Abstract
Sorafenib is currently used to treat advanced and/or unresectable hepatocellular carcinoma (HCC), but the increase of the median survival was only 3 months. Moreover, sorafenib has severe side effects and patients develop resistance quickly. Epigenetic alterations such as DNA methylation play a decisive role in the development and progression of HCC. To our knowledge, there are no studies that analysed the global DNA methylation changes in HCC cells treated with sorafenib. Using MeDip-chip technologies, we found 1230 differentially methylated genes in HA22T/VGH cells treated with sorafenib compared to untreated cells. Gene ontology and pathway analysis allowed identifying several enriched signaling pathways involved in tumorigenesis and cancer progression. Among the genes differentially methylated we found genes related to apoptosis, angiogenesis and invasion, and genes belonging to pathways known to be deregulated in HCC such as RAF/MEK/ERK, JAK-STAT, PI3K/AKT/mTOR and NF-κB. Generally, we found that oncogenes tended to be hypermethylated and the tumor suppressor genes tended to be hypomethylated after sorafenib treatment. Finally, we validated MeDip-chip results for several genes found differentially methylated such as BIRC3, FOXO3, MAPK3, SMAD2 and TSC2, using both COBRA assay and direct bisulfite sequencing and we evaluated their mRNA expression. Our findings suggest that sorafenib could affect the methylation level of genes associated to cancer-related processes and pathways in HCC cells, some of which have been previously described to be directly targeted by sorafenib.
Keywords: sorafenib, hepatocellular carcinoma, epigenomics, DNA methylation, HA22T/VGH, MeDip-chip, human promoter 1.0R array, gene ontology, COBRA assay, direct bisulfite sequencing
Introduction
Hepatocellular carcinoma (HCC) is the most frequent primary malignancy of the liver. Globally, it is the fifth most common cancer in men and seventh among women. There are over half a million new HCC cases diagnosed annually worldwide, with Mediterranean countries such as Italy, Spain, and Greece that have intermediate incidence rates of 10-20 per 100,000 individuals (1). The standard treatment for HCC patients with good prognosis is surgical resection, with the rate of five-year survival of 89-93% (2). However, for the majority of patients with an advanced and/or unresectable HCC the only therapeutic option is the oral multikinase inhibitor sorafenib (3). Sorafenib was developed in 1995. This molecule is a small drug developed by Bayer and Onyx that is able to inhibit strongly the activity of CRAF that was later named sorafenib. Besides affecting CRAF, sorafenib also inhibits BRAF, VEGFR-1, -2 and -3, PDGFR-β, FGFR1, c-Kit, Flt-3 and RET (4). Sorafenib reduces proliferation by inhibiting the activity of RAF in the MAPK pathway, induces apoptosis by blocking elF4E phosphorylation and the initiation of Mcl-1 translation, and reduces the tumor microvasculature targeting PDGFR-β and VEGFR-2/3 (5). In 2008, the SHARP clinical trial showed that the median overall survival increased from 7.9 months to 10.7 months in the group treated with sorafenib compared to placebo (6). Although HCC patients treated with sorafenib have an increase in survival time, sorafenib still presented very low rate of tumor response, suggesting the existence of some acquired mechanism of primary resistance. Some studies have suggested several mechanisms underlying the onset of sorafenib resistance in HCC such as the up regulation of Akt, the increased expression of EGFR and HER-3, the activation of epithelial-mesenchymal transition (EMT) process and cell autophagy (7).
It is well known that hepatocarcinogenesis is a complex process, not only favoured by the accumulation of genetic alterations, but also by epigenetic alterations. In fact, there are many studies indicating the importance of epigenetic processes that have largely changed the view of HCC. These studies indicated that HCC presents many alterations of its epigenome such as global genomic DNA hypomethylation, gene-specific DNA hyper- or hypomethylation, abnormal expression of DNA methyltransferases and histone modifying enzymes, altered histone modification patterns, and aberrant expression of microRNAs and long non-coding RNA (8).
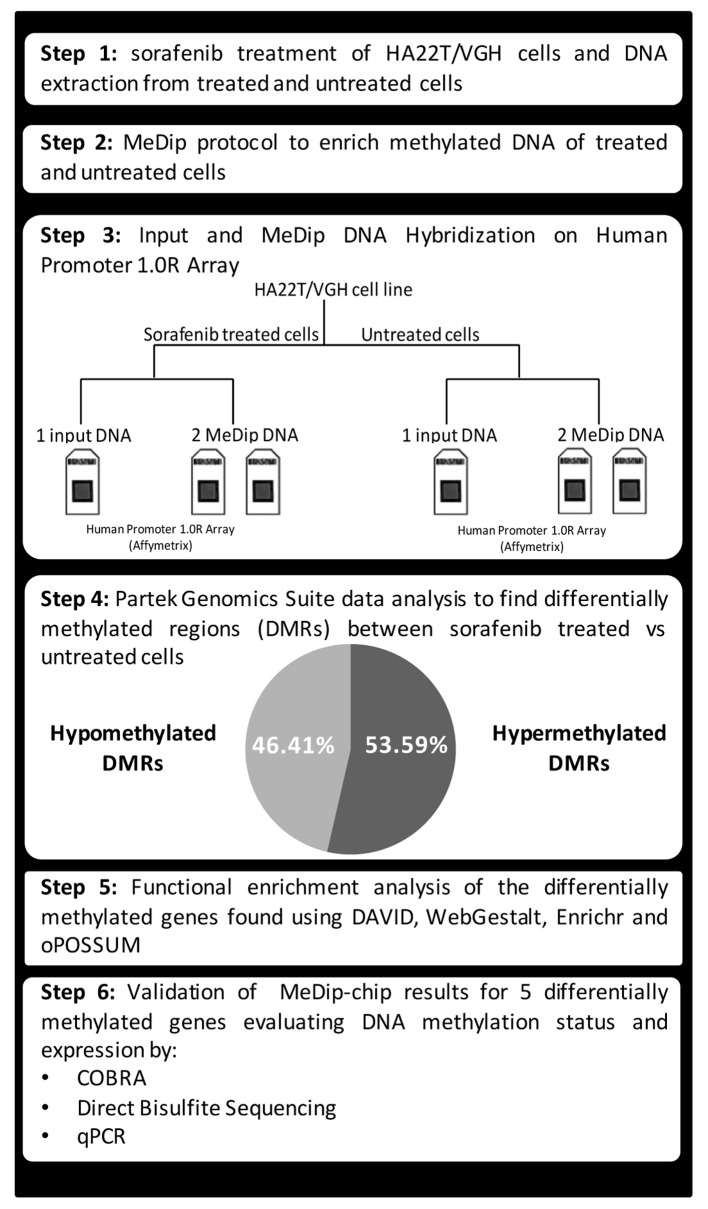
To our knowledge there are few studies in the literature regarding the epigenetic effects of sorafenib in cancer cells. In one study, A549 adenocarcinoma alveolar cancer epithelial cells were treated simultaneously with TGF-β1 and sorafenib and global changes to histone modification were determined. The authors demonstrated that sorafenib largely reversed the changes in histone modifications that occurred during EMT in A549 cells (9). In another study, it was demonstrated that the antiproliferative effect of sorafenib on HCC cells could be due to the degradation of the histone methyltransferase EZH2 promoted by sorafenib through the way of proteasome. Overexpression of EZH2 reduced the effects of sorafenib, while the siRNA against EZH2 increased the effect of the drug. It is assumed, therefore, that the epigenetic modifications mediated by EZH2 may be another molecular mechanism crucial for the acquisition of resistance to sorafenib (10). Furthermore, the interaction of sorafenib with microRNAs has been demonstrated elsewhere (11). In our previous study, we found that the combined treatment of HA22T/VGH HCC cells with miR-193a and sorafenib led to further inhibition of proliferation compared to sorafenib treatment alone (11,12). However, DNA methylation has not been studied on global level in HCC cells treated with sorafenib. Herein, we evaluated the changes on DNA methylation profile in HCC cells treated with sorafenib. In particular, we used MeDip-chip technology to profile the DNA methylation variation in HA22T/VGH cells treated with sorafenib compared to untreated cells. The genes found differentially methylated by bioinformatics tools were then subjected to functional enrichment analysis and we found an enrichment of many gene ontology terms and molecular signalling pathways likely promoted by sorafenib treatment. Finally, we validated MeDip-chip results for several genes using both COBRA assay and direct bisulfite sequencing and then we evaluated their mRNA expression (Fig. 1).
Figure 1.
Schematic flow chart of the experimental design followed.
Materials and methods
Cell culture, sorafenib treatment and DNA and RNA extraction
HA22T⁄VGH undifferentiated cells, kindly provided by N. D'Alessandro (University of Palermo, Palermo, Italy), were maintained in RPMI-1640 (Life Technologies) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 incubator. SKHep1Clone3 (SKHep1C3), selected from human HCC-derived cells (SKHep1: ATCC HTB-52), was maintained in Earle's MEM (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies) at 37°C in a 5% CO2 incubator. Sorafenib (Nexavar®, BAY43-9006) was synthesized at Bayer Corporation and was dissolved in 100% DMSO (Sigma-Aldrich) and diluted in RPMI-1640 (Life Technologies) to the required concentration with a final DMSO concentration of 0.1%. Cells were cultured in 10 cm Ø plates up to almost confluence and were treated with sorafenib (5 µM) or solvent (0.1% DMSO) for 24 h. Genomic DNA and total RNA were extracted from treated and untreated cells using Wizard Genomic DNA purification kit (Promega) and TRIzol reagent (Life Technologies), respectively, according to the manufacturer's instructions, and the nucleic acid quantifications were performed using NanoDrop (Thermo Fisher Scientific).
MeDip-chip
The global DNA methylation profiles of cells treated with sorafenib or left untreated were obtained by methylated DNA immunoprecipitation coupled with Affymetrix Human Promoter 1.0R Tiling Arrays (MeDip-chip) using a modification of the Affymetrix chromatin immunoprecipitation assay protocol. Genomic DNA from sorafenib treated and untreated cells was fragmented using micrococcal nuclease (New England Biolabs) to obtain DNA fragments ranging from 200 to 500 nucleotides and used them as input (IN) DNA. Agilent Bioanalyzer with the RNA 6000 Nano LabChip kit was used to check the size of fragmented DNA. Part of IN DNA (4 µg) was immunoprecipitated (IP) with 10 µl of anti-5-methylcytosine Antibody (Eurogentec, BI-MECY-0100) using the MeDip protocol (13), with minor modifications. The antibody-DNA complexes were immunoprecipitated using Dynabeads® Protein G immunoprecipitation kit (Life Technologies) and the methylated enriched DNA was purified by standard phenol/chloroform procedure and precipitated with isopropanol. The MeDip was performed in duplicate for each DNA sample starting with initial fragmentation step. A total of 200 ng of IN and IP DNA was amplified with the Affymetrix Chromatin Immunoprecipitation Assay Protocol. Hybridization on Human Promoter 1.0R array was performed using the reagents contained in GeneChip® Hybridization, Wash, and Stain kit (Affymetrix) using GeneChip hybridization oven 640 (Affymetrix). Array washing and staining was performed with the use of the Fluidics Station 450 (Affymetrix). Arrays were scanned with the GeneChip Scanner 3000 7G (Affymetrix) and raw data were extracted with GeneChip Operating System (GCOS) software from Affymetrix. In total 3 arrays per each cellular condition (2 IP DNA and 1 IN DNA) were performed. IP and IN DNA data have been deposited in the National Center for Biotechnology Information (NCBI) Gene 97 Expression Omnibus (GEO) data repository, and are accessible (GSE72257).
