Abstract
Arterioles and sinusoids of the bone marrow (BM) are accompanied by stromal cells that express nerve/glial antigen 2 (NG2) and leptin receptor (LepR), and constitute specialised niches that regulate quiescence and proliferation of haematopoietic stem cells (HSCs). However, how niche cells differentially regulate HSC functions remains unknown. Here, we show that the effects of cytokines regulating HSC functions are dependent on the producing cell sources. Deletion of chemokine C-X-C motif ligand 12 (Cxcl12) or stem cell factor (Scf) from all perivascular cells marked by Nestin-GFP dramatically depleted BM HSCs. Selective Cxcl12 deletion from arteriolar NG2+ cells, but not from sinusoidal LepR+ cells, caused HSC reductions and altered HSC localisation in BM. By contrast, deletion of Scf in LepR+ cells, but not NG2+ cells, led to reductions in BM HSC numbers. These results uncover distinct contributions of cytokines derived from perivascular cells in separate vascular niches to HSC maintenance.
Introduction
Haematopoietic stem cells (HSCs) self-renew and differentiate into all blood types in response to various demands during life. HSC functions are finely regulated by specialized “niche” cells in the bone marrow1–4. The location of the HSC niches in the bone marrow remains controversial. Recent analyses with improved surface marker identification and bone marrow imaging have suggested that HSCs are largely perivascular5–7. Knock-in mice of GFP in the chemokine C-X-C motif ligand 12 (Cxcl12) locus reveal that the brightest GFP-expressing stromal cells (commonly referred to as Cxcl12-abundant reticular — CAR— cells) are distributed around sinusoids6. Cxcl12 and other niche factors are expressed by perivascular cells marked by Nes-GFP, which contain all mesenchymal stem cell (MSC) activity in the bone marrow, and are physically associated with HSCs5. Nes-GFP+ cells thus overlap with CAR cells as both stromal cell types differentiate into adipocytic and osteoblastic mesenchymal lineages8. Perivascular cells marked by constitutive expression of Cre driven by the LepR9, 10, Osterix or Prx-1-cre-derived cells11 have also been shown to contribute to HSC maintenance via synthesis of Cxcl12 and Scf, whereas the deletion of the same factors in committed osteoblasts (using Osteocalcin-cre) did not reveal a significant HSC phenotype11. Knock-in reporter mice for Cxcl12 and Scf revealed a major (>95%) overlap in the perivascular stromal cells expressing these niche factors9, 10. Additionally, no significant alterations in HSC numbers were observed upon genetic deletion of Cxcl12 or Scf using Nestin-creER transgenic mice9, 10, but the significance of these results remains unclear since Cre expression, even if driven by the same promoter, is low among Nes-GFP+ cells12. Thus, the exact functional contribution of Nes-GFP+ cells in niche activity remains unclear.
Recent whole-mount tridimensional (3D) imaging of the bone marrow revealed two major subsets of Nes-GFP cells where stromal cells with bright GFP signals are exclusively associated with arterioles of the bone marrow whereas Nes-GFP+ cells with lower GFP levels are distributed ubiquitously around sinusoids. The latter subset largely corresponds to LepR-cre-marked cells, whereas the former is labelled by NG2 pericyte marker13. The role of arteriole-associated stromal cells in regulation of HSC quiescence is suggested by significant changes in HSC associations with arterioles, compared to randomly assigned virtual HSCs, upon recovery after chemotherapy, after the administration of polyinosinic:polycytidylic acid, or in animals genetically deficient of Pml, all of which lead to HSC proliferation13. Schwann cells (GFAP+) in the bone marrow, which are exclusively associated with arterioles, activate TGF-β that promotes HSC quiescence14. However, it has recently been argued that HSCs are randomly distributed in the bone marrow and that arteriole-associated NG2+ cells do not contribute to niche activity15. Other studies using HoxB5-marked HSCs have also suggested a uniform distribution in the bone marrow16, whereas other recent reports have argued for distinct vascular niche contributions17, 18. Thus, whether vascular niches differentially contribute to HSC behaviour is controversial.
RESULTS
NG2-cre targets virtually all peri-vascular Nestin-GFP+ cells in the bone marrow
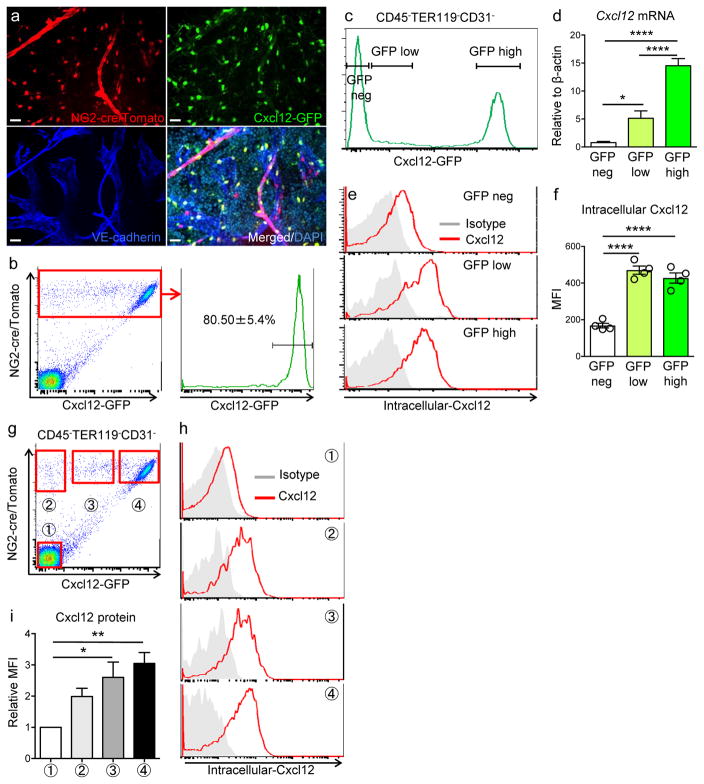
To evaluate the contributions of NG2+ cells in the HSC niche, we crossed NG2-cre transgenic mice with ROSA26-loxP-stop-loxP-tdTomato reporter (iTdTomato) and Nes-GFP transgenic mice. Whole-mount imaging analyses of the bone marrow revealed that constitutive NG2-driven Cre expression efficiently labelled Nes-GFP+ stromal cells including both peri-arteriolar Nes-GFPbright and homogenously distributed peri-sinusoidal Nes-GFPdim cells (Fig. 1a, b). FACS analyses of digested bone marrow nucleated cells confirmed that 96.9 ± 1.3% of CD45− TER119− CD31− Nes-GFP+ stromal cells were marked by NG2-cre/iTdTomato (Fig. 1c), suggesting that NG2-cre recombines in the entire Nes-GFP+ stromal cell population of the adult bone marrow. Consistent with the trilineage mesenchymal features of NG2-cre-targeted cells, we found labelling in osteocytes, chondrocytes and adipocytes (Supplementary Fig. 1a–c). However, we found that a small fraction (~10%) of endothelial cells was labelled (Supplementary Fig. 1d, e). As LepR+ stromal cells represent a large subset (~80%) of Nes-GFP+ cells located around sinusoids13, 19, we examined the relationships among NG2-cre targeted cells, arteriolar NG2+ and sinusoidal LepR+ cells. Staining of bone marrow from NG2-cre/iTdTomato/Nes-GFP mice with anti-NG2 and anti-LepR antibodies revealed that a high proportion of TdTomato+ cells (88.5 ± 1.6%) expressed LepR (Fig. 1d, e). While LepR-cre marked a small portion of Nest-GFPbright cells, NG2-cre labelled all the Nes-GFPbright cells (Supplementary Fig. 1f, g). Immunoreactive NG2+ cells around arterioles were also targeted by NG2-cre (Supplementary Fig. 1h). These data thus indicate that NG2-cre exclusively targets the non-endothelial perivascular stromal Nes-GFP+ cell population.
Figure 1. NG2-cre labels peri-vascular niche cells.
