRNPs, only hnRNP A1 was obviously decreased in BxPC3KdFyn cells compared with control cells. In addition, the level of hnRNP A1 was decreased in the hnRNP E1 complex in BxPC3KdFyn cells when the SF2/ASF level was increased (Fig. 4B). These results suggested that hnRNP A1 and SF2/ASF may serve important roles in the hnRNP E1 spliceosome complex.
To explore the functions of hnRNP A1 and SF2/ASF in the spliceosome, target-specific siRNAs were used to knock down their expression in BxPC3KdFyn cells. As shown in Fig. 4E and F, knockdown of hnRNP A1 or SF2/ASF in BxPC3KdFyn cells completely eliminated the construction of the hnRNP E1 complex and ultimately prevented the expression of integrin β1C. By contrast, knockdown of SRp20 did not affect the interaction of hnRNP E1 with hnRNP A1 and SF2/ASF, and did not alter the splicing of integrin β1 in BxPC3KdFyn cells. Therefore, in the spliceosome complex of BxPC3KdFyn cells, hnRNP A1, hnRNP E1 and SF2/ASF were key proteins responsible for integrin β1 splicing.
To further determine whether hnRNP A1, hnRNP E1 and SF2/ASF have direct roles in the post-transcriptional modification of integrin β1 pre-mRNA, endogenous complexes of RNA with hnRNP A1, E1 and SF2/ASF were extracted from BxPC3KdFyn cells to analyze the presence of integrin β1 pre-mRNA. hnRNP A1 and SF2/ASF, but not hnRNP E1, were directly bound to the pre-mRNA of integrin β1 (Fig. 4G). These results suggested that hnRNP E1 spliceosome regulated integrin β1 splicing by using hnRNP A1 and SF2/ASF to recognize the pre-mRNA of integrin β1 in pancreatic cancer cells.
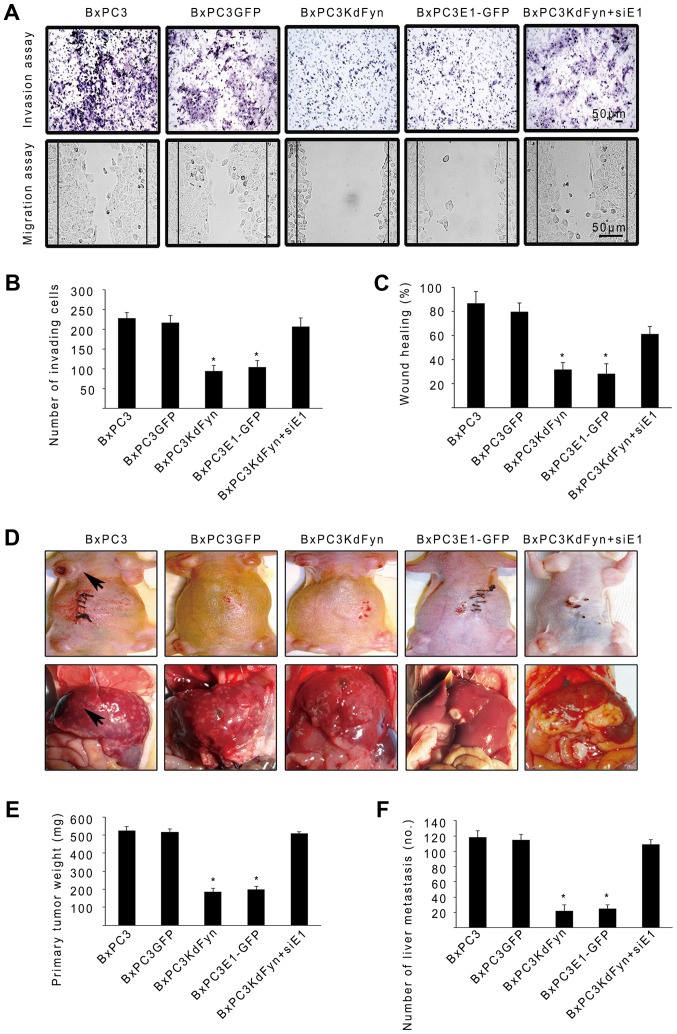
Effect of Fyn and hnRNP E1 on the invasion and metastasis of pancreatic cancer cells in vitro and in vivo
Transwell and wound-healing assays were employed to determine the effect of Fyn and hnRNP E1 on the invasion and migration of pancreatic cancer cells in vitro. As shown in Fig. 5A–C, inhibition of the activity of Fyn or overexpression of hnRNP E1 significantly decreased the number of invading cells and the migration of BxPC3 pancreatic cancer cells. By contrast, knockdown of the expression of hnRNP E1 in the BxPC3KdFyn cells promoted the invasion and migration of tumor cells. Subsequently, the effects of Fyn and hnRNP E1 on pancreatic cancer cell tumorigenesis and metastasis were investigated in vivo by subcutaneous or intrasplenic injection of 2×105 tumor cells into nude mice. After 5–8 weeks, the mice were sacrificed to detect subcutaneous tumors and liver metastases, the findings revealed that inhibition of Fyn activity