Data analysis
The raw data were imported into Partek Genomics Suite (PGS) software version 6.6, normalized with the RMA algorithm and converted to log2 values. Hierarchical clustering and principal component analysis were performed on the raw data to assess the quality and reproducibility of the sample. For each cellular condition, the two IP CEL files were normalized through subtraction of log2 IN signal intensity of each of 4.6 million array probes to log2 IP signal. The differences between sorafenib-treated and untreated cells were analyzed using ANOVA. Significant differentially methylated regions (DMR) were obtained using the MAT algorithm (model-based analysis of tiling arrays algorithm) (14) with the following parameters: P-value threshold <0.001, sliding window = 2,000 bp, minimum number of probes in a region = 10. DMR with a positive or negative MAT score were considered hypermethylated or hypomethylated, respectively, in sorafenib-treated cells compared to untreated cells. Using PGS function, each DMR was associated to its nearest gene of the human genome. The genomic coordinates of each DMR were calculated based on UCSC human genomic assembly version 18. The ratio between CpG observed and CpG expected for each DMR was calculated using the following formula: Observed/expected CpG = number of CpG x (DMR length) / (number of C x number of G). In order to study the effect of DNA methylation changes on promoters and gene bodies only DMR between −5,000 bp from TSS and +5,000 bp from the end of the annotated gene were included for the next analyses. For functional enrichment analysis of gene ontology terms and molecular signaling pathway the following tools were used: DAVID (https://david.ncifcrf.gov/), WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/), Enrichr (http://amp.pharm.mssm.edu/Enrichr/) and oPOSSUM 3.0 (http://opossum.cisreg.ca/oPOSSUM3/). A more stringent cut-off (P-value <0.0008) associated to DMR was applied to remove non-relevant genes and reduced the list of DMR and corresponding genes used for performing the following functional enrichment analyses.
COBRA and direct bisulfite sequencing quantification of CpG methylation
In order to validate the DNA methylation status in a set of genomic regions detected as hypermethylated (positive MAT score) or hypomethylated (negative MAT score) by the MeDip-chip profiling we performed combined bisulfite restriction analysis (COBRA) and direct bisulfite sequencing for quantitative DNA methylation analysis, and using the original DNA samples for sorafenib treated and untreated cells that were used in the MeDip-chip experiment. The gene specific bisulfite primers were designed to overlap five MAT detected DMR using the MethPrimer Software (BIRC3: forward, 5′-TTAGTTTGTTTGGTTGTGAAGAGAA-3′, reverse, 5′-ACTCCCTAACCCATACCCTATACAC-3′; FOXO3 forward, 5′-GGGTTGTTTTTTGAGGATT-3′, reverse, 5′-ACCAACCCCTTACCCTAAA-3′; MAPK3 forward, 5′-TTAGTTTTTTAG GAGGTTAAGGTGG-3′, reverse, 5′-CCACAAACTATAAACAAAAAATCTCC-3′; SMAD2 forward, 5′-GGGGTTTGTAGGGGTTTGT-3′, reverse, 5′-TCCCAACCATTAAAAAACATC-3′; TSC2 forward, 5′-GTTTTTGTGTTATGATTTTGGGAA-3′, reverse, 5′-ACATCCATCCACTACAAAACAAAAT-3′). Bisulfite treatment of genomic DNA was performed using EZ DNA Methylation kit (Zymo Research). For COBRA analysis the restriction enzymes TaqI and HpyCH4IV (New England Biolabs) were used to examine the methylation status of the amplified regions. In brief, 100–150 ng of the amplified products were digested with the restriction enzymes, which digest methylated DNA but not unmethylated DNA. The digested DNA was electrophoresed on 3% agarose gels in 1X TBE, and visualized by ethidium bromide staining. We used a UV image analyzer to quantify the integrated optical density (IOD) of the bands. Methylation level (%) for a specific cytosine was defined as the sum of the IOD of the shifted bands divided by the IOD of all bands in each lane. All experiments were repeated twice to assess the reproducibility of results.
For direct bisulfite sequencing analysis the amplified products obtained previously were purified and sequenced on an Applied Biosystems 3130xl Genetic Analyzer. The sequencing reactions were performed using Bigdye Terminator v1.1 Cycle Sequencing kit (Life Technologies) and purified with Performa Ultra 96-Well Plate (EdgeBio, Gaithersburg, MD, USA). The abi files were analyzed with Accelrys DS Gene 1.5 (Accelrys). Since bisulfite treatment does not convert methylated cytosine to uracil, the cytosine chromatogram peaks at each CpG location will correspond to the degree of methylation at that site within a given sample. Likewise, the thymine chromatogram peaks will correspond to the degree of cytosines that were not methylated. Furthermore, the relative size of these peaks will be proportionally related to the total percentage of cytosine bases that were methylated. Therefore, according to Parrish et al (15), methylation levels for each CpG site within the DNA amplicon can be quantified by measuring the ratio between peak height values of cytosine (C) and thymine (T), yielding the basic equation for the methylation percentage to be [C/(C+T) ×100]. If the reverse primer was used, the guanine (G) and adenine (A) peak heights should be used instead, yielding the equation [G/(G+A) ×100]. Each amplicon was sequenced in triplicate to assess the reproducibility of results.
RT-qPCR expression analysis
In order to investigate the relationship between DNA methylation changes and gene expression in sorafenib-treated and untreated cells (HA22T/VGH and SKHep1C3), RT-qPCR was performed to determine the gene expression of several genes whose DNA methylation status was validated previously with COBRA and direct bisulfite sequencing. An amount of 1 µg of total RNA was retrotranscribed using M-MLV-RT enzyme (Life Technologies) and pre-designed primers used for qPCR validation were purchased from Bio-Rad (Hercules, CA, USA). GoTaq qPCR Master Mix was supplied by Promega.
The mRNA expression of BIRC3, FOXO3, MAPK3, SMAD2 and TSC2 genes was evaluated by qPCR using PrimePCR™ SYBR® Green Assay (Bio-Rad, BIRC3: qHsaCID0013381; FOXO3: qHsaCID0023235; MAPK3: qHsaCID0010939; SMAD2: qHsaCID0022031; TSC2: qHsaCID0018594; YWHAZ: qHsaCID0013897). The relative quantification of the expression in sorafenib-treated HCC cells compared to untreated cells was based on ΔΔCt method. YWHAZ was used as housekeeping gene and untreated cells were used as a reference sample. We chose YWHAZ because it has been shown to have a constant expression during the process of apoptosis in tumor cell lines (16). qPCR experiment was performed twice. For each qPCR reaction, we used 20 ng of cDNA. Each gene in the two cellular conditions was assayed in triplicate. After qPCR reaction, denaturation curve was performed to ensure the specificity of the primers.
Results
Detection of global DNA methylation changes in HCC sorafenib-treated cells by using Human Promoter array technology
We previously demonstrated that HA22T/VGH cells were sensitive to sorafenib (11). To assess whether these undifferentiated HCC cells treated with sorafenib displayed genome wide changes in DNA methylation, we performed methylated DNA immunoprecipitation (MeDip) followed by hybridization on tiling array. We used MeDip in combination with Human Promoter 1.0 Tiling array platform to identify changes of hypo- and hypermethylation with greater sensitivity in HA22T/VGH cells treated with sorafenib in comparison to cells treated with vehicle (DMSO) only.
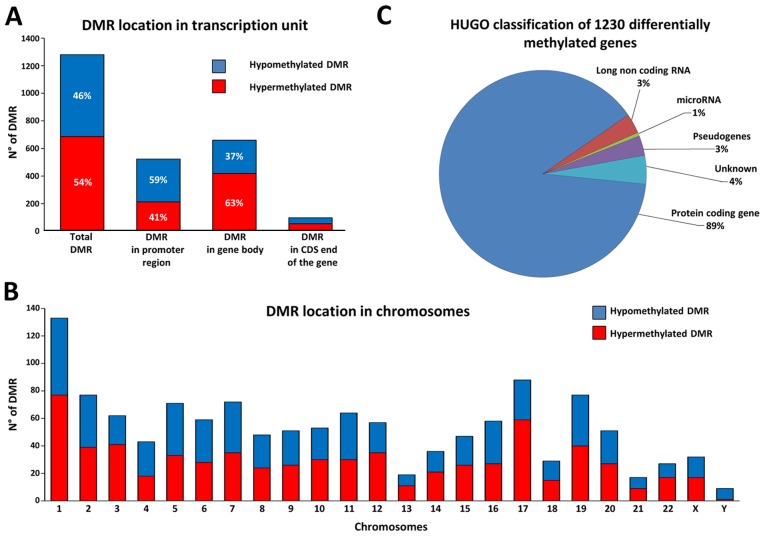
We detected the differentially methylated regions (DMR) in HA22T/VGH treated with sorafenib using untreated cells as baseline. Significant DMR were obtained by PGS using ANOVA and MAT algorithm with specific parameters (see Materials and methods). We considered only DMR between −5,000 bp from the TSS and +5,000 bp from the end of the coding DNA sequence (CDS) of the genes. From the comparison, we obtained 1,280 DMR corresponding to 1,230 distinct annotated genes, with 46.4% of hypomethylated DMR and 53.6% of hypermethylated DMR (Fig. 2A). To have a general view of the post-sorafenib genomic landscape we considered the distribution of DMR respect to TSS and CpG density, and the frequency of DMR in the different chromosomes.
Figure 2.
Differentially methylated regions (DMR) and associated genes in HA22T/VGH cells treated with sorafenib. Distribution of the 1,280 DMR based on the position respect the TSS of the gene (A) and chromosomes (B). (C) The graph shows the class of the genes catalogued with online repository of HGNC (HUGO gene nomenclature committee) and the corresponding percentage.
