Abstract
A laboratory strain (GY) of Helicoverpa armigera (Hübner) was established from surviving larvae collected from transgenic cotton expressing a Bacillus thuringiensis var. kurstaki insecticidal protein (Bt cotton) in Gaoyang County, Hebei Province, People's Republic of China, in 2001. The GYBT strain was derived from the GY strain through 28 generations of selection with activated Cry1Ac delivered by diet surface contamination. When resistance to Cry1Ac in the GYBT strain increased to 564-fold after selection, we detected high levels of cross-resistance to Cry1Aa (103-fold) and Cry1Ab (>46-fold) in the GYBT strain with reference to those in the GY strain. The GYBT strain had a low level of cross-resistance to B. thuringiensis var. kurstaki formulation (Btk) (5-fold) and no cross-resistance to Cry2Aa (1.4-fold). Genetic analysis showed that Cry1Ac resistance in the GYBT strain was controlled by one autosomal and incompletely recessive gene. The cross-resistance pattern and inheritance mode suggest that the Cry1Ac resistance in the GYBT strain of H. armigera belongs to “mode 1,” the most common type of lepidopteran resistance to B. thuringiensis toxins. A cadherin gene was cloned and sequenced from both the GY and GYBT strains. Disruption of the cadherin gene by a premature stop codon was associated with a high level of Cry1Ac resistance in H. armigera. Tight linkage between Cry1Ac resistance and the cadherin locus was observed in a backcross analysis. Together with previous evidence found with Heliothis virescens and Pectinophora gossypiella, our results confirmed that the cadherin gene is a preferred target for developing DNA-based monitoring of B. thuringiensis resistance in field populations of lepidopteran pests.
The cotton bollworm Helicoverpa armigera (Hübner) is one of the most serious insect pests of cotton in Asia, Australia, and Africa (10). It has developed resistance to almost all groups of chemical insecticides because of their intense use (12, 20). The failure of insecticides to control H. armigera has been a strong incentive for the development and adoption of transgenic cotton expressing a Bacillus thuringiensis insecticidal protein (Bt cotton) in the People's Republic of China, Australia, and India. Bt cotton has been a popular alternative for lepidopteran pest control since 1996. Yields increased, and the use of insecticides and thus farmers' production costs decreased in the United States, the People's Republic of China, South Africa, and Mexico (17). The Bt cotton area in the People's Republic of China was expanded to 2.1 million ha in 2002, equivalent to 51% of the total cotton area of 4.1 million ha (9).
The evolution of resistance to both B. thuringiensis subspecies and individual toxins has been demonstrated in the laboratory for many insects and has been found in field populations of Plutella xylostella (3, 22). Laboratory selection in Australia, India, and the People's Republic of China has demonstrated the capacity of H. armigera to develop high levels of resistance to Cry1Ac (1, 11, 14). Large-scale planting of transgenic Bt cotton creates intensive selection pressure on cotton bollworms in the field. The development of resistance to B. thuringiensis toxin by H. armigera is now considered the major threat to the long-term effectiveness of environmentally benign Bt cotton and will eventually compromise the benefit of transgenic Bt cotton. Effective resistance management programs will be needed to preserve the long-term utility of Bt cotton. Consequently, it is urgent to understand the cross-resistance pattern, inheritance mode, and mechanism of Cry1Ac resistance in H. armigera in order to design an effective resistance management program for Bt cotton.
In this paper, we report the cross-resistance pattern and inheritance mode of a laboratory-selected H. armigera strain with a very high level of resistance to Cry1Ac and the disruption of a cadherin gene which was found to be associated with Cry1Ac resistance in this strain.
MATERIALS AND METHODS
Insects.
A laboratory strain (GY) of H. armigera was established from 300 surviving larvae collected from transgenic Bt cotton (in late growth stage) in Gaoyang County, Hebei Province, People's Republic of China, in August of 2001. This GY strain was then reared on an artificial diet in the laboratory without exposure to any B. thuringiensis toxins or other insecticides. A resistant strain named GYBT was derived from the original GY strain by 28 generations of selection with activated Cry1Ac by surface contamination bioassay. The Oxford strain was supplied by the Institute of Virology in Oxford, United Kingdom. This strain originated from Africa and has been kept in laboratories for more than 20 years without any exposure to insecticides.
Larvae of H. armigera were reared on an artificial diet based on wheat germ and soybean powder at 27 ± 1°C with a 16-h-light-8-h-dark photoperiod. Adults were held under the same temperature and light conditions at an rH of 60% and supplied with a 10% sugar solution.
Toxins.