or overexpression of exogenous hnRNP E1 within BxPC3 pancreatic cancer cells significantly decreased the primary tumor mass and liver metastases. Furthermore, knockdown of hnRNP E1 in the BxPC3KdFyn cells promoted the growth of the primary tumor mass and liver metastases (Fig. 5D–F). These results suggested that Fyn/hnRNP E1 signaling regulated the invasion and metastases of pancreatic cancer cells in vitro and in vivo.
Figure 5.
Effect of Fyn and hnRNP E1 on the invasion and metastasis of pancreatic cancer cells. (A–C) Transwell and wound-healing assays revealed that the activity of Fyn and/or the expression of hnRNP E1 significantly affected the invasion and migration abilities of BxPC3 pancreatic cancer cells in vitro. (D–F) An assay of the subcutaneous tumor mass and liver metastatic nodules revealed that the activity of Fyn and/or the expression of hnRNP E1 significantly affected the tumorigenesis and metastasis of BxPC3 pancreatic cancer cells in vivo. Each group of xenograft experiments included 5 mice. *P<0.05. Data are presented as the mean ± standard deviation from ≥3 independent experiments.
Expression of hnRNP E1 and integrin β1 are associated with metastasis of pancreatic cancer
Our data showed that Fyn, hnRNP E1 and integrin β1 were involved in the metastases of pancreatic cancer cells. Therefore, we next analyzed the expression of these proteins in pancreatic cancer using the Human Protein Atlas database and pancreatic cancer tissues.
In the Human Protein Atlas database, the expression of hnRNP E1 was positive in pancreatic cancer and the malignant cells exhibited moderate to strong cytoplasmic positivity (http://www.proteinatlas.org/ENSG00000169564-PCBP1/cancer/tissue/pancreatic+cancer). In pancreatic cancer tissues obtained from our institution, the mRNA expression of integrin β1A was correlated with lymph node metastasis (P=0.001), hepatic metastasis (P=0.005) and the tumor stage (P=0.002) of pancreatic cancer. By contrast, only cytoplasmic and not nuclear, expression of hnRNP E1 was correlated with lymph node metastasis (P=0.004), hepatic metastasis (P=0.011) and the tumor stage of pancreatic cancer (P=0.001) (Table III). In addition, the cytoplasmic expression of hnRNP E1 was positively correlated with the mRNA expression of integrin β1A (Table IV). Therefore, the expression of hnRNP E1 and integrin β1A were associated with metastasis of pancreatic cancer.
Table III.
Associations between the expression of integrin β1A, hnRNP E1 and the categorical clinicopathological parameters of pancreatic carcinoma.