Considering the position of DMR regarding TSS we found 40.9% of DMR localized in promoter region, 51.6% in gene body and 7.6% in CDS end of the gene. In particular, DMR in promoter regions presented more hypomethylated DMR, whereas DMR in gene bodies presented the opposite trend with more hypermethylated DMR (Fig. 2A). Calculating the CpG density for each DMR with the observed to expected CpG ratio (obs/exp CpG) formula we found that 22.8% of DMR have an obs/exp CpG >60%, 28.4% of DMR an obs/exp CpG between 60–30%, and 48.8% of DMR with an obs/exp CpG <30% (Table I). In particular, we found that the DMR with an obs/exp CpG >60% were almost exclusively subjected to a hypomethylation phenomenon, whereas the DMR with an obs/exp CpG <30% were preferentially subjected to a hypermethylation phenomenon. This trend was repeated regardless whether the DMR was in the promoter region, in the gene body or in the CDS end of the gene (Table I). Then, observing the frequency of distribution in the different chromosomes, we found that the 1,280 DMR were distributed quite uniformly along the 24 chromosomes according with their gene density (Fig. 2B). However, some chromosomes showed more hypermethylated DMR (chr 1, chr 12, chr 17 and chr 22) whereas others showed more hypomethylated DMR (chr 4 and chr Y). Finally, to have a general view of the gene classes involved we catalogued the 1,230 differentially methylated genes by using the online repository of HGNC (HUGO gene nomenclature committee), and we found that the majority of DMR were close to a protein coding gene (Fig. 2C).
Table I.
Distribution of the 1,280 DMR based on CpG density.
| DMR | Observed to expected CpG ratio (Obs/Exp CpG)a | No. of DMR | Hypermethylated DMR (%) | Hypomethylated DMR (%) |
|---|---|---|---|---|
| Total DMR | Obs/Exp CpG ≥60% | 292 | 2.7 | 97.3 |
| 60% < Obs/Exp CpG <30% | 364 | 55.5 | 44.5 | |
| Obs/Exp CpG ≤30% | 624 | 76.3 | 23.7 | |
| DMR in promoter region | Obs/Exp CpG ≥60% | 215 | 1.9 | 98.1 |
| 60% < Obs/Exp CpG <30% | 114 | 50.9 | 49.1 | |
| Obs/Exp CpG ≤30% | 194 | 77.8 | 22.2 | |
| DMR in gene body | Obs/Exp CpG ≥60% | 60 | 6.7 | 93.3 |
| 60% < Obs/Exp CpG <30% | 223 | 58.7 | 41.3 | |
| Obs/Exp CpG ≤30% | 377 | 75.3 | 24.7 | |
| DMR in CDS end of the gene | Obs/Exp CpG ≥60% | 17 | 0 | 100 |
| 60% < Obs/Exp CpG <30% | 27 | 48.1 | 51.9 | |
| Obs/Exp CpG ≤30% | 53 | 77.4 | 22.6 |
Observed to expected CpG ratio = number of CpG x (DMR length) / (number of C x number of G).
Functional enrichment analysis of the 1,230 genes found differentially methylated in the sorafenib-treated cells
In order to assign a biological meaning to the 1,230 genes found differentially methylated in sorafenib-treated cells compared to untreated cells in MeDip-chip analysis, we conducted a functional enrichment analysis using different online tools.
DAVID identified 18 enriched annotation terms associated to our gene list. In particular, five BIOCARTA signaling pathways were related to receptor tyrosine kinase (RTK) signal transduction (Table II). Of note were the changes in DNA methylation in the genes of these pathways. For example, among the 10 genes found differentially methylated in the EGF pathway, 7 oncogenes (OG) were hypermethylated and 2 tumor suppressor genes (TSG) were hypomethylated (Table III). This occurred in a similar way also in the other 4 pathways. The results of this analysis may indicate that sorafenib modified the level of DNA methylation of HA22T/VGH cells in key genes of RTK signal transduction pathways on which sorafenib acts directly.
Table II.
Molecular signaling pathways found enriched by DAVID.
| Category | Term | Count | P-value | Adjusted P-value Benjamini | Genes |
|---|---|---|---|---|---|
| BIOCARTA | EGF signaling pathway | 10 | 1.02E-05 | 2.06E-03 | CSNK2A3, FOS, HRAS, JAK1, MAPK3, MAPK8, PIK3CA, SHC1, STAT3, STAT5A |
| BIOCARTA | PDGF signaling pathway | 9 | 4.57E-05 | 4.58E-03 | CSNK2A3, FOS, HRAS, JAK1, MAPK3, MAPK8, SHC1, STAT3, STAT5A |
| BIOCARTA | IL 2 signaling pathway | 8 | 1.42E-04 | 9.49E-03 | CSNK2A3, FOS, HRAS, JAK1, MAPK3, MAPK8, SHC1, STAT5A |
| BIOCARTA | EPO signaling pathway | 7 | 4.44E-04 | 2.21E-02 | CSNK2A3, FOS, HRAS, MAPK3, MAPK8, SHC1, STAT5A |
| BIOCARTA | TPO signaling pathway | 7 | 8.39E-04 | 3.32E-02 | CSNK2A3, FOS, HRAS, MAPK3, MAPK8, SHC1, STAT5A |
The hypomethylated genes (negative MAT-score) are in bold.
Table III.
Genes of EGF signaling pathway found differently methylated.
| Gene | MAT score | OG/TSG |
|---|---|---|
| HRAS | −9.91 | OG |
| MAPK8 (JNK1) | −9.62 | TSG |
| SHC1 | −9.34 | TSG/OG |
| STAT3 | 9.84 | OG |
| STAT5A | 9.94 | OG |
| FOS | 10.04 | OG |
| JAK1 | 10.18 | OG |
| PIK3CA | 10.32 | OG |
| CSNK2A3 | 11.02 | – |
| MAPK3 (ERK) | 12.42 | OG |
OG/TSG = evidence that a given gene acts as oncogene (OG), tumor suppressor gene (TSG) or neither (−) according with literature data.
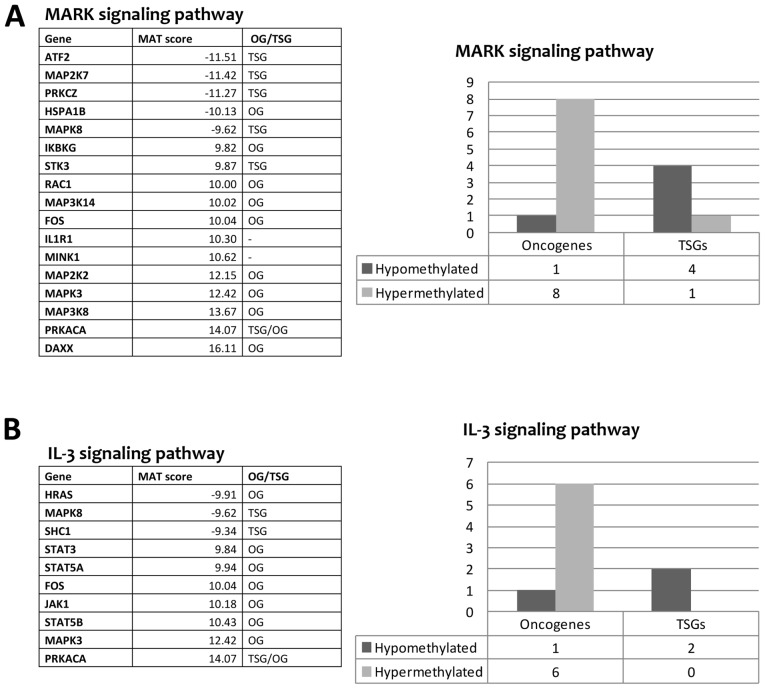
Gene ontology analysis performed by WebGestalt found 16 terms significantly enriched. In particular, the following 3 terms had a lower level of abstraction and were more specific: 'Sequence-specific DNA binding RNA polymerase II transcription factor activity', 'Structure-specific DNA binding' and 'Double-stranded DNA binding'. The first term was related to transcription factors that bind the RNA polymerase II (29 genes). The second and third term (son of the second term) are related to transcription factors that bind directly to DNA (24 and 20 genes, respectively). Among these genes, we found several transcription factors relevant in tumorigenesis and/or cancer progression such as FOXO3, STAT3 and STAT5A that displayed a change in DNA methylation level following sorafenib treatment. Pathway analysis performed by WebGestalt found 10 Wikipathways in our list of genes (Table IV). The genes that occur most often in these pathways were ATF2, CDKN1A, FOS, FOXO3, HRAS, IKBKG, JAK1, MAP2K2, MAP2K7, MAP3K14, MAP3K8, MAPK3, MAPK8, PIK3CA, PRKCZ, RAC1, SHC1, SHC2, STAT3, STAT5A, and STAT5B. In the literature, there is evidence that these genes have a role in tumorigenesis and/or cancer progression. We found a strong tendency of OG to be hypermethylated and TSG to be hypomethylated after sorafenib treatment (Fig. 3). This may indicate that sorafenib could trigger the hypermethylation of several OG and the hypomethylation of other TSG as downstream effect. In addition, the pathway analysis revealed that many genes belonging to MAPK cascade and JAK-STAT signaling pathway (IL-2/3/5 signaling pathways), known to be involved in sorafenib mechanism of action, had a change in the level of methylation after sorafenib treatment (Fig. 3). The MAPK signaling pathway showed a MEK gene (MAP2K2) and an ERK gene (MAPK3) hypermethylated after sorafenib treatment. In contrast, genes belonging to the JNK signaling pathway (MAP2K7, MAPK8) were hypomethylated indicating a possible activation of this pathway (Fig. 3A). Regarding the JAK-STAT pathway, the genes JAK1, STAT3, STAT5A and STAT5B were hypermethylated with probable inhibition of this pathway (Fig. 3B).
Table IV.
