Abstract
Site-specific attachment of paramagnetic spin labels to biomolecules causes distance-dependent line-broadening effects, which can be exploited to study the structure and dynamics of these molecules in solution. This protocol describes how to attach nitroxide spin labels to proteins and how to collect and analyze NMR data using these labeled samples. We also explain how to derive distance restraints for paramagnetic relaxation enhancement nuclear magnetic resonance (PRE-NMR) studies.
Keywords: Paramagnetic relaxation enhancement, Large protein NMR, Nuclear magnetic resonance spectroscopy, Nitroxide spin label, Distance restraints
Background
This protocol describes how to attach nitroxide spin labels to proteins and how the modified proteins can be employed to derive distance restraints using paramagnetic relaxation enhancement nuclear magnetic resonance (PRE-NMR) methods. Site-specific attachment of paramagnetic spin labels to proteins enhances the transverse relaxation rates of nearby nuclei leading to line-broadening effects that can be used to derive distance restraints (Battiste and Wagner, 2000; Iwahara et al., 2004 ; Clore and Iwahara, 2009). PRE-derived distance restraints have been used to characterize the structures of various molecules, including amongst others membrane proteins ( Roosild et al., 2005 ), multi-domain proteins displaying inter-domain dynamics ( Sjodt et al., 2016 ), single domain proteins (Battiste and Wagner, 2000), protein-DNA complexes (Clore and Iwahara, 2009), transient protein-protein interactions ( Tang et al., 2007 ; Villareal et al., 2011 ), and intrinsically disordered proteins ( Bertoncini et al., 2005 ). Two approaches are generally used to obtain PRE-derived distance restraints using proteins that are labeled with paramagnetic probes. The first method quantitatively measures the probe’s effects on the transverse relaxation rates of nearby protein nuclei by determining Γ2 (described in detail by Iwahara and Clore) ( Iwahara et al., 2004 ; Clore and Iwahara, 2009). The second method is less quantitative, but in practice it is easier to implement. It was originally employed by Battiste and Wagner, and measures the probe’s effects by comparing cross-peak intensity ratios in diamagnetic- and paramagnetic-spectra of the labeled protein (Battiste and Wagner, 2000). The nitroxide spin label MTSL is frequently used as a paramagnetic probe in PRE-NMR studies of proteins because it is readily attached via a disulfide bond to cysteine residues, and also relatively small, inexpensive, and commercially available. The aim of this protocol is to describe how to attach MTSL nitroxide spin labels to proteins and how to derive PRE-distance restraints following the approach described by Battiste and Wagner (2000).
Materials and Reagents
Amber vial
Desalting spin column, for example, a ZebaTM spin desalting column, 7k MWCO, 2 ml (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 89889)
15 ml conical test tube
Aluminum foil
A centrifugal filter, such as Amicon Ultra-15 centrifugal filter (EMD Millipore, catalog number: UFC900308)
Standard 5 mm NMR tube (SP Industries, catalog number: 535-PP-7)
2D [1H-15N]-HSQC spectrum of the native protein acquired using standard methods ( Cavanagh et al., 1995 )
Dithiothreitol (DTT); > 99% purity (Gold Bio, catalog number: DTT50)
Deuterium oxide (D2O); ≥ 99.8% purity (Sigma-Aldrich, catalog number: 617385)
Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl); ≥ 99% purity (Fisher Scientific, catalog number: BP153)
Sodium chloride (NaCl); ≥ 99% purity (Fisher Scientific, catalog number: S271)
S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl (MTSL) methanesulfonothioate (Toronto Research Chemicals, catalog number: O875000)
Acetonitrile; ≥ 99.9% purity (Fisher Scientific, catalog number: A996)
Sodium phosphate monobasic (NaH2PO4); ≥ 98% purity (Fisher Scientific, catalog number: S369)
Sodium azide; ≥ 99% purity (Fisher Scientific, catalog number: S227I)
Sodium ascorbate; ≥ 98% purity (Sigma-Aldrich, catalog number: A7631)
Labeling buffer (see Recipes)
200 mM MTSL stock solution (see Recipes)
Example NMR buffer (see Recipes)
250 mM sodium ascorbate stock solution (see Recipes)
Note: These reagents were used to obtain the PRE data according to Sjodt et al., 2016. However, other brands of these materials may be used if necessary.