(a,b) Whole-mount images of sternums from NG2-cre/ iTdTomato/ Nes-GFP transgenic mice stained with anti-VE-cadherin antibody. Dashed lines delineate the borders between bone and bone marrow. Representative image from 3 mice. Scale bars, 100 μm in (a), 20 μm in (b). NG2-cre targeted cells overlap with both peri-arteriolar Nes-GFP+ cells (arrows) and peri-sinusoidal Nes-GFP+ cells (arrowheads). (c) Representative FACS plots showing the percentage of NG2-cre/ iTdTomato positive cells within CD45− TER119− CD31− Nes-GFP+ bone marrow stromal cells isolated from NG2-cre/ iTdTomato / Nes-GFP transgenic mice. n=4 mice. (d) Whole-mount images of sternum from NG2-cre/ iTdTomato transgenic mice stained with anti-LepR and anti-VE-cadherin antibodies. Scale bars, 20μm. All panels show the same area for different channels (NG2-cre, LepR, VE-cadherin and merged fluorescence images). (e) Representative FACS plots of CD45− TER119− CD31− bone marrow stromal cells stained with anti-LepR antibody isolated from NG2-cre/ iTdTomato mice. n=3 mice. Data are represented as mean ± SEM. Statistics Source Data are available in Supplementary Table1.
NG2-cre-marked cells are main source of Cxcl12 in the bone marrow
Since Cxcl12 production in mature osteolineage cells has been shown to be dispensable for HSC maintenance10, 11, NG2-cre transgenic mice give the opportunity to define functionally the contribution of the entire population of Nes-GFP+ cells in HSC maintenance. We crossed NG2-cre/iTdTomato or LepR-cre/iTdTomato with Cxcl12-GFP knock-in mice6 to interrogate the relationships with CAR cells. As previously reported20, the vast majority of stromal cells marked by LepR-cre (~90%) were CAR cells (Supplementary Fig. 2a, b). Likewise, a large proportion of NG2-marked cells (~80%) were CAR cells (Fig. 2a, b). We sorted GFP-negative, low and bright cells from Cxcl12-GFP knock-in mice and found, as expected, that GFP expression positively correlated with endogenous Cxcl12 mRNA (Fig. 2c, d). However, staining for intracellular Cxcl12 protein in the same subpopulations revealed, unexpectedly, that GFP low stromal cells expressed as much Cxcl12 as GFP-bright canonical CAR cells (Fig. 2e, f.). To confirm the staining specificity, we crossed NG2-cre/Cxcl12-GFP mice with Cxcl12fl/fl mice, and evaluated intracellular Cxcl12 level in each population. We found that the positive signal was completely abrogated in stromal cells in NG2-cre+ Cxcl12fl/Gfp mice, indicating that the intracellular staining was specific (Supplementary Fig. 2c). Interestingly, all NG2-cre-targeted stromal cells, whether from the GFP bright classical CAR cell gate (Fig. 2g, ➃), the GFP low (Fig. 2g, ➂), or GFP-negative gate (Fig. 2g, ➁), expressed similar levels of Cxcl12 protein (Fig. 2h, i). These results indicate that all NG2-marked cells in flushed bone marrow produce high levels of Cxcl12 protein.
Figure 2. NG2-cre-marked cells are main source of Cxcl12 in the bone marrow.
(a) Whole-mount images of sternal bone marrow from NG2-cre/ iTdTomato/ Cxcl12-GFP mice stained with anti-VE-cadherin antibody and DAPI. Representative images from 3 mice. All panels show the same area for different channels (NG2-cre, Cxcl12-GFP and merged fluorescence images). Scale bars, 20 μm. (b) Representative FACS plots showing the percentage of Cxcl12-GFP+ cells within CD45− TER119− CD31− NG2-cre/ iTdTomato+ cells. Data are represented as mean ± SEM. n=5 mice. (c) Representative histogram showing the gating scheme for isolation of CD45− TER119− CD31− cell subpopulations from Cxcl12-GFP mice used in (d). (d) Gene expression analysis of Cxcl12 in sorted CD45− TER119− CD31− cells. n=6 mice. Statistical analysis was done by one-way ANOVA. (e,f) FACS analyses of intracellular Cxcl12 protein in CXCL12-GFP mice. (e) Histograms showing intracellular Cxcl12 protein level of each cell subpopulation in CD45− TER119− CD31− cells. (f) Quantification of intracellular Cxcl12 protein level. n=4 mice from two independent experiments. Statistical analysis was done by one-way ANOVA. (g–i) Analyses of intracellular Cxcl12 protein in NG2-cre/ iTdTomato/ Cxcl12-GFP mice. (g) FACS plots showing gating scheme in CD45− TER119− CD31− stromal cells of bone marrow. Tomato−GFP− (➀), Tomato+GFP− (➁), Tomato+GFPmid (➂), Tomato+GFPhi (➃). (h) Representative histograms showing intracellular Cxcl12 protein level in each population. (i) Quantification of intracellular Cxcl12 protein level. Summary of median fluorescent intensity (MFI) of intracellular Cxcl12 relative to that of TdTomato− GFP− cells. Statistical analysis was done by one-way ANOVA. n=7 mice. Data are represented as ± S.E.M. (b, d, f, i). Statistics Source Data are available in Supplementary Table1.
Cxcl12 from distinct peri-vascular niche cells contributes differentially to HSC functions
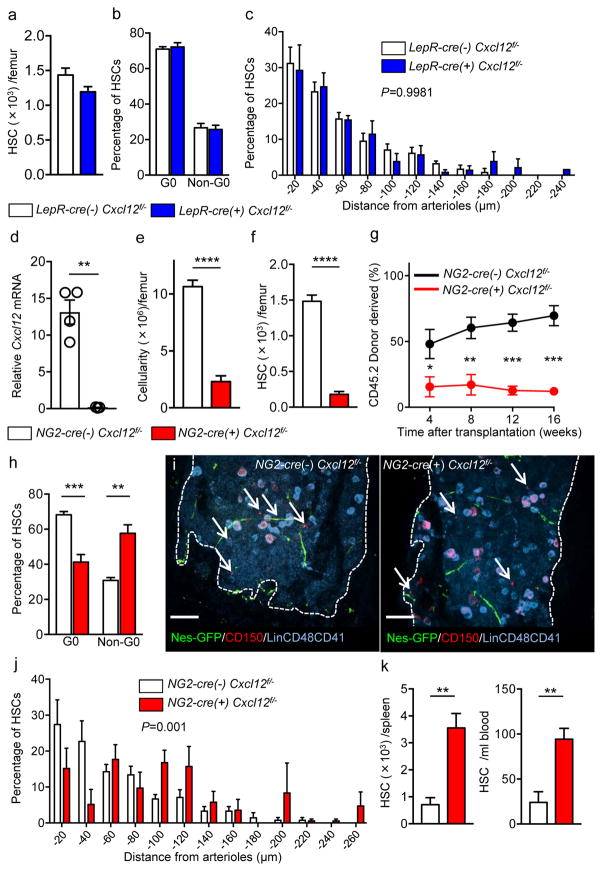
We then crossed Cxcl12fl/fl mice with CMV-cre transgenics to generate animals with one germline knockout allele (Cxcl12fl/−) and then with NG2-cre animals (or LepR-cre mice for comparison) to determine the contribution of all Nes-GFP+ cells in the synthesis of this niche factor (Supplementary Fig. 3a). There was a ~80% reduction of Cxcl12 expression in LepR-expressing cells identified by anti-LepR antibody (which stains 82.2 ± 2.1 % of LepR-marked cells), suggesting a high deletion efficiency in this population (Supplementary Fig. 3b). Consistent with a prior study10, deletion of Cxcl12 in LepR-cre-targeted cells led to mobilization of HSCs into blood and spleen, but no significant reduction of HSCs in the bone marrow (Supplementary Fig. 3c, and Fig. 3a). We investigated the functional relevance of the Cxcl12 deletion in LepR+ cells and observed no significant alteration in HSC cycling profile and distribution in the bone marrow relative to arterioles by 3D imaging (Fig. 3b, c). Deletion of Cxcl12 in NG2-marked cells drastically reduced Cxcl12 expression in Nes-GFP+ stromal cells without affecting the expression levels of other niche factors including Scf, vascular cell adhesion molecule-1 (Vcam1), and angiopoietin-1 (Angpt1) (Fig. 3d and Supplementary Fig. 3d). Whereas the absolute number of Nes-GFP+ stromal cells was not altered by the deletion of Cxcl12 (Supplementary Fig. 3e), the bone marrow cellularity was significantly reduced (Fig. 3e). Importantly, the number of phenotypic HSCs in the bone marrow was markedly decreased in NG2-cre/ Cxcl12fl/− mice, a finding confirmed by competitive repopulation assays (Fig. 3f, g and Supplementary Fig. 3f). Committed progenitors were also decreased in Cxcl12-depleted bone marrow (Supplementary Fig. 3g). Interestingly, Cxcl12 deletion in NG2-cre-marked stromal cells forced HSCs to exit from quiescence (Fig. 3h and Supplementary Fig. 3h) and led to redistribution of remaining HSCs away from arterioles. (Fig. 3i, j). In addition, a robust HSC mobilization was observed in NG2-cre/ Cxcl12fl/− mice (Fig. 3k). Since NG2+ Nes-GFP+ cells associated to portal vessels form an HSC niche in the foetal liver21, we also evaluated the impact of constitutive NG2-driven Cre deletion in the neonatal liver to rule out a contribution from defective embryonic HSC specification or foetal HSC proliferation. We found that the cellularity and the number of phenotypic HSCs in the liver of newborn NG2-cre/ Cxcl12fl/− mice were comparable to those of control animals (Supplementary Fig. 3i), which is consistent with prior studies showing that Cxcl12 is dispensable for HSCs in foetal liver22. These data strongly suggest that Cxcl12 derived from Nes-GFP+ cells play essential roles for HSC maintenance and retention in the bone marrow.
