Abstract
We developed a pentachlorophenol (PCP)-degrading, methanogenic fixed-film reactor by using broken granular sludge from an upflow anaerobic sludge blanket reactor. This methanogenic consortium was acclimated with increasing concentrations of PCP. After 225 days of acclimation, the reactor was performing at a high level, with a PCP removal rate of 1,173 μM day−1, a PCP removal efficiency of up to 99%, a degradation efficiency of approximately 60%, and 3-chlorophenol as the main chlorophenol residual intermediate. Analyses by PCR-denaturing gradient gel electrophoresis (DGGE) showed that Bacteria and Archaea in the reactor stabilized in the biofilms after 56 days of operation. Important modifications in the profiles of Bacteria between the original granular sludge and the reactor occurred, as less than one-third of the sludge DGGE bands were still present in the reactor. Fluorescence in situ hybridization experiments with probes for Archaea or Bacteria revealed that the biofilms were composed mostly of Bacteria, which accounted for 70% of the cells. With PCR species-specific primers, the presence of the halorespiring bacterium Desulfitobacterium hafniense in the biofilm was detected very early during the reactor acclimation period. D. hafniense cells were scattered in the biofilm and accounted for 19% of the community. These results suggest that the presence of PCP-dehalogenating D. hafniense in the biofilm was crucial for the performance of the reactor.
Widespread use of pentachlorophenol (PCP), mostly as a wood-preserving agent, has led to contamination of the environment. PCP and other chlorophenols (CPs) are very toxic molecules and are common environmental pollutants. PCP can be transformed into less toxic compounds by anaerobic microorganisms (22) such as reductive dehalogenating bacteria (14). Anaerobic processes can be used to decontaminate PCP-containing waste or groundwater and when coupled with chemical extraction of PCP can also decontaminate soils and sediments (25).
Anaerobic-biofilm-based processes, such as upflow anaerobic sludge blanket (UASB) reactors and fixed-film (FF) bioreactors, have been tested for the degradation of PCP and other halogenated compounds (see reference 41 for a review). FF bioreactors have many advantages over other types of bioreactors, such as a shorter start-up period, no requirement for bed expansion or fluidization, and no risk of accidental washout of the biomass.
In contrast to their aerobic counterparts, the structure of anaerobic biofilms has been studied in only a few instances. Juteau et al. (24) examined the biofilm present in anaerobic PCP-degrading bioreactors by scanning electron microscopy. They observed that the biofilm surface was populated by various morphologically different species, namely, cocci, rods such as Methanosaeta sp., and filamentous microorganisms, whereas the bottom of the biofilm was principally dominated by rods including Methanosaeta-like organisms. Major drawbacks of scanning electron microscopy are that it provides information limited to the morphology of the microorganisms and that the drastic dehydration steps in the preparation of biofilm samples cause loss of structure. These can be overcome by the use of fluorescence in situ hybridization (FISH) coupled with confocal laser scanning microscopy (CLSM), which allows the detection of specific microorganisms in a biofilm by hybridization of specific fluorescent oligonucleotide probes targeting the 16S rRNA sequences without disruption of the three-dimensional structure of the biofilm (46).
Looking at UASB reactors, the FISH technique has been used to localize specific bacterial strains such as halorespiring bacteria within granules. For instance, Desulfomonile tiedjei cells in a 3-chlorobenzoate-fed reactor colonized the granules as inner as well as surface microcolonies (1). In a perchloroethylene-fed reactor inoculated with Sulfurospirillum (formerly Dehalospirillum) multivorans, microcolonies were detected mainly at the surface of the granules (20). However, once added to autoclaved granules of a PCP-fed reactor, Desulfitobacterium hafniense DCB-2 showed a uniform distribution in the granules (11). Finally, we have previously demonstrated in a PCP-fed UASB bioreactor augmented with D. hafniense (formerly D. frappieri) PCP-1 that strain PCP-1 cells colonized the outer section of granules (27). This spatial organization suggests that strain PCP-1 protects other members of the consortium from PCP toxicity, thus supporting our observation of enhanced efficiency when PCP-1 bacteria are present.
To our knowledge, no study reporting the spatial arrangement in anaerobic FF reactors of a specific anaerobic, halogenated compound degrader, or any anaerobic xenobiotic degrader, has been published. Characterization of biofilms from anaerobic FF reactors has been performed mostly for sulfate-reducing bacteria (3, 36, 37) and methanogens (38). A better understanding of biofilm structure in anaerobic FF reactors may allow the design of improved bioreactors. Here we describe the biofilm of a methanogenic, PCP-fed FF reactor containing D. hafniense. This report combines analytical and biodegradation data on the reactor with the microbial profiles, the biofilm structure, and the spatial arrangement of D. hafniense within the biofilm.
MATERIALS AND METHODS
Anaerobic FF bioreactor.
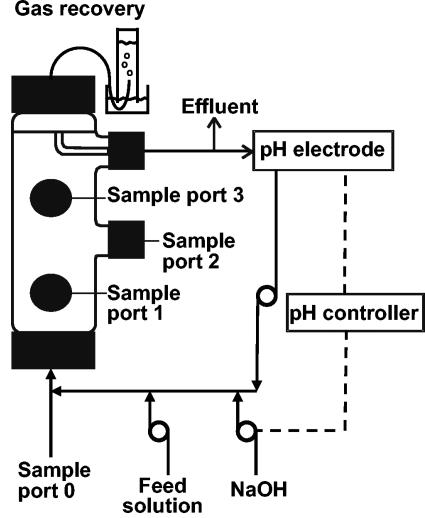
The bioreactor (Fig. 1) was custom manufactured by Lasalle Scientifique, Inc. (Montreal, Québec, Canada). The bioreactor had a total volume of 505 ml and an effective volume of 375 ml. It was packed with approximately 275 g of Raschig rings (8.0 by 8.0 mm) (Ace Glass Inc., Vineland, N.J.) to allow biofilm colonization. Since biofilm cannot be examined on rings by CLSM, small pieces of microscope glass slides (0.5 to 1.0 by 1.0 cm) were included in the reactor. The bioreactor was made of glass cylinders (height, 25 cm; outside diameter, 7 cm) with large plastic caps (GL-45 threads) on the ends. It had three screw-cap lateral openings (outside diameter, 24 mm) that allowed sampling of the packing material. Each of these openings was also plugged with a butyl stopper that was pierced with a needle for liquid sampling. Another lateral opening was used as an effluent port. The liquid was recirculated from the effluent port to the influent port with a peristaltic pump and Pharmed tubing (Norton Performance Plastics, Akron, Ohio). A sampling port was also located in the recirculating loop.