Equipment
Centrifuge (Beckman Coulter, model: Allegra X-14R)
SX4750A swinging bucket rotor (Beckman Coulter, model: SX4750A ARIESTM Roter)
-
MALDI-TOF mass spectrometer (Thermo Fisher Scientific, Applied BiosystemsTM, model: Voyager-DE-STR)
Note: This product has been discontinued. Examples of other MALDI-TOF instruments include Shimadzu, model: AXIMA Assurance Linear MALDI-TOF; Bruker, model: microflex LT/SH.
-
NMR experiments system
Note: NMR experiments used in this protocol are part of the Bruker standard pulse sequence library and were performed on Bruker Avance spectrometers equipped with triple-resonance cryogenic probes (Bruker Corporation).
Software
-
NMRPipe ( Delaglio et al., 1995 )
-
Bruker TopSpinTM
-
Sparky (Goddard and Kneller, 2008)
-
Microsoft Excel
-
XPLOR-NIH ( Schwieters et al., 2003 )
Procedure
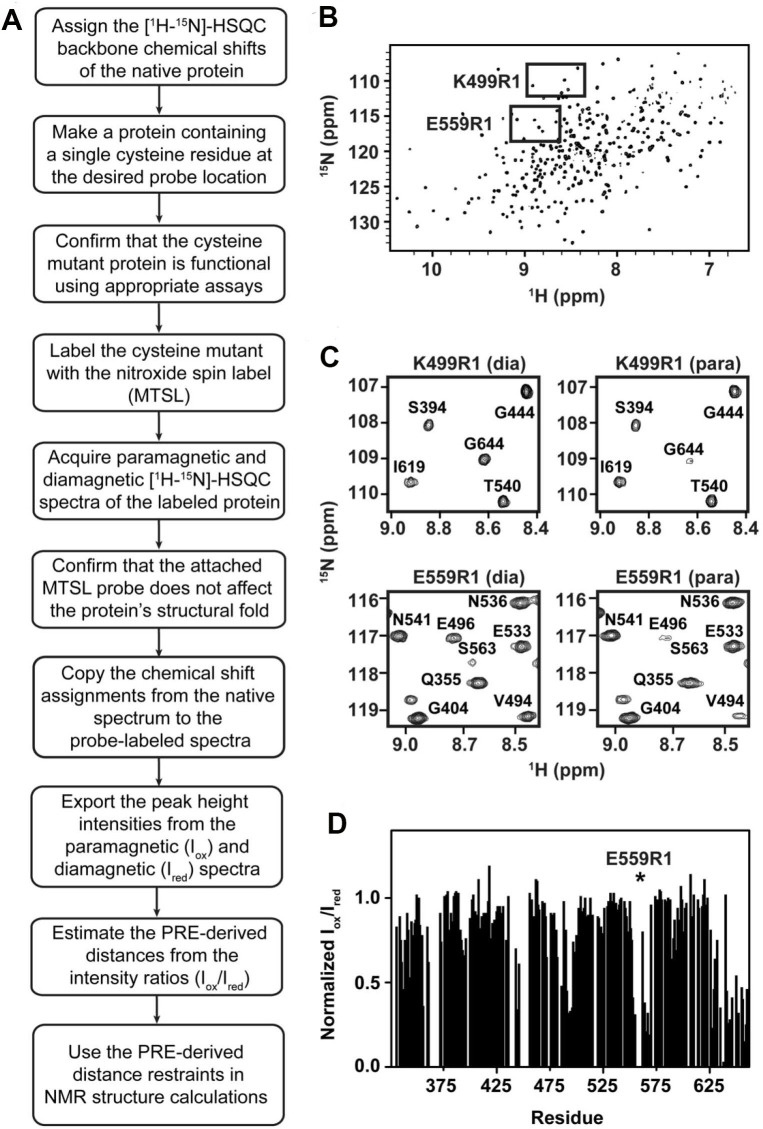
Figure 1A shows a flow chart of the steps that need to be performed in order to label a protein with MTSL and how PRE-NMR distance restraints can be measured. In the procedure, the MTSL probe is attached via a disulfide bond to a protein containing a single cysteine residue. NMR spectra are then acquired and interpreted to extract distance restraints. In order to interpret the NMR spectra, the backbone chemical shifts of the protein must first be obtained.