Molecular signaling pathways found enriched by WebGestalt tool.
| Category | Molecular signaling pathway | Genes count | Raw P-value | Adjusted P-value | Genes |
|---|---|---|---|---|---|
| Wikipathway | Insulin signaling pathway | 20 | 4.65E-08 | 6.88E-06 | FOS, FOXO3, GRB10, HRAS, MAP2K2, MAP2K7, MAP3K14, MAP3K8, MAPK3, MAPK8, MINK1, PIK3CA, PRKAA2, PRKCZ, RAC1, RPS6KA4, SHC1, SHC2, TRIB3, TSC2 |
| Wikipathway | Kit receptor signaling pathway | 12 | 3.16E-07 | 1.65E-05 | FOS, FOXO3, GRB10, HRAS, MAP2K2, MAPK3, MAPK8, SHC1, STAT3, STAT5A, STAT5B |
| Wikipathway | Prolactin signaling pathway | 14 | 3.35E-07 | 1.65E-05 | FOS, HRAS, JAK1, MAP2K2, MAPK3, MAPK8, PIK3CA, RAC1, SHC1, STAT3, STAT5A, STAT5B, TEC, ZAP70 |
| Wikipathway | AGE-RAGE pathway | 12 | 1.53E-06 | 5.66E-05 | ATF2, LGALS3, MAPK3, MAPK8, MMP7, PRKCZ, RAC1, SHC1, SMAD2, STAT3, STAT5A, STAT5B |
| Wikipathway | IL-2 signaling pathway | 10 | 2.13E-06 | 6.29E-05 | FOS, FOXO3, HRAS, JAK1, MAP2K2, MAPK3, SHC1, STAT3, STAT5A, STAT5B |
| Wikipathway | IL-3 signaling pathway | 10 | 2.55E-06 | 6.29E-05 | FOS, HRAS, JAK1, MAPK3, MAPK8, PRKACA, SHC1, STAT3, STAT5A, STAT5B |
| Wikipathway | MAPK signaling pathway | 17 | 5.33E-06 | 0.0001 | ATF2, DAXX, FOS, HSPA1B, IKBKG, IL1R1, MAP2K2, MAP2K7, MAP3K14, MAP3K8, MAPK3, MAPK8, MINK1, PRKACA, PRKCZ, RAC1, STK3 |
| Wikipathway | IL-5 signaling pathway | 9 | 6.07E-06 | 0.0001 | FOXO3, FOS, JAK1, MAP2K2, MAPK3, SHC1, STAT3, STAT5A, STAT5B |
| Wikipathway | AMPK signaling pathway | 10 | 1.65E-05 | 0.0002 | TSC2, CAB39, CDKN1A, CPT1A, FASN, PFKFB3, PIK3CA, PLCB1, PRKAA2, PRKACG |
| Wikipathway | TCR signaling pathway | 13 | 1.03E-05 | 0.0002 | ATF2, FOS, HRAS, IKBKG, IRF4, MALT1, MAP2K2, MAP3K14, MAP3K8, MAPK3, MAPK8, SHC1, ZAP70 |
Figure 3.
Examples of molecular signaling pathways found enriched by WebGestalt. (A) MAPK signaling pathway. (B) IL-3 signaling pathway. The tables report the genes whose level of methylation was changed after the treatment of HA22T/VGH with sorafenib. Negative and positive MAT-scores indicate a given gene hypomethylated or hypermethylated, respectively. The column 'OG/TSG' indicates the presence of evidence in published reports that a given gene acts as oncogene (OG), tumor suppressor gene (TSG) or neither (−). Histograms indicate the number of OG or TSG that were hypomethylated or hypermethylated in our study.
In summary, among the 1,230 differentially methylated genes identified by functional enrichment analysis, several genes had a role in tumorigenesis and/or cancer progression described in literature. Some of these genes were associated with functions linked to apoptosis, invasion and angiogenesis. Moreover, we found that sorafenib affected the methylation level of genes belonging to important signaling pathways such as RAF/MEK/ERK, JNK, JAK-STAT, PI3K/AKT/mTOR and NF-κB.
Non-coding genes found differentially methylated in sorafenib-treated cells
Among the 1,230 differentially methylated genes, 47 were non-coding genes (Table V) with 16 antisense RNAs, 20 lncRNAs including 15 lincRNAs, 7 microRNAs, three small nucleolar RNAs, and one small nucleolar RNA. In particular, MALAT1, HOTTIP, miR-149 and miR-675 are known to be associated with the tumorigenesis and/or cancer progression.
Table V.
Non-coding genes found differently methylated and associated with the tumorigenesis and/or cancer progression.
| Gene | DMR length | Num. CpG | CpG observed/CpG expected | P-value | MAT score | DMR position | Note |
|---|---|---|---|---|---|---|---|
| ATXN8OS | 2110 | 11 | 0.18 | 3.69E-04 | −10.15 | Gene body | Antisense RNA. Implicated in localization and activity of splicing factors |
| C1RL-AS1 | 3076 | 34 | 0.16 | 5.16E-04 | 10.24 | Gene body | Antisense RNA |
| CACNA1C-AS1 | 2453 | 117 | 0.58 | 1.66E-04 | −11.01 | Promoter region | Antisense RNA |
| CECR5-AS1 | 2036 | 26 | 0.24 | 3.87E-04 | 10.50 | Gene body | Antisense RNA |
| EPHA1-AS1 | 985 | 9 | 0.25 | 5.16E-04 | 10.21 | Gene body | Antisense RNA. Diseases associated with EPHA1-AS1 include Alzheimer's disease |
| FAM41C | 2699 | 53 | 0.36 | 1.47E-04 | −11.08 | Gene body | lncRNA class |
| FAM66D | 2279 | 42 | 0.41 | 2.58E-04 | 11.10 | Gene body | Antisense RNA |
| GATA6-AS1 | 2895 | 188 | 0.88 | 4.06E-04 | −10.07 | Promoter region | Antisense RNA |
| HOTTIP | 1434 | 20 | 0.39 | 2.21E-04 | −10.65 | Promoter region | Antisense RNA. Role in cancer (48) |
| HOXB-AS3 | 4175 | 222 | 0.61 | 1.66E-04 | −10.87 | Gene body | Antisense RNA. Diseases associated with HOXB-AS3 include obesity |
| LINC00029 | 2382 | 105 | 0.47 | 7.01E-04 | −9.49 | Gene body | Long intergenic non-protein coding |
| LINC00174 | 1674 | 34 | 0.42 | 4.24E-04 | −10.00 | Gene body | Long intergenic non-protein coding |
| LINC00261 | 1054 | 9 | 0.19 | 3.69E-05 | −13.06 | Gene body | Long intergenic non-protein coding |
| LINC00261 | 2135 | 104 | 0.66 | 7.01E-04 | −9.41 | Gene body | Long intergenic non-protein coding |
| LINC00277 | 3609 | 52 | 0.28 | 2.58E-04 | 11.03 | Promoter region | Long intergenic non-protein coding |
| LINC00319 | 3036 | 124 | 0.42 | 9.22E-05 | −12.22 | Promoter region | Long intergenic non-protein coding |
| LINC00539 | 2157 | 77 | 0.59 | 3.32E-04 | −10.20 | Gene body | Long intergenic non-protein coding |
| LINC00539 | 964 | 14 | 0.22 | 3.87E-04 | 10.53 | Gene body | Long intergenic non-protein coding |
| LINC00628 | 2253 | 33 | 0.24 | 5.53E-05 | 13.09 | CDS end of the gene | Long intergenic non-protein coding |
| LINC00637 | 2399 | 213 | 0.94 | 7.56E-04 | −9.35 | Gene body | Long intergenic non-protein coding |
| LINC00643 | 679 | 5 | 0.24 | 5.53E-05 | 13.24 | Gene body | Long intergenic non-protein coding |
| LINC00663 | 379 | 7 | 0.45 | 1.84E-05 | −13.75 | CDS end of the gene | Long intergenic non-protein coding |
| LINC00857 | 3166 | 179 | 0.67 | 7.01E-04 | −9.42 | Gene body | Long intergenic non-protein coding |
| LINC00957 | 2385 | 142 | 0.55 | 1.47E-04 | −11.09 | Gene body | Long intergenic non-protein coding |
| LINC00999 | 2549 | 109 | 0.41 | 1.84E-05 | −14.02 | Gene body | Long intergenic non-protein coding |
| LINC01002 | 2343 | 114 | 0.46 | 9.22E-05 | −12.24 | Gene body | Long intergenic non-protein coding |
| LINC01056 | 663 | 45 | 0.68 | 4.06E-04 | −10.10 | Gene body | Long intergenic non-protein coding |
| MALAT1 | 2205 | 73 | 0.51 | 1.84E-05 | 16.14 | CDS end of the gene | IncRNA. Role in cancer (47) |
| MIR149 | 3820 | 308 | 0.64 | 1.29E-04 | −11.22 | Gene body | miRNA. Role in cancer (49) |
| MIR3939 | 1740 | 10 | 0.15 | 4.98E-04 | 10.26 | CDS end of the gene | miRNA |
| MIR4635 | 2310 | 128 | 0.55 | 1.29E-04 | −11.27 | Gene body | miRNA |
| MIR4683 | 2321 | 281 | 0.96 | 4.06E-04 | −10.05 | Promoter region | miRNA |
| MIR5091 | 1362 | 106 | 0.86 | 1.11E-04 | −11.65 | Gene body | miRNA |
| MIR5187 | 2601 | 31 | 0.17 | 4.42E-04 | 10.39 | Gene body | miRNA |
| MIR675 | 3245 | 150 | 0.51 | 7.37E-05 | −12.38 | Promoter region | miRNA. Role in cancer (50) |
| MRPL23-AS1 | 2003 | 85 | 0.42 | 7.74E-04 | −9.28 | Promoter region | Antisense RNA |
| MYLK-AS1 | 2516 | 138 | 0.70 | 1.84E-05 | −14.32 | Gene body | Antisense RNA |
| RNF219-AS1 | 2572 | 108 | 0.64 | 3.69E-04 | −10.14 | Gene body | Antisense RNA. SNP associated with the regulation of blood pressure and alcohol consuming |
| SNHG18 | 2171 | 182 | 0.75 | 7.74E-04 | −9.26 | Gene body | Small nucleolar RNA |
| SNORA51 | 2385 | 52 | 0.33 | 6.82E-04 | 9.97 | Gene body | Small nucleolar RNA |