FIG. 1.
Schematic representation of the FF reactor.
The bioreactor was operated at 35°C, with a hydraulic retention time of 28 to 35 h. The pH was adjusted to 7.3 ± 0.1 with a 5.0-g/liter NaOH solution. Recirculation in the reactor provided a linear upflow liquid velocity of 0.79 m day−1. The reactor was fed with a solution containing the following (in grams per liter): PCP, 0.0013 to 0.364; sucrose, 1.99; n-butyrate, 0.63; yeast extract (Gibco Laboratories), 0.05; ethanol (95%), 0.46; KH2PO4, 0.04; K2HPO4, 0.05; NaHCO3, 0.88; KHCO3, 1.24; NH4HCO3 0.45; resazurin (BDH Chemicals Ltd., Poole, England), 0.001. A trace mineral solution was also added to this feeding solution. The final concentration of each mineral was as follows (in milligrams per liter): FeSO4 · 7H2O, 5.71; H3BO3, 0.42; ZnSO4 · 7H2O, 1.47; CuSO4, 0.49; MnSO4 · H2O, 4.55; CoSO4 · 7H2O, 1.82; NiSO4 · 6H2O, 0.84; (NH4)6Mo7O24 · 4H2O, 1.47; AlK(SO4)2 · 12H2O, 0.18; Na2EDTA, 5.26; MgSO4 · 7H2O, 9.00; Na2SeO4, 0.14; Na2WO4, 0.07 (27, 44). The mean chemical oxygen demand (COD) of the feeding solution was 3,500 mg/liter.
Before use, the nutrient solution was sparged for 10 min with a gas mixture containing H2-CO2-N2 (10:10:80; Praxair, Mississauga, Ontario, Canada), and then butyrate, ethanol, and PCP (99% pure; Aldrich Chemical Company, Inc., Milwaukee, Wis.) were added. Growth of microorganisms in the feeding solution was limited by keeping it at 4°C.
Anaerobic granular sludge was obtained from an industrial reactor treating wastewater from an apple-processing plant (Rougemont, Québec, Canada). The reactor was inoculated with granules (equivalent to 523 mg of volatile suspended solids) that were homogenized mechanically and diluted 1:4 with anoxic phosphate-buffered saline (PBS; 130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4). In order to facilitate bacterial attachment to the surface of the packing material prior to feeding, the reactor was left in recirculation mode for 3 days following inoculation. The PCP load started at 5 μM day−1 and was gradually increased to 1,173 μM day−1 after 225 days of operation.
Chemical analyses of the reactor.
Gas production was monitored daily at the effluent port by water displacement in an inverted cylinder (15) as illustrated in Fig. 1. Samples were taken weekly from the bioreactor influent and effluent ports and analyzed for volatile fatty acids (VFA) and CPs. CODs were analyzed every 3 to 5 weeks. For VFA and CP analyses, suspended solids were removed by centrifugation at 6,000 × g for 2 min.
For acetate, propionate, and n-butyrate (VFA) monitoring, 300-μl samples were mixed with 100 μl of a heptanoic acid solution (final concentration, 153 mg liter−1) as an internal standard and 200 μl of 3% formic acid (final concentration, 1%) to increase VFA volatilization. One microliter was manually injected into a Hewlett-Packard 6890 gas chromatograph (Agilent Technologies, Montreal, Québec, Canada) equipped with a flame ionization detector (SPB-1000; 0.25-μm thickness; Supelco, Bellefonte, Pa.). The sample was injected in splitless mode with helium as the carrier gas (4.0 ml min−1) and run at 120°C for 4 min and then at 135, 150, 165, and 180°C for 1 min each with a ramp of 15°C min−1 between temperatures. The flame ionization detector was heated at 220°C with an H2 flow of 40.0 ml min−1, an airflow of 450 ml min−1, and an N2 flow of 35 ml min−1. A 3% formic acid sample was analyzed between samples to prevent carryover.
CPs were analyzed by high-performance liquid chromatography as described before (23, 24). COD was analyzed by the closed-reflux method (12).
DNA extraction, PCR, and denaturing gradient gel electrophoresis (DGGE).
Sampling of Raschig rings and glass slides coated with biofilm was performed in an anaerobic chamber through the sampling ports (Fig. 1). DNA was extracted from two Raschig rings for the biofilm samples. Glass beads (300 mg, 0.4- to 0.5-mm diameter) and a volume of 1 ml of extraction buffer (Tris-HCl at 50 mM [pH 8.0], EDTA at 10 mM, polyvinylpolypyrrolidone at 1%) were added, and then samples were homogenized in a bead beater (Fast Prep FP120, Bio 101 ThermoSavant; Qbiogene, Carlsbad, Calif.) twice for 20 s at speed 4 and centrifuged at 16,000 × g for 15 min. DNA in the supernatant was purified by successive phenol-chloroform-isoamyl alcohol and chloroform-isoamyl alcohol extractions and ethanol precipitation (40).