Figure 3. Cxcl12 from distinct peri-vascular niche cells contributes differentially to HSC functions.
(a–c) Analyses of LepR-cre/ Cxcl12fl/− mice. (a) Absolute numbers of HSCs in BM. n=6 mice for each group. (b) FACS analyses of cell cycle of HSCs with Ki-67 and Hoechst 33342 staining. n=5 mice per group. (c) HSC localization relative to arterioles. Error bars: n=3 mice. The p value has been calculated using n=129 HSCs for cre (−), 160 HSCs for cre (+), pooled from 3 mice per group. P=0.9981. (d–k) Analyses of NG2-cre/ Cxcl12flox/−. mice (d) Cxcl12 mRNA expression relative to β-actin in CD45−TER119−CD31−Nes-GFP+ cells from NG2-cre(−) Cxcl12f/− and NG2-cre(+) Cxcl12f/− mice. n=4 mice for cre (−), n=3 mice for cre (+), from two independent experiments. (e,f) Bone marrow cellularity (e) and absolute numbers of phenotypic CD150+CD48−Lineage−Sca-1+c-kit+ (LSK) HSCs (f) per one femur. n=10 mice. (g) Percentages of donor-derived cells after competitive reconstitution. n=5 mice per group. (h) Quantification of cell cycle of HSCs with Ki-67 and Hoechst 33342 staining. n=5 mice for cre (−), n=7 mice for cre (+). (i) Representative images of whole-mount immunofluorescent staining of the sternal bone marrow from 3 mice. Arrows indicate CD150+CD48−CD41−Lineage− HSCs. Dashed lines depict the border between bone and bone marrow. Scale bars, 100 μH. (j) HSC localization relative to arterioles. Error bars: n=3 mice for cre (−), n=4 mice for cre (+). The p value has been calculated using n=139 HSCs pooled from 3 mice for cre (−), 105 HSCs pooled from 4 mice for cre (+). P=0.001. (k) Absolute numbers of HSCs in the spleen (left) and blood (right). n=6 mice (cre-), 8 mice (cre+) for spleen. n=5 mice (cre-), n=7 mice (cre+) for blood. Data are represented as mean ± S.E.M. Data were analysed with two-tailed t-test (a, b, d–f, h, k) and Two-sample Kolmogorov-Smirnov test (c, j).
Cxcl12 from peri-arteriolar cells contributes to HSC maintenance
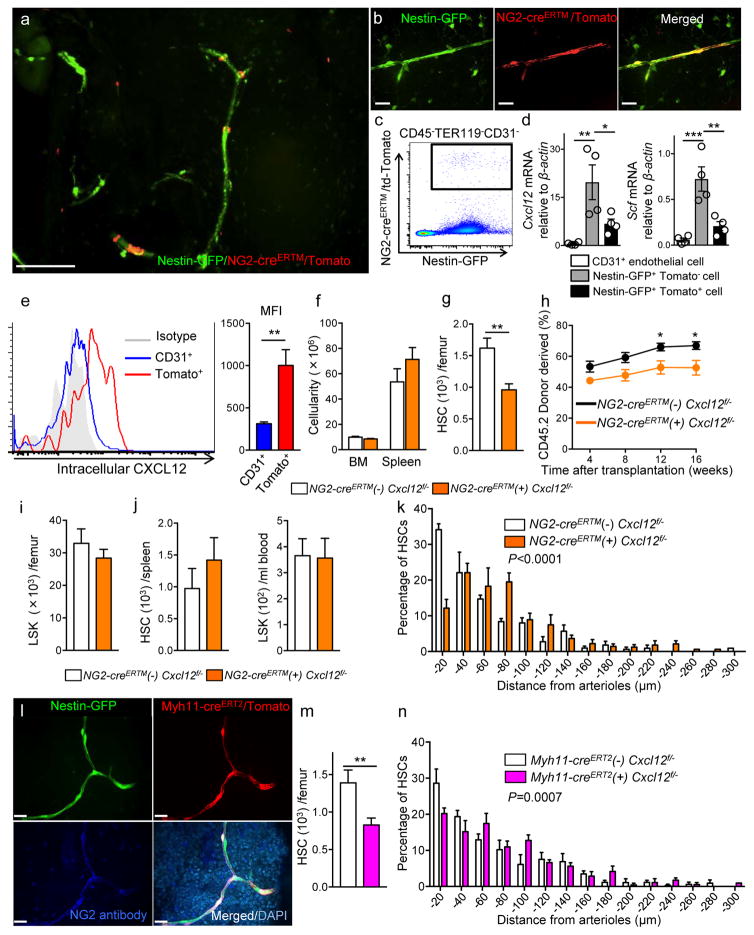
The difference in the phenotype of NG2-cre- or LepR-cre-induced deletion, combined with the significant Cxcl12 expression in NG2-marked cells outside of the canonical CAR cell population, raised the possibility that adult stromal cells expressing NG2 significantly contributed to Cxcl12 production in the bone marrow. In triple-transgenic NG2-creERTM /Nes-GFP/ iTdTomato mice, TdTomato expression was observed in peri-arteriolar Nes-GFP+ stromal cells, as well as in cells of the mature osteolineage (Fig. 4a, b). No labelling was detected in peri-sinusoidal Nes-GFP+ cells or endothelial cells (Fig. 4a, b). We next examined the expression level of the niche factors in TdTomato+ bone marrow stromal cells which exclusively expressed Nes-GFP using q-PCR analyses of Nes-GFP+ TdTomato+ stromal cells isolated from digested flushed bone marrow cells after 8 weeks of tamoxifen administration (Supplementary Fig. 4a and Fig. 4c) and found that the expression level of Cxcl12 and Scf in Nes-GFP+ TdTomato+ stromal cells was equal to or above CD31+ endothelial cells which have been reported to contribute to niche activity4,5 (Fig. 4d). Using platelet-derived growth factor receptor β (PDGFRβ), expressed in pericytes but not in osteolineage cells, we were able to enrich for a cell population exhibiting gene expression profiles similar to NG2-creERTM targeted cells from NG2-DsRed mice (Supplementary Fig. 4b, c). Using Cxcl12-GFP mice as reporter in NG2-creERTM/ iTdTomato or NG2-DsRed transgenics, GFP was not detectable in NG2+ cells compared to the bright NG2-negative canonical CAR cells (Supplementary Fig. 4d, e). Since significant endogenous Cxcl12 mRNA is expressed in NG2+ cells, we assessed intracellular Cxcl12 protein level in NG2-creERTM/ iTdTomato cells, which revealed that NG2+ cells expressed higher Cxcl12 protein than CD31+ bone marrow endothelial cells (Fig. 4e). These results thus suggest that all stromal cells marked by NG2 produce Cxcl12. It remains unclear whether, and if so how, intracellular content of Cxcl12 or GFP content in Cxcl12 knock-in mice, reflects Cxcl12 secretion which is highly regulated, notably via cell-cell contact, glycan presentation, and gap junctions23.