The lyophilized powder of four δ-endotoxins (Cry1Aa, Cry1Ab, Cry1Ac, and Cry2Aa) was kindly provided by Jean-Michel Vassal, CIRAD-AMIS, Montpellier, France. The Cry1Aa, Cry1Ab, Cry1Ac, and Cry2Aa used for bioassays were all in activated form. A standard sample of B. thuringiensis var. kurstaki formulation (Btk; 15,000 IU mg−1) was obtained from the Bt Research and Development Centre in the Agriculture Science Academy of Hubei Province, People's Republic of China.
Bioassays and selection.
The toxicity of the various B. thuringiensis proteins to H. armigera was determined by a surface contamination bioassay. The toxin suspensions of the proteins were diluted with 0.01 M phosphate-buffered saline (pH 7.4) to generate five to seven serial dilutions. Phosphate-buffered saline was used as a control. A disk of an artificial diet (diameter, 1.6 cm) was put into a 24-well plate and made to fit into the inner wall and bottom of the plate by gentle pressure. One hundred microliters of the toxin solution was applied to the diet surface and allowed to air dry. One second-instar larva, starved for 4 h, was placed in each well of the plate and covered with two layers of nylon net to prevent escape. Forty-eight larvae were tested for each concentration. The mortality and body masses of the survivors were measured after 5 days of keeping them at 26 ± 1°C with a 16-h-light-8-h-dark photoperiod and 60% rH. Bioassay data were analyzed using the Poloplus software (LeOra Software).
For the selection experiments, more than 1,000 second-instar larvae were challenged each generation with a dose of activated Cry1Ac with the protocols described above. After 5 days, the largest 50% larvae (i.e., those with a body mass of >10 mg) were transferred to a normal diet to complete development.
Cross-resistance.
To examine the cross-resistance pattern of the GYBT strain, the toxicity of Cry1Aa, Cry1Ab, Cry1Ac, Cry2Aa, and Btk to the GYBT strain was compared with that to the control (GY) strain. The resistance ratio was expressed as the ratio of the 50% lethal concentration (LC50) of the GYBT strain to that of the GY strain.
Inheritance of resistance.
Female or male pupae of the GYBT and GY strains were separated. Virgin female moths from the GYBT strain were mass-crossed with male moths of the GY strain and vice versa. The responses of F1 hybrids from the two reciprocal crosses to activated Cry1Ac were determined. The F1 hybrids were backcrossed to the GYBT strain, because the resistance was incompletely recessive.
The dominance of resistance was calculated using the method of Stone (21). The degree of dominance (D) values ranged from −1 (completely recessive) to +1 (completely dominant).
If the resistance is monogenic, backcrossing of F1 hybrids (resistant heterozygote [rs]) with GYBT (resistant homozygote [rr]) would produce 50% rr and 50% rs progeny (26). The expected percentages of mortality in the backcross were calculated from the regression lines of the two parental strains as described by Georghiou (6). The values of χ2 for the expected and observed number of dead larvae at each concentration were calculated. A χ2 test of the discrepancy between the observations and the predictions from the assumption of monofactorial inheritance was made to determine goodness of fit.
Reverse transcription-PCR of the cotton bollworm cadherin gene.
Total RNA of the midguts from sixth-instar larvae was extracted with the SV total RNA isolation system (Promega) according to the manufacturer's instructions and reverse transcribed with the Moloney murine leukemia virus reverse transcriptase (Promega). The cDNA of the midgut of the susceptible GY strain was amplified by PCR using specific primers based on a cadherin cDNA sequence of H. armigera in GenBank under the accession number AF519180. Four pairs of primers used to amplify the cotton bollworm cadherin gene are summarized in Table 1. The 3′ end of the cadherin gene of H. armigera was obtained by using rapid amplification of cDNA ends (RACE). The 3′-RACE cDNA was synthesized by using an oligo(dT)15 adapter primer [5′-AGTGGTAACAACGCAGAGTA(T15)-3′]. PCR products of the expected size were excised and purified by using the Wizard DNA purification system (Promega) and cloned by using the pGEM-T easy vector system (Promega). At least three clones for each fragment were fully sequenced.
TABLE 1.
Primers used to amplify the cotton-bollworm cadherin-gene (Ha-BtR)
| Primer name | Primer sequence | cDNA positions (bp) |
|---|---|---|
| Hacad-1F | 5′-ATCGGCTGGTGGATTG(CT)TGTTC-3′ | −179 to −158 |
| Hacad-1R | 5′-CGCTCTGGTCTGGGTATTGCTA-3′ | 86 to 107 |
| Hacad-2F | 5′-GTAGCAATACCCAGACCAGAGC-3′ | 85 to 106 |
| Hacad-2R | 5′-GCTTACTGCGTTACCCAT(AT)AGAG-3′ | 1676 to 1698 |
| Hacad-3F | 5′-TAATGGGTAACGCAGT-3′ | 1679 to 1694 |
| Hacad-3R | 5′-CGACAATGCTGTAATAGGTG-3′ | 3864 to 3883 |
| Hacad-4F | 5′-AAGGAGCGAGCAGTAGT-3′ | 3424 to 3440 |
| Hacad-adp | 5′-AGTGGTAACAACGCAGAGTA-3′ | —a |
Position of the 3′ adapter.