Figure 1. A flow chart of nitroxide labeling and representative PRE data.
A. A flow chart describing the steps taken when site-specifically labeling proteins with nitroxide spin labels for PRE-derived structural studies. B. Representative [1H-15N]-HSQC spectrum of IsdHN2N3 (see Sjodt et al., 2016 for more information). Boxed regions indicate the representative sections of each PRE probe’s spectra shown in (C) (K499R1 and E559R1). C. Magnified regions showing the selective distance dependent line-broadening for the K499R1 (top) and E559R1 (bottom) probes. For each probe the diamagnetic and paramagnetic spectra are shown on the left and right panels, respectively. D. Representative PRE profile of the E559R1 probe data. Normalized intensity ratios (Iox/Ired) are shown as a function of residue number. The asterisk depicts the location of the probe. Errors of the ratio measurements are approximately 10-15% based on signal to noise of the NMR spectra, which can lead to values in excess of 1. Errors also can occur as a result of manipulating the sample (adding ascorbate to reduce the probe) and instrument instability. These errors are partially accounted for by adding ± 5 Å to the distance restraints that are obtained from the ratio data (Battiste and Wagner, 2000).
-
Assign the backbone chemical shifts of the native protein
Acquire a high quality 2D [1H-15N]-HSQC spectrum of the native protein to be used as a reference (i.e., a spectrum containing the appropriate number of resolved cross-peaks based on the amino acid sequence of the protein).
Assign the backbone amide chemical shifts in the native protein using standard NMR procedures (for standard NMR experiments see Cavanagh et al., 1995 ).
-
Generate a protein mutant with a single cysteine residue
For these studies, the protein should contain a single cysteine residue. Check the protein’s primary sequence to determine if it contains native cysteine residues that could be inadvertently labeled by the nitroxide probe. If necessary, make conservative mutations to these cysteine residues to prevent non-specific labeling. If biochemical assays are available for the protein, they should be performed to ensure these mutations do not impair its function.
Choose the desired location in the protein to which the nitroxide probe will be attached. This location needs to be at a residue whose sidechain is solvent-exposed. The residue should be located in a structured loop or within defined secondary structure. This is important because if the probe is located in a structurally disordered region of the protein, the increased flexibility of the polypeptide backbone can reduce the quantitative strength of the technique.
Perform site-directed mutagenesis to incorporate a single cysteine residue at the desired location to which the nitroxide probe will be attached.
Purify 15N-labeled protein containing the cysteine mutation. Use buffers that are supplemented with 2.5 mM DTT to prevent formation of inter-molecular disulfide bonds that can lead to protein aggregation (see Note 1). For a review on expression and purification of proteins see Gräslund et al. (2008) .
Prior to labeling the purified cysteine mutant with MTSL, verify that the cysteine mutation does not affect the structure of the protein. This can be accomplished by comparing the [1H-15N]-HSQC spectra of the mutant and native proteins. The chemical shift position of the cross-peaks in the spectra should be similar, with only localized changes occurring near the site of the mutation.
-
Site-specific labeling the protein
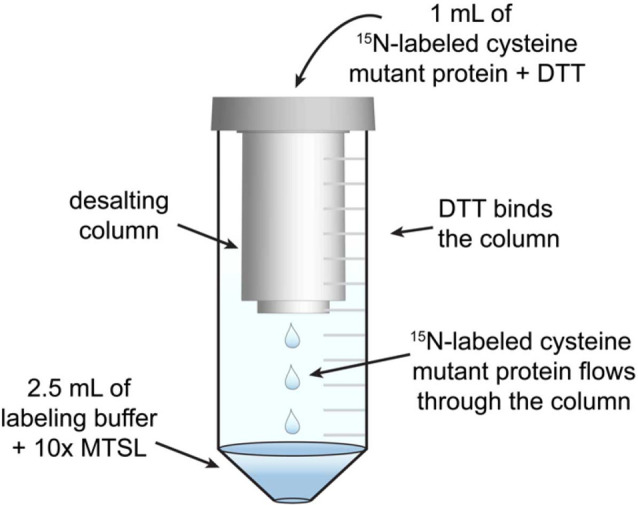
The aim of the following steps is to label the 1H-15N labeled purified cysteine mutant of the protein with MTSL. Care is taken to maintain the cysteine mutant under reducing conditions until it is modified with MTSL. The apparatus used to label the protein is shown in Figure 2, and consists of a conical tube and desalting column.
