Abstract
Signal-transducing functions driven by the cytoplasmic domain of membrane type-1 matrix metalloproteinase (MT1-MMP) are believed to regulate many inflammation-associated cancer cell functions including migration, proliferation, and survival. Aside from upregulation of the inflammation biomarker cyclooxygenase-2 (COX-2) expression, MT1-MMP’s role in relaying intracellular signals triggered by extracellular pro-inflammatory cues remains poorly understood. Here, we triggered inflammation in HT1080 fibrosarcoma cells with phorbol-12-myristate-13-acetate (PMA), an inducer of COX-2 and of MT1-MMP. To assess the global transcriptional regulatory role that MT1-MMP may exert on inflammation biomarkers, we combined gene array screens with a transient MT1-MMP gene silencing strategy. Expression of MT1-MMP was found to exert both stimulatory and repressive transcriptional control of several inflammasome-related biomarkers such as interleukin (IL)-1B, IL-6, IL-12A, and IL-33, as well as of transcription factors such as EGR1, ELK1, and ETS1/2 in PMA-treated cells. Among the signal-transducing pathways explored, the silencing of MT1-MMP prevented PMA from phosphorylating extracellular signal–regulated kinase, inhibitor of κB, and p105 nuclear factor κB (NF-κB) intermediates. We also found a signaling axis linking MT1-MMP to MMP-9 transcriptional regulation. Altogether, our data indicate a significant involvement of MT1-MMP in the transcriptional regulation of inflammatory biomarkers consolidating its contribution to signal transduction functions in addition to its classical hydrolytic activity.
Keywords: Inflammation, inflammasome, MT1-MMP, cancer, gene transcription
Introduction
Membrane type 1-matrix metalloproteinase (MT1-MMP) is a transmembrane matrix metalloprotease which degrades interstitial collagens and extracellular matrix (ECM) components under both normal and pathophysiological processes including atherosclerosis, muscular diseases, and cancer.1–3 It also functions as a signal-transducing intermediate as numerous lines of evidence suggest that MT1-MMP relays signals linking cancer to cancer-related inflammation.4–7 Overexpression of MT1-MMP was found to trigger phosphorylation of the signal transducers and activators of transcription 3 (STAT3)8 and of a MT1-MMP/nuclear factor-κ-light-chain-enhancer of an activated B cell (NF-κB) signaling axis found to also act as a checkpoint controller of cyclooxygenase-2 (COX-2) expression in CD133+ U87 glioblastoma cells.9 JAK/STAT3 and NF-κB are involved in major pathways regulating inflammation, are constitutively active in most cancers, and are often activated by cancer risk factors.10 Moreover, hypoxia and acidic conditions, as are found within solid tumors, are known to activate NF-κB11,12 and to correlate with increased MT1-MMP expression.13–15 Given that numerous gene products linked to inflammation, survival, proliferation, invasion, angiogenesis, and metastasis are regulated by NF-κB,16 a better understanding of MT1-MMP’s contribution to pro-inflammatory pathway regulation may provide opportunities for both prevention and treatment of cancer. The MT1-MMP has been well characterized as a cell surface pro-MMP-2 activator.17 Along with other MT-MMPs, it has a common domain structure consisting of a signal peptide, a pro-domain, a catalytic domain, a hinge, a hemopexin-like domain, a transmembrane domain, and a stalk region.18 The MT1-MMP, MT2-MMP, MT3-MMP, and MT5-MMP are further characterized by a short cytoplasmic domain, whereas glycosylphosphatidylinositol anchors characterize MT4-MMP and MT6-MMP.3 Gene silencing, deletions, or point mutations within the cytoplasmic domain of MT1-MMP have been shown to alter RhoA/ROK expression by attenuating thrombin-triggered RhoA and Rac1 activation in endothelial cells,19 or RhoA-mediated CD44 shedding in glioma cells,20 and to change the phosphorylation status of extracellular signal–regulated kinase (Erk),21 signal transducers and activators of transcription 3 (STAT3),8,15 and Akt.22 The MT1-MMP gene silencing has also provided evidence regarding its involvement in transcriptional regulation as it prevented concanavalin-A–induced STAT3-mediated and NF-κB–mediated regulation of inflammation, autophagy, and angiogenesis in numerous cell models.23,24
Recent studies have suggested cooperative links between inflammasomes, which are cytoplasmic protein complexes that sense and assemble in response to diverse inflammatory stimuli such as infection-associated or stress-associated stimuli,25 with many downstream cellular processes such as autophagy,26 angiogenesis,27 and carcinogenesis.28 The inflammasome components, on complex scaffold protein assembly,29 trigger caspase activity and lead to the cleavage of precursors and release of the pro-inflammatory cytokines interleukin 1β (IL-1β) and IL-18.30 The MT1-MMP knockdown suppressed angiogenesis in glioma tumors through decreased production of pro-angiogenic factors, such as vascular endothelial growth factor and IL-8,31 and was also found to regulate the transcription of colony-stimulating factors 2 and 3 in mesenchymal stromal cells.8 In addition, MT1-MMP was also demonstrated to regulate caspase activity,32 angiogenesis,33 and cell migration.34 Whether any MT1-MMP intracellular signaling functions are linked to inflammasome regulation remains to be determined.