Four overlapping fragments of the cadherin gene from the GY strain were cloned and sequenced from positions −179 (in the 5′ untranslated region) to 107, 85 to 1698, 1679 to 3883, and 3424 to the poly(A) tail. The full amino acid sequence of the cadherin gene from the GY strain is more than 98% identical to that of the cadherin gene of H. armigera with the GenBank accession number AF519180.
Linkage analysis.
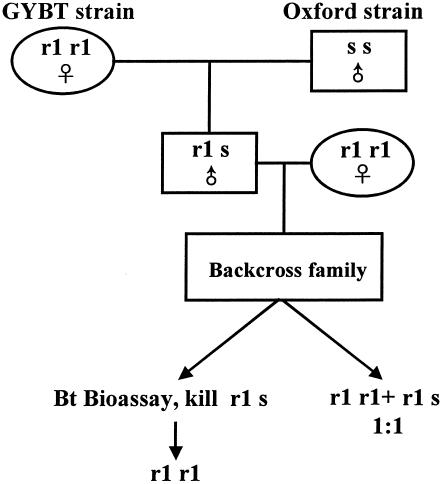
To test whether Cry1Ac resistance was genetically linked with the cadherin locus, we conducted a linkage analysis experiment. Figure 1 shows the schematic program for backcross analysis, which was adapted from the work of Heckel et al. (8). A male from the resistant GYBT strain was crossed with a female from the susceptible Oxford strain to produce a family of hybrid F1 offspring. An F1 male was backcrossed to a female from GYBT to produce the backcross family. Some of the larvae from the backcross family were treated with 1 μg of Cry1Ac/cm2 for 5 days, and the survivors were then reared to the final instar on a normal artificial diet. Untreated control larvae from the backcross family were reared to the final instar on a normal artificial diet.
FIG. 1.
Experimental design for linkage analysis of Cry1Ac resistance in H. armigera. ♀, females; ♂, males.
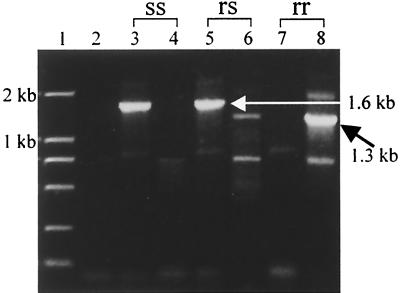
The cDNA made from sixth-instar larval midguts of the treated surviving larvae and the untreated control larvae from the backcross family was individually synthesized using the same oligo(dT)15 adapter primer as that for 3′ RACE. Two PCRs with two pairs of primers (Hacad-2F-Hacad-2R and Hacad-2F-Hacad-adp) were made for each sample. The amplification reaction mixture (50 μl) contained 1 μl of cDNA template, a 1 μM concentration each of 5′ and 3′ primers, a 150 μM concentration of the deoxynucleoside triphosphates, 2 mM MgCl2, and 1 U of Taq polymerase. PCR was performed for 30 cycles of 30 s at 94°C, 1 min at 64°C, and 2 min at 72°C. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining. Genotypes of the cadherin gene were discriminated depending on banding patterns as shown in Fig. 2. When the primer pair Hacad-2F-Hacad-2R was used for PCR amplification, the susceptible homozygote (ss) and heterozygote (rs) shared a 1.6-kb fragment, while the resistant homozygote did not have this fragment. However, when the primer pair Hacad-2F-Hacad-adp was used, the resistant homozygote (rr) and the heterozygote (rs) shared a 1.3-kb fragment, while the susceptible homozygote did not have this fragment.
FIG. 2.
Banding patterns of three cadherin genotypes from H. armigera. The primer pair Hacad-2F-Hacad-2R was used for lanes 3, 5, and 7, and the primer pair Hacad-2F-Hacad-adp was used for lanes 4, 6, and 8. Lane 1, DNA marker; lane 2, negative control (no cDNA template); lanes 3 and 4, homozygous susceptible allele cDNA template (ss); lanes 5 and 6, heterozygous allele cDNA template (rs); lanes 7 and 8, homozygous resistant allele cDNA template (rr).
RESULTS
Cross-resistance.