Figure 4. Cxcl12 deletion in NG2-creERTM targeted cells alters HSC numbers and location in the BM.
(a,b) Whole-mount images of sternum from NG2-creERTM/iTdTomato/Nes-GFP mice. Representative images from 3 mice. Scale bars, 100 μm in (a) and 20 μm in (b). (c) Representative FACS plot of bone marrow stromal cells isolated from NG2-creERTM/iTdTomato /Nes-GFP mice. (d) Quantitative real-time PCR of Cxcl12 and Scf in CD45−TER119−CD31+, CD45−TER119−CD31−Nes-GFP+, and CD45−TER119−CD31− Nes-GFP+NG2-creERTMTdTomato+ cells 8 weeks of tamoxifen administration. n=4 mice. (e) Representative histogram showing intracellular Cxcl12 levels in NG2-creERTMTdTomato+ cells (left). MFI of intracellular Cxcl12 protein (right). n=7 mice. (f–k) Analyses of NG2-creERTM/Cxcl12fl/− mice. (f) Cellularity in the bone marrow and spleen. BM; n=10 mice for cre (−), n=13 mice for cre (+), Spleen; n=5 mice for cre (−), n=8 mice for cre (+). (g) Numbers of CD150+ CD48− LSK HSCs in BM. n=10 mice for cre (−), n=13 mice for cre (+). (h) Percentages of donor-derived cells after competitive reconstitution. n=8 mice for cre (−), n=13 mice for cre (+). (i) LSK cells in BM. n=10 mice for cre (−), n=13 mice for cre (+). (j) HSC numbers in the spleen (left) and LSK cells in blood (right). n=5 mice for cre (−), n=8 mice for cre (+). (k) HSC localization relative to arterioles. Error bars: n=3 mice. The p value has been calculated using n=254 HSCs for cre (−), 238 HSCs for cre (+) pooled from 3 mice per group. P<0.0001. (l) Whole-mount sternal images from Myh11-creERT2/Nes-GFP/iTdTomato mice stained with anti-NG2 antibody. Representative images from 3 mice. Scale bars, 20 μm. (m, n) Analyses of Myh11-creERT2/Cxcl12fl/− mice. (m) Numbers of CD150+CD48− LSK HSCs in BM. n=5 mice for cre (−), n=8 mice for cre (+). (n) HSC localization relative to arterioles. Error bars: n=3 mice. The p value has been calculated using n=220 HSCs pooled from 3 mice for cre (−), 239 HSCs pooled from 4 mice for cre (+). P=0.0007. Data are represented as ± S.E.M. (d–k, m, n). Statistical significance was assessed using two-tailed t-test (e–j, m), one-way ANOVA (d), and Two-sample Kolmogorov-Smirnov test (k, n).
To evaluate Cxcl12 functions in NG2 expressing cells of postnatal bone marrow, we generated NG2-creERTM/ Cxcl12fl/− mice (Supplementary Fig.4f). Since recombination efficiency in NG2-creERTM mice is ~30% when after tamoxifen administration in adult mice, we injected tamoxifen at 2–3 week age and then analysed 6–8 weeks after treatment to maximize the recombination in this compartment. To evaluate the NG2 promoter activity in TdTomato+ cells after tamoxifen treatment, we confirmed Cspg4 (NG2) expression in TdTomato+ cells 7–8 weeks after tamoxifen injection (Supplementary Fig. 4g). Because both Cxcl12 and Gt(ROSA)26Sortm14(CAG-tdTomato)Hze alleles are closely linked (2.08cM apart) on Chromosome 6, we were not able to generate NG2-creERTM/Cxcl12fl/−/iTdTomato mice to verify the deletion efficiency of Cxcl12 in NG2-creERTM targeted cells. Targeted deletion of Cxcl12 in NG2+ cells led to a significant reduction of HSC numbers in bone marrow, which was confirmed by competitive repopulation assays, without affecting bone marrow cellularity, LSK cells in the bone marrow, HSCs in the spleen, LSK in the blood or composition of mature cells in the blood (Fig. 4f–j and Supplementary Fig. 4h, i). In addition, we found that HSCs were located further away from arterioles in NG2-creERTM/Cxcl12fl/− marrow (Fig. 4k). To confirm these results, we intercrossed Myh11-creERT2 transgenic mice, reported to target vascular smooth muscle cells, with iTdTomato and Nes-GFP transgenics24. Whole mount immunofluorescence analysis of the bone marrow of triple-transgenic mice revealed strong and selective recombination with arteriole-associated stromal cells (Fig. 4l), which was also confirmed by the staining with anti-NG2 antibody (Supplementary Fig. 4j). Myh11-creERT2-labeled cells were uniformly Nes-GFP+ (Supplementary Fig. 4k) and also expressed Cxcl12 as determined by intracellular staining (Supplementary Fig. 4l). We then bred Myh11-cre ERT2 mice with Cxcl12fl/− animals to evaluate the impact of Cxcl12 deletion in this subpopulation of arteriole-associated Nes-GFP+ cells using the same tamoxifen regimen. We also confirmed that TdTomato+ cells expressed Myh11 at the time of analyses (Supplementary Fig. 4m). We found that the number of HSCs in the bone marrow was significantly reduced (by 40%) (Fig. 4m) and the distribution of HSCs was altered in Myh11-creERT2-deleted BM (Fig. 4n), confirming an important contribution of arteriole-associated stromal cells in the synthesis of Cxcl12 for HSC maintenance.
Scf from ubiquitously distributed LepR+, but not peri-arteriolar niche cells promotes HSC maintenance
The distinct phenotypes of Cxcl12 deletion from NG2+ arteriolar and LepR+ sinusoidal niches suggest differential functions of these stromal cells in HSC maintenance. To investigate roles of Scf, the other cytokine shown to be critical for HSC maintenance25, we assessed the expression of Scf in these distinct niches. We analysed NG2-cre/iTdTomato/Scf-GFP mice and confirmed Scf-GFP+ cells ~100% overlapped with NG2-cre targeted cells (Fig. 5a, b). Q-PCR analyses showed that NG2+ peri-arteriolar niche cells marked with NG2-creERTM or NG2DsRed+PDGFRβ+ expressed modest level of Scf (Fig. 4d and Supplementary Fig. 4c), which was confirmed by immunofluorescence imaging of the bone marrow from NG2-creERTM/iTdTomato/Scf-GFP mice (Supplementary Fig. 5a).
Figure 5. NG2-cre, but not NG2-creERTM, targeted cells are the source of Scf in the bone marrow.