Inflammasomes are believed to be promising therapeutic targets in cancer-related clinical conditions as their inhibition, or the neutralization of their products, profoundly affects carcinogenesis and tumor progression.35 Loss of MT1-MMP leads to abnormal proteolytic processing of ECM components in Mmp14-deficient mice which consequently triggered signaling events that alter the nuclear lamina and cytoskeleton structure, ultimately leading to nuclear envelope abnormalities, increased DNA damage, altered gene transcription, and activation of cell senescence.36 Here, we designed a proof-of-concept study in which we sought to further address whether MT1-MMP exerts any transcriptional regulation of inflammasome-related biomarkers on pro-inflammatory stimulus in an HT1080 fibrosarcoma cell model.
Materials and Methods
Materials
Sodium dodecylsulfate (SDS) and bovine serum albumin were purchased from Sigma (Oakville, ON, USA). Electrophoresis reagents were purchased from Bio-Rad (Mississauga, ON, USA). Micro bicinchoninic acid protein assay reagents were from Pierce (Rockford, IL, USA). The monoclonal anti-MT1-MMP catalytic domain antibody clone 3G4.2 was from EMD Millipore (Billerica, MA, USA). The polyclonal antibodies against inhibitor of κB (IκB), p105, phosphorylated p105, Erk, and phosphorylated Erk were purchased from Cell Signaling (Danvers, MA, USA). The monoclonal antibody against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was from Advanced Immunochemical Inc. (Long Beach, CA, USA). Horseradish peroxidase–conjugated donkey anti-rabbit and anti-mouse IgG secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). All other reagents were from Sigma-Aldrich (Oakville, ON, Canada).
Cell culture and small interfering RNA transfection
The human HT1080 fibrosarcoma cell line was purchased from American Type Culture Collection and maintained in Eagle’s Minimum Essential Medium containing 10% (v/v) fetal calf serum (HyClone Laboratories, Logan, UT, USA), 2 mM glutamine, 100 units/mL penicillin, and 100 µg/mL streptomycin and was cultured at 37°C under a humidified atmosphere containing 5% CO2. Gene silencing of MT1-MMP was performed through cell transfection of 20-nM MT1-MMP–specific small interfering RNA (siRNA) (Hs_MMP14_6 HP validated siRNA; QIAGEN, Valencia, CA, USA; SI03648841) or with a scrambled siRNA sequence (QIAGEN; AllStar Negative Control siRNA, SI03650318) as a control using lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Small interfering RNA and mismatch siRNA were synthesized by QIAGEN and annealed to form duplexes. About 24 hours after transfection, cells were treated with up to 30-nM phorbol-12-myristate-13-acetate (PMA) or vehicle in a serum-free medium for 18 hours.
Immunoblotting procedures
Following treatments or transfection, HT1080 cells were washed with phosphate-buffered saline and lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, 120 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 0.1% Triton) in the presence of phosphatase and protease inhibitors on ice for 30 minutes. Cell debris was pelleted by centrifugation for 10 minutes at high speed. Protein concentration was quantified using a micro bicinchoninic acid protein assay kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). Proteins from control and treated cells were separated by SDS-polyacrylamide gel electrophoresis (PAGE). After electrophoresis, proteins were electrotransferred to polyvinylidene difluoride membranes, which were then blocked overnight at 4°C with 5% nonfat dry milk in Tris-buffered saline (150 mM NaCl, 20 mM Tris-HCl, pH 7.5) containing 0.3% Tween-20 (TBST). Membranes were further washed in TBST and incubated with primary antibodies directed against MT1-MMP (1/10 000), p105, phosphorylated p105, Erk, phosphorylated Erk, IκB, phosphorylated IκB (1/1000), or GAPDH (1/1500). Washing was then performed in TBST, followed by a 1-hour incubation with horseradish peroxidase–conjugated anti-rabbit IgG (1/10 000) or anti-mouse IgG (1/5000) in TBST containing 5% nonfat dry milk. Immunoreactive material was visualized by Western Lightning Enhanced Chemiluminescence Pro (Perkin Elmer, Waltham, MA, USA).