The GYBT strain was derived from the GY strain by 28 generations of selection with activated Cry1Ac. When resistance to Cry1Ac in the GYBT strain increased to 564-fold after this selection, high levels of cross-resistance to Cry1Aa (103-fold) and Cry1Ab (>46-fold) were detected in the GYBT strain, while the GYBT strain had a low level of cross-resistance to Btk (5-fold) but no cross-resistance to Cry2Aa (1.4-fold) (Table 2).
TABLE 2.
Cross-resistance patterns of the Cry1Ac-selected strain of H. armigera (GYBT)
| Strain | Toxin | LC50 (μg/cm2) | 95% FLb | Slope ± SE | RRa |
|---|---|---|---|---|---|
| GY | Cry1Aa | 1.44 | 0.80-2.38 | 1.17 ± 0.24 | |
| Cry1Ab | 3.23 | 2.15-5.28 | 1.42 ± 0.31 | ||
| Cry1Ac | 0.1 | 0.08-0.12 | 1.60 ± 0.17 | ||
| Cry2Aa | 1.02 | 0.57-1.67 | 1.21 ± 0.30 | ||
| Btk | 624 | 492-768 | 2.09 ± 0.25 | ||
| GYBT | Cry1Aa | 148 | 89-459 | 1.16 ± 0.32 | 103 |
| Cry1Ab | >150 | >46 | |||
| Cry1Ac | 56.4 | 31.4-217 | 1.0 ± 0.23 | 564 | |
| Cry2Aa | 1.41 | 0.97-2.01 | 1.61 ± 0.26 | 1.4 | |
| Btk | 3,102 | 1.67 ± 0.23 | 5 |
RR, the resistance ratio determined by dividing the LC50 of the GYBT strain by the LC50 of the reference GY strain.
FL, fiducial limit.
Inheritance of resistance.
Responses of the resistant GYBT isolates, the susceptible GY isolates, and their F1 progeny to Cry1Ac toxin are summarized in Table 3. The responses to Cry1Ac of F1 progeny from reciprocal crosses between the GYBT and GY strains were not statistically different from each other. This result indicates that Cry1Ac resistance is not sex linked. By using the method of Stone (21), the D values in two reciprocal crosses were calculated to be −0.49 and −0.55 at the LC50, suggesting that Cry1Ac resistance was incompletely recessive.
TABLE 3.
Response of the resistant GYBT, susceptible GY, and F1 progeny of H. armigera to activated Cry1Ac toxin
| Strain or progeny | LC50 (μg/cm2) | 95% FLa | Slope ± SE | RRb | Dc | No. of larvae tested |
|---|---|---|---|---|---|---|
| GY | 0.10 | 0.08-0.12 | 1.60 ± 0.17 | 1 | 437 | |
| GYBT | 56.4 | 31.4-217 | 1.00 ± 0.23 | 564 | 233 | |
| F1 progeny | ||||||
| GYBT females × GY males | 0.51 | 0.30-0.83 | 2.03 ± 0.24 | 5.1 | −0.49 | 235 |
| GY females × GYBT males | 0.41 | 0.32-0.52 | 1.78 ± 0.25 | 4.1 | −0.55 | 204 |
FL, fiducial limit.
RR, the resistance ratio determined by dividing the LC50 of the resistant strain by the LC50 of the reference GY strain.
D was calculated using the method of Stone (21).
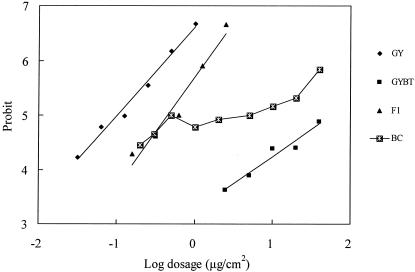
If the resistance is assumed to be controlled by a single gene, progeny of the backcrossing of the F1 hybrids (rs) with GYBT (rr) should consist of 50% rr and 50% rs progeny. In the log dosage-probit curve of mortality due to Cry1Ac of the backcross progeny, there was a distinct plateau corresponding to 50% mortality (probit 5) (Fig. 3). This plateau suggests that Cry1Ac resistance may be controlled by one gene. The χ2 test of the goodness of fit between the observations and predictions under the assumption of monofactorial inheritance (Σχ2 = 10.771 < χ2 [P = 0.05]; df = 7) further supports the single-gene hypothesis.
FIG. 3.
Dosage-mortality regression lines for the progeny of the susceptible strain (GY), resistant strain (GYBT), F1 (GY × GYBT), and backcrossed strain (BC) (F1 × GYBT). Pooled data from the reciprocal crosses were combined to draw the F1 line.
It is concluded that Cry1Ac resistance in the GYBT strain of H. armigera is controlled by a single, autosomal, incompletely recessive locus.