PCR mixtures of 50 μl contained 10 mM Tris-HCl buffer (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 200 μM deoxynucleoside triphosphates, 2.5 U of Taq polymerase (Amersham Biosciences, Baie d'Urfé, Québec, Canada), 10 pmol of each primer (Table 1), 0.5 μg of bovine serum albumin per μl, and approximately 10 ng of DNA. Amplifications were carried out with a model 480 Thermal Cycler (Perkin-Elmer) starting with a 5-min step at 80°C during which DNA was added, and then the reaction went to 94°C for 5 min and 55°C for 5 min, followed by 30 to 35 cycles at 72°C for 2 min, 94°C for 40 s, and 55°C for 1 min and a final extension at 72°C for 10 min.
TABLE 1.
Oligonucleotides used in this study
| Common name | Generic name | Target(s) | Sequence (5′-3′) | Position | Reference |
|---|---|---|---|---|---|
| Primers for PCR-DGGE | |||||
| PRBA341fa | S-D-Bact-0341-a-A-18 | Bacteria | CCTACGGGAGGCAGCAG | 341-357 | 34 |
| PRUN518r | S-D-Bact-0518-a-A-18 | Bacteria | ATTACCGCGGCTGCTGG | 518-534 | 34 |
| ARC344fa | S-D-Arch-0344-a-A-20 | Archaea | ACGGGGAGCAGCAGGCGCGA | 344-363 | 39 |
| ARC915r | S-D-Arch-0915-a-A-20 | Archaea | GTGCTCCCCCGCCAATTCCT | 915-934 | 39 |
| Specific primers for Desulfitobacterium | |||||
| PCP1G | S-S-D.frapp-0056-(D. frappieri PCP-1)-a-S-18 | D. hafniense | CGAACGGTCCAGTGTCTA | 56-73 | 28 |
| PCP4D | S-S-D.frapp-0580-(D. frappieri PCP-1)-a-A-20 | D. hafniense | AGGTACCGTCATGTAAGTAC | 599-580 | 28 |
| De2 | S-G-Desulfito-704-(D. frappieri PCP-1)-a-A-22 | Desulfitobacteriumb | CCTAGGTTTTCACACCAGACTT | 725-704 | 28 |
| De1 | S-G-Desulfito-322-(D. frappieri PCP-1)-a-S-20 | Desulfitobacteriumb | GCTATCGTTARTRGATGGAT | 322-341 | 28 |
| Fluorescent probes | |||||
| ARC915-Cy3 | S-D-Arch-0915-a-A-20 | Archaea | GTGCTCCCCCGCCAATTCCT | 934-915 | 42 |
| EUB338-Cy3 | S-D-Bact-0338-a-A-18 | Bacteria | GCTGCCTCCCGTAGGAGT | 355-338 | 2 |
| NONEUB338-Cy3 | Negative control | ACTCCTACGGGAGGCAGC | 47 | ||
| PCP-1-4-Cy3 | S-S-D.frapp-0327-(D. frappieri PCP-1)-a-A-19 | D. hafniense, D. chlororespirans | GCGGATCCATCTACTAACG | 345-327 | 27 |
| PCP-1-8-Cy5 | S-S-D.frapp-0576-(D. frappieri PCP-1)-a-A-19 | D. hafniense | CCGTCATGTAAGTACATTA | 594-576 | 27 |
| Helpers for PCP-1-8-Cy5 | |||||
| h3 | S-S-D.frapp-0555-(D. frappieri PCP-1)-a-A-21 | TTTACATACTTACCGTTCGTC | 575-555 | 27 | |
| h4 | S-S-D.frapp-0595-(D. frappieri PCP-1)-a-A-18 | GGGCTTCCTCCTCAGGTA | 612-595 | 27 |
GC clamp (CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG).
Except D. metallireducens, two-nucleotide difference.
PCR amplification of 16S rRNA gene sequences of Bacteria for DGGE profiles was performed with the PRBA341f and PRUN518r primers (Table 1). However, for the 16S rRNA gene sequences of Archaea, a nested PCR was necessary to obtain a detectable amount of amplicons. The first PCR was performed as described above, with the ARC344f and ARC915r primers (with a hybridization temperature of 50°C), and the nested PCR was performed with the ARC344f and PRUN518r primers (Table 1) and 1 to 2 μl of the first PCR product. DGGE was performed as previously described (26), with a linear urea-formamide gradient of 20 to 70%.
Detection of Desulfitobacterium bacteria in anaerobic granular sludge.
Anaerobic enrichment of the indigenous flora of Rougemont sludge was performed as described by Lanthier et al. (28). Briefly, the granular sludge was broken mechanically with an industrial blender and diluted 1:4 in anaerobic sterile mineral salt medium (8). A volume of 2.8 ml of diluted sludge was used to inoculate 35 ml of liquid medium. Two series of enrichments were done, with and without 1.5 ppm of 2,4,6-trichlorophenol (Sigma-Aldrich). Noninoculated controls were also prepared, as well as extraction-negative controls. The sludge was incubated at 30°C for 14 days. DNA extraction was performed as described above, and PCR amplifications with Desulfitobacterium-specific primers (Table 1) were done as previously described (28).
FISH of biofilm samples.
Biofilm samples grown on small glass slides were washed for 2 min in ice-cold filtered PBS and fixed for 45 min in paraformaldehyde-4% (wt/vol) PBS (pH 7.2) in 15-ml conic tubes. They were then washed again in PBS for 5 min and stored at 4°C. For hybridization, biofilm samples were first dehydrated in a graded series of 50, 80, and 95% ethanol solutions for 5 min each and then soaked in an acetylation solution as described previously (27). The slides were then mounted on microscope slides for hybridization experiments. Hybridization solution (200 to 500 μl of 30% deionized formamide, 0.9 M NaCl, 0.02 M Tris-HCl, 0.01% sodium dodecyl sulfate [final pH 7.2]) containing 2.5 ng of fluorescent probe per μl−1 and helper oligonucleotides (if required) (Table 1) was added. For D. hafniense detection, biofilm samples were simultaneously hybridized with probe S-S-D.frapp-576 (D. frappieri)-a-A-19 (PCP-1-8) and helper probes S-S-D.frapp-555 (D. frappieri)-a-A-21 (h3) and S-S-D.frapp-595 (D. frappieri)-a-A-18 (h4) as described by Lanthier et al. (27). Autofluorescence controls were prepared with hybridization buffer containing no fluorescent probe.