Total RNA isolation, complementary DNA synthesis, and real-time quantitative real-time polymerase chain reaction
Total RNA was extracted from HT1080 cell monolayers using TRIzol reagent (Life Technologies, Gaithersburg, MD, USA). For complementary DNA (cDNA) synthesis, 1 µg of total RNA was reverse-transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). Complementary DNA was stored at −80°C prior to polymerase chain reaction (PCR). Gene expression was quantified by real-time quantitative PCR using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). DNA amplification was conducted using an Icycler iQ5 (Bio-Rad), and product detection was performed by measuring binding of the fluorescent dye SYBR Green I to double-stranded DNA. The following primer sets were provided by QIAGEN: IL-1B (Hs_ IL1B_1_SG, QT00021385), IL-6 (Hs_IL6_1_SG, QT00083720), IL-12A (Hs_IL12A_1_SG, QT00000357), IL-33 (Hs_IL33_1_SG, QT00041559), EGR1 (Hs_ EGR1_1_SG, QT0026505), ETS1 (Hs_ETS1_1_SG, QT00049133), ETS2 (Hs_ETS2_1_SG, QT00086198), ELK1 (Hs_ELK1_1_SG, QT00034104), β-actin (Hs_ ACTB_2_SG, QT01680476), GAPDH (Hs_GAPDH_1_ SG, QT00079247), and Peptidylprolyl Isomerase A (PPIA) (Hs_PPIA_4_SG, QT01866137). The relative quantities of target gene messenger RNA (mRNA) against an internal control, β-actin/GAPDH/PPIA RNA, were measured by following a ΔCT method employing an amplification plot (fluorescence signal vs cycle number). The difference (ΔCT) between the mean values in the triplicate samples of target gene and those of β-actin/GAPDH/PPIA RNA was calculated by iQ5 Optical System Software version 2.0 (Bio-Rad), and the relative quantified value was expressed as 2−∆CT.
Gelatin zymography
Gelatin zymography was used to assess the extent of pro-MMP-2 gelatinolytic activity and activation status as previously described.37 Briefly, a 20 µL aliquot of the culture medium was subjected to SDS-PAGE in a gel containing 0.1 mg/mL gelatin, a substrate that is efficiently hydrolyzed by pro-MMP-9, pro-MMP-2, and MMP-2. The gels were then incubated in 2.5% Triton X-100 and rinsed in nanopure distilled H2O. Gels were further incubated at 37°C for 20 hours in 20 mM NaCl, 5 mM CaCl2, 0.02% Brij-35, 50 mM Tris-HCl buffer, pH 7.6 and then stained with 0.1% Coomassie Brilliant Blue R-250 and destained in 10% acetic acid and 30% methanol in H2O. Gelatinolytic activity was detected as unstained bands on a blue background.
Human transcription factors and inflammasome PCR arrays
The human inflammasomes (PAHS-097Z) and transcription factors (PAHS-075Z) RT2 Profiler PCR Array screens (SA Biosciences, Frederick, MD, USA) were performed according to the manufacturer’s protocol. The detailed list of these key transcription factor genes can be found on the manufacturer’s Web site (http://www.sabiosciences.com/ArrayList.php). Using real-time quantitative PCR, we analyzed expression of a focused panel of genes related to transcription factor genes. Relative gene expressions were calculated using the 2−ΔΔCT method, in which CT indicates the fractional cycle number where the fluorescent signal reaches detection threshold. The ‘delta-delta’ method uses the normalized ΔCT value of each sample, calculated using a total of 5 endogenous control genes (B2M, HPRT1, RPL13A, GAPDH, and ACTB). Fold change values are then presented as average fold .. for genes in siScrambled PMA-treated HT1080 cells relative to siMT1-MMP PMA-treated cells. Detectable PCR products were obtained and defined as requiring <35 cycles. The resulting raw data were then analyzed using the PCR Array Data Analysis Template (http://www.sabiosciences.com/pcrarraydataanalysis.php). This integrated Web-based software package automatically performs all ΔΔCT-based fold-change calculations from our uploaded raw threshold cycle data.