Cadherin alleles from susceptible and resistant H. armigera.
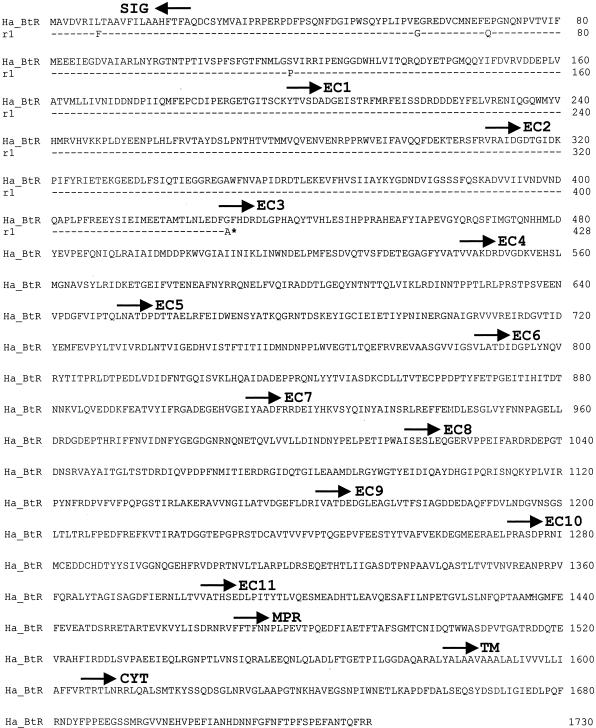
Cadherin cDNA isolated from the susceptible GY strain had 5,537 bp encoding a predicted protein of 1,730 amino acids, which we named Ha-BtR (GenBank accession no. AY647974). The amino acid sequence of Ha-BtR was 80% identical to the cadherin-like protein from Heliothis virescens (HevCaLP; GenBank accession no. AF367362), 60% identical to BtR175 from Bombyx mori (GenBank accession no. BAA77212), 58% identical to BT-R1 from Manduca sexta (GenBank accession no. AF319973), and 53% identical to BtR from Pectinophora gossypiella (GenBank accession no. AY198374). Similar to the structures of other lepidopteran cadherins, the proposed structure of Ha-BtR included a putative membrane signal sequence, 11 extracellular cadherin repeats, a membrane-proximal extracellular domain, a transmembrane domain, and a cytoplasmic domain (Fig. 4).
FIG. 4.
Amino acid sequence of Ha-BtR and resistance allele r1 deduced from cDNA. Identical residues are designated by dashes. An asterisk shows the premature stop codon in r1. Protein sequence analysis was done by using the Simple Modular Architecture Research Tool (http://smart.ox.ac.uk). Horizontal arrows specify start sites of putative domains. SIG, signal peptide predicted by using SignalP 3.0; EC, cadherin repeat; MPR, membrane-proximal region; TM, transmembrane domain; CYT, cytoplasmic domain.
In the resistant GYBT strain, we identified only one short allele (r1) of the Ha-BtR gene. The r1 allele had a premature stop codon at position 429 of the Ha-BtR gene (GenBank accession no. AY647975) (Fig. 4). All tested individuals (>30) from the GYBT strain had homozygous r1 alleles; however, all tested individuals (>30) from the GY strain were homozygous for the Ha-BtR gene. This finding indicates that the GYBT strain is homozygous for the r1 allele.
Linkage analysis.
Two hundred eleven second-instar larvae from the backcross family were treated with 1 μg of Cry1Ac/cm2. After 5 days, 107 larvae (50.7%) had survived (body mass, >10 mg), and the others (49.3%) were dead or seriously inhibited (body mass, <5 mg). Twenty-one larvae from the 107 survivors were randomly chosen for cadherin genotyping, and all 21 were found to be homozygous for the r1 allele of the Ha-BtR gene (r1r1). It can be deduced that all survivors (50.7%) were homozygous for the r1 allele, and no recombinations of Cry1Ac resistance and the cadherin locus were observed (crossing over occurred in an F1 male).
Thirty untreated control larvae from the backcross family were also randomly selected for cadherin genotyping. As expected, nearly 50% of the individual larvae (16 in 30) were homozygous for the r1 allele (r1r1) and nearly 50% of them (14 in 30) were heterozygous for the r1 allele (r1s).
The results from backcross analysis confirm that the cadherin locus is tightly linked with Cry1Ac resistance in the GYBT strain.