| Parameters | Integrin β1A mRNA expression
|
Cytoplasmic hnRNP E1 protein expression
|
|||
|---|---|---|---|---|---|
| Median expression (range) | P-value | Low | High | P-value | |
| Age (years) | 0.183 | 0.805 | |||
| <65 | 44.343 (24.355–82.093) | 27 | 29 | ||
| ≥65 | 28.273 (17.489–58.197) | 9 | 11 | ||
| Sex | 0.847 | 0.092 | |||
| Male | 37.853 (18.741–74.376) | 5 | 12 | ||
| Female | 28.671 (23.728–79.674) | 31 | 28 | ||
| Tumor size (cm) | 0.517 | 0.961 | |||
| ≤2 | 55.170 (23.557–80.806) | 16 | 18 | ||
| >2 | 29.847 (18.257–74.357) | 20 | 22 | ||
| Tumor location | 0.601 | 0.108 | |||
| Head | 35.482 (21.230–74.376) | 34 | 33 | ||
| Body and tail | 64.364 (27.496–79.674) | 2 | 7 | ||
| Lymph node metastasis | 0.001 | 0.004 | |||
| Negative | 28.051 (10.770–57.022) | 27 | 17 | ||
| Positive | 70.630 (34.876–96.331) | 9 | 23 | ||
| Vascular invasion | 0.150 | 0.258 | |||
| Negative | 36.222 (25.467–77.838) | 30 | 29 | ||
| Positive | 29.816 (5.563–64.032) | 6 | 11 | ||
| Neural invasion | 0.147 | 0.426 | |||
| Negative | 29.361 (9.563–56.324) | 11 | 9 | ||
| Positive | 40.934 (25.618–80.162) | 25 | 31 | ||
| Duodenal invasion | 0.328 | 0.417 | |||
| Negative | 27.934 (9.758–63.527) | 8 | 6 | ||
| Positive | 37.038 (24.087–78.756) | 28 | 34 | ||
| Hepatic metastases | 0.005 | 0.011 | |||
| Negative | 29.847 (16.785–69.192) | 35 | 31 | ||
| Positive | 72.487 (64.115–106.964) | 1 | 9 | ||
| Differentiation | 0.56 | 0.404 | |||
| Well | 49.798 (25.015–69.084) | 1 | 4 | ||
| Moderate | 35.482 (18.741–80.892) | 26 | 25 | ||
| Poor | 28.189 (25.769–30.836) | 9 | 11 | ||
| T-stage (UICC) | 0.419 | 0.077 | |||
| T1, 2 | 35.482 (25.467–77.188) | 26 | 21 | ||
| T3, 4 | 39.998 (12.114–76.003) | 10 | 19 | ||
| Tumor stage (UICC) | 0.002 | 0.001 | |||
| I | 28.632 (13.919–44.015) | 21 | 8 | ||
| II | 46.717 (18.839–82.530) | 14 | 22 | ||
| III, IV | 76.003 (54.517–117.941) | 1 | 10 | ||
The bold values indicate P-values <0.05, UICC Union for International Cancer Control.
Table IV.
Correlation between the expressions of β1A (mRNA) and hnRNP E1 (protein).
| n | Spearman rank correlation analysis
|
||
|---|---|---|---|
| Correlation coefficient | P-value | ||
| hnRNP E1 protein expression β1A mRNA expression | 76 | 0.556 | <0.01 |
Discussion
The present study revealed a novel mechanism by which Fyn/hnRNP E1 signaling regulates pancreatic cancer metastasis by affecting the alternative splicing of integrin β1. The results demonstrated that inhibition of Fyn activity upregulated the expression of PAK1 and promoted the phosphorylation and nuclear localization of hnRNP E1, which affected the alterative splicing of integrin β1. Subsequently, an hnRNP E1 spliceosome complex including SR proteins and hnRNPs was shown to regulate the alternative splicing of integrin β1. In this spliceosome complex, hnRNP A1, hnRNP E1 and SF2/ASF were key proteins responsible for integrin β1 splicing. Importantly, hnRNP A1 and SF2/ASF directly bound to the pre-mRNA of integrin β1 to achieve post-transcriptional regulation. In vivo and in vitro studies demonstrated that suppression of Fyn activity and/or overexpression of hnRNP E1 significantly decreased the invasion and metastasis of pancreatic cancer cells. Finally, the expression of hnRNP E1 and integrin β1A were associated with the metastasis of pancreatic cancer.