Statistical data analysis
Data are representative of 3 or more independent experiments. Statistical significance was assessed using Student’s unpaired t test. Probability values of less than .05 were considered significant and an asterisk (*) identifies such significance in the figures.
Results
PMA triggers Erk, p105, and IkB transient phosphorylation in HT1080 fibrosarcoma cells
We first addressed the responsiveness of HT1080 fibrosarcoma cells to PMA, which is known to trigger inflammation and to prime cell migration and proliferation38 in part through a NF-κB–transducing pathway.39,40 More specifically, we assessed the phosphorylation status of IκB, of the p105 precursor protein of NF-κB1, as well as of the Erk. HT1080 cells were treated with 30 nM PMA for up to 60 minutes, and the phosphorylation status of the above molecules was determined by immunoblotting as described in the “Materials and Methods” section (Figure 1A). We found that IκB optimal phosphorylation occurred at 10 minutes, whereas those of p105 and Erk were delayed and occurred later at 30 minutes (Figure 1B).
Figure 1.
PMA triggers Erk, p105, and IκB transient phosphorylation in HT1080 fibrosarcoma cells. Serum-starved HT1080 fibrosarcoma cells were treated for up to 60 minutes with vehicle or 30 nM PMA, following which cell lysates were isolated, electrophoresed via SDS-PAGE, and immunodetected for (A) P-IκB, IκB, P-p105, p105, P-Erk, and Erk proteins as described in the “Materials and Methods” section. (B) Quantification was performed by scanning densitometry of a representative autoradiogram. Erk indicates extracellular signal–regulated kinase; IκB, inhibitor of κB; PMA, phorbol-12-myristate-13-acetate; SDS-PAGE, sodium dodecylsulfate-polyacrylamide gel electrophoresis.
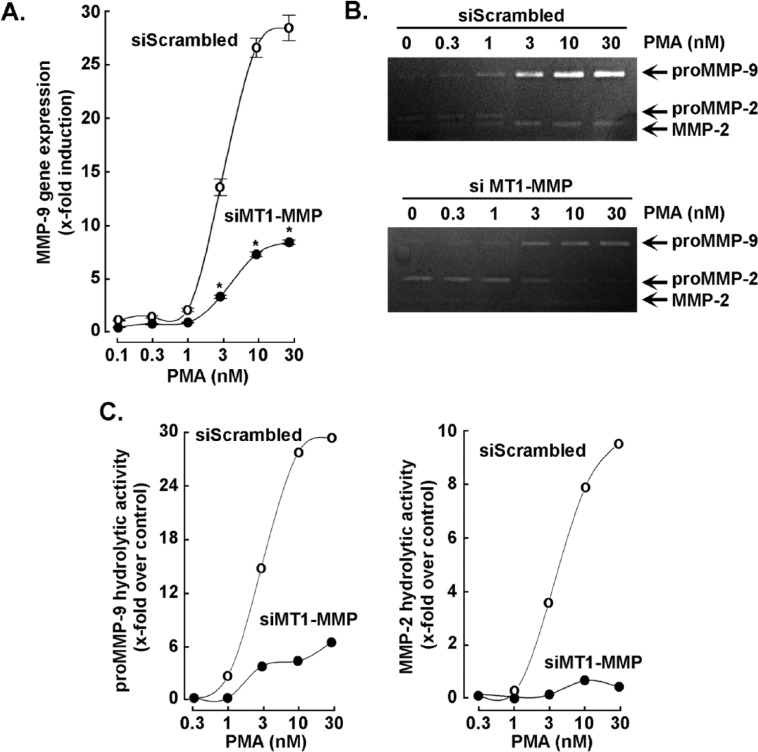
MT1-MMP gene silencing abrogates PMA-induced Erk, p105, and IkB phosphorylation in HT1080 fibrosarcoma cells
Given the hypothesized role of MT1-MMP as an intermediate in several signal-transducing pathways particularly involved in the regulation of cell invasion and inflammation,8,13,15,23 we next transiently silenced MT1-MMP gene expression in HT1080 fibrosarcoma cells and evaluated the impact of this gene silencing on PMA-induced signaling. We found that although MT1-MMP expression was significantly reduced (Figure 2, bottom panels), the PMA-induced phosphorylation status of IκB, p105, and Erk were all significantly abrogated. This reinforces the claim that MT1-MMP can modulate downstream end-product expression, potentially through transcriptional regulation.