Buffer exchange the 15N-labeled cysteine mutant into labeling buffer that has been supplemented with fresh 2.5 mM DTT. The protein concentration should be at least 250-300 µM.
Make a stock solution of MTSL by adding acetonitrile directly to the amber vial in which the MTSL was shipped. The final concentration of the stock solution should be 200 mM MTSL. Store the MTSL stock solution at -20 °C (see Notes 2 and 3).
Immediately before starting the labeling reaction, dilute the protein with labeling buffer to a final concentration of 250-300 µM protein. The final volume of the diluted protein solution should be 1 ml. The protein should be diluted with labeling buffer that does not contain DTT. From this step forward, all buffers used in this procedure should not contain DTT.
Equilibrate a Zeba spin desalting column with labeling buffer as per the manufacturer’s instructions.
Construct the MTSL solution that will be used to label the protein. Pipette 2.5 ml of labeling buffer into a clean 15 ml conical tube. Add MTSL from the stock solution to the conical tube. The amount added should be sufficient to achieve a final concentration of MTSL that is 10x the molar amount of protein that will be labeled (e.g., if the final concentration of protein in the reaction will be 300 µM then the concentration of MTSL should be 3 mM). Make sure to shield all MTSL solutions from light by covering the tube with foil first.
Construct the apparatus that will be used to label the protein (Figure 2). Place the equilibrated desalting column into the conical tube from step C5. The column will rest above the MTSL-containing solution and will leave room for the protein solution to flow through.
Slowly pipette the protein onto the center of the column’s resin bed (Figure 2).
Centrifuge as per the manufacturer’s instructions. The protein will flow through the column and into the labeling solution, while DTT present in the protein solution will remain on the column. The process is considered complete after the elution of all the protein solution into the MTSL solution at the bottom of the collection tube. This will produce a solution located at the bottom of the conical tube that has a total volume of ~3.5 ml. Discard the desalting column.
Cover the conical tube containing the modification reaction with foil and let it incubate under agitation for 15 min at room temperature.
After 15 min, add additional MTSL from the stock solution to the modification reaction. The amount of added MTSL should yield a final concentration that is 20x greater than the protein’s concentration in the conical tube.
Place tube back onto a rotating device and let the protein:MTSL mixture incubate under agitation overnight at room temperature (~16 h).
Buffer exchange the MTSL-labeled protein sample into the desired NMR buffer using a fresh Amicon Ultra-15 centrifugal filter. This will remove any excess (unligated) MTSL (see Notes 4-6). As mentioned above, the NMR buffer should not contain any DTT.
Verify that the protein is labeled with MTSL by MALDI-TOF mass spectrometry. As compared to the unligated protein, the mass of the modified protein should increase by ~186 Da (the weight of the attached probe). Although MALDI-TOF is not quantitative, in the mass spectrum > 90% of the protein should be labeled with MTSL. If biochemical assays are available for the protein, they should be performed on labeled protein to verify that MTSL attachment does not impair its function.
For NMR studies, the MTSL modified protein should have a final concentration of ~300 µM (see Note 7). The NMR buffer should contain 8-10% D2O, but be devoid of any DTT. Place the sample in a standard NMR tube and shield from light using foil.
-
NMR data acquisition
Use a standard 2D- [1H-15N]-HSQC pulse sequence to acquire a spectrum of the freshly MTSL-labeled protein. Typically, the 1H and 15N dimensions are defined by 2,048 and 256 complex points, respectively. At this time, the sample is in its paramagnetic state. Its spectrum should be well dispersed, indicating that the modified protein is folded and not aggregating.
Make a stock solution of 250 mM sodium ascorbate by dissolving it in NMR buffer.