Alternative splicing of the mRNA encoding certain integrin subunits increases diversity within the integrin family, and alters the function of the specific integrin (22). Previous studies have shown that splicing of integrin αv affected the proliferation and pro-angiogenic properties of human endothelial cells (21). Four splice variants (A, B, C and D) have been identified in the integrin β1 family, and the β1A isoform is known to be widely expressed among all mammalian species tested to date. Splice variants B, C and D which inhibit β1A-mediated focal adhesion formation, focal adhesion kinase phosphorylation, fibronectin matrix assembly, cell spreading and motility, have been shown to be downregulated or even absent in many types of tumor tissues and may be involved in tumor progression (22,46–49). However, the formation of splice variants of integrin β1 has not been fully described. In the present study, inhibition of Fyn activity and/or overexpression of hnRNP E1 was found to significantly promote the splicing of the integrin β1C variant in pancreatic cancer cells and decrease the invasion and migration of tumor cells in vitro, thus eventually suppressing tumorigenesis and metastasis of pancreatic cancer cells in vivo. In human pancreatic cancer tissues obtained from our institution, the expression of hnRNP E1 and integrin β1A were associated with the metastasis of pancreatic cancer. These results suggest that Fyn and hnRNP E1 affect pancreatic cancer metastasis by participating in the regulation of integrin β1 splicing.
As a member of the SFKs, Fyn has been shown to affect many cellular processes, including mRNA splicing (50,51). However, the mechanism by which Fyn regulates integrin β1 alternative splicing has not been reported. In the present study, the activity of Fyn was demonstrated to regulate the alternative splicing of integrin β1 via hnRNP E1. Inhibition of Fyn activity significantly promoted the phosphorylation and nuclear localization of hnRNP E1, which affected the alterative splicing of integrin β1 in pancreatic cancer cells. In addition, overexpression of hnRNP E1 significantly promoted the splicing of integrin β1C. By contrast, knockdown of hnRNP E1 expression eliminated integrin β1C splicing induced by KdFyn. These results suggest that Fyn acts though hnRNP E1 to regulate the alternative splicing of integrin β1. Additionally, inhibition of Fyn activity upregulated PAK1, which phosphorylated hnRNP E1 through direct interaction. Therefore, Fyn functions through PAK1 to induce phosphorylation and nuclear localization of hnRNP E1, which regulates integrin β1 alternative splicing.
The different components of the spliceosome determine the splicing of individual pre-mRNAs and assist in the accurate recognition of splice sites (52,53). The hnRNPs are involved in processing heterogeneous nuclear RNAs into mature mRNAs, as well as functioning as trans-acting factors in the regulation of gene expression under the phosphorylation of serine and threonine residues or methylation of arginine residues (54). The SR proteins, which are phosphorylated by the SR-specfic protein kinase family, including cdc2-like kinase, are key determinants of exon identity, and function as molecular adaptors to link the pre-mRNA to the splicing machinery (55–58). In the present study, hnRNP E1 was found to form a complex with SR proteins and hnRNPs, which coordinately regulated the alternative splicing of integrin β1 in pancreatic cancer cells. In this spliceosome complex, hnRNP E1, hnRNP A1 and SF2/ASF served key roles in the regulation of integrin β1 splicing. Firstly, the amounts of hnRNP A1 and SF2/ASF in the hnRNP E1 spliceosome complex were demonstrated to be inversely correlated with one another among BxPC3 cells with different Fyn activity levels, suggesting that hnRNP A1 and SF2/ASF may have opposing roles in integrin β1 splicing. This phenomenon is consistent with a previous report, which showed that these two proteins have competing roles in regulation of alternative splicing of the Ron gene (59). Additionally, in the present study, knockdown of either hnRNP A1 or SF2/ASF completely eliminated the formation of the hnRNP E1 spliceosome complex and ultimately prohibited the alternative splicing of integrin β1 in pancreatic cancer cells, suggesting that hnRNP A1 and SF2/ASF were the key proteins responsible for integrin β1 splicing. Furthermore, RNA immunoprecipitation revealed that hnRNP A1 and SF2/ASF, but not hnRNP E1, directly bound to the pre-mRNA of integrin β1, suggesting that the hnRNP E1 spliceosome complex acts via binding of hnRNP A1 and SF2/ASF to the pre-mRNA of integrin β1. Therefore, the hnRNP E1 spliceosome regulates integrin β1 splicing via hnRNP A1- and SF2/ASF-mediated recognition of the pre-mRNA of integrin β1.