Figure 2.
MT1-MMP gene silencing abrogates PMA-induced Erk, p105, and IkB phosphorylation in HT1080 fibrosarcoma cells. Transient MT1-MMP gene silencing was performed in HT1080 cells as described in the “Materials and Methods” section. Serum-starved HT1080 fibrosarcoma cells were then treated for either 10 minutes to monitor Erk phosphorylation or 30 minutes to monitor p105 and IκB phosphorylation within vehicle or 30 nM PMA-treated cells. Cell lysates were isolated, electrophoresed via SDS-PAGE, and immunodetected for P-IκB, IκB, P-p105, p105, P-Erk, Erk, MT1-MMP, and GAPDH proteins as described in the “Materials and Methods” section. Erk indicates extracellular signal–regulated kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IκB, inhibitor of κB; PMA, phorbol-12-myristate-13-acetate; SDS-PAGE, sodium dodecylsulfate-polyacrylamide gel electrophoresis.
Stimulatory and repressive regulatory impacts on transcription factor and cytokine transcription on MT1-MMP gene silencing in PMA-stimulated HT1080 fibrosarcoma cells
To more broadly assess the global transcriptional impact of MT1-MMP on PMA-stimulated cells, we used a gene array approach to survey some downstream transcription factors (Figure 3) and the inflammasome response (Figure 4). Among the 84 transcription factor genes in each array which were assessed downstream of PMA stimulation, 61% were induced by PMA (Figure 3A) in cells where MT1-MMP was silenced, suggesting that basal MT1-MMP exerted a repressive transcriptional control on these genes. Among these, several were involved in cell proliferation, survival, and inflammation. However, expression of 29% of the transcription factors assessed was decreased under these same conditions (Figure 3B), suggesting MT1-MMP bestowed stimulatory effects on these genes following PMA treatment. These genes involved similar proliferation and inflammation processes as in those inhibited by MT1-MMP (Table 1), confirming the dynamic balance that the signaling contribution of MT1-MMP exerts within the cell. A similar screen was performed regarding inflammasome-associated biomarkers. Interestingly, 17% of the genes tested were upregulated in cells where MT1-MMP was silenced (Figure 4A), whereas 83% were inhibited (Figure 4B). The latter observation suggests that MT1-MMP exerts a significant role in the positive transcriptional regulation of inflammasome-related genes in PMA-stimulated cells (Table 1) (refer to the appended supplemental data section for the list of all genes screened and effects).
Figure 3.
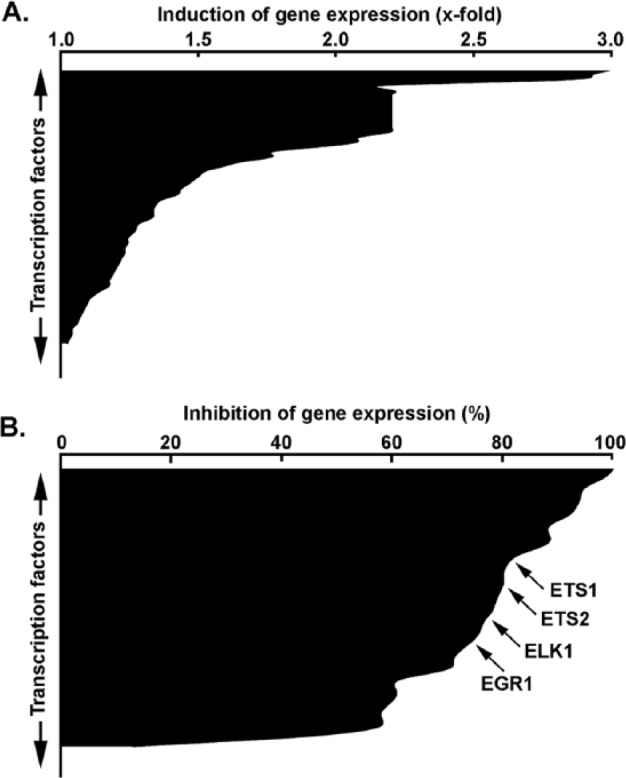
Stimulative and repressive regulatory impacts on transcription factors transcription on MT1-MMP gene silencing in PMA-stimulated HT1080 fibrosarcoma cells. Gene silencing was performed with either siScrambled or siMT1-MMP in HT1080 fibrosarcoma cells. Cells were then serum-starved in the presence or absence of 30 nM PMA for 24 hours. Total RNA isolation and qRT-PCR were performed as described in the “Materials and Methods” section to assess expression of a subset of 84 different transcription factor genes using human PCR arrays as described in the “Materials and Methods” section. (A) PMA stimulatory response expressed as the ratio of siMT1-MMP–transfected cells over siScrambled-transfected cells (x-fold induction). (B) PMA repressive response expressed as the ratio of siMT1-MMP–transfected cells over siScrambled-transfected cells (% inhibition). PMA indicates phorbol-12-myristate-13-acetate; SDS-PAGE, sodium dodecylsulfate-polyacrylamide gel electrophoresis; qRT-PCR, quantitative real-time polymerase chain reaction.