After the [1H-15N]-HSQC spectrum has been acquired of the paramagnetic protein (step D1), remove the NMR tube from the magnet. Using a long pipette that fits into the NMR tube, add sodium ascorbate to a final concentration that is 5x greater than the concentration of the labeled protein. This step should be performed carefully, by pipetting the solution up and down slowly so as to mix the solution, while preventing any sample loss. Addition of sodium ascorbate will reduce MTSL’s unpaired electron to form the diamagnetic state. Let the sample reduce for a minimum of 3 h at room temperature (no agitation is needed).
Acquire a [1H-15N]-HSQC spectrum of the reduced diamagnetic MTSL:protein sample using acquisition parameters that are identical to those used to acquire the paramagnetic spectrum described in step D1.
Figure 2. Schematic of the apparatus used to label proteins with MTSL.
Data analysis
-
Process and analyze the paramagnetic and diamagnetic NMR data
Process the acquired NMR data using standard software (e.g., NMRPipe [ Delaglio et al., 1995 ] or Bruker TopSpinTM software). Both the paramagnetic and diamagnetic datasets should be processed identically. Representative [1H-15N]-HSQC spectra of diamagnetic and paramagnetic samples are shown in Figures 1B and 1C.
Analyze the NMR spectra using standard software (e.g., Sparky) (Goddard and Kneller, 2008). The diamagnetic spectrum should be analyzed first, as it is most closely related to the previously assigned spectrum of the native protein. Assign the cross-peaks in the diamagnetic spectrum using the known chemical shifts of the native protein. This can readily be accomplished by overlaying the [1H-15N]-HSQC spectra of the native and labeled proteins, and then transferring the chemical shift assignments. The cross peaks in the spectra should overlay well, with only localized differences occurring for cross peaks that originate from residues that are located near the attached probe. Considerable care must be taken to make sure that the assignments are correctly assigned.
Measure the intensities of the cross peaks in the assigned spectrum of the diamagnetic protein. Care should be taken to make sure that the cross peaks that are analyzed are well resolved (see Note 8). When making the intensity measurement, make sure that it is made at the peak’s maximum. In our experience, measuring peak height is sufficient for extracting distance restraints. However, measuring cross peak volumes is also acceptable. It is critical to extensively check the sequence-specific chemical shift assignments of the diamagnetic spectrum. Generate a list of cross peak intensities using the analysis software. The list should contain the cross peak’s intensity and the identity of the residue.
-
Label the cross peaks in the paramagnetic spectrum using the curated chemical shift list that was used to analyze the diamagnetic spectrum. Using the procedures outlined in Data analysis A3, measure the peak intensities of isolated cross peaks in the paramagnetic spectrum. If a cross-peak in the paramagnetic spectrum is significantly broadened such that its intensity is near the noise level (e.g., Figure 1C, residue S563 in the E559R1 paramagnetic panel), accurate measurement of its intensity is not possible. Therefore, these cross peaks should not be employed to quantitatively relate the NMR data to inter-atomic distances, and intensities from these cross peaks should not be analyzed further. However, the identity of residues exhibiting significant line broadening in the paramagnetic spectrum should be noted. This is because distance restraints to these amino acids can still be employed in the structure calculations as complete broadening indicates that they are within ~12 Å of the probe. Use the software to generate a list of cross peak intensities using the analysis software. The list should contain the cross peak’s intensity and the identity of the residue. A sample table depicting a list of cross peak intensities is shown in Table 1.
aNote: This data was adapted from the NMR study of IsdHN2N3 (Sjodt et al., 2016).
Calculate the intensity ratios using standard analysis software (e.g., Microsoft Excel). Import both the diamagnetic and paramagnetic intensity lists (generated from Data analysis A3 and A4) into a single spreadsheet. For each residue, use the cross peak intensity data to calculate the ratio of its intensity in the paramagnetic (Iox) and diamagnetic (Ired) states. A representative plot of the intensity ratios versus protein sequence is shown in Figure 1D.