| SNORD52 | 2303 | 69 | 0.50 | 2.77E-04 | −10.33 | Gene body | Small nucleolar RNA |
| SPANXA2-OT1 | 2060 | 15 | 0.20 | 5.16E-04 | 10.21 | Gene body | Antisense RNA |
| SPATA41 | 2102 | 13 | 0.19 | 1.66E-04 | −11.01 | Promoter region | lncRNA class |
| TOB1-AS1 | 2251 | 209 | 0.83 | 7.74E-04 | −9.31 | Gene body | Antisense RNA |
| TTC28-AS1 | 2534 | 47 | 0.45 | 2.40E-04 | 11.15 | Gene body | Antisense RNA. Associated with obesity |
| ZNRD1-AS1 | 1415 | 8 | 0.11 | 7.74E-04 | 9.83 | Gene body | Antisense RNA. Associated with Graves' disease and systemic lupus erythematosus |
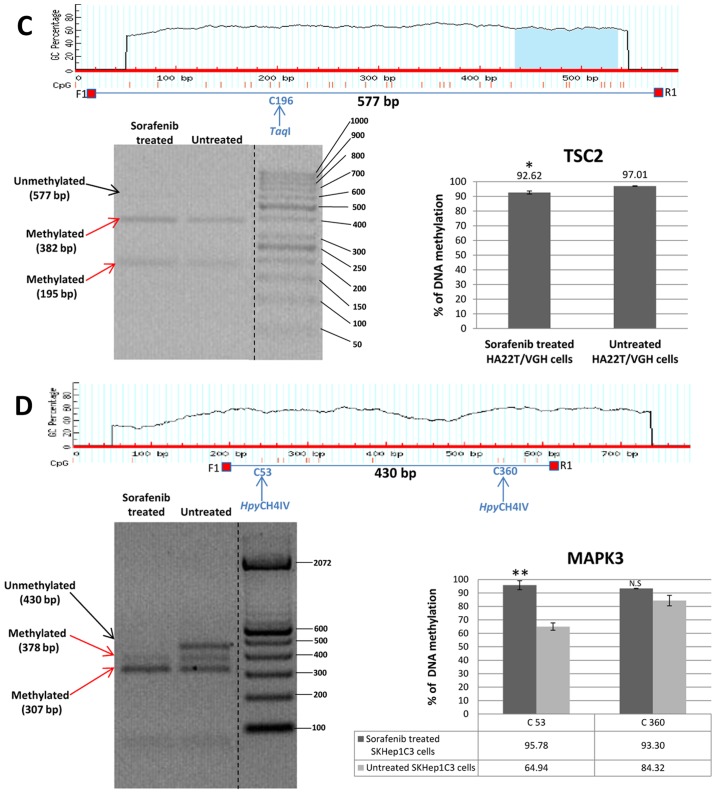
Validation of MeDip-chip results by COBRA assay and direct bisulfite sequencing of the genes BIRC3, FOXO3, MAPK3, SMAD2 and TSC2
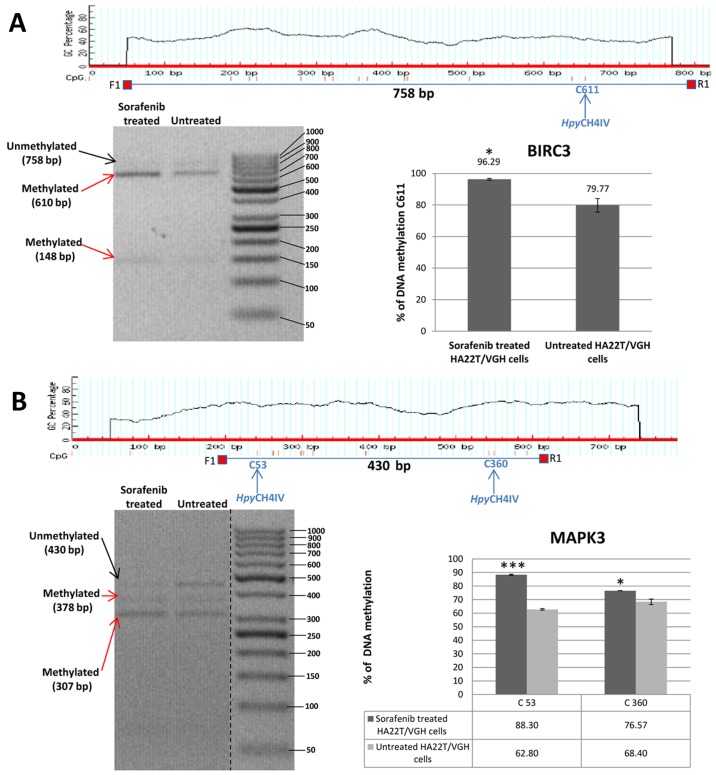
In order to validate the methylation status of the genes found hypomethylated or hypermethylated in MeDip-chip experiment, we performed quantitative analysis of CpGs by COBRA assay and direct bisulfite sequencing. Among the OGs and TSGs found differentially methylated, we chose to validate the following genes: BIRC3, FOXO3, MAPK3, SMAD2 and TSC2 (Table VI). These 5 genes were chosen for their biological significance, for the location of the DMR in the associated gene and for the number of CpGs contained in the DMR. Our aim was to better understand the relationship between DNA methylation in regions with high or low density of CpGs located in different portions of the gene (promoter or intragenic) and gene expression. In HA22T/VGH cells the DMR relative to BIRC3 and MAPK3 were found hypermethylated in sorafenib-treated cells by MeDip-chip experiment. COBRA assay results are in line with these findings and confirmed that the level of DNA methylation in this DMR was higher in sorafenib treated cells compared to the untreated cells (Fig. 4A and B).
Table VI.
Genes validated by quantitative COBRA assay and direct bisulfite sequencing.
| Gene | Chr | DMR start | DMR end | DMR length | No. of CpG in DMR | CpG observed/CpG expected | P-value | MAT score | DMR position |
|---|---|---|---|---|---|---|---|---|---|
| BIRC3 | chr11 | 101693878 | 101695822 | 1944 | 25 | 0.27 | 1.29E-04 | +12.10 | Gene body: Intronic |
| FOXO3 | chr 6 | 108987593 | 108990713 | 3120 | 312 | 0.87 | 2.77E-04 | −10.33 | Promoter region |
| MAPK3 | chr16 | 30037080 | 30039335 | 2255 | 29 | 0.21 | 1.11E-04 | +12.42 | Gene body: Intronic and exonic |
| SMAD2 | chr18 | 43709687 | 43712269 | 2582 | 228 | 0.94 | 1.66E-04 | −10.86 | Promoter region |
| TSC2 | chr16 | 2043747 | 2047150 | 3403 | 121 | 0.40 | 3.69E-05 | −13.04 | Gene body: Intronic and exonic |
Figure 4.
Validation of MeDip-chip results by COBRA assay in sorafenib treated and untreated HCC cells. (A) The DMR associated to BIRC3 gene was 1,944 bp long and the primers were designed to amplify a 758 bp fragment from nucleotides 1,032 to 1,790 relative to the DMR start. The digested bands (610 and 148 bp) indicate the presence of methylation (red arrows). The band at the top (758 bp) corresponds to undigested DNA (black arrow). The histograms indicate the level of methylation in cytosine 611 in sorafenib treated cells and untreated cells by COBRA analysis. The results indicated a hypermethylation of cytosine 611 in sorafenib treated cells. (B) The DMR associated to MAPK3 gene was 2,255 bp long and the primers were designed to amplify a 430 bp fragment from nucleotides 1,660 to 2,090 relative to the DMR start. The digested bands (378 and 307 bp) indicate the presence of methylation (red arrows). The band at the top (430 bp) corresponds to undigested DNA (black arrow). The bands of 52 and 71 bp were not visible on the gel. The histograms indicate the level of methylation in cytosine 53 and cytosine 360 in sorafenib treated cells and untreated cells by COBRA analysis. The results indicated a hypermethylation of cytosine 53 and cytosine 360 in sorafenib treated cells. All the assays were performed using 3.0% gel in 1X TBE. Error bars of the histograms are referred to standard deviation of the means of two experiment performed. *P<0.05, **P<0.01 and ***P<0.001 indicate that expression difference between the two cellular conditions was statistically significant. Results of one representative experiment of two independent experiments are shown. Validation of MeDip-chip results by COBRA assay in sorafenib treated and untreated HCC cells. (C) The DMR associated to TSC2 gene was 3,403 bp long and the primers were designed to amplify a 577 bp fragment from nucleotides 1,107 to 1,684 relative to the DMR start. The digested bands (382 and 195 bp) indicate the presence of methylation (red arrows). The band at the top (577 bp) corresponds to undigested DNA (black arrow). The histograms indicate the level of methylation in cytosine 196 in sorafenib-treated cells and untreated cells by COBRA analysis. The results indicated a slight hypomethylation of cytosine 196 in sorafenib treated cells. All the assays were performed using 3.0% gel in 1X TBE. The COBRA analysis was also conducted in sorafenib treated and untreated SKHep1C3 cells. In SKHep1C3 (D) cells the DMR associated to MAPK3 gene was found hypermethylated in sorafenib treated cells compared to untreated cells. In particular, the results indicated a hypermethylation of cytosine 53 in sorafenib treated cells. All the assays were performed using 3.0% gel in 1X TBE. Error bars of the histograms are referred to standard deviation of the means of two experiment performed. *P<0.05, **P<0.01 and ***P<0.001 indicate that expression difference between the two cellular conditions was statistically significant. N.S means that the results was not statistically significant. Results of one representative experiment of two independent experiments are shown.
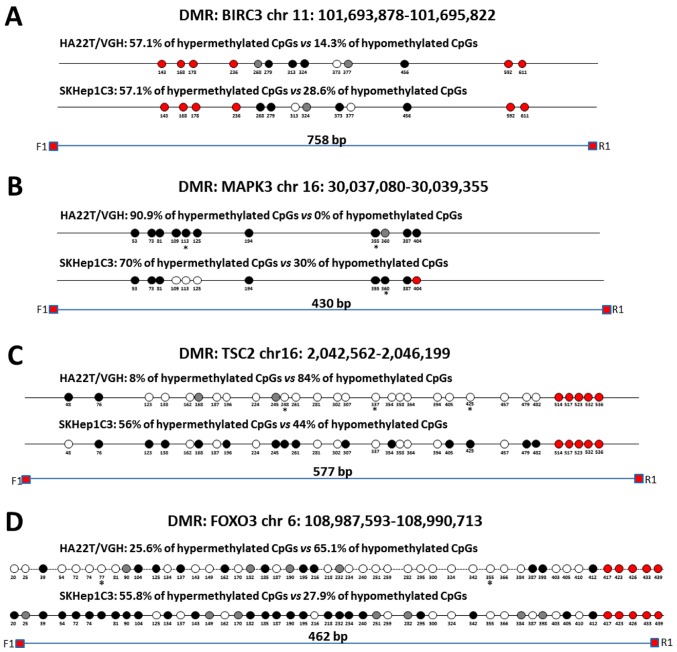
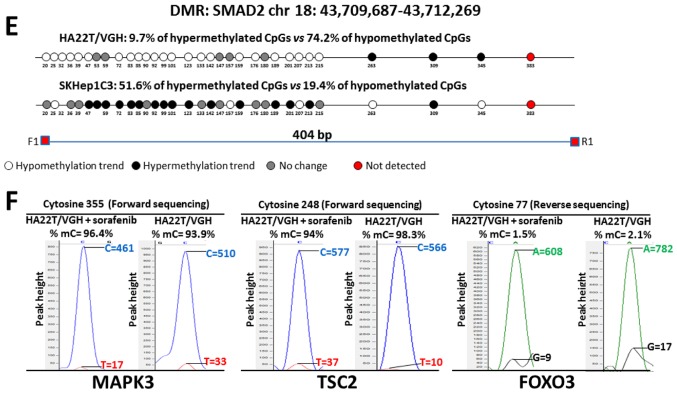
Direct bisulfite sequencing on BIRC3 DMR found that 57.14% of CpGs analysed showed a hypermethylation trend in sorafenib-treated compared to -untreated cells and 14.29% of CpGs showed a hypomethylation trend (Fig. 5A). Direct bisulfite sequencing of MAPK3 DMR displayed that 90.91% of CpGs analysed showed a hypermethylation trend in sorafenib-treated compared to untreated cells, and 0% of CpGs showed a hypomethylation trend (Fig. 5B). The DMR relative to TSC2, FOXO3 and SMAD2 were found hypomethylated in MeDip-chip experiment. COBRA assay carried out on TSC2 DMR confirmed that the level of DNA methylation in this DMR was lower in sorafenib-treated cells than -untreated cells (Fig. 4C). Moreover, direct bisulfite sequencing found that 84% of CpGs analysed showed a hypomethylation trend in sorafenib-treated cells, and 8% of CpGs analysed showed a hypermethylation trend (Fig. 5C and F) confirming the hypomethylation trend of TSC2 gene found by MeDip-chip experiment. For DMR relative to FOXO3 and SMAD2, direct bisulfite sequencing displayed 65.12% of CpGs analysed with a hypomethylation trend in sorafenib-treated cells and 25.58% of CpGs analysed with a hypermethylation trend for FOXO3 DMR (Fig. 5D and F), and for SMAD2 DMR, 74.19% of CpGs analysed showed a hypomethylation trend in sorafenib-treated cells, and 9.68% of CpGs analysed showed a hypermethylation trend (Fig. 5E). Therefore, the assay confirmed the hypomethylation trend of FOXO3, SMAD2 and TSC2 DMRs found by MeDip-chip experiment.