(a) Whole-mount sternum from NG2-cre/ iTdTomato/ Scf-GFP mice, anti-VE-cadherin. Representative images from 3 mice. Scale bars, 20 μm. (b) Representative FACS plot showing percentage of NG2-cre/ iTdTomato+ cells within CD45−TER119−CD31−Scf-GFP+ cells. n=3 mice. (c–e) Analyses of LepR-cre/ Scffl/− mice. (c) Numbers of HSCs (left) in BM and LSK cells in spleen (right). n=4 mice for cre (−), n=3 mice for cre (+). (d) FACS analyses of HSC (CD150+CD48−LSK) cell cycle with Ki-67 and Hoechst 33342 staining. n=5 mice for cre (−), n=6 mice for cre (+). (e) HSC localization relative to arterioles. Error bars: n=3 mice. P value has been calculated using n=272 HSCs for cre (−), 293 HSCs for cre (+) pooled from 3 mice per group. P=0.3402. (f–i) Analyses of NG2-cre/ Scffl/− mice. (f) Numbers of total BM cells (left) and CD150+CD48−LSK HSCs (right) in BM. n=5 mice for cre (−), n=7 mice for cre (+). (g) Percentages of donor-derived cells after competitive reconstitution. n=5 mice for cre (−), n=7 mice for cre (+). (h) FACS analyses of HSC cell cycle with Ki-67 and Hoechst 33342 staining. n=6 mice for cre (−), n=7 mice for cre (+). (i) HSC localization relative to arterioles. Error bars: n=3 mice. P value has been calculated using n=224 HSCs for cre (−), 274 HSCs for cre (+) pooled from 3 mice per group. P=0.2872. (j–l) Analyses of NG2-creERTM/ Scffl/− mice. (j) Absolute numbers of total cells (left) and HSCs (right) per one femur BM. n=8 mice for cre (−), n=6 mice for cre (+). (k) Percentages of donor-derived cells after competitive reconstitution. n=9 mice for cre (−), n=7 mice for cre (+). (l) HSC localization relative to arterioles. Error bars: n=3 mice. P value has been calculated using n=161 HSCs for cre (−), 152 HSCs for cre (+) pooled from 3 mice per group. P=0.0868. Data are represented as ± S.E.M. (b–l). Statistical significance was assessed using two-tailed t-test (c, d, f–h, j, k) and Two-sample Kolmogorov-Smirnov test (e, i. l). Statistics Source Data are available in Supplementary Table1.
We, therefore, assessed the functions of Scf in these distinct niches by analysing LepR-cre, NG2-cre, or NG2-creERTM/Scffl/− lines. Deletion of Scf in LepR-cre targeted cells which marked broadly distributed Nes-GFP+, LepR expressing stromal cells (Supplementary Fig. 5b) led to reductions in HSC numbers in the bone marrow and increased LSK cells in the spleen (Fig. 5c), which is consistent with previous studies9. LepR-cre/Scffl/− mice showed no effects on cell cycle or localization of HSCs in the bone marrow (Fig. 5d, e). Deletion of Scf in NG2-cre targeted cells led to a reduction of cellularity and HSC numbers in the bone marrow (Fig. 5f). Competitive transplantation assays have confirmed the reduction of long-term reconstitution activity (Fig. 5g and Supplementary Fig. 5c). The numbers of HSCs in spleen and LSK cells in blood were comparable to those of littermate control mice (Supplementary Fig. 5d). As was the case with LepR-cre/Scffl/− mice, neither cell cycle of HSCs nor distribution was altered in the NG2-cre/Scffl/− marrow (Fig. 5h, i). In addition, there was a reduction observed in the numbers of myeloid cell lineage cells in peripheral blood while NG2-cre/Scffl/− mice had normal haematopoietic lineage composition in the spleen (Supplementary Fig. 5e). Scf has been shown to be required for foetal liver haematopoiesis, and Scf deletion in LepR-cre targeted cells showed no effect on HSC number in the liver of newborn mice9. We found that HSC number in NG2-cre/ Scffl/− newborn liver was decreased without a significant change in cellularity (Supplementary Fig. 5f), suggesting that NG2-cre targeted cells are important source of Scf essential for HSCs not only in the marrow but also foetal liver26.
We next examined whether Scf from peri-arteriolar niche cells played roles in HSC maintenance using NG2-creERTM/Scffl/− mice. After 6–8 weeks of tamoxifen administration, NG2-creERTM/Scffl/− mice showed no significant changes in the numbers of total cells and HSCs in the bone marrow, spleen or blood, or in the location of HSCs in the bone marrow compared to Scffl/− littermates treated with tamoxifen (Fig. 5j–l, and Supplementary Fig. 5g, h). Although it is possible that Scf supplied from the neighbouring peri-sinusoidal cells or peri-arteriolar stromal cells that escaped Cre recombination may have compensated the deletion of Scf in NG2-creERTM/Scfflox/− mice, these results confirm the importance of LepR+ peri-sinusoidal cells rather than NG2+ peri-arteriolar niche cells as a major source of Scf essential for maintenance of HSCs in the bone marrow.
Distinct contributions of vascular-associated cells in niche activity
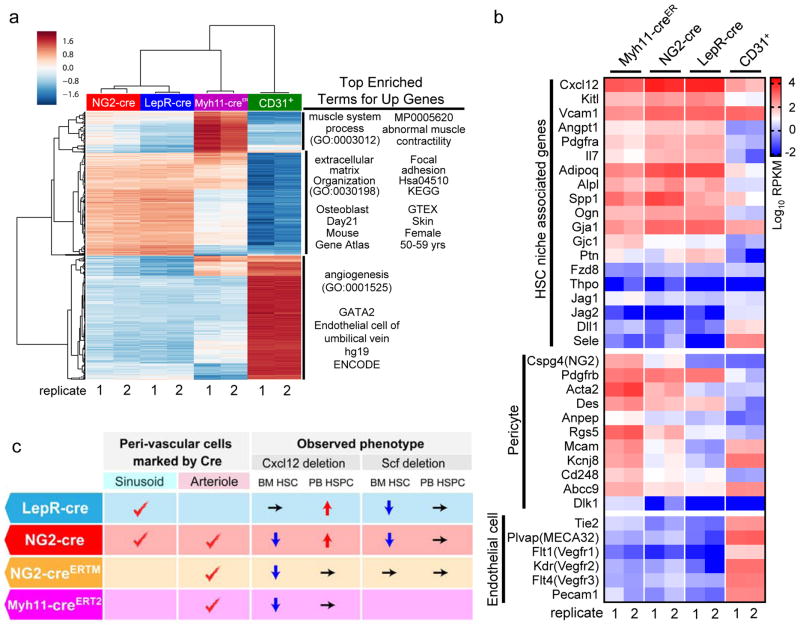
To further explore the differential genetic makeup of various peri-vascular niche cell populations, we analysed the transcriptomes by RNA-seq of Myh11-creERT2 /TdTomato+ (peri-arteriolar), LepR-cre /TdTomato+ (peri-sinusoidal), NG2-cre/TdTomato+ (combined arteriolar and sinusoidal stroma), and CD31+ endothelial cells. Consistent with the high degree of overlapping function, NG2- and LepR-marked stromal cells expression profiles closely clustered by various unsupervised clustering analyses, whereas Myh11-marked and CD31+ endothelial cells showed distinct expression vectors, while repeats were highly reproducible (Fig. 6a). Enrichment analyses for differentially expressed gene sets across the cell types revealed enrichment for muscle-related terms in Myh11-creER- marked cells, including muscle system process (GO:0003012, adjusted p-value < 9.13e-8, proportion test) and abnormal muscle contraction phenotype based on MGI’s Mammalian Phenotype ontology (MP0005620, adjusted p < 4.16e-10). Ng2-cre- and LepR-cre-labelled cells were enriched for extracellular matrix components (GO:0030198, adjusted p < 2.12e-25), cell adhesion pathway members (hsa04510, KEGG, adjusted p < 0.000004), and genes highly expressed in osteoblasts (osteoblast day21, Mouse Gene Atlas, adjusted p < 8.819e-28) and human skin (GTEX, skin, female, 50–59 yrs; p < 1.67e-51). Enriched terms in the genes up-regulated in the endothelial cells are angiogenesis (GO:0001525, adjusted p < 4.14e-14) and targets of GATA2 as determined by ChIP-seq experiments from the ENCODE project conducted in endothelial cell of umbilical vein (p < 2.52e-21). Generally, all non-endothelial stromal cell populations expressed more genes associated with the HSC niche, except for Vcam1, E-selectin (Sele) or Delta-like ligand-1 (Dll1) which were more selectively expressed in endothelial cells (Fig. 6b). Since we utilized Myh11-creERT2 cells as peri-arteriolar stromal cells, the RNA-sequence data does not include the NG2-creERTM cells. These results suggest distinct contributions of vascular-associated cells in niche activity.