DISCUSSION
The most common type of lepidopteran resistance to B. thuringiensis toxins is called “mode 1.” This type is characterized by high resistance (over 500-fold) to at least one Cry1A toxin, recessive inheritance, little or no cross-resistance to Cry1C, and reduced binding of at least one Cry1A toxin (24). Mode 1 resistance has been reported for at least one strain each from the diamondback moth, Plutella xylostella (23), the tobacco budworm, Heliothis virescens (5), the Indian meal moth, Plodia interpunctella (13, 29), and the pink bollworm, Pectinophora gossypiella (15). Cry1Ac resistance in the GYBT strain of H. armigera was more than 500-fold and incompletely recessive. This strain also shows strong cross-resistance to two other Cry1A toxins (Cry1Aa and Cry1Ab). Although lack of Cry1Ac binding was not tested, the GYBT strain may possess mode 1 resistance.
Previous work suggested that aminopeptidases (digestive enzymes) or cadherins (cell adhesion proteins) serve as receptors for B. thuringiensis toxins in the insect midgut. Genetic mapping experiments with the resistant YHD2 strain of Heliothis virescens showed tight linkage between resistance to Cry1Ac and a cadherin gene (called BtR-4 or HevCaLP) but not to genes encoding aminopeptidases. Insertion of a retrotransposon disrupts BtR-4 in the YHD2 strain, which has more than 10,000-fold resistance to Cry1Ac (5). Later, three mutant alleles of a cadherin-encoding gene associated with high resistance to Cry1Ac, each with a unique major deletion, were identified in the AZP-R strain of Pectinophora gossypiella (15).
The product of the cadherin gene can serve as a receptor for B. thuringiensis toxins in the midguts of lepidopteran insects (16, 28). The stop codon in the r1 allele of the cadherin gene is expected to block the production of the binding region and make the B. thuringiensis toxin lose the specific binding receptor in the midgut of H. armigera. Thus, it is not surprising to find that disruption of the Ha-BtR gene by a premature stop codon is tightly linked with Cry1Ac resistance in the GYBT strain. So far, at least five different mutations of the cadherin gene that result in failure to produce a full-length protein are associated with Cry1Ac resistance in lepidoptera.
For our resistant GYBT strain, the initial population was collected from a Bt cotton field in 2001 and selected with activated Cry1Ac, which is similar to the protein expressed in Bt cotton. It is reasonable to presume that the mechanism we picked up in the laboratory was possibly present in the field. However, field populations may harbor other resistance mutations of the same cadherin gene as in the pink bollworm, a possibility which may complicate any future DNA-based screening. Nonetheless, the cadherin locus is the most likely target to develop a DNA-based screening of resistance to Bt cotton in field populations of H. armigera.
Mode 1 resistance is the most common type of B. thuringiensis resistance found in both the laboratory- and field-selected strains of insects, but it is not the only type (25). A resistant strain of H. armigera (BKBT) was selected with activated Cry1Ac from a laboratory strain (BK77) that originated from Cote d'Ivoire in 1977 (27). The BKBT strain showed 160-fold resistance to Cry1Ac and strong cross-resistance to Cry1Aa and Cry1Ab but not to Cry2Aa. Cry1Ac resistance in the BKBT strain was completely dominant (27). Subsequent genetic mapping results indicated that Cry1Ac resistance in the BKBT strain is linked with five informative AFLP markers which are all in one linkage group (Yidong Wu and Jean-Michel Vassal, unpublished data). This suggests that the cotton bollworm can use different defense mechanisms to produce similar phenotypic patterns of resistance.
One requirement for a “stacked” gene resistance management strategy to work is that the stacked toxins should have different modes of action (4). A recent model of the interaction of B. thuringiensis toxins with larval midgut binding sites in H. armigera showed that Cry1Aa, Cry1Ab, and Cry1Ac competed for common binding sites but that Cry2A did not share the binding sites with Cry1Ac (2). The GYBT strain has strong cross-resistance to Cry1A toxins but none to Cry2Aa. An Australian strain of H. armigera selected with Cry1Ac showed a cross-resistance pattern similar to that of the GYBT strain, and the Australian strain was shown to have lost binding of Cry1Ac (1). These results are in agreement with the binding data and model of Estela et al. (2). Second-generation Bt cotton producing both Cry1Ac and Cry2Ab toxins has been approved for commercial planting in Australia since 2002 (9). If Cry2Ab is not cross-resistant with the Cry1Ac toxin, this kind of two-gene Bt cotton may not only broaden the pest control spectrum but also be used as a resistance management strategy for both toxins.