Hybridization was carried out in an Omnislide in situ thermal cycler (Thermo Electron Corporation, Waltham, Mass.) for 2 h at 46°C. The humidity chamber was filled with towels soaked with 30 ml of hybridization buffer with no fluorescent probe. After this step, slides were washed twice for 20 min at 48°C in 10 ml of the washing buffer. The slides were then rinsed with deionized water, counterstained in the dark with 200 μl of 1 μM YOYO-1 (Molecular Probes, Eugene, Oreg.) for 15 min, and rinsed with deionized water. Slides were mounted with antifade (ProLong Antifade Kit; Molecular Probes). Slides were then left overnight at room temperature in the dark and then stored at 4°C until microscopic examination. Autofluorescence was very weak in the red channel and absent in the far red channel and therefore did not interfere with the fluorescence signals from the different probes tested.
Slides were examined on a Nikon Eclipse E800 confocal microscope (Bio-Rad, Mississauga, Ontario, Canada) equipped with a Nikon 60× Plan Apo oil immersion objective (numerical aperture of 1.4 and 2.6× zoom) and a Radiance 2000 digital camera (Bio-Rad). The confocal microscope was equipped with a krypton-argon dual laser (488 and 568 nm) and a diode laser (638 nm). For quantification analysis, pictures of at least 20 random microscope fields at different depths were taken with the 60× objective (1.9× zoom). These images (1,280 by 1,024 pixels, 12 bits) corresponded to biofilm sections of 110 by 88 μm (9,680-μm2 area; the total area analyzed was equal to 193,600 μm2) and had a pixel size of 0.39 μm. Images were averaged by Kalman filtration with eight running scans per image (32). The acquisition software used was LaserSharp 2000 (version 4.3).
Cell areas of Bacteria, Archaea, and D. hafniense were quantified with the MetaMorph imaging system (version 4.5r0) (Universal Imaging Corporation, Downingtown, Pa.). The boundary of the biofilm to be quantified was defined by setting the background threshold of the images and transforming them into binary images. Application of a single cycle of two different filters (dilatation and erosion), followed by another filter to remove single pixels, helped to increase the signal-to-noise ratio. The biofilm area on binary images was then quantified for each fluorescent probe and then compared to the total biofilm area stained with YOYO-1 (29, 32).
RESULTS
Monitoring of the PCP-degrading, methanogenic FF reactor.
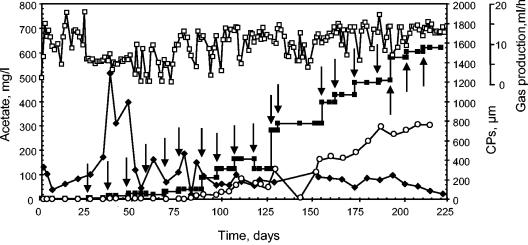
A methanogenic FF reactor was exposed to an increasing PCP load (Fig. 2). It reached a PCP-degrading rate of 1,173 μM day−1 after 225 days of operation, with a PCP removal efficiency of up to 99% and a degradation efficiency of approximately 60%. Only 3-CP was observed as a CP intermediate throughout the experiment.
FIG. 2.
Reactor monitoring. The PCP load (closed squares; micromolar per day) was determined at the influent port of the reactor. The hydraulic retention times were 28 to 35 h. Concentrations of acetate (lozenges) and 3-CP (open circles) were determined at the effluent port. Gas production (open squares) was monitored by water displacement. Arrows indicate the times when the PCP load in the influent was changed.
Gas production and VFA degradation were used as indicators of methanogenic activity (35). The reactor reached an average gas production of 9.6 ml h−1 after 3 to 5 days following start-up and was mostly stable throughout the experiment (Fig. 2). Analysis of the composition of gas from a similar anaerobic FF reactor showed that methane accounted for 69%. Acetate gradually increased after start-up and then stabilized to less than 100 ppm after 60 days of operation (Fig. 2). Butyrate (0.63 g liter−1 at start-up) decreased after start-up and reached trace level after 20 days of operation. Propionate, generated during the process, also was detected at a trace level during the time of operation. To determine the organic matter degradation capacity of the reactor, the influent and effluent port CODs were compared. COD reduction varied between 56 and 72% during the time of operation.
We used PCR primers specific to desulfitobacteria and D. hafniense to determine the presence of such bacteria (Table 1). Specific signals were detected with both sets of primers after 14 days of operation (data not shown). To determine if desulfitobacteria were present in the original sludge, PCRs with the same primers were performed on DNA extracted from the original granular sludge. No specific signal was detected. The granular sludge was then cultured under conditions promoting desulfitobacterial growth. Positive PCR signals were obtained with this enrichment, suggesting the presence of endogenous desulfitobacteria in the original sludge.
Determination of Bacteria and Archaea profiles by PCR-DGGE.
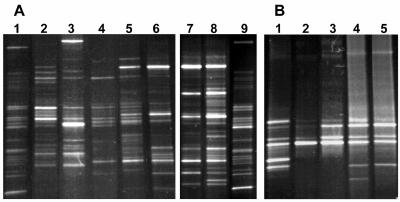
Monitoring of the changes in the microbiota during the acclimation process was performed by PCR-DGGE. Total DNA was extracted from biofilm samples, as well as from the original granular sludge, and 16S rRNA sequences were amplified for Bacteria or Archaea. We observed that Bacteria and Archaea were mostly stable from day 56 in the biofilm (Fig. 3A). Important modifications occurred in Bacteria between the reactor biofilm and the original granular sludge used as the inoculum. Although both types of samples contained approximately 25 DGGE bands, only 3 to 6 bands were common. These differences can be attributed to the reactor type (FF versus granular) and to the feeding medium. The Archaea profiles were less complex than the Bacteria profiles, with 8 to 10 bands in the granular sludge (Fig. 3B). The most prominent bands were also present in the biofilm. This result suggests that methanogenesis was occurring with the same type of Archaea in both reactors.