Figure 6. Distinct contributions of vascular-associated cells in niche activity.
(a) RNA-seq analysis of peri-vascular niche cells. Heatmap of unsupervised hierarchical clustering of significant enriched genes for sorted CD45−TER119−CD31−NG2-cre/TdTomato+, CD45−TER119−CD31−LepR-cre/TdTomato+, CD45−TER119−CD31−Myh11-creERT2/TdTomato+, and CD45−TER119−CD31+ cells was created with clustergrammer. 2 mice for each group. Enrichment analysis for each cluster as determined by hierarchical clustering of the rows was performed with Enrichr40. (b) Heatmap expression levels of selected genes defined by previous studies for HSC niche cell4, pericyte34, and endothelial cells6. The values of log transformed reads per kilobase per million mapped reads (RPKM) obtained from RNA-seq were visualized using GraphPad Prism7. Genes with absent expression were assigned the lowest value of the gene pool for visualization. 2 mice for each group. (c) Summary of phenotypes of niche factor deleted mice in peri-vascular stromal cells.
DISCUSSION
Recent evidence indicates that HSCs are uniformly perivascular in the bone marrow5–7. However, whether specific regions or structures of the bone marrow provide defined microenvironments for distinct HSCs remains controversial. Results herein argue that such selected microenvironment exists and highlight the possibility of heterogeneity among niche factor-producing perivascular cells (Fig. 6c and Supplementary Fig. 6).
While knock-in reporter mice have suggested stromal cells synthesizing Scf and Cxcl12 nearly completely overlap with each other10, the differential contributions of perivascular cell subsets argue that GFP reporters may not faithfully reflect the complexity of regulated secretion and presentation of niche factors in the local environment. That HSCs are not randomly distributed in the bone marrow, is also supported by the association and regulation of an HSC subset by the megakaryocyte27–29, and a recent study that has found that quiescent HSCs with low reactive oxygen species (ROS) are localized near arterioles and megakaryocytes30. Notch signalling expands a CD31+ arteriole-ending endothelial cell population reduced in the ageing bone marrow18. As lineage-biased HSCs are identified31–33, further studies will determine the extent by which HSC heterogeneity is matched by niche heterogeneity.
METHODS
Animals
B6;FVB-Tg(Cspg4-cre)1Akik/J (NG2-cre), B6.129-Leprtm2(cre)Rck/J (LepR-cre), B6.Cg-Tg(Cspg4-cre/Esr1*)BAkik/J (NG2-creERTM), B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (iTdTomato), Cspg4-DsRed.T1 (NG2DsRed), Kitltm1.1Sjm/J (SCF-GFP), B6.FBV-Tg(Myh11-cre/ERT2)1Soff/J mice were purchased from Jackson Laboratory, Cxcl12-GFP mice and Cxcl12fl/fl mice were a gift from Dr. Takashi Nagasawa (Osaka University, Japan), and Scffl/fl mice were gift from Dr. Sean Morrison. C57BL/6-CD45.1/2 congenic strains were purchased from the National Cancer Institute. All mice were maintained in pathogen-free conditions under a 12:12-h light/dark cycle and fed ad libitum. All experiments were carried out using littermates except for Myh11-cre/ERT2 mice. Since all males have the transgene in Myh11-cre/ERT2 mice, we used age- matched Cxcl12flox/− mice that were grown and maintained in the same room as control. All mice were analysed at 6–12 weeks of age. Gender was not selected in all experiments except for the experiments using Myh11-cre/ERT2 animals. Peripheral blood collection was carried out by retro-orbital plexus bleeding with heparinized microcapillary tube under deep anaesthesia. All experimental procedures were approved by the Animal Care and Use Committees of Albert Einstein College of Medicine.
In vivo treatment
For induction of CreERTM or CreERT2 mediated recombination, two-week old mice were injected intraperitoneally with 0.5mg tamoxifen (Sigma) dissolved in corn oil (Sigma) twice a day for two round of 5 consecutive days with a 7-day interval.
Flow cytometry and cell sorting
For the analyses of hematopoietic cells, bone marrow cells were flushed and dissociated by gently passing through 21G needle. Spleen cells were obtained by grinding and passing through 70 μm nylon filter. Ammonium chloride was used for red blood cell (RBC) lysis. For analysis of stromal cells, bone marrow cells were flushed and digested with 1mg ml−1 collagenase IV (Gibco) and 2mg ml−1 dispase (Gibco) in Hank’s balanced salt solution (Gibco) for 30–45 min at 37°C. After lysing RBCs with ammonium chloride, cells were filtered through 100 μm nylon mesh. For analysis of newborn liver cells, livers were minced into small pieces with scissors and digested with 3mg ml−1 type 1 collagenase (Sigma) in HBSS with shaking for 5 min. Digested cells were dissociated by passing through 18G needle and 21G needle. After lysing RBCs with ammonium chloride, cells were filtered through 70 μm nylon mesh. For FACS analysis, cells were stained with antibodies in PEB (PBS containing 0.5% BSA and 2mM EDTA) buffer for 20–60 min on ice. For cell sorting, we used L-15 FACS buffer 5 as a staining buffer. The following antibodies were used: Allophycocyanin (APC)-anti-Gr-1 (eBioscience, cat No. 17-5931, clone RB6-8C5), phycoerythrin (PE)-anti-CD11b (eBioscience, cat No. clone M1/70), APC-eFluor780-anti-CD45R (eBioscience, cat No. 47-0452, clone RA3-6B2), PerCP-Cyanine5.5-anti-CD3e (eBioscience, cat No. 45-0031, clone 145-2C11), biotin-anti-Lineage (TER119, RB6-8C5, RA3-6B2, M1/70, 145-2C11, BD Biosciences, cat No. 559971), fluorescein isothiocyanate (FITC)-anti-CD45.2 (eBioscience, cat No. 11-0454, clone 104), FITC-anti-Ly6A/E (eBioscience, cat No. 11-5981, clone D7), PE-Cy7-anti-CD117 (eBioscience, cat No. 25-1171, clone 2B8), PerCP-eFluor710-anti-CD48 (eBioscience, cat No. 46-0481, clone HM48-1), FITC-anti-Ki67 (eBioscience, cat No. 11-5698, clone SolA 15) eFluor660-anti-Ki67 (eBioscience, cat No. 50-5698, clone SolA 15), PE-Cy7-anti-CD31 (eBioscience, cat No.25-0311, clone 390), PE-Cy7-anti-CD24 (eBioscience, cat No. 25-0242, clone M1/69), APC-eFluor780-anti-CD45 (eBioscience, cat No. 47-0451, clone 30-F11), APC-eFluor780-anti-TER119 (eBioscience, cat No. 47-5921, clone TER119), APC-anti-CD140b (eBioscience, cat No. 17-1402, clone APB5), PerCP-eFluor710-anti-CD16/32 (eBioscience, cat No. 46-0161, clone 93), eFluor660-anti-CD34 (eBioscience, cat No. 50-0341, clone RAM34), PE-anti-CD135 (eBioscience, cat No. 12-1351, clone A2F10), PE-anti-CD127 (eBioscience, cat No. 12-1271, clone A7R34), Streptavidin APC-eFluor780 (eBioscience, cat No. 47-4317), PE-anti-CD150 (Biolegend, cat No. 115904, clone TC15-12F12.2), Alexa Fluor 647-anti-CD43 (Biolegend, cat No. 121212, clone 1B11), Alexa Fluor 647-anti-CD144 (Biolegend, cat No. 138006, clone BV13), Alexa Fluor 647-anti-CD31 (Biolegend, cat No. 102516, clone MEC13.3). All antibodies above were used in 1:100 dilution. For intracellular CXCL12 staining, cells were fixed and permeabilised using Cytofix/Cytoperm kit (BD Biosciences) according manufacturer’s protocol. Isotype control (IC002A, R&D systems, 1;10 dilution) and antii-Cxcl12 antibody conjugated APC (IC350A, R&D systems, 1:10 dilution) were used for labelling. Flow cytometric analyses were carried out using an LSR II flow cytometer equipped with FACS Diva 6.1 software (BD Biosciences). Dead cells and debris were excluded by FSC, SSC and 4′,6-diamino-2-phenylindole (DAPI) (Sigma) staining. Cell sorting was performed on FACSAria Cell Sorter (BD Biosciences). Data were analysed with FlowJo (Tree Star) software.