Figure 4.
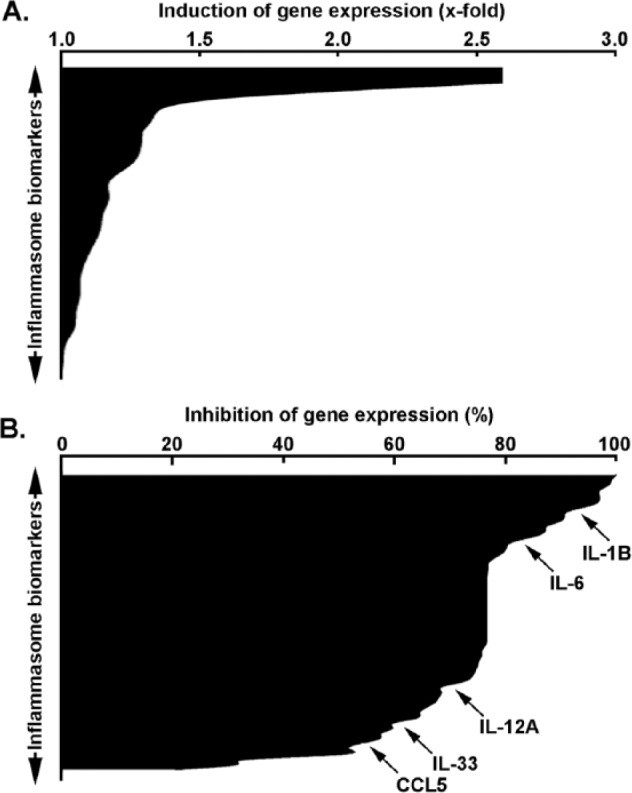
Stimulative and repressive regulatory impact on inflammasome-related transcription on MT1-MMP gene silencing in PMA-stimulated HT1080 fibrosarcoma cells. Gene silencing was performed with either siScrambled or siMT1-MMP in HT1080 fibrosarcoma cells. Cells were then serum-starved in the presence or absence of 30 nM PMA for 24 hours. Total RNA isolation and qRT-PCR were performed as described in the “Materials and Methods” section to assess expression of a subset of 84 different inflammasome-related genes using human PCR arrays as described in the “Materials and Methods” section. (A) PMA stimulatory response expressed as the ratio of siMT1-MMP–transfected cells over siScrambled-transfected cells (x-fold induction). (B) PMA repressive response expressed as the ratio of siMT1-MMP–transfected cells over siScrambled-transfected cells (% inhibition). PMA indicates phorbol-12-myristate-13-acetate; SDS-PAGE, sodium dodecylsulfate-polyacrylamide gel electrophoresis; qRT-PCR, quantitative real-time polymerase chain reaction.
Table 1.
Suppressive transcriptional role of MT1-MMP silencing in PMA-treated cells.