In conclusion, the results of the present study provide a potential explanation for how the Fyn/hnRNP E1 spliceosome signaling regulates pancreatic cancer metastasis. Inhibition of Fyn activity promoted the phosphorylation and nuclear localization of hnRNP E1, leading to construction of the hnRNP E1 spliceosome complex, thus resulting in alterative splicing of integrin β1. In the hnRNP E1 spliceosome complex, hnRNP A1 and SF2/ASF are responsible for binding to the pre-mRNA of integrin β1 to achieve posttranscriptional modification. Suppression of Fyn and/or overexpression of hnRNP E1 significantly decreased the invasion and metastasis of pancreatic cancer cells. Thus, by altering integrin β1 splicing, the Fyn/hnRNP E1 spliceosome signal modulates the metastasis of pancreatic cancer.
Acknowledgments
The authors would like to thank Professors Lei Cai and Xiaobin Feng for their assistance with the writing assistance and proofreading of the present study. This work was supported by the National Natural Science Foundation of China under the grant no. 81430063; and the Natural Science Foundation of Southwest Hospital under the grant no. SWH2016JCYB-46.
References
- 1.Binker MG, Binker-Cosen MJ, Binker-Cosen AA, Cosen-Binker LI. Microenvironmental factors and extracellular matrix degradation in pancreatic cancer. JOP. 2014;15:280–285. doi: 10.6092/1590-8577/2638. [DOI] [PubMed] [Google Scholar]
- 2.Erkan M. Understanding the stroma of pancreatic cancer: Co-evolution of the microenvironment with epithelial carcinogenesis. J Pathol. 2013;231:4–7. doi: 10.1002/path.4213. [DOI] [PubMed] [Google Scholar]
- 3.Lunardi S, Muschel RJ, Brunner TB. The stromal compartments in pancreatic cancer: Are there any therapeutic targets? Cancer Lett. 2014;343:147–155. doi: 10.1016/j.canlet.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 4.Grzesiak JJ, Ho JC, Moossa AR, Bouvet M. The integrin-extracellular matrix axis in pancreatic cancer. Pancreas. 2007;35:293–301. doi: 10.1097/mpa.0b013e31811f4526. [DOI] [PubMed] [Google Scholar]
- 5.Grzesiak JJ, Tran Cao HS, Burton DW, Kaushal S, Vargas F, Clopton P, Snyder CS, Deftos LJ, Hoffman RM, Bouvet M. Knockdown of the beta(1) integrin subunit reduces primary tumor growth and inhibits pancreatic cancer metastasis. Int J Cancer. 2011;129:2905–2915. doi: 10.1002/ijc.25942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh N, Clynes M, Crown J, O'Donovan N. Alterations in integrin expression modulates invasion of pancreatic cancer cells. J Exp Clin Cancer Res. 2009;28:140. doi: 10.1186/1756-9966-28-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao H, Zeng ZZ, Fay KS, Veine DM, Staszewski ED, Morgan M, Wilder-Romans K, Williams TM, Spalding AC, Ben-Josef E, et al. Role of α5β1 integrin up-regulation in radiation-induced invasion by human pancreatic cancer cells. Transl Oncol. 2011;4:282–292. doi: 10.1593/tlo.11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilbig A. Src kinase and pancreatic cancer. Recent Results Cancer Res. 2008;177:179–185. doi: 10.1007/978-3-540-71279-4_19. [DOI] [PubMed] [Google Scholar]
- 9.Je DW, O YM, Ji YG, Cho Y, Lee DH. The inhibition of SRC family kinase suppresses pancreatic cancer cell proliferation, migration, and invasion. Pancreas. 2014;43:768–776. doi: 10.1097/MPA.0000000000000103. [DOI] [PubMed] [Google Scholar]
- 10.Ma YC, Shi C, Zhang YN, Wang LG, Liu H, Jia HT, Zhang YX, Sarkar FH, Wang ZS. The tyrosine kinase c-Src directly mediates growth factor-induced Notch-1 and Furin interaction and Notch-1 activation in pancreatic cancer cells. PLoS One. 2012;7:e33414. doi: 10.1371/journal.pone.0033414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagathihalli NS, Merchant NB. Src-mediated regulation of E-cadherin and EMT in pancreatic cancer. Front Biosci (Landmark Ed) 2012;17:2059–2069. doi: 10.2741/4037. [DOI] [PubMed] [Google Scholar]