FIG. 3.
Bacteria and Archaea profiles by PCR-DGGE. DNA was extracted from reactor biofilm samples. 16S rRNA gene sequences were PCR amplified with primers specific for Bacteria or Archaea, and PCR products were transferred onto a 20 to 70% urea-formamide DGGE gel. (A) Bacteria. Lanes: 1 and 9, original granular sludge; 2 to 8, biofilm samples taken after 7, 14, 35, 56, 81, 116, and 180 days of operation, respectively. Lanes 7 to 9 are from another DGGE gel and were adjusted on the basis of the profiles of the original sludge. (B) Archaea. Lane 1, original granular sludge; lanes 2 to 5, biofilm samples taken after 7, 14, 56, and 81 days of operation, respectively.
Biofilm structure determined by FISH.
Biofilm sampling was carried out after stabilization of the Bacteria and Archaea as determined by PCR-DGGE. As observed by phase-contrast microscopy, the area colonized by the biofilm increased with time although a number of areas were devoid of biofilm. The biofilm was irregular, having a thickness varying between 10 and 30 μm.
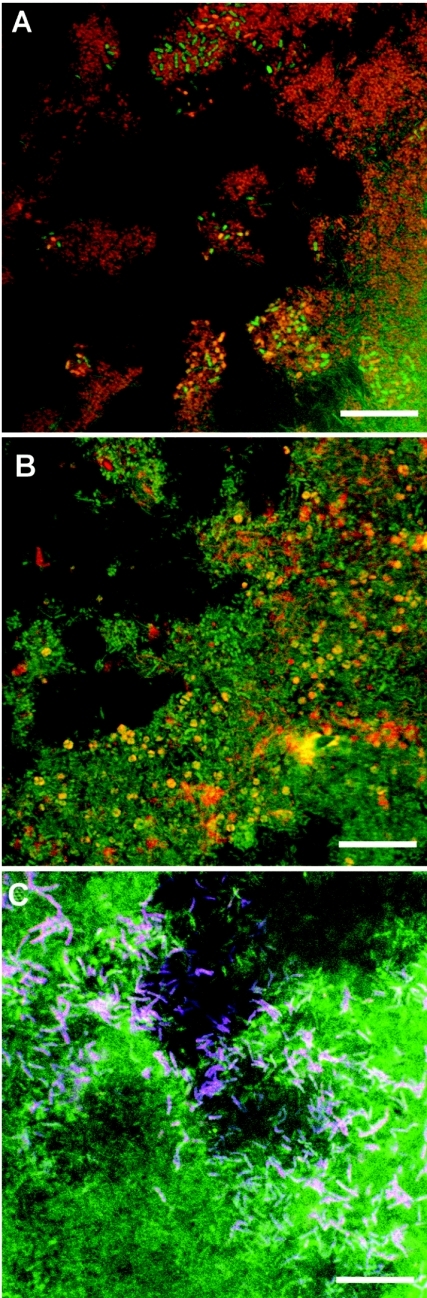
Several biofilm samples were hybridized with fluorescent probes at different time points. Representative results are illustrated in Fig. 4. Bacteria were more abundant than Archaea and were composed of different types of cocci, coccobacilli, and rods (Fig. 4A). Archaea were less diversified, mostly Methanosarcina-like microorganisms, but some rods were also observed (Fig. 4B). D. hafniense cells were detected in high numbers scattered throughout the biofilm, sometimes as loose microcolonies. The hybridization signals of two specific probes (PCP-1-4 and PCP-1-8) coupled to different fluorochromes and both targeting D. hafniense in the biofilm gave almost identical patterns, thus confirming the specificity of D. hafniense detection (Fig. 4C).
FIG. 4.
Spatial arrangement of Bacteria, Archaea, and D. hafniense in the anaerobic FF reactor. Biofilm samples were fixed, permeabilized, hybridized with fluorescently labeled oligonucleotides, and then counterstained with YOYO-1 before being examined by CLSM. FISH was performed with EUB338-Cy3 (Bacteria, panel A), ARC915-Cy3 (Archaea, panel B), and PCP-1-4-Cy3 and PCP-1-8-Cy5 (D. hafniense, panel C). Panels A, B, and C are biofilms taken after 56, 116, and 225 days of operation, respectively. Bars correspond to 20 μm.
Archaea, Bacteria, and D. hafniense were quantified in the biofilms by taking at least 20 cross-sectional images of the biofilm (Table 2). This quantitative analysis confirmed the predominance of Bacteria, with a proportion of 70%, over Archaea, with a proportion of 10%. Quantitative analysis also showed that D. hafniense cells covered 19% of the total surface of the biofilm.
TABLE 2.
Proportions of Bacteria, Archaea, and D. hafniense in the reactor biofilms
Percentage of the total surface of the biomass revealed by the probe as determined by staining with YOYO-1.
n, number of microscopic fields examined by CLSM.