Evidence and validation of MT1-MMP requirement in PMA-induced cytokine and transcription factor transcription
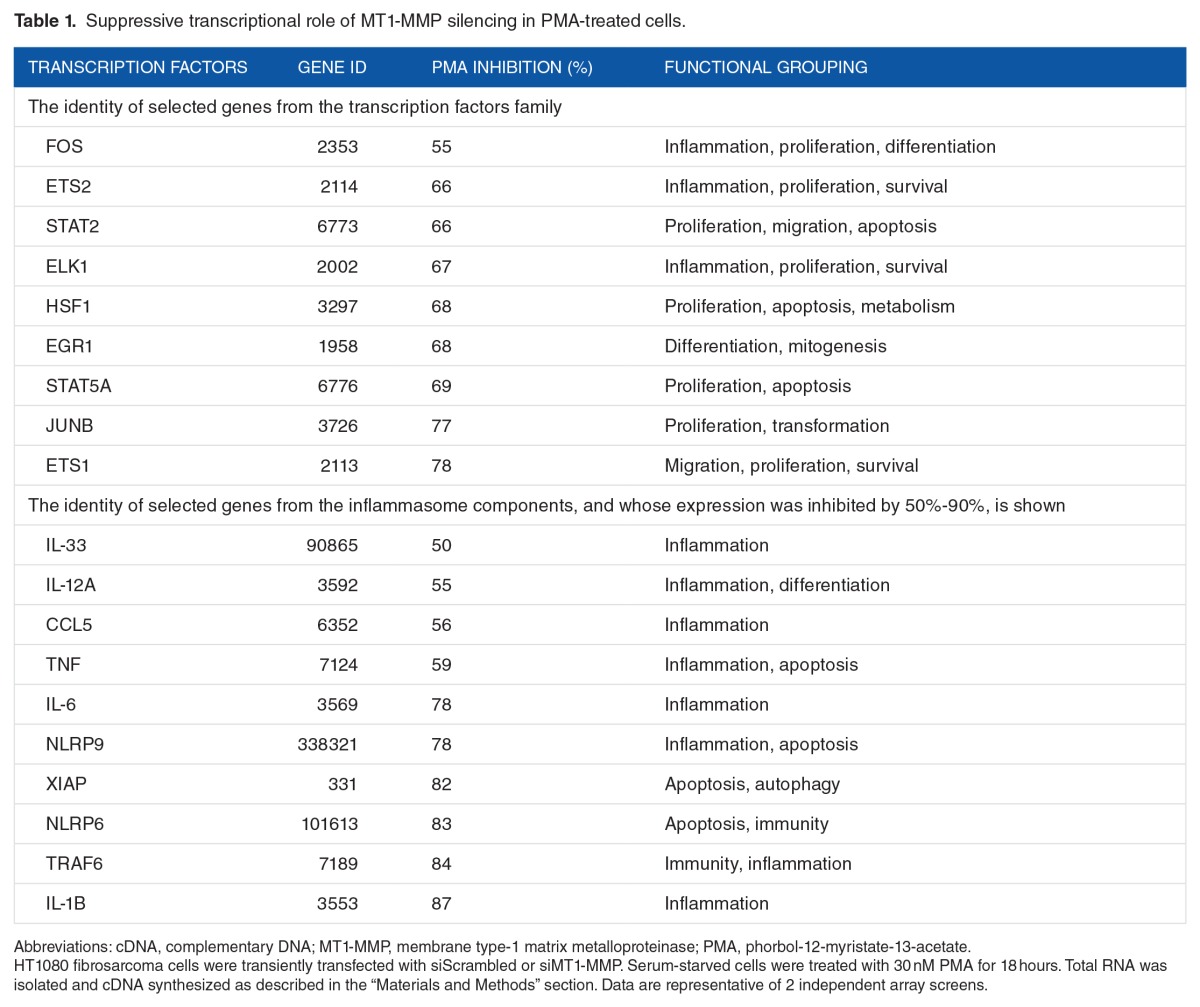
Having established that MT1-MMP plays a role in the regulation of inflammasome, we next focused on the potential stimulatory roles that MT1-MMP originally exerts on gene transcription, which had been repressed by PMA stimulation in MT1-MMP-silenced cells (Figures 3B and 4B). Although of interest, the repressive transcriptional role of MT1-MMP, as seen by MT1-MMP silencing in PMA-treated cells (Figures 3A and 4A) will be explored in a subsequent study. Dose-response treatments of PMA were applied to siScrambled or siMT1-MMP-transfected cells, and qRT-PCR was performed for specific inflammasome-related (Figure 5) and transcription factor (Figure 6) target genes. We confirmed that PMA was able to trigger gene expression of IL-1B, IL-6, IL-12, and IL-33 in control (siScrambled) cells, whereas MT1-MMP gene silencing prevented this PMA-induced expression (Figure 5). Similarly, transcription of ELK1, early growth response protein 1 (EGR1), ETS1, and ETS2 were also induced by PMA in conditions where MT1-MMP was present, whereas MT1-MMP silencing prevented these increases (Figure 6).
Figure 5.