Figure 5.
Validation of MeDip-chip results by direct bisulfite sequencing in sorafenib-treated and -untreated HCC cells. Forward sequencing chromatogram peaks for thymine (unmethylated) and cytosine (methylated) or reverse sequencing chromatogram peaks for adenine (unmethylated) and guanine (methylated) were compared in bisulfite treated DNA to determine the average level of methylation for each CpG within a given sample. The white and black circles indicate that the mean level of CpG methylation in sorafenib-treated cells was lower (hypomethylation) or higher (hypermethylation) respect to untreated cells. Grey circles indicate that the mean level of CpG methylation was the same in the two cellular conditions (difference <0.1% was not considered relevant). Red circles indicate undetected CpGs. *P<0.05 and **P<0.01 indicate that methylation difference between the two cellular conditions was statistically significant. (A) For BIRC3 in HA22T/VGH, 4 out 7 CpGs showed a hypermethylation trend in sorafenib treated cells, while 1 of 7 CpGs showed a hypomethylation trend. For BIRC3 in SKHep1C3, 4 out 7 CpGs showed a hypermethylation trend in sorafenib-treated cells, while 2 of 7 CpGs showed a hypomethylation trend. The primer set was designed to amplify a 758 bp fragment from nucleotides 1,032 to 1,790 relative to the DMR start. (B) For MAPK3 in HA22T/VGH, 10 out 11 CpGs showed a hypermethylation trend in sorafenib-treated cells, while 0 out 11 CpGs showed a hypomethylation trend. For MAPK3 in SKHep1C3, 7 out 10 CpGs showed a hypermethylation trend in sorafenib-treated cells, while 3 out 10 CpGs showed a hypomethylation trend. The primer set was designed to amplify a 430 bp fragment from nucleotides 1,660 to 2,090 relative to the DMR start. (C) For TSC2 in HA22T/VGH, 21 out 25 CpGs showed a hypomethylation trend in sorafenib-treated cells, while 2 out 25 CpGs showed a hypermethylation trend. For TSC2 in SKHep1C3, 11 out 25 CpGs showed a hypomethylation trend in sorafenib-treated cells, while 14 out 25 CpGs showed a hypermethylation trend. The primer set was designed to amplify a 577 bp fragment from nucleotides 1,107 to 1,684 relative to the DMR start. (D) For FOXO3 in HA22T/VGH, 28 out 43 CpGs showed a hypomethylation trend in sorafenib-treated cells, while 11 of 43 CpGs showed a hypermethylation trend. For FOXO3 in SKHep1C3, 12 out 43 CpGs showed a hypomethylation trend in sorafenib-treated cells, while 24 of 43 CpGs showed a hypermethylation trend. The primer set was designed to amplify a 430 bp fragment from nucleotides 1,780 to 2,242 relative to the DMR start. Validation of MeDip-chip results by direct bisulfite sequencing in sorafenib treated and untreated HCC cells. (E) For SMAD2 in HA22T/VGH, 23 out 31 CpGs showed a hypomethylation trend in sorafenib-treated cells, while 3 out 31 CpGs showed a hypermethylation trend. For SMAD2 in SKHep1C3, 6 out 31 CpGs showed a hypomethylation trend in sorafenib-treated cells, while 16 out 31 CpGs showed a hypermethylation trend. The primer set was designed to amplify a 404 bp fragment from nucleotides 2,118 to 2,581 relative to the DMR start. (F) Examples of chromatogram peaks for thymine (unmethylated) and cytosine (methylated; forward sequencing) and for adenine (unmethylated) and guanine (methylated; reverse sequencing) relative to CpGs whose mean differences in DNA methylation were statistically significant between the two cellular conditions.
To compare the results obtained in other sorafenib-sensitive HCC cell lines, we extended some validation analysis by COBRA assay and direct bisulfite sequencing to SKHep1C3 cells BIRC3 and MAPK3 resulted hypermethylated in line with HA22T/VGH cells (Figs. 4D and 5A and B). TSC2 displayed substantially unchanged DNA methylation level (Fig. 5C); finally, FOXO3 and SMAD2 showed an opposite DNA methylation trend respect to HA22T/VGH cells (Fig. 5D–E).
RT-qPCR expression analysis of the genes BIRC3, FOXO3, MAPK3, SMAD2 and TSC2 in sorafenib-treated and untreated HCC cells
To evaluate the relationship between DNA methylation changes and gene expression in sorafenib-treated and untreated cells, we evaluated the mRNA expression of the genes whose DNA methylation status was previously validated by COBRA and direct bisulfite sequencing (BIRC3, FOXO3, MAPK3, SMAD2 and TSC2) by RT-qPCR. The results showed that the 5 genes studied were differentially expressed in cells treated with sorafenib compared to untreated cells (Fig. 6A). In particular, BIRC3, FOXO3 and SMAD2 were upregulated while MAPK3 and TSC2 were downregulated. FOXO3, MAPK3 and SMAD2 showed an inverse relationship between gene expression and DNA methylation level with FOXO3 and SMAD2 being hypomethylated and upregulated and MAPK3 was hypermethylated and downregulated in sorafenib-treated compared to untreated cells. The DMR of FOXO3 (MAT score = −10.33; RQ=3.24) and SMAD2 (MAT score = −10.86; RQ=1.95) were located in the promoter region of their associated genes, and their hypomethylation in sorafenib-treated cells were associated with an upregulation of their mRNA. These results were expected because of the well-known effect on gene expression of inhibition of DNA methylation in promoter regions. BIRC3 and TSC2 showed a direct relationship between DNA methylation level and gene expression. BIRC3 was hypermethylated and upregulated and TSC2 was hypomethylated and downregulated. We performed the RT-qPCR analysis in SKHep1C3 cells to compare the results obtained in HA22T/VGH cell line. SKHep1C3 cells displayed the same expression trend found in HA22T/VGH cells for BIRC3, FOXO3, SMAD2 and TSC2 and the inverse expression trend for MAPK3 (Fig. 6B).
Figure 6.
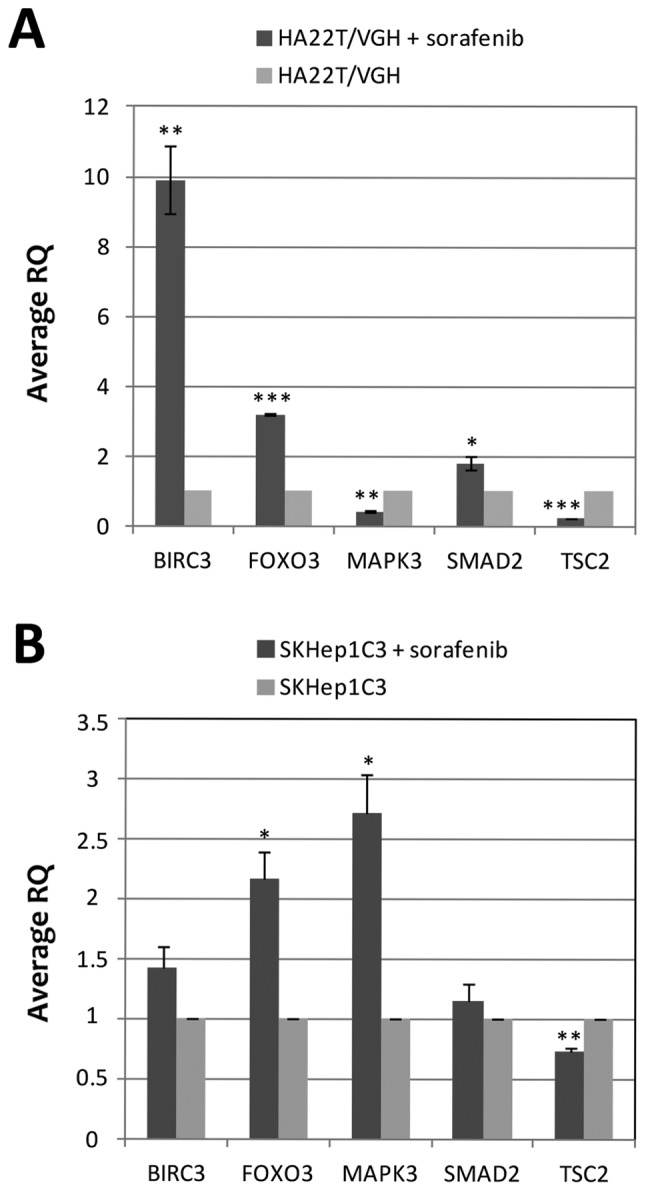
Evaluation of mRNA expression by RT-qPCR in sorafenib treated and untreated HCC cells. The mRNA expression of the genes BIRC3, FOXO3, MAPK3, SMAD2 and TSC2 was evaluated in HA22T/VGH (A) and SKHep1C3 (B) sorafenib-treated and untreated cells by RT-qPCR, to establish the relationship between DNA methylation changes and gene expression. Relative expression was calculated using ΔΔCt method and an untreated cell sample as a reference. The housekeeping gene YWHAZ was used. Error bars of the RQ panels refer to standard deviation of the RQ means of the two experiment performed. *P<0.05, **P<0.01 and ***P<0.001 indicate that the expression difference between the two cellular conditions was statistically significant.