DISCUSSION
The reactor had to be gradually exposed to increasing amounts of PCP to allow adaptation of the anaerobic consortium, more specifically, the xenobiotic-sensitive methanogens (4). It has been shown that low PCP concentrations (0.75 to 11.3 μM) completely inhibit methanogenesis (13, 49). For instance, Tartakovsky et al. (44, 45) demonstrated with a UASB bioreactor that when the PCP concentration is increased too rapidly, accumulation of more toxic intermediates occurs (i.e., trichlorophenols), leading to reactor failure. Accumulation of acetate after start-up can be explained by the lower growth rate of methanogenic Archaea compared to fermentative and acetogenic bacteria, which degraded organic matter, propionate, and butyrate rapidly following start-up. However, aceticlastic methanogens, which are responsible for the transformation of acetate into CH4 and CO2, could have been inhibited by the toxicity of PCP or its intermediates, such as 3-CP, since methanogens are particularly sensitive to toxic compounds (4). Also, only trace levels of butyrate and propionate were found in the effluents, thus demonstrating that acetogenic bacteria were not affected by the toxicity in the reactor and were readily transforming these VFA. These results are different from those obtained by Wu et al. (49), who tested the effect of PCP on the VFA degradation of PCP-acclimatized and nonacclimatized anaerobic granular sludge. They found that all VFA-degrading populations were affected by PCP toxicity. More specifically, propionate degraders were the most sensitive to the presence of PCP, followed by acetate-utilizing methanogens and then by butyrate degraders. These differences from our system could be attributed to the dissimilar inocula used, which probably contained different microorganisms.
The degradation rate achieved by our reactor (1,173 μM day−1) compares advantageously to those achieved in other studies (17, 18). For instance, Juteau et al. (24) obtained 99% PCP removal with a PCP load of 60 μM day−1 with their FF bioreactor. They observed mostly 3,4-DCP and 3-CP as major intermediates and no significant 3-CP degradation. Beaudet et al. (6) obtained complete dechlorination of PCP with a PCP loading rate of up to 68 μM day−1 in an anaerobic FF reactor fed with PCP extracted from contaminated wood chips.
More studies on PCP degradation with UASB anaerobic bioreactors have been performed and have demonstrated that these reactors could degrade higher PCP loads than FF bioreactors (13, 17, 19, 48). For instance, Wu et al. (49) reported the highest PCP load to be degraded by an anaerobic bioreactor. The maximum PCP load tested was 330 to 364 μM day−1, with removal of more than 99% of the PCP and no detection of CP intermediates. In the present study, our reactor achieved a threefold higher PCP removal rate. However, 3-CP, the main PCP degradation intermediate, was still present in the effluent. These differences can be attributed to a distinct granular sludge inoculum. We used granular sludge treating wastewater from an apple-processing plant, whereas the UASB reactor studied by Wu et al. (49) was inoculated with both (i) granules previously enriched with a mixture of VFA and (ii) contaminated soils enriched anaerobically in the presence of PCP.
Our FF reactor also achieved better degradation than UASB reactors inoculated with other desulfitobacteria. Inoculation of a UASB bioreactor with sterilized granular sludge with D. hafniense DCB-2 resulted in 99% transformation of PCP into 3,4,5-trichlorophenol when the reactor was fed a PCP load of up to 158 μM day−1 (11), whereas UASB reactors inoculated with strain PCP-1 reached a PCP load of 300 μM day−1 with 99% PCP degradation and less than 5% residual 3-CP was detected in the effluent (44).
Aerobic systems are also very efficient for the degradation of PCP. For example, Stinson et al. (43) were able to degrade PCP loads of up to 1,352 μM day−1 in an aerobic FF bioreactor, which is comparable to what has been obtained with our reactor. Jarvinen et al. (21) obtained 99.9% PCP degradation with a PCP load of 2,780 μM day−1 with an aerobic fluidized bed reactor.
D. hafniense was detected in the reactor biofilm and was present at a low level in the granular sludge. The conditions that prevailed in the reactor probably allowed this bacterium to grow and colonize the biofilm. Enrichment of desulfitobacteria from anaerobic microflora exposed to halogenated compounds has been frequently observed (7, 9, 10, 30). Lanthier et al. (28) demonstrated by PCR that desulfitobacteria are widely distributed in different soils and sediments from the Province of Québec, Canada. Desulfitobacteria use halogenated compounds as electron acceptors in a process named “halorespiration,” which allows them to generate energy (16).
Stabilization of Bacteria and Archaea as observed by PCR-DGGE correlates with the acetate profile, which was only stable after 60 days. Tartakovsky et al. (44) showed stabilization of Bacteria after 17 days of operation in a PCP-fed UASB reactor. Liu et al. (31) demonstrated stabilization of the Bacteria and Archaea in two anaerobic acidogenic bioreactors after 13 days of operation. The biofilm in our bioreactor had an uneven surface ranging in thickness from 10 to 30 μm and was devoid of clear mushroom-like structures or channels. This result is consistent with the slow-growing nature of anaerobic microorganisms (33) and also with the lower biomass production in anaerobic bioreactors compared to aerobic systems (41). The general aspect of the biofilms was similar to what has been reported with other types of anaerobic biofilm. For example, Amann et al. (3) looked at the structure of anaerobic biofilms on glass coverslips in anaerobic FF sulfidogenic reactors and observed a 5- to 10-μm-thick and irregular biofilm after 3 weeks of reactor operation. Araujo et al. (5) examined an anaerobic biofilm growing in a chemostat containing a Robbins device and inoculated with disaggregated granular sludge. They observed that the biofilm rapidly colonized the polypropylene surface and formed an unequal-thickness layer of 0 to 9 μm after 11 days of operation.