MT1-MMP requirement in PMA-induced inflammasome-related transcription. Gene silencing was performed with either siScrambled (open circles) or siMT1-MMP (closed circles) in HT1080 fibrosarcoma cells. Cells were then serum-starved in the presence or absence of up to 30 nM PMA for 24 hours. Total RNA isolation and qRT-PCR were performed as described in the “Materials and Methods” section to assess expression of MT1-MMP, IL-6, IL-12, IL-33, or IL-1B.
Figure 6.
MT1-MMP requirement in PMA-induced transcription factors transcription. Gene silencing was performed with either siScrambled (open circles) or siMT1-MMP (closed circles) in HT1080 fibrosarcoma cells. Cells were then serum-starved in the presence or absence of up to 30 nM PMA for 24 hours. Total RNA isolation and qRT-PCR were performed as described in the “Materials and Methods” section to assess expression of ELK1, EGR1, ETS1, or ETS2. PMA indicates phorbol-12-myristate-13-acetate; qRT-PCR, quantitative real-time polymerase chain reaction.
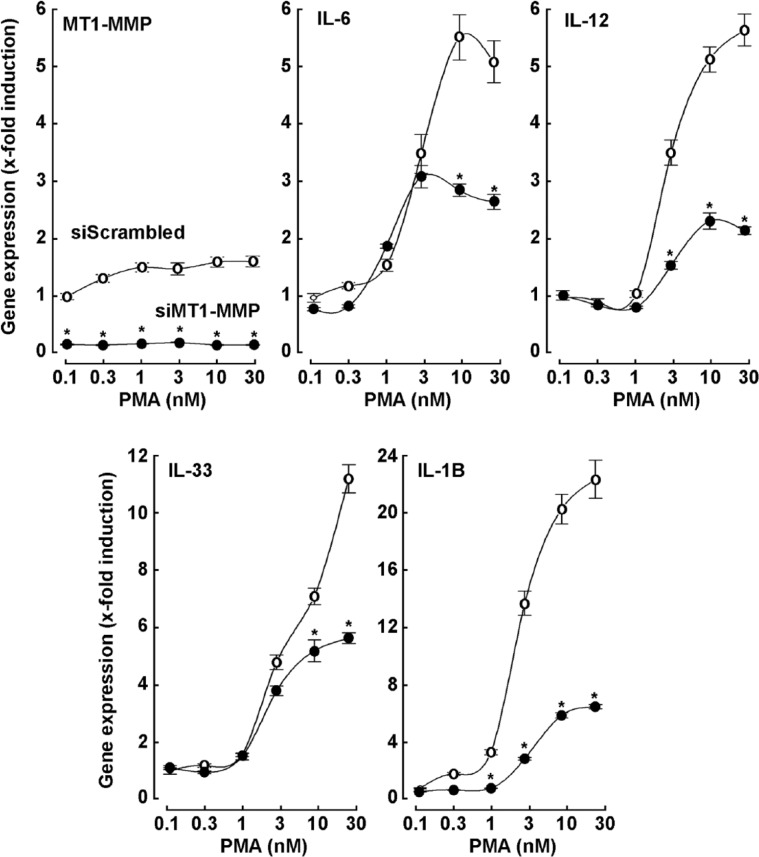
MT1-MMP gene silencing abrogates PMA-induced MMP-9 expression and function in HT1080 fibrosarcoma cells
Parallel stimulation by MT1-MMP of biomarkers for cell proliferation/invasion and for inflammation phenotypes associated with cancer cell invasiveness suggests the possibility of a common element downstream of MT1-MMP. We assessed the expression of MMP-9, a secreted MMP known to be highly inducible by inflammation-mediating factors such as tumor necrosis factor (TNF) and PMA.38,41,42 The MT1-MMP was again transiently silenced and this was shown to prevent PMA-mediated MMP-9 induction (Figure 7A). This was further confirmed at the protein level using zymography to assess the extracellular hydrolytic activity of pro-MMP-9 and of pro-MMP-2 (Figure 7B). Although pro-MMP-9 hydrolytic activity was increased by PMA, this was largely prevented by MT1-MMP gene silencing (Figure 7C). A detectable activation of latent pro-MMP-2 to active MMP-2 was observed on PMA treatment, but this too was repressed in MT1-MMP–silenced cells confirming the functional repression of MT1-MMP at the cell surface.
Figure 7.
MT1-MMP gene silencing abrogates PMA-induced MMP