Immunofluorescence imaging
Preparation of whole-mount sternum for immunofluorescence staining: Alexa Fluor 647-anti-CD144 (Biolegend, cat No. 138006, clone BV13, 5μg/mouse) and/or Alexa Fluor 647-anti-CD31 (Biolegend, cat No. 102516, clone MEC13.3, 5μg/mouse) antibodies were injected via retro-orbital plexus 10 minutes prior to euthanize mice as needed, then deeply anesthetized mice were perfused with ice-cold phosphate buffer saline (PBS) followed by ice-cold 4% paraformaldehyde (PFA). Sternal bones were collected and transected with a surgical blade into 2–3 fragments. The fragments were bisected sagitally for the bone marrow cavity to be exposed, and then fixed with 4% PFA for 30 min. After rinse with PBS, bone pieces were blocked/permeabilised in PBS containing 20% normal goat serum and 0.5% TritonX-100. The tissues were incubated with primary antibodies for overnight. The primary antibodies used were anti-LepR (R&D systems, AF497, 1:100), anti-NG2 (Milipore, AB5320, 1:100). After rinsing in PBS, tissues were incubated with secondary antibodies for 2 hours. The secondary antibodies used were Alexa Fluor 647 donkey anti-goat IgG (Molecular Probes, A-21447), Alexa Fluor 488 (Molecular Probes, A-11034) or Alexa Fluor 568 goat anti-rabbit IgG (Molecular Probes, A-11011). DAPI was used for nuclear staining. For LepR staining, fixed samples were blocked in TNB blocking buffer (PerkinElmer), and primary antibody was incubated in TNB blocking buffer.
Endogenous HSC staining in sternum bone marrow was performed as previously described 13. Briefly, sternal bone marrow fragments were fixed with 4%PFA for 30 min, then rinsed with PBS, were blocked/permeabilised in PBS containing 20% normal goat serum and 0.5% TritonX-100. Primary antibodies were incubated for 2 overnights at room temperature. After rinsing the tissue with PBS, the tissues were incubated with secondary antibodies for 2h. The primary antibodies used were biotin-anti-Lineage (TER119, RB6-8C5, RA3-6B2, M1/70, 145-2C11, BD Biosciences, cat No. 559971, 1:100), biotin-anti-CD48 (eBioscience, cat No. 13-0481, HM48-1, 1:100), biotin-anti-CD41 (eBioscience, cat No. 13-0411, MWReg30, 1:3000), Alexa Fluor 647-anti-CD144 (Biolegend, cat No. 138006, clone BV13, 1:100), PE-anti-CD150 (Biolegend, cat No. 115904, clone TC15-12F12.2, 1:100) The secondary antibody used was Streptavidin eFluor 450 (eBioscience, cat No. 48-4317, 1:100).
Preparation of frozen sections of long bones: femoral bones were perfusion-fixed and then post-fixed with 4% PFA for 30 min. Bones were decalcified in 10% EDTA (pH7.4) for 3 days at room temperature, and were incubated in 30 % sucrose/PBS for cryoprotection and embedded in SCEM embedding medium (SECTION-LAB). Twenty-μm thick frozen sections were cut with Cryostat (CM3050 S, Leica) using Kawamoto’s tape transfer method34. For immunofluorescence staining of femoral bone sections, after rinsing with PBS, slides were blocked with PBS containing species-matched serum. Slides were incubated with a primary antibody for overnight at 4°C, visualized by fluorescent-conjugated secondary antibodies, and mounted in Vectashield Mounting Medium with DAPI (Vector Laboratories). The primary antibodies used were anti-Aggrecan (Abcam, AB1031, 1:400), anti-Osteocalcin (TAKARA, M188, clone R21C-01A, 1:400), and anti-Perilipin (Cell Signaling Technology, cat No. 9349, clone D1D8, 1:1000). The secondary antibodies used were Alexa Fluor 647 anti-rat IgG (Molecular Probes, A-21247), Alexa Fluor 647 anti-rabbit IgG (Molecular Probes, A32733). All images were acquired using a ZEISS AXIO examiner D1 microscope (Zeiss) with a confocal scanner unit, CSUX1CU (Yokogawa), and reconstructed in three dimensions with Slide Book software (Intelligent Imaging Innovations).
Blood cell count
Blood cells were analysed on an ADVIA 120 haematology system (SIEMENS).
RNA isolation and quantitative PCR
Sorted cells were collected in lysis buffer. Messenger RNA isolation was performed using Dynabeads® mRNA DIRECTTM Micro Kit (Life Technologies) and reverse transcriptions were performed using RNA to cDNA EcoDry Premix (Clonetech) according to the manufacturer’s protocols. Quantitative PCR was performed with SYBR Green (Roche) on ABI PRISM 7900HT Sequence Detection System or QuantStudio 6 Flex Real-time PCR System (all from Applied Biosystems). The PCR protocol consisted of one cycle at 95°C (10 min) followed by 40 cycles of 95°C (15 s) and 60°C (1 min). Expression of β-actin was used as a control. The average of threshold cycle number (Ct) for each tested mRNA was used to quantify the relative expression of each gene: 2-(Ct(gene) - Ct(control)). Primers used include: β-actin forward, 5′-GCTTCTTTGCAGCTCCTTCGT-3′, β-actin reverse, 5′-ATCGTCATCCATGGCGAACT-3′, Cxcl12 forward, 5′-CGCCAAGGTCGTCGCCG-3′, Cxcl12 reverse, 5′-TTGGCTCTGGCGATGTGGC-3′, Scf forward, 5′-CCCTGAAGACTCGGGCCTA-3′, Scf reverse, 5′-CAATTACAAGCGAAATGAGAGCC-3′, Vcam-1 forward, 5′-GACCTGTTCCAGCGAGGGTCTA-3′, Vcam-1 reverse, 5′-CTTCCATCCTCATAGCAATTAAGGTG-3′, Ang-1 forward, 5′-CTCGTCAGACATTCATCATCCAG-3′, Ang-1 reverse, 5′-CACCTTCTTTAGTGCAAAGGC-3′. Cspg4 (NG2) forward, 5′-AGGACCTAACATTCCGGGTCA-3′, reverse, 5′-CTGTGTTGTGGAGGATCTGTATG-3′ Myh11 forward, 5′-CAGCTGGAAGAGGCAGAGGAGG-3′ reverse. 5′-AACAAATGAAGCCTCGTTTCCTCTC-3′.
RNA preparation and next-generation sequencing
Total RNA from sorted NG2-cre/TdTomato+, LepR-cre/TdTomato+, Myh11-creERT2/TdTomato+ bone marrow stromal cells, and CD31+ endothelial cells was extracted using the RNAeasy Plus Micro kit (Qiagen). The integrity and purity of total RNA were assessed using an Aligent 2100 Bioanalyzer (Agilent Technologies) Complementary DNA was generated using SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech Laboratories) from 1ng of total RNA. Nextera XT DNA Sample preparation Kit (Illumina) was used for preparation of DNA libraries. The libraries were then submitted for Illumina HiSeq2500 sequencing (Illumina) according to the standard operation procedure.