At present, the most promising B. thuringiensis crop resistance management measure is based on the high dose-refuge strategy (7, 19). To be effective, the following conditions should exist. First, the Bt cotton plants should be highly toxic against the target pest, and B. thuringiensis resistance should be recessive, so that the plants kill heterozygous resistant larvae. Furthermore, the frequency of B. thuringiensis-resistant organisms should be less than 10−3, and there are very few resistant homozygotes. Third, there should be enough susceptible insects to dilute homozygous resistant insects. In the United States and Australia, formal nontransgenic refugium requirements have been implemented. However, the People's Republic of China has not pursued this route, partly in the expectation that small field sizes and agronomic heterogeneity would restrain the development of resistance, at least for the polyphagous pests, and partly in acceptance of the impracticability of regulating the implementation of refugia (18, 30). These factors may account for the lack of unequivocal field evidence of enhanced resistance to Bt cotton containing Cry1Ac in the People's Republic of China. However, it must be noted that the efficacy of Bt cotton against the cotton bollworm decreases with plant aging and thus there are some survivors in B. thuringiensis fields in the late season (31). This lower efficacy of Bt cotton later in the season may favor the survival of heterozygotes (rs). It is very important to test the relative survival rates of the three genotypes of H. armigera (rr, rs, and ss) in the early, middle, and late growth stages of Bt cotton to see whether heterozygote survival in the late season will indeed pose a threat to the continuing efficacy of Bt cotton. DNA-based screening for mutations in the cadherin locus may help to monitor changes in resistance gene frequency.
Acknowledgments
We thank Jean-Michel Vassal (CIRAD, Montpellier, France) for kindly providing B. thuringiensis toxins used in this study and D. Russell (NRI, Chatham, United Kingdom) and G. Lövei (DIAS, Slagelse, Denmark) for correcting the manuscript.
This work was funded by an EU-Framework 5 Inco-Dev project (ICA4-CT-2001-10069) and partially supported by the Teaching and Research Award Program for Outstanding Young Teachers in Higher Education Institutions of MOE, Beijing, People's Republic of China (TRAPOYT), and a grant from the National High Technology Research and Development Program of China (program 863) to Y. Wu.
REFERENCES
- 1.Akhurst, R. J., W. J. James, L. J. Bird, and C. Beard. 2003. Resistance to the Cry1Ac δ-endotoxin of Bacillus thuringiensis in the cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J. Econ. Entomol. 96:1290-1299. [DOI] [PubMed] [Google Scholar]
- 2.Estela, A., B. Escriche, and J. Ferré. 2004. Interaction of Bacillus thuringiensis toxins with larval midgut binding sites of Helicoverpa armigera (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 70:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferré, J., B. Escriche, Y. Bel, and J. Van Rie. 1995. Biochemistry and genetics of insect resistance to Bacillus thuringiensis insecticidal crystal proteins. FEMS Microbiol. Lett. 132:1-7. [Google Scholar]
- 4.Ferré, J., and J. Van Rie. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47:501-533. [DOI] [PubMed] [Google Scholar]
- 5.Gahan, L. J., F. Gould, and D. G. Heckel. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857-860. [DOI] [PubMed] [Google Scholar]
- 6.Georghiou, G. P. 1969. Genetics of resistance to insecticides in house flies and mosquitoes. Exp. Parasitol. 26:224-255. [DOI] [PubMed] [Google Scholar]
- 7.Gould, F. 1998. Sustainability of transgenic insecticidal cultivars: integrating pest genetics and ecology. Annu. Rev. Entomol. 43:701-726. [DOI] [PubMed] [Google Scholar]
- 8.Heckel, D. G., L. J. Gahan, Y. Liu, and B. Tabashnik. 1999. Genetic mapping of resistance to Bacillus thuringiensis toxins in diamondback moth using biphasic linkage analysis. Proc. Natl. Acad. Sci. USA 96:8373-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James, C. 2002. Preview: global status of commercialized transgenic crops: 2002. ISAAA brief no. 27. International Service for the Acquisition of Agri-biotech Applications, Ithaca, N.Y.
- 10.King, A. B. S. 1994. Heliothis/Helicoverpa (Lepidoptera: Noctuidae), p. 39-106. In G. A. Matthews and J. P. Tunstall (ed.), Insect pests of cotton. CAB International, Wallingford, United Kingdom.