-
Estimating distance restraints from the NMR data
The aim of this section is to describe how to relate the cross-peak intensity ratio data calculated in the previous section to distance restraints. A detailed description describing the effects of paramagnetic relaxation on cross peak intensity, and its relationship to interatomic distance has been described in detail elsewhere (Battiste and Wagner, 2000; Clore, 2015). The reader is encouraged to consult these references to understand the underlying theory. Briefly, the effect can be described by the following equation:
( 1)
where, Iox and Ired are the measured intensities of the cross peaks for the paramagnetic and diamagnetic forms of the protein, respectively, t is the total evolution time of the transverse proton magnetization during the NMR experiment. The values of R correspond to the rate of the transverse relaxation of the amide proton, R2 is the intrinsic relaxation of the proton, R2sp, is the contribution to the relaxation caused by the paramagnetic probe. The value of R2sp is dependent upon r, the distance between the amide proton and the probe. This relationship is described using a modified version of the Solomon-Bloembergen as shown in Equation 2 (Solomon and Bloembergen, 1956; Battiste and Wagner, 2000)
( 2)
where, K is a constant (1.23 x 10-32 cm6 sec-2) that describes the spin properties of the MTSL spin label (Battiste and Wagner, 2000), ωh is the Larmor frequency of the proton spin, τC is the apparent PRE correlation time ( Simon et al., 2010 ). As ωh is known, and values of τC can be estimated based on the molecular weight of the protein ( Cavanagh et al., 1995 ), a calibration curve can be generated using Equations 1 and 2. The curve relates the intensity ratio for a given cross peak to the distance between the amide proton and the probe. For MTSL probes, Iox/Ired ratios less than 1 indicate that the probe and proton atom are within 13-25 Å, whereas Iox/Ired ratios equal to ~0 and ~1 indicate that the probe and proton are separated by less than ~12 Å, or more than ~30 Å, respectively. It is critical to note, that the calibration curves only provide an estimate of the interatomic distances as the probe exhibits significant flexibility. As a result, the distance restraints obtained from the intensity data are imprecise and structure calculation strategies represent the probe using multiple conformers ( Iwahara et al., 2004 ; Clore, 2015).
Construct an intensity to distance calibration curve (Iox/Ired versus r). This is done using Equations 1 and 2, and estimated values of τC and the average amide proton linewidth (R2/π) for structured residues in the protein. A representative calibration curve is shown in Figure 3 and employs values of 23 Hz and 16.4 ns for R2/π and τC, respectively.
Convert the experimental intensity ratios into distance separations using the calibration curve generated in Data analysis B1.
-
Construct a distance restraint table to be used in XPLOR-NIH calculations (Figure 4). The distance restraints should be made between the nitrogen atom of the MTSL ring and the affected amide proton using standard XPLOR-NIH restraint table format ( Schwieters et al., 2003 ). It should be stressed that these distances boundaries are estimates and are not to be used as accurate distance measurements. Two types of distance restraints are employed in the calculations:
If Iox/Ired is < 0.80, the restraint is called ‘attractive’ and is given the assigned distance from Data analysis B2 with an error of ± 5 Å (see Note 9).
If Iox/Ired ≥ 0.80, the restraint is called ‘repulsive’ and is given a lower bound distance of 20 Å with no upper bound distance (Battiste and Wagner, 2000; Reckel et al., 2011 ; Gottstein et al., 2012 ).
Table 1. Sample list of peak intensities and ratiosa .
| Chemical Shift Assignment |
15N-Chemical Shift (ppm) |
1H-Chemical Shift (ppm) |
Paramagnetic Peak Height | Diamagnetic Peak Height |
Peak Height Ratio (Paramagnetic: Diamagnetic) |
| E343 | 128.26 | 9.05 | 827155 | 787464 | 1.05 |
| H344 | 119.68 | 8.92 | 259998 | 744839 | 0.35 |
| A346 | 126.94 | 8.37 | 423217 | 1671520 | 0.25 |
| N348 | 116.84 | 7.98 | 686791 | 1164493 | 0.59 |
| R350 | 120.95 | 9.23 | 169304 | 377806 | 0.45 |
Figure 3. Calibration curve for the estimation of PRE-derived distances.