Discussion
Sorafenib is the first multi-kinase inhibitor designed to inhibit the activity of RAF kinase, and is currently the only treatment option for advanced and/or unresectable hepatocellular carcinoma. Although SHARP (3) and other clinical trials confirmed the effectiveness of sorafenib compared to placebo, the median survival was nearly 3 months longer for patients treated with sorafenib than for those given placebo. Moreover, sorafenib had severe side effects and patients developed resistance quickly (7). It is well known that epigenetic alterations in addition to genetic mutations play an additive role in the development and progression of cancer.
To our knowledge, there are no studies about sorafenib effects on global DNA methylation changes in cancer cells. In this novel study, conducted on HCC derived cells (the undifferentiated HA22T/VGH cells) the data obtained showed that sorafenib mediated DNA methylation variations in 1280 regions corresponding to 1230 unique genes. We consider this the main novelty of our work. Data obtained with functional enrichment analysis suggested for the first time that sorafenib in HA22T/VGH cells affected the methylation level of different genes known to be associated with tumorigenesis and/or cancer progression (i.e. in apoptosis, invasion and angiogenesis and important signaling pathways deregulated in HCC).
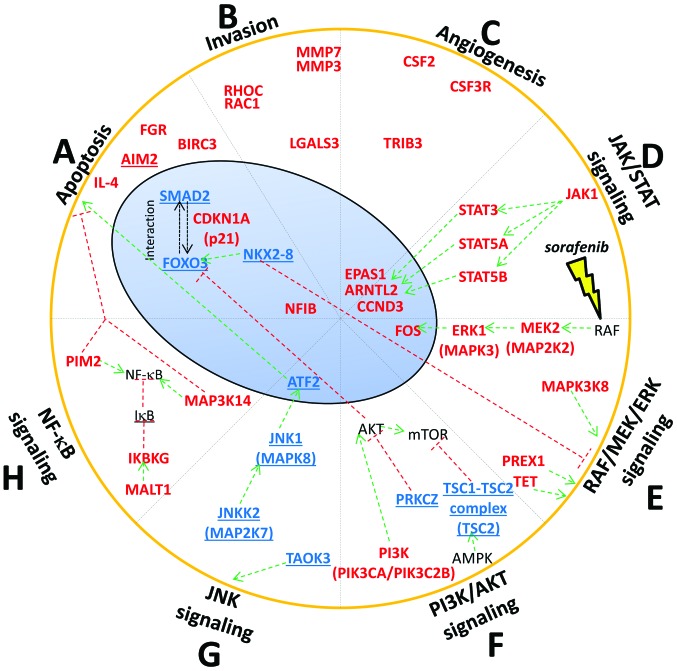
Apoptosis
Several studies showed that sorafenib induced apoptosis in different types of cancer cell lines, including HCC cells (6). Our results showed that different genes involved in apoptosis were differentially methylated after the treatment with sorafenib. We found hypomethylation of FOXO3, SMAD2 and hypermethylation of CDKN1A (p21) (Fig. 7A). FOXO3 is a transcription factor with pro-apoptotic function. Lu et al found that FOXO3 was downregulated in HCC tissue (17). Moreover, Wehler et al found that sorafenib-sensitive colorectal cancer cells were defined by medium-strong FOXO3 protein expression (18). We examined FOXO3 gene expression in 2 sorafenib HCC treated cell lines. We have found hypomethylation of FOXO3 and a correspondent upregulation of its mRNA in HA22T/VGH cells likely leading to increased protein expression levels. In SKHep1C3, we found hypermethylation of FOXO3 by bisulfite sequencing and upregulation of mRNA. In this regard, we sequenced a small region (462 bp) of the DMR (differential methylated region) that was 3120 bp long. For this reason, we do not exclude to find hypomethylation within the DMR in other portions; however, other molecular mechanisms may be responsible of FOXO3 mRNA increasing in SKHep1C3.
Figure 7.
Genes associated to tumorigenesis and/or cancer progression found differentially methylated in HA22T/VGH sorafenib treated cells. The genes are grouped based on their role: apoptosis (A), invasion (B) and angiogenesis (C) as well as genes belonging to several signaling pathways such as JAK-STAT (D), RAF/MEK/ERK (E), PI3K/AKT (F), JNK (G) and NF-κB (H). Generally, we found a trend where oncogenes were hypermethylated and TSG were hypometh-ylated after sorafenib treatment. In light blue and red are reported genes found hypomethylated or hypermethylated, respectively. Genes known to be TSG are underlined while genes known to be oncogenes are not underlined. Green and red lines indicate the activation or inhibition of a gene product or a signaling pathway, respectively.
We observed the hypomethylation of FOXO3 and the upregulation of FOXO3 mRNA in 2 sorafenib-treated cell lines and these events may determine FOXO3 protein up regulation. The role of SMAD2 as TSG or OG is still controversial and depends on the cellular context (19). When Smad proteins are activated by TGF-β, they form a complex with FOXO proteins and turn on the growth inhibitory genes p15INK4B and p21Cip1 (19). We found that SMAD2 was hypomethylated in sorafenib-treated cells and its mRNA was upregulated in both cell lines analyzed. Conversely, we found p21 (CDKN1A) hypermethylated. This result is intriguing because p21 is a pleiotropic protein and can also have anti-apoptotic activity (20). Weiss et al found that sorafenib decreased p21 expression both in renal cell carcinoma and in HCC cell lines, suggesting that the decrease of p21 protein is at least in part responsible for the high cytotoxicity of sorafenib (20).
Invasion
Sorafenib is known to inhibit invasion in HCC cells. Ha et al reported that sorafenib inhibits migration and invasion of HCC cells through suppression of different MMP expression (21). Moreover, Chiang et al demonstrated that the invasion promoted by TPA (12-O-tetradecanoylphorbol-13-acetate) that induced MMP-9 and VEGF expression was inhibited by sorafenib via the suppression of ERK/NF-κB pathway in HCC cells (22). We found that sorafenib promoted the hypermethylation of crucial genes implicated in invasion process such as MMP3, MMP7, RAC1, RHOC and LGALS3 (Fig. 7B). MMP3 and MMP7 are well known proteases implicated in the degradation of ECM and invasion process. RAC1 is known to be involved in various cellular functions such as cell growth, division, morphology, polarity and migration, and its overexpression was correlated with poor prognosis in HCC (23). RHOC was found overexpressed in HCC and it had an important role in invasion and metastasis (24). Finally, downregulation of LGALS3 was demonstrated to cause a decrease of uPAR levels and inhibits the proliferation, migration and invasion of HCC (25).
Angiogenesis
Anti-angiogenic activity of sorafenib is well documented in HCC (6). Our results showed that the proangiogenic factor EPAS1 was hypermethylated in our study. EPAS1 is an oncogene that works in response to hypoxic conditions and is overexpressed in HCC (26). Cervello et al found that EPAS1 mRNA resulted downregulated after treating HCC cells with sorafenib (27).
JAK-STAT pathway
In mammals, the JAK/STAT pathway is the principal signaling mechanism for a wide array of cytokines and growth factors. JAK activation stimulates cell proliferation, differentiation, cell migration and apoptosis (28). Yang et al found that sorafenib caused dephosphorylation of STAT3 and downregulation of cyclin D3 (CCND3) in glioblastomas (29). The downregulation of cyclin D3 mRNA after sorafenib treatment was reported by Cervello et al in HCC cells (27). We found the hypermethylation of the following component of JAK-STAT signaling after sorafenib treatment: JAK1, STAT3, STAT5A, STAT5B and CCND3 (Fig. 7D). Noteworthy, CCND3, a key component for G1/S phase transition, is induced by the transcription factor STAT3 and STAT5 here both were found hypermethylated.
MAPK pathway: RAF/MEK/ERK cascade
Sorafenib was initially designed to inhibit the kinase activity of RAF and consequently the RAF/MEK/ERK cascade involved in different processes of survival and cellular growth. Liu et al demonstrated that sorafenib blocked the RAF/MEK/ERK pathway by inhibiting the phosphorylation of MEK and ERK and it downregulated cyclin D1 levels in two HCC cell lines (6). We observed MAPK3 (ERK1) and MAP2K2 (MEK2) hypermethylated in sorafenib treated HA22T/VGH cells. We also found FOS, one of the effectors of RAF/MEK/ERK cascade, hypermethylated (Fig. 7E). Walter et al found the downregulation of FOS in human osteosarcoma cells after sorafenib treatment (30).
PI3K/AKT pathway
PI3K/AKT signaling is an important survival/proliferative pathway involving various growth factors, cytokines, and activation of receptors and it is one of the most commonly activated signaling pathway in human cancer. However, the role of this pathway in the mechanisms of action of sorafenib is not fully elucidated. Gedaly et al showed that sorafenib enhance the level of p-AKT in HCC HuH7 cells (31). However, Zhang et al found opposite results using Human SMMC-7721 hepatic carcinoma cells. They found that sorafenib inhibited hepatic tumor growth by reducing PI3K/AKT signaling pathway (32). Furthermore, Cervello et al found no variation of p-AKT in HCC HepG2 cell line treated with sorafenib (27). In this context, we found DNA methylation changes in different component of PI3K/AKT pathway, but with no clear tendency to turn on or turn off this pathway in HA22T/VGH treated with sorafenib. We found PIK3CA and PIK3C2B, subunits of PI3K, hypermethylated (Fig. 7F). We found TSC2, inhibitor of mTOR, hypomethylated (Fig. 7F), although the mRNA validation showed a downregulation of the TSC2 mRNA in the 2 cell lines analyzed.
Concerning the correlation between DNA methylation and mRNA expression, our results indicated that among the 5 selected genes BIRC3 and TSC2 displayed a direct trend between DNA methylation and mRNA expression and FOXO3, MAPK3 and SMAD2 showed an inverse trend in HA22T/VGH cells.