- 11.Kranthi, K. R., S. Kranthi, S. Ali, and S. K. Banerjee. 2000. Resistance to Cry1Ac δ-endotoxin of Bacillus thuringiensis in a laboratory selected strain of Helicoverpa armigera (Hübner). Curr. Sci. 78:1001-1004. [Google Scholar]
- 12.McCaffery, A. R. 1998. Resistance to insecticides in heliothine lepidoptera: a global view. Philos. Trans. R. Soc. Lond. Ser. B 353:1735-1750. [Google Scholar]
- 13.McGaughey, W. H. 1985. Insect resistance to the biological insecticide Bacillus thuringiensis. Science 229:193-195. [DOI] [PubMed] [Google Scholar]
- 14.Meng, F., J. Shen, W. Zhou, and H. Cen. 2003. Long-term selection for resistance to transgenic cotton expressing Bacillus thuringiensis toxin in Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae). Pest Manag. Sci. 60:167-172. [DOI] [PubMed] [Google Scholar]
- 15.Morin, S., R. W. Biggs, M. S. Sisterson, L. Shriver, C. Ellers-Kirk, D. Higginson, D. Holley, L. J. Gahan, D. G. Heckel, Y. Carrière, T. J. Dennehy, J. K. Brown, and B. E. Tabashnik. 2003. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc. Natl. Acad. Sci. USA 100:5004-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngamatsu, Y., T. Koike, K. Sasaki, A. Yoshimoto, and Y. Furukawa. 1999. The cadherin-like protein is essential to specificity determination and cytotoxic action of the Bacillus thuringiensis insecticidal CryIAa toxin. FEBS Lett. 460:385-390. [DOI] [PubMed] [Google Scholar]
- 17.Pray, C. E., J. Huang, R. Hu, and S. Rozelle. 2002. Five years of Bt cotton in China—the benefits continue. Plant J. 31:423-430. [DOI] [PubMed] [Google Scholar]
- 18.Russell, D. 2004. Integrated pest management for insect pests of cotton in less developed countries, p. 141-179. In A. R. Horowitz and I. Ishaaya (ed.), Insect pest management: field and protected crops. Springer-Verlag, Berlin, Germany.
- 19.Shelton, A. M., J. D. Tang, R. T. Roush, T. D. Metz, and E. D. Earle. 2000. Field tests on managing resistance to Bt-engineered plants. Nat. Biotechnol. 18:339-342. [DOI] [PubMed] [Google Scholar]
- 20.Shen, J., and Y. Wu. 1995. Resistance of Helicoverpa armigera to insecticides and its management. China Agricultural Press, Beijing, People's Republic of China.
- 21.Stone, B. F. 1968. A formula for determining degree of dominance in case of monofactorial inheritance of resistance to chemicals. Bull. W. H. O. 38:325-326. [PMC free article] [PubMed] [Google Scholar]
- 22.Tabashnik, B. E. 1994. Evolution of resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 39:47-79. [Google Scholar]
- 23.Tabashnik, B. E., Y. B. Liu, N. Finson, L. Masson, and D. G. Heckel. 1997. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc. Natl. Acad. Sci. USA 94:1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabashnik, B. E., Y. B. Liu, T. Malvar, D. G. Heckel, L. Masson, and J. Ferré. 1998. Insect resistance to Bacillus thuringiensis: uniform or diverse? Philos. Trans. R. Soc. Lond. Ser. B 353:1751-1756. [Google Scholar]
- 25.Tabashnik, B. E. 2001. Breaking the code of resistance. Nat. Biotechnol. 19:922-924. [DOI] [PubMed] [Google Scholar]
- 26.Tsukamoto, M. 1963. The log dosage-probit curve in genetic researches of insect resistance to insecticides. Botyu-Kagaku 28:91-98. [Google Scholar]
- 27.Uraichuen, S. 2002. Comparative study of the toxicity and receptors of Bacillus thuringiensis Berliner δ-endotoxins against two major pests of cotton, Helicoverpa armigera and Heliothis virescens: relation with resistance. Ph.D. thesis. National Superior Agronomic School of Montpellier, Montpellier, France.
- 28.Vadlamudi, R. K., E. Weber, I. Ji, T. H. Ji, and L. A. Bulla, Jr. 1995. Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J. Biol. Chem. 270:5490-5494. [DOI] [PubMed] [Google Scholar]
- 29.Van Rie, J., W. H. McGaughey, D. E. Johnson, M. D. Barnett, and H. Van Mellaert. 1990. Mechanism of insect resistance to the microbial insecticide Bacillus thuringiensis. Science 247:72-74. [DOI] [PubMed] [Google Scholar]
- 30.Wu, K., Y. Guo, and S. Gao. 2002. Evaluation of the natural refuge function for Helicoverpa armigera (Hübner) within Bt transgenic cotton growing areas in north China. J. Econ. Entomol. 95:832-837. [DOI] [PubMed] [Google Scholar]
- 31.Wu, K., Y. Guo, N. Lv, J. T. Greenplate, and R. Deaton. 2003. Efficacy of transgenic cotton containing a cry1Ac gene from Bacillus thuringiensis against Helicoverpa armigera (Lepidoptera: Noctuidae) in northern China. J. Econ. Entomol. 96:1322-1328. [DOI] [PubMed] [Google Scholar]