- 12.Chaimowitz NS, Falanga YT, Ryan JJ, Conrad DH. Fyn kinase is required for optimal humoral responses. PLoS One. 2013;8:e60640. doi: 10.1371/journal.pone.0060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman NM, Yoder AN, Houtman JC. Non-catalytic functions of Pyk2 and Fyn regulate late stage adhesion in human T cells. PLoS One. 2012;7:e53011. doi: 10.1371/journal.pone.0053011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du CP, Tan R, Hou XY. Fyn kinases play a critical role in neuronal apoptosis induced by oxygen and glucose deprivation or amyloid-β peptide treatment. CNS Neurosci Ther. 2012;18:754–761. doi: 10.1111/j.1755-5949.2012.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yadav V, Denning MF. Fyn is induced by Ras/PI3K/Akt signaling and is required for enhanced invasion/migration. Mol Carcinog. 2011;50:346–352. doi: 10.1002/mc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen ZY, Cai L, Bie P, Wang SG, Jiang Y, Dong JH, Li XW. Roles of Fyn in pancreatic cancer metastasis. J Gastroenterol Hepatol. 2010;25:293–301. doi: 10.1111/j.1440-1746.2009.06021.x. [DOI] [PubMed] [Google Scholar]
- 17.Kapp TG, Rechenmacher F, Sobahi TR, Kessler H. Integrin modulators: a patent review. Expert Opin Ther Pat. 2013;23:1273–1295. doi: 10.1517/13543776.2013.818133. [DOI] [PubMed] [Google Scholar]
- 18.Landowski TH, Gard J, Pond E, Pond GD, Nagle RB, Geffre CP, Cress AE. Targeting integrin α6 stimulates curative-type bone metastasis lesions in a xenograft model. Mol Cancer Ther. 2014;13:1558–1566. doi: 10.1158/1535-7163.MCT-13-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun C, Zargham R, Shao Q, Gui X, Marcus V, Lazaris A, Salman A, Metrakos P, Qu X, Gao Z. Association of CD98, integrin β1, integrin β3 and Fak with the progression and liver metastases of colorectal cancer. Pathol Res Pract. 2014;210:668–674. doi: 10.1016/j.prp.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Moro L, Perlino E, Marra E, Languino LR, Greco M. Regulation of beta1C and beta1A integrin expression in prostate carcinoma cells. J Biol Chem. 2004;279:1692–1702. doi: 10.1074/jbc.M307857200. [DOI] [PubMed] [Google Scholar]
- 21.Gauck S, Schultheiss HP, Rauch U, Eisenreich A. Modulation of the isoform expression of Cyr61 and Integrin-αv in human microvascular endothelial cells. Cardiovasc Syst. 2013;1:8. doi: 10.7243/2052-4358-1-8. [DOI] [Google Scholar]
- 22.de Melker AA, Sonnenberg A. Integrins: Alternative splicing as a mechanism to regulate ligand binding and integrin signaling events. BioEssays. 1999;21:499–509. doi: 10.1002/(SICI)1521-1878(199906)21:6<499::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 23.Fornaro M, Steger CA, Bennett AM, Wu JJ, Languino LR. Differential role of β1C and β1A integrin cytoplasmic variants in modulating focal adhesion kinase, protein kinase B/AKT, and Ras/Mitogen-activated protein kinase pathways. Mol Biol Cell. 2000;11:2235–2249. doi: 10.1091/mbc.11.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moro L, Greco M, Maiorano E, Selvaggi L, Marra E, Perlino E. Transcriptional regulation of beta1 integrin expression in the physio/pathological states of human endometrial tissues. Int J Oncol. 2005;26:457–465. [PubMed] [Google Scholar]