RNA-Seq analysis
RNA-Seq data generated from Illumina HiSeq 2500 were processed following a recently developed reproducible pipeline36. Briefly, paired-end sequencing reads were aligned to the mouse genome using Spliced Transcripts Alignment to a (STAR)37. We next used featureCount38 to assign aligned reads to genes. Count per Million (CPM) was used as the expression quantification method. The CPM matrix was log2 transformed and Z-score scaled before performing Principal Component Analysis (PCA) and Hierarchical Clustering (HC).
To identify signature genes for each cell populations, we normalized the read count matrix using Voom39 and performed gene wise moderated F-tests to test if a gene is differentially expressed across the four cell populations. Nominal p-values were corrected using Benjamini-Hochberg procedure to adjust for multiple hypothesis testing. Signature genes for each cell populations were identified using the following criteria: adjusted p-value < 0.001 and log2 fold change over at least 2 other cell types. Enrichment analyses for signature genes were performed using Enrichr40.
Competitive transplantation
Competitive transplantation assays were performed using CD45.1/CD45.2 congenic system. Equivalent volumes of bone marrow cells collected from gene deleted mice or control mice (CD45.2) were transplanted into lethally irradiated (12Gy) CD45.1 recipients with 0.3×106 competitor CD45.1 cells. CD45.1/CD45.2 chimerism of recipients’ blood was analysed up to 4 months after transplantation.
Cell cycle analysis
Cell cycle analysis was performed as described previously13. Briefly, bone marrow cells were stained with surface markers, fixed in 2% PFA in PBS for 20 min, washed, permeabilised with 0.1% Triton X-100 in PBS for 15 min, and stained with anti-Ki67 antibody and Hoechst 33342 (Sigma) at 20 μg ml−1 for 30 min.
CFU-C assay
CFU-C assay was performed by bleeding animals retro-orbitally into EDTA-containing tubes and subsequently centrifuging the whole blood over lympholyte-M (CEDARLANE) to separate red blood cells from mononuclear cells. The mononuclear cell fraction was collected and plated in CFU-C media (Stem cell technologies, 3534). After one week, colonies were counted and CFU-C/ml blood were calculated per animal.
Statistics and Reproducibility
All data are represented as mean ± s.e.m. of at least three independent experiments, unless otherwise noted in the figure legends. We used two-tailed Student’s t tests for evaluating the significance of difference unless otherwise indicated. Two-sample Kolmogorov-Smirnov tests were used for comparisons of distribution patterns. Statistical analyses were performed using GraphPad Prism 6 or 7 software. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
No statistical method was used to predetermine sample size. The experiments were not randomized and investigators were not blinded to allocation during experiments and outcome analyses.
Data availability
RNA sequencing data have been deposited in Gene Expression Omnibus under accession number GSE89811. Statistical source data supporting Figs 1c, e, 2b, 5b, and Supplementary Figs 1d, e, f, g, 4a have been provided in Supplementary Table 1. All data supporting the conclusion of this paper are available from the authors on request.
Supplementary Material
Acknowledgments
We thank C. Prophete, P. Ciero and C. Cruz for technical assistance and L. Tesfa, Y. Wang, and D. Sun for help with cell sorting. We thank Drs. Takashi Nagasawa and Sean Morrison for providing reagents. This work was supported by R01 grants from the National Institutes of Health (NIH) (DK056638, HL116340, HL097819 to P.S.F), New York Stem Cell Foundation and NIH’s Common Fund (U54HL127624, U54CA189201 to AM). We are also grateful to the New York State Department of Health (NYSTEM Program) for shared facility (C029154) and research support (N13G-262) and the Leukaemia and Lymphoma Society’s Translational Research Program. Y.K. is supported by JSPS Grant-in Aid for Scientific Research (B)(15H04859) and the Takeda Science Foundation. N.A. is supported by JSPS Postdoctoral Fellowships for Research Abroad.
Footnotes
Contributions
N.A. performed most of the experiments and analysed data; H.P. performed CFU-C experiment; Z.W., N.F.F, and A.M. analysed RNA-seq data; A.B. bred Myh11-creERT2 mice; P.S.F initiated and directed the study. N.A, Y.K., and P.S.F. interpreted data and wrote manuscript. All of the authors contributed to the design of experiments, discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Boulais PE, Frenette PS. Making sense of hematopoietic stem cell niches. Blood. 2015;125:2621–2629. doi: 10.1182/blood-2014-09-570192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu VW, Scadden DT. Heterogeneity of the bone marrow niche. Curr Opin Hematol. 2016;23:331–338. doi: 10.1097/MOH.0000000000000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvi LM, Link DC. The hematopoietic stem cell niche in homeostasis and disease. Blood. 2015;126:2443–2451. doi: 10.1182/blood-2015-07-533588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20:833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 8.Omatsu Y, et al. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenbaum A, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenette PS, Pinho S, Lucas D, Scheiermann C. Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu Rev Immunol. 2013;31:285–316. doi: 10.1146/annurev-immunol-032712-095919. [DOI] [PubMed] [Google Scholar]
- 13.Kunisaki Y, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki S, et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- 15.Acar M, et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526:126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JY, et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530:223–227. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itkin T, et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016;532:323–328. doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusumbe AP, et al. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature. 2016;532:380–384. doi: 10.1038/nature17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizoguchi T, et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan JA, et al. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science. 2016;351:176–180. doi: 10.1126/science.aad0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ara T, et al. A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J Immunol. 2003;170:4649–4655. doi: 10.4049/jimmunol.170.9.4649. [DOI] [PubMed] [Google Scholar]
- 23.Schajnovitz A, et al. CXCL12 secretion by bone marrow stromal cells is dependent on cell contact and mediated by connexin-43 and connexin-45 gap junctions. Nature immunology. 2011;12:391–398. doi: 10.1038/ni.2017. [DOI] [PubMed] [Google Scholar]
- 24.Wirth A, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 25.Broudy VC. Stem cell factor and hematopoiesis. Blood. 1997;90:1345–1364. [PubMed] [Google Scholar]
- 26.Bowie MB, Kent DG, Copley MR, Eaves CJ. Steel factor responsiveness regulates the high self-renewal phenotype of fetal hematopoietic stem cells. Blood. 2007;109:5043–5048. doi: 10.1182/blood-2006-08-037770. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura-Ishizu A, Takubo K, Kobayashi H, Suzuki-Inoue K, Suda T. CLEC-2 in megakaryocytes is critical for maintenance of hematopoietic stem cells in the bone marrow. The Journal of experimental medicine. 2015;212:2133–2146. doi: 10.1084/jem.20150057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao M, et al. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat Med. 2014;20:1321–1326. doi: 10.1038/nm.3706. [DOI] [PubMed] [Google Scholar]
- 29.Bruns I, et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat Med. 2014;20:1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itkin T, et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016 doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notta F, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351:aab2116. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanjuan-Pla A, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502:232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto R, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Akasaka T, et al. Relation of phasic coronary flow velocity characteristics with TIMI perfusion grade and myocardial recovery after primary percutaneous transluminal coronary angioplasty and rescue stenting. Circulation. 2000;101:2361–2367. doi: 10.1161/01.cir.101.20.2361. [DOI] [PubMed] [Google Scholar]
- 35.Kawamoto T, Shimizu M. A method for preparing 2- to 50-micron-thick fresh-frozen sections of large samples and undecalcified hard tissues. Histochem Cell Biol. 2000;113:331–339. doi: 10.1007/s004180000149. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Ma’ayan A. An open RNA-Seq data analysis pipeline tutorial with an example of reprocessing data from a recent Zika virus study. F1000Res. 2016;5:1574. doi: 10.12688/f1000research.9110.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 39.Law CW, et al. Genome Biology. 2014;15:1–17. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited in Gene Expression Omnibus under accession number GSE89811. Statistical source data supporting Figs 1c, e, 2b, 5b, and Supplementary Figs 1d, e, f, g, 4a have been provided in Supplementary Table 1. All data supporting the conclusion of this paper are available from the authors on request.