Targeting specific microbial species in multispecies anaerobic biofilms has not been reported frequently in the literature. For instance, Amann et al. (3) and Raskin et al. (37) were the first to report the detection of specific microorganisms in anaerobic biofilms by FISH. With specific hybridization probes targeting specific 16S rRNA sequences, they visualized two sulfate-reducing bacterial populations, named population types 1 and 2, in sulfidogenic biofilms established in anaerobic fixed-bed bioreactors. These populations were related to Desulfuromonas acetoxidans and Desulfovibrio vulgaris, respectively, and were arranged as microcolonies. In this study, D. hafniense cells were found scattered throughout the multispecies methanogenic biofilm and accounted for almost 20% of the biofilm in the reactor. Scattering could have occurred by cell motility. For instance, D. hafniense DCB-2 is motile as it has one or two flagella (10), and even if D. hafniense PCP-1 was reported as a nonmotile microorganism (7), motile cells have been observed in pure culture (unpublished results). Scattering could have also been the result of biofilm detachment and reattachment elsewhere on the slide. Distribution of D. hafniense throughout the biofilm is probably important for the community, as this bacterium would dehalogenate PCP in less toxic compounds, as does D. hafniense PCP-1 (7). In return, syntrophic associations with other microorganisms in the biofilm must occur as D. hafniense, when grown as a pure culture, needs specific carbon sources such as pyruvate to grow and specific electron donors to perform PCP dechlorination. In this respect, D. hafniense could have used chemotaxis toward pyruvate producers to scatter throughout the biofilm. We previously demonstrated that D. hafniense PCP-1 forms microcolonies and is mostly present at the surface of PCP-fed UASB reactors (27). However, in this particular case, strain PCP-1 was added to already formed granules. The presence of strain PCP-1 at the surface would then protect the other microorganisms inside the granules from PCP toxicity. Knowing more about the syntrophic associations between D. hafniense and Bacteria or Archaea would allow a better understanding of PCP degradation in these reactors.
Acknowledgments
This research was supported by a grant to R.V. from the Natural Sciences and Engineering Research Council of Canada (NSERC grant OGP0155558), the Fonds Québécois de la Recherche sur la Nature et les Technologies, and the Foundation Armand-Frappier.
We greatly appreciate the expert technical assistance of S. Milot, R. Alary, and M. Desrosiers at INRS—Institut Armand-Frappier and thank the laboratory of Serge Guiot at the Biotechnological Research Institute of Montreal for providing anaerobic sludge. We also thank Nathalie Arbour for kindly revising the English style and grammar.
REFERENCES
- 1.Ahring, B. K., N. Christiansen, I. Mathrani, H. V. Hendriksen, A. J. L. Macario, and E. C. de Macario. 1992. Introduction of a de novo bioremediation ability, aryl reductive dechlorination, into anaerobic granular sludge by inoculation of sludge with Desulfomonile tiedjei. Appl. Environ. Microbiol. 58:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., J. Stromley, R. Devereux, R. Key, and D. A. Stahl. 1992. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl. Environ. Microbiol. 58:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angelidaki, I., L. Ellegaard, and B. K. Ahring. 2003. Applications of the anaerobic digestion process. Adv. Biochem. Eng. Biotechnol. 82:1-33. [DOI] [PubMed] [Google Scholar]
- 5.Araujo, J. C., G. Brucha, J. R. Campos, and R. F. Vazoller. 2000. Monitoring the development of anaerobic biofilms using fluorescent in situ hybridization and confocal laser scanning microscopy. Water Sci. Technol. 41(12):69-77. [Google Scholar]
- 6.Beaudet, R., G. McSween, F. Lépine, S. Milot, and J.-G. Bisaillon. 1997. Anaerobic biodegradation of pentachlorophenol in a liquor obtained after extraction of contaminated chips and wood powder. J. Appl. Microbiol. 82:186-190. [DOI] [PubMed] [Google Scholar]
- 7.Bouchard, B., R. Beaudet, R. Villemur, G. McSween, F. Lépine, and J.-G. Bisaillon. 1996. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int. J. Syst. Bacteriol. 46:1010-1015. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, S. A., D. R. Shelton, D. Berry, and J. M. Tiedje. 1983. Anaerobic biodegradation of phenolic compounds in digested sludge. Appl. Environ. Microbiol. 46:50-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitenstein, A., A. Saano, M. Salkinoja-Salonen, J. R. Andreesen, and U. Lechner. 2001. Analysis of a 2,4,6-trichlorophenol-dehalogenating enrichment culture and isolation of the dehalogenating member Desulfitobacterium frappieri TCP-A. Arch. Microbiol. 175:133-142. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen, N., and B. K. Ahring. 1996. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int. J. Syst. Bacteriol. 46:442-448. [Google Scholar]
- 11.Christiansen, N., and B. K. Ahring. 1996. Introduction of a de novo bioremediation activity into anaerobic granular sludge using the dechlorinating bacterium DCB-2. Antonie Leeuwenhoek 69:61-66. [DOI] [PubMed] [Google Scholar]
- 12.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.) 1999. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, D.C.