- 25.Eisenreich A. Regulation of vascular function on posttranscriptional level. Thrombosis. 2013;2013:948765. doi: 10.1155/2013/948765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barash Y, Garcia JV. Predicting alternative splicing. Methods Mol Biol. 2014;1126:411–423. doi: 10.1007/978-1-62703-980-2_28. [DOI] [PubMed] [Google Scholar]
- 27.Kotlajich MV, Crabb TL, Hertel KJ. Spliceosome assembly pathways for different types of alternative splicing converge during commitment to splice site pairing in the A complex. Mol Cell Biol. 2009;29:1072–1082. doi: 10.1128/MCB.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schor IE, Gómez Acuña LI, Kornblihtt AR. Coupling between transcription and alternative splicing. Cancer Treat Res. 2013;158:1–24. doi: 10.1007/978-3-642-31659-3_1. [DOI] [PubMed] [Google Scholar]
- 29.Verbeeren J, Niemelä EH, Turunen JJ, Will CL, Ravantti JJ, Lührmann R, Frilander MJ. An ancient mechanism for splicing control: U11 snRNP as an activator of alternative splicing. Mol Cell. 2010;37:821–833. doi: 10.1016/j.molcel.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Zhang T, Huang XH, Dong L, Hu D, Ge C, Zhan YQ, Xu WX, Yu M, Li W, Wang X, et al. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol Cancer. 2010;9:72. doi: 10.1186/1476-4598-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barboro P, Ferrari N, Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352:152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 32.Dery KJ, Gaur S, Gencheva M, Yen Y, Shively JE, Gaur RK. Mechanistic control of carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM1) splice isoforms by the heterogeneous nuclear ribonuclear proteins hnRNP L, hnRNP A1, and hnRNP M. J Biol Chem. 2011;286:16039–16051. doi: 10.1074/jbc.M110.204057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garayoa M, Man YG, Martínez A, Cuttitta F, Mulshine JL. Downregulation of hnRNP A2/B1 expression in tumor cells under prolonged hypoxia. Am J Respir Cell Mol Biol. 2003;28:80–85. doi: 10.1165/rcmb.4880. [DOI] [PubMed] [Google Scholar]
- 34.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang HY, Dou KF. PCBP1 is an important mediator of TGF-β-induced epithelial to mesenchymal transition in gall bladder cancer cell line GBC-SD. Mol Biol Rep. 2014;41:5519–5524. doi: 10.1007/s11033-014-3428-7. [DOI] [PubMed] [Google Scholar]
- 36.Chen ZY, Cai L, Zhu J, Chen M, Chen J, Li ZH, Liu XD, Wang SG, Bie P, Jiang P, et al. Fyn requires HnRNPA2B1 and Sam68 to synergistically regulate apoptosis in pancreatic cancer. Carcinogenesis. 2011;32:1419–1426. doi: 10.1093/carcin/bgr088. [DOI] [PubMed] [Google Scholar]
- 37.Meng Q, Rayala SK, Gururaj AE, Talukder AH, O'Malley BW, Kumar R. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci USA. 2007;104:5866–5871. doi: 10.1073/pnas.0701065104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lian WX, Yin RH, Kong XZ, Zhang T, Huang XH, Zheng WW, Yang Y, Zhan YQ, Xu WX, Yu M, et al. THAP11, a novel binding protein of PCBP1, negatively regulates CD44 alternative splicing and cell invasion in a human hepatoma cell line. FEBS Lett. 2012;586:1431–1438. doi: 10.1016/j.febslet.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Hodson MJ, Hudson AJ, Cherny D, Eperon IC. The transition in spliceosome assembly from complex E to complex A purges surplus U1 snRNPs from alternative splice sites. Nucleic Acids Res. 2012;40:6850–6862. doi: 10.1093/nar/gks322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie DB, Schellenberg MJ, MacMillan AM. Spliceosome structure: Piece by piece. Biochim Biophys Acta. 2009;1789:624–633. doi: 10.1016/j.bbagrm.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Shcherbakova I, Hoskins AA, Friedman LJ, Serebrov V, Corrêa IR, Jr, Xu MQ, Gelles J, Moore MJ. Alternative spliceosome assembly pathways revealed by single-molecule fluorescence microscopy. Cell Rep. 2013;5:151–165. doi: 10.1016/j.celrep.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Feltz C, Anthony K, Brilot A, Pomeranz Krummel DA. Architecture of the spliceosome. Biochemistry. 2012;51:3321–3333. doi: 10.1021/bi201215r. [DOI] [PubMed] [Google Scholar]