In our study the DMR of BIRC3, MAPK3 and TSC2 were located in intragenic regions and the analysis of gene expression of associated genes returned both positive and negative relationship between transcription and intragenic DNA methylation. Some authors have already described that DNA methylation in intragenic regions can lead to both positive and negative relationship between transcription and intragenic DNA methylation (33). BIRC3 (MAT score = +12.10; RQ=9.22) and TSC2 (MAT score = −13.04; RQ=0.27) showed the same direct trend, while MAPK3 (MAT score = +12.42; RQ=0.41) showed an inverse trend between gene expression and DNA methylation. Moreover, the DMR of BIRC3 and MAPK3 contained fewer CpGs and were located near CpG islands. In literature, the regions that flank CpG islands with less CG density have been recently described as 'CpG island shores'. Different studies have described the role of CpG island shore methylation in cancer and how it can affect gene expression. For example, Irizarry et al reported that most of the methylation differences between normal and cancer samples occur in CpG island shores (34). To our knowledge this is the first time that the association between CpG island shore methylation and changes of gene expression in HCC cells treated with sorafenib has been reported although further studies are needed to confirm these findings.
Since the methylome of SKHep1C3 was not analyzed, we do not assume that sorafenib determines the same global DNA methylation changes observed in HA22T/VGH cells even if this cell line is sorafenib-sensitive. It would be of interest in the future to identify the DMRs in different HCC cell lines in order to know the common DMRs and those depending on their own malignant characteristics and on their differentiation state. In this regard, the DNA methylation variations of the 5 selected genes in SKHep1C3 cells displayed a partial overlap compared to those obtained in HA22T/VGH cells, but importantly their expression levels were affected. In SKHep1C3 the trend of the gene expression dysregulation was concordant in 4 out 5 genes in respect to HA22T/VGH cells although not statistically significant for BIRC3 and SMAD2. Further, in SKHep1C3, we found hypermethylation of FOXO3 by bisulfite sequencing and upregulation of mRNA. In this regard, we outline that we sequenced a small region (462 bp) of the DMR (differential methylated region) that was 3120 bp long. For this reason, we do not exclude to find hypomethylation within the DMR in other portions; however, some other mechanisms of gene expression control, other than DNA methylation, such as histone modifications, chromatin remodelling and post-transcriptional regulation (i.e. ncRNAs) should not to be excluded in determining mRNA dysregulation following sorafenib treatments.
In summary, this is the first study that analyzed the global DNA methylation changes induced by sorafenib in cancer cells in particular in the HCC sorafenib sensitive HA22T/VGH cells. Our data show that sorafenib affected the DNA methylation status of numerous genes involved in tumorigenesis and cancer progression, with a general trend of oncogenes to be hypermethylated and tumor suppressor genes to be hypomethylated. These genes are involved in processes such as apoptosis, invasion and angiogenesis, and different signaling pathways deregulated in HCC.
Acknowledgments
We thank Dr M. Crosatti (University of Leicester; Leicester, UK) for the linguistic revision of the manuscript. We are grateful to Professor Marina Colombi and Professor Massimo Gennarelli, responsible for the Affymetrix platform at the Division of Biology and Genetics, Department of Molecular and Translational Medicine, Brescia. This study was supported by Lega Italiana per la Lotta contro i Tumori (LILT), by Comitato Nazionale Universitario (CNU) Brescia, by the Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) local funds of the University of Brescia. E.A. was supported by a Postdoctoral fellowship from Associazione Davide Rodella Onlus, Brescia.
Glossary
Abbreviations
- COBRA
combined bisulfite restriction analysis
- DMR
differentially methylated region
- IN DNA
input DNA
- IP DNA
immunoprecipitated DNA
- MAT algorithm
model-based analysis of tiling arrays algorithm
- MeDip
methylated DNA immunoprecipitation
- PGS
partek genomics suite
- 5 mC
5-methylcytosine
- hmC
5-hydroxymethylcytosine
References
- 1.Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: Consider the population. J Clin Gastroenterol. 2013;47(Suppl):S2–S6. doi: 10.1097/MCG.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 4.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 5.Carlomagno F, Anaganti S, Guida T, Salvatore G, Troncone G, Wilhelm SM, Santoro M. BAY 43-9006 inhibition of oncogenic RET mutants. J Natl Cancer Inst. 2006;98:326–334. doi: 10.1093/jnci/djj069. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, Wilhelm S, Lynch M, Carter C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 7.Gauthier A, Ho M. Role of sorafenib in the treatment of advanced hepatocellular carcinoma: An update. Hepatol Res. 2013;43:147–154. doi: 10.1111/j.1872-034X.2012.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014;342:223–230. doi: 10.1016/j.canlet.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Chen YL, Ji G, Fang W, Gao Z, Liu Y, Wang J, Ding X, Gao F. Sorafenib inhibits epithelial-mesenchymal transition through an epigenetic-based mechanism in human lung epithelial cells. PLoS One. 2013;8:e64954. doi: 10.1371/journal.pone.0064954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, Zhu Y, He H, Liu J, Xu L, Zhang H, Liu H, Liu W, Liu Y, Pan D, et al. Sorafenib suppresses growth and survival of hepatoma cells by accelerating degradation of enhancer of zeste homolog 2. Cancer Sci. 2013;104:750–759. doi: 10.1111/cas.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salvi A, Conde I, Abeni E, Arici B, Grossi I, Specchia C, Portolani N, Barlati S, De Petro G. Effects of miR-193a and sorafenib on hepatocellular carcinoma cells. Mol Cancer. 2013;12:162. doi: 10.1186/1476-4598-12-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossi I, Arici B, Portolani N, De Petro G, Salvi A. Clinical and biological significance of miR-23b and miR-193a in human hepatocellular carcinoma. Oncotarget. 2017;8:6955–6969. doi: 10.18632/oncotarget.14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 14.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parrish RR, Day JJ, Lubin FD. Direct bisulfite sequencing for examination of DNA methylation with gene and nucleotide resolution from brain tissues. Curr Protoc Neurosci Chapter 7. 2012 doi: 10.1002/0471142301.ns0724s60. Unit 7.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira E, Cronjé MJ. Selection of suitable reference genes for quantitative real-time PCR in apoptosis-induced MCF-7 breast cancer cells. Mol Biotechnol. 2012;50:121–128. doi: 10.1007/s12033-011-9425-3. [DOI] [PubMed] [Google Scholar]
- 17.Lu M, Ma J, Xue W, Cheng C, Wang Y, Zhao Y, Ke Q, Liu H, Liu Y, Li P, et al. The expression and prognosis of FOXO3a and Skp2 in human hepatocellular carcinoma. Pathol Oncol Res. 2009;15:679–687. doi: 10.1007/s12253-009-9171-z. [DOI] [PubMed] [Google Scholar]
- 18.Wehler TC, Hamdi S, Maderer A, Graf C, Gockel I, Schmidtmann I, Hainz M, Berger MR, Theobald M, Galle PR, et al. Single-agent therapy with sorafenib or 5-FU is equally effective in human colorectal cancer xenograft - no benefit of combination therapy. Int J Colorectal Dis. 2013;28:385–398. doi: 10.1007/s00384-012-1551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seoane J, Le HV, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/S0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 20.Wettersten HI, Hee Hwang S, Li C, Shiu EY, Wecksler AT, Hammock BD, Weiss RH. A novel p21 attenuator which is structurally related to sorafenib. Cancer Biol Ther. 2013;14:278–285. doi: 10.4161/cbt.23374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha TY, Hwang S, Moon KM, Won YJ, Song GW, Kim N, Tak E, Ryoo BY, Hong HN. Sorafenib inhibits migration and invasion of hepatocellular carcinoma cells through suppression of matrix metalloproteinase expression. Anticancer Res. 2015;35:1967–1976. [PubMed] [Google Scholar]
- 22.Chiang IT, Liu YC, Wang WH, Hsu FT, Chen HW, Lin WJ, Chang WY, Hwang JJ. Sorafenib inhibits TPA-induced MMP-9 and VEGF expression via suppression of ERK/NF-κB pathway in hepatocellular carcinoma cells. In Vivo. 2012;26:671–681. [PubMed] [Google Scholar]
- 23.Yang W, Lv S, Liu X, Liu H, Yang W, Hu F. Up-regulation of Tiam1 and Rac1 correlates with poor prognosis in hepatocellular carcinoma. Jpn J Clin Oncol. 2010;40:1053–1059. doi: 10.1093/jjco/hyq086. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Yang LY, Yang ZL, Huang GW, Lu WQ. Expression and significance of RhoC gene in hepatocellular carcinoma. World J Gastroenterol. 2003;9:1950–1953. doi: 10.3748/wjg.v9.i9.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng D, Hu Z, He F, Gao C, Xu L, Zou H, Wu Z, Jiang X, Wang J. Downregulation of galectin-3 causes a decrease in uPAR levels and inhibits the proliferation, migration and invasion of hepatocellular carcinoma cells. Oncol Rep. 2014;32:411–418. doi: 10.3892/or.2014.3170. [DOI] [PubMed] [Google Scholar]
- 26.Bangoura G, Liu ZS, Qian Q, Jiang CQ, Yang GF, Jing S. Prognostic significance of HIF-2alpha/EPAS1 expression in hepatocellular carcinoma. World J Gastroenterol. 2007;13:3176–3182. doi: 10.3748/wjg.v13.i23.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cervello M, Bachvarov D, Lampiasi N, Cusimano A, Azzolina A, McCubrey JA, Montalto G. Molecular mechanisms of sorafenib action in liver cancer cells. Cell Cycle. 2012;11:2843–2855. doi: 10.4161/cc.21193. [DOI] [PubMed] [Google Scholar]
- 28.Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, Factor VM, Thorgeirsson SS. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Yang F, Brown C, Buettner R, Hedvat M, Starr R, Scuto A, Schroeder A, Jensen M, Jove R. Sorafenib induces growth arrest and apoptosis of human glioblastoma cells through the dephosphorylation of signal transducers and activators of transcription 3. Mol Cancer Ther. 2010;9:953–962. doi: 10.1158/1535-7163.MCT-09-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter I, Wolfesberger B, Miller I, Mair G, Burger S, Gallè B, Steinborn R. Human osteosarcoma cells respond to sorafenib chemotherapy by downregulation of the tumor progression factors S100A4, CXCR4 and the oncogene FOS. Oncol Rep. 2014;31:1147–1156. doi: 10.3892/or.2013.2954. [DOI] [PubMed] [Google Scholar]
- 31.Gedaly R, Angulo P, Chen C, Creasy KT, Spear BT, Hundley J, Daily MF, Shah M, Evers BM. The role of I3K/mTOR inhibition in combination with sorafenib in hepatocellular carcinoma treatment. Anticancer Res. 2012;32:2531–2536. [PubMed] [Google Scholar]
- 32.Zhang CZ, Wang XD, Wang HW, Cai Y, Chao LQ. Sorafenib inhibits liver cancer growth by decreasing mTOR, AKT, and I3K expression. J BUON. 2015;20:218–222. [PubMed] [Google Scholar]
- 33.Kulis M, Queirós AC, Beekman R, Martín-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829:1161–1174. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]