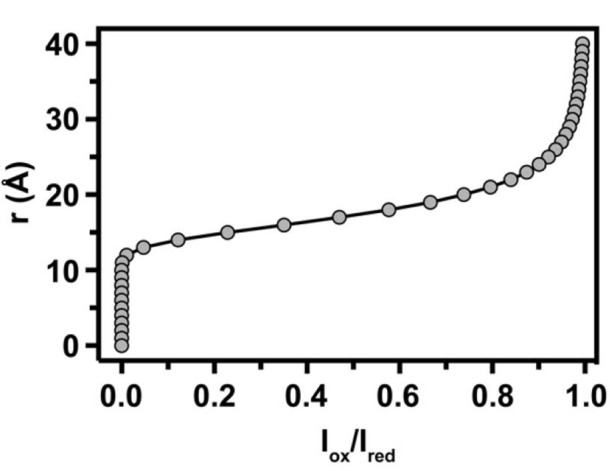
A representative calibration curve used for the conversion of intensity ratios into distances for the NMR study of IsdHN2N3 ( Sjodt et al., 2016 ). The curve was generated according to Equations 1 and 2 using an average linewidth (R2/π) and correlation time (τC) of 23 Hz and 16.4 ns, respectively.
Figure 4. Example of XPLOR-NIH restraint table.
An example of PRE-distance restraint input table in XPLOR-NIH format. This table was adapted from the NMR study of IsdHN2N3 ( Sjodt et al., 2016 ). The ‘segid ALT’ portion identifies the MTSL molecule that was modeled onto the starting structure in three randomized orientations. (For more information on generating a restraint table, the reader is encouraged to refer to Schwieters et al., 2003 ; Iwahara et al., 2004 ; Sjodt et al., 2016 ).
Notes
Make fresh DTT solution before each use.
MTSL will precipitate at high concentrations of acetonitrile, so keep the stock concentrations below 200 mM.
MTSL is light- and air-sensitive.
Make sure to extensively wash the protein in step C11. Excess unligated MTSL will lead to artifacts in the NMR experiments.
The NMR buffer should be optimized for the protein of interest. For a review on buffers suitable for NMR see Kelly et al. (2002) .
The pH of the buffer should be chosen based on the stability of the protein as the MTSL radical is quite stable once ligated to the protein. However, at basic pH it is possible for some of the free MTSL groups to be hydrolyzed to thiols. These groups could react with the remaining free MTSL molecules and lead to formation of disulfide linked biradicals, which would decrease the labeling efficiency. Therefore, buffers that are above neutral values should be avoided. To account for this possibility, a 20-fold molar excess of MTSL is used during the labeling process to ensure efficient labeling of the protein.
The protein concentration should be kept low to prevent non-specific inter-molecular PRE effects.
It has been reported that the tail of a cross-peak that is completely broadened by PRE effects can reduce the height of a neighboring peak by up to 15%, giving rise to false positive broadening effects (Battiste and Wagner, 2000).
The large error bounds placed on the attractive restraints are provided to account for the inherent flexibility of the MTSL probe.
Recipes
-
Labeling buffer
50 mM Tris-HCl (pH 7.8)
50 mM NaCl
-
200 mM MTSL stock solution
Dissolve 10 mg MTSL into 18.92 μl acetonitrile
-
Example NMR buffer
20 mM NaH2PO4 (pH 6.0)
50 mM NaCl
0.01% NaN3
-
250 mM sodium ascorbate stock solution
Dissolve 49.5 mg sodium ascorbate into deionized water to a final volume of 1 ml
Acknowledgments
This protocol was adapted from Sjodt et al. (2016). This work was supported by National Institutes of Health Grants AI52217 (to R.T.C.) and the National Institutes of Health Award F31GM101931 (to M.S.).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Battiste J. L. and Wagner G.(2000). Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry 39(18): 5355-5365. [DOI] [PubMed] [Google Scholar]
- 2.Bertoncini C. W., Jung Y. S., Fernandez C. O., Hoyer W., Griesinger C., Jovin T. M. and Zweckstetter M.(2005). Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci U S A 102(5): 1430-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh J., Fairbrother W. J., Palmer A. G. and Skelton N. J.(1995). Protein NMR Spectroscopy: Principles and Practice. Academic Press pp: 587 [Google Scholar]
- 4.Clore G. M.(2015). Practical aspects of paramagnetic relaxation enhancement in biological macromolecules. Methods Enzymol 564: 485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clore G. M. and Iwahara J.(2009). Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev 109(9): 4108-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J. and Bax A.(1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6(3): 277-293. [DOI] [PubMed] [Google Scholar]