- 13.Duff, S. J. B., K. J. Kennedy, and A. J. Brady. 1995. Treatment of dilute phenol/PCP wastewaters using the upflow anaerobic sludge blanket (UASB) reactor. Water Res. 29:645-651. [Google Scholar]
- 14.El Fantroussi, S., H. Naveau, and S. N. Agathos. 1998. Anaerobic dechlorinating bacteria. Biotechnol. Prog. 14:167-188. [DOI] [PubMed] [Google Scholar]
- 15.Fang, H. H., H. Liu, and T. Zhang. 2002. Characterization of a hydrogen-producing granular sludge. Biotechnol. Bioeng. 78:44-52. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 17.Hendriksen, H. V., and B. Ahring. 1993. Anaerobic dechlorination of pentachlorophenol in fixed-film and upflow anaerobic sludge blanket reactors using different inocula. Biodegradation 3:399-408. [Google Scholar]
- 18.Hendriksen, H. V., S. Larsen, and B. K. Ahring. 1991. Anaerobic degradation of PCP and phenol in fixed-film reactors: the influence of an additional substrate. Water Sci. Technol. 24(3-4):431-436. [Google Scholar]
- 19.Hendriksen, H. V., S. Larsen, and B. K. Ahring. 1992. Influence of a supplemental carbon source on anaerobic dechlorination of pentachlorophenol in granular sludge. Appl. Environ. Microbiol. 58:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hörber, C., N. Christiansen, E. Arvin, and B. K. Ahring. 1998. Improved dechlorinating performance of upflow anaerobic sludge blanket reactors by incorporation of Dehalospirillum multivorans into granular sludge. Appl. Environ. Microbiol. 64:1860-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarvinen, K. T., E. S. Melin, and J. A. Puhakka. 1994. High-rate bioremediation of chlorophenol-contaminated groundwater at low temperatures. Environ. Sci. Technol. 28:2387-2392. [DOI] [PubMed] [Google Scholar]
- 22.Jensen, J. 1996. Chlorophenols in the terrestrial environment. Rev. Environ. Contam. Toxicol. 146:25-51. [DOI] [PubMed] [Google Scholar]
- 23.Juteau, P., R. Beaudet, G. McSween, F. Lépine, and J.-G. Bisaillon. 1995. Study of the reductive dechlorination of pentachlorophenol by a methanogenic consortium. Can. J. Microbiol. 41:862-868. [DOI] [PubMed] [Google Scholar]
- 24.Juteau, P., R. Beaudet, G. McSween, F. Lépine, S. Milot, and J.-G. Bisaillon. 1995. Anaerobic biodegradation of pentachlorophenol by a methanogenic consortium. Appl. Microbiol. Biotechnol. 44:218-224. [Google Scholar]
- 25.Khodadoust, A. P., M. T. Suidan, G. A. Sorial, and D. D. Dionysiou. 1999. Desorption of pentachorophenol from soil using mixed solvents. Environ. Sci. Technol. 33:4483-4491. [Google Scholar]
- 26.Labbé, N., P. Juteau, S. Parent, and R. Villemur. 2003. Bacterial diversity in a marine methanol-fed denitrification reactor at the Montreal Biodome, Canada. Microb. Ecol. 46:12-21. [DOI] [PubMed] [Google Scholar]
- 27.Lanthier, M., B. Tartakovsky, R. Villemur, G. DeLuca, and S. R. Guiot. 2002. Microstructure of anaerobic granules bioaugmented with Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 68:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanthier, M., R. Villemur, F. Lépine, J. Bisaillon, and R. Beaudet. 2001. Geographic distribution of Desulfitobacterium frappieri PCP-1 and Desulfitobacterium spp. in soils from the province of Québec, Canada. FEMS Microbiol. Ecol. 36:185-191. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence, J. R., D. R. Korber, G. M. Wolfaardt, and D. E. Caldwell. 1997. Analytical imaging and microscopy techniques, p. 29-51. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Setzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. American Society for Microbiology, Washington, D.C.
- 30.Lecouturier, D., J. J. Godon, and J. M. Lebeault. 2003. Phylogenetic analysis of an anaerobic microbial consortium deiodinating 5-amino-2,4,6-triiodoisophthalic acid. Appl. Microbiol. Biotechnol. 62:400-406. [DOI] [PubMed] [Google Scholar]
- 31.Liu, W. T., O. C. Chan, and H. H. Fang. 2002. Microbial community dynamics during start-up of acidogenic anaerobic reactors. Water Res. 36:3203-3210. [DOI] [PubMed] [Google Scholar]
- 32.Manz, W., K. Wendt-Potthoff, T. R. Neu, U. Szewzyk, and J. R. Lawrence. 1999. Phylogenetic composition, spatial structure, and dynamics of lotic bacterial biofilms investigated by fluorescent in situ hybridization and confocal laser scanning microscopy. Microb. Ecol. 37:225-237. [DOI] [PubMed] [Google Scholar]
- 33.Morris, J. G. 1994. Obligately anaerobic bacteria in biotechnology. Appl. Biochem. Biotechnol. 48:75-106. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pind, P. F., I. Angelidaki, B. K. Ahring, K. Stamatelatou, and G. Lyberatos. 2003. Monitoring and control of anaerobic reactors. Adv. Biochem. Eng. Biotechnol. 82:135-182. [DOI] [PubMed] [Google Scholar]
- 36.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raskin, L., R. I. Amman, L. K. Poulsen, B. E. Rittmann, and D. A. Stahl. 1995. Use of ribosomal RNA-based molecular probes for characterization of complex microbial communities in anaerobic biofilms. Water Sci. Technol. 31(1):261-272. [Google Scholar]
- 38.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Skiadas, I. V., H. N. Gavala, J. E. Schmidt, and B. K. Ahring. 2003. Anaerobic granular sludge and biofilm reactors. Adv. Biochem. Eng. Biotechnol. 82:35-67. [DOI] [PubMed] [Google Scholar]
- 42.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stinson, M. K., H. S. Skovronek, and T. J. Chresand. 1991. EPA site demonstration of BioTrol aqueous treatment system. J. Air Waste Manage. Assoc. 41:228-233. [DOI] [PubMed] [Google Scholar]
- 44.Tartakovsky, B., M.-J. Levesque, R. Dumortier, R. Beaudet, and S. R. Guiot. 1999. Biodegradation of pentachlorophenol in a continuous anaerobic reactor augmented with Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 65:4357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tartakovsky, B., M. F. Manuel, D. Beaumier, C. W. Greer, and S. R. Guiot. 2001. Enhanced selection of an anaerobic pentachlorophenol-degrading consortium. Biotechnol. Bioeng. 73:476-483. [DOI] [PubMed] [Google Scholar]
- 46.Wagner, M., P. Hutzler, and R. Amann. 1998. Three-dimensional analysis of complex microbial communities by combining confocal laser scanning microscopy and fluorescence in situ hybridization. .In M. H. F. Wilkinson and F. Schut (ed.), Digital image analysis of microbes: imaging, morphometry, fluorometry and motility techniques and applications. John Wiley & Sons, Ltd., Chichester, England.
- 47.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 48.Woods, S. L., J. F. Ferguson, and M. M. Benjamin. 1989. Characterization of chlorophenol and chloromethoxybenzene biodegradation during anaerobic treatment. Environ. Sci. Technol. 23:62-68. [Google Scholar]
- 49.Wu, W.-M., L. Bhatnagar, and J. G. Zeikus. 1993. Performance of anaerobic granules for degradation of pentachlorophenol. Appl. Environ. Microbiol. 59:389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]