- 43.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:3. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svineng G, Fässler R, Johansson S. Identification of beta1C-2 a novel variant of the integrin beta1 subunit generated by utilization of an alternative splice acceptor site in exon C. Biochem J. 1998;330:1255–1263. doi: 10.1042/bj3301255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manzotti M, Dell'Orto P, Maisonneuve P, Fornaro M, Languino LR, Viale G. Down-regulation of β1C integrin in breast carcinomas correlates with high proliferative fraction, high histological grade, and larger size. Am J Pathol. 2000;156:169–174. doi: 10.1016/S0002-9440(10)64716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meredith JE, Jr, Kiosses WB, Takada Y, Schwartz MA. Mutational analysis of cell cycle inhibition by integrin beta1C. J Biol Chem. 1999;274:8111–8116. doi: 10.1074/jbc.274.12.8111. [DOI] [PubMed] [Google Scholar]
- 48.Moro L, Greco M, Ditonno P, Battaglia M, Marra E, Perlino E. Transcriptional regulation of the beta1C integrin splice variant in human prostate adenocarcinoma. Int J Oncol. 2003;23:1601–1606. [PubMed] [Google Scholar]
- 49.Perlino E, Lovecchio M, Vacca RA, Fornaro M, Moro L, Ditonno P, Battaglia M, Selvaggi FP, Mastropasqua MG, Bufo P, et al. Regulation of mRNA and protein levels of beta1 integrin variants in human prostate carcinoma. Am J Pathol. 2000;157:1727–1734. doi: 10.1016/S0002-9440(10)64809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito YD, Jensen AR, Salgia R, Posadas EM. Fyn: A novel molecular target in cancer. Cancer. 2010;116:1629–1637. doi: 10.1002/cncr.24879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mattioli C, Pianigiani G, Pagani F. Cross talk between spliceosome and microprocessor defines the fate of pre-mRNA. Wiley Interdiscip Rev RNA. 2014;5:647–658. doi: 10.1002/wrna.1236. [DOI] [PubMed] [Google Scholar]
- 53.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: Past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 55.Eisenreich A, Boltzen U, Poller W, Schultheiss HP, Rauch U. Effects of the Cdc2-like kinase-family and DNA topoisomerase I on the alternative splicing of eNOS in TNF-alpha-stimulated human endothelial cells. Biol Chem. 2008;389:1333–1338. doi: 10.1515/BC.2008.152. [DOI] [PubMed] [Google Scholar]
- 56.Eisenreich A, Bogdanov VY, Zakrzewicz A, Pries A, Antoniak S, Poller W, Schultheiss HP, Rauch U. Cdc2-like kinases and DNA topoisomerase I regulate alternative splicing of tissue factor in human endothelial cells. Circ Res. 2009;104:589–599. doi: 10.1161/CIRCRESAHA.108.183905. [DOI] [PubMed] [Google Scholar]
- 57.Eisenreich A, Zakrzewicz A, Huber K, Thierbach H, Pepke W, Goldin-Lang P, Schultheiss HP, Pries A, Rauch U. Regulation of pro-angiogenic tissue factor expression in hypoxia-induced human lung cancer cells. Oncol Rep. 2013;30:462–470. doi: 10.3892/or.2013.2413. [DOI] [PubMed] [Google Scholar]
- 58.Howard JM, Sanford JR. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA. 2015;6:93–110. doi: 10.1002/wrna.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, Biamonti G. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 2013;41:8665–8679. doi: 10.1093/nar/gkt579. [DOI] [PMC free article] [PubMed] [Google Scholar]