- 7.Gräslund S., Nordlund P., Weigelt J., Hallberg B. M., Bray J., Gileadi O., Knapp S., Oppermann U., Arrowsmith C., Hui R., Ming J., S. dhe-Paganon, Park H. W., Savchenko A., Yee A., Edwards A., Vincentelli R., Cambillau C., Kim R., Kim S. H., Rao Z., Shi Y., Terwilliger T. C., Kim C. Y., Hung L. W., Waldo G. S., Peleg Y., Albeck S., Unger T., Dym O., Prilusky J., Sussman J. L., Stevens R. C., Lesley S. A., Wilson I. A., Joachimiak A., Collart F., Dementieva I., Donnelly M. I., Eschenfeldt W. H., Kim Y., Stols L., Wu R., Zhou M., Burley S. K., Emtage J. S., Sauder J. M., Thompson D., Bain K., Luz J., Gheyi T., Zhang F., Atwell S., Almo S. C., Bonanno J. B., Fiser A., Swaminathan S., Studier F. W., Chance M. R., Sali A., Acton T. B., Xiao R., Zhao L., Ma L. C., Hunt J. F., Tong L., Cunningham K., Inouye M., Anderson S., Janjua H., Shastry R., Ho C. K., Wang D., Wang H., Jiang M., Montelione G. T., Stuart D. I., Owens R. J., Daenke S., Schütz A., Heinemann U., Yokoyama S., Büssow K. and Gunsalus K. C.(2008). Protein production and purification. Nat Methods 5(2): 135-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goddard T. D. and Kneller D. G.(2008). Sparky NMR analysis software.
- 9.Gottstein D., Reckel S., Dotsch V. and Guntert P.(2012). Requirements on paramagnetic relaxation enhancement data for membrane protein structure determination by NMR. Structure 20(6): 1019-1027. [DOI] [PubMed] [Google Scholar]
- 10.Iwahara J., Schwieters C. D. and Clore G. M.(2004). Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule . J Am Chem Soc 126(18): 5879-5896. [DOI] [PubMed] [Google Scholar]
- 11.Kelly A. E., Ou H. D., Withers R. and Dotsch V.(2002). Low-conductivity buffers for high-sensitivity NMR measurements. J Am Chem Soc 124(40): 12013-12019. [DOI] [PubMed] [Google Scholar]
- 12.Reckel S., Gottstein D., Stehle J., Lohr F., Verhoefen M. K., Takeda M., Silvers R., Kainosho M., Glaubitz C., Wachtveitl J., Bernhard F., Schwalbe H., Guntert P. and Dotsch V.(2011). Solution NMR structure of proteorhodopsin. Angew Chem Int Ed Engl 50(50): 11942-11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roosild T. P., Greenwald J., Vega M., Castronovo S., Riek R. and Choe S.(2005). NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science 307(5713): 1317-1321. [DOI] [PubMed] [Google Scholar]
- 14.Schwieters C. D., Kuszewski J. J., Tjandra N. and Clore G. M.(2003). The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160(1): 65-73. [DOI] [PubMed] [Google Scholar]
- 15.Simon B., Madl T., Mackereth C. D., Nilges M. and Sattler M.(2010). An efficient protocol for NMR-spectroscopy-based structure determination of protein complexes in solution. Angew Chem Int Ed Engl 49(11): 1967-1970. [DOI] [PubMed] [Google Scholar]
- 16.Sjodt M., Macdonald R., Spirig T., Chan A. H., Dickson C. F., Fabian M., Olson J. S., Gell D. A. and Clubb R. T.(2016). The PRE-Derived NMR model of the 38.8-kDa Tri-Domain IsdH protein from staphylococcus aureus suggests that it adaptively recognizes human hemoglobin. J Mol Biol 428(6): 1107-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solomon I. and Bloembergen N.(1956). Nuclear magnetic interactions in the HF molecule. J Chem Phys 25: 261. [Google Scholar]
- 18.Tang C., Schwieters C. D. and Clore G. M.(2007). Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature 449(7165): 1078-1082. [DOI] [PubMed] [Google Scholar]
- 19.Villareal V. A., Spirig T., Robson S. A., Liu M., Lei B. and Clubb R. T.(2011). Transient weak protein-protein complexes transfer heme across the cell wall of Staphylococcus aureus. J Am Chem Soc 133(36): 14176-14179. [DOI] [PMC free article] [PubMed] [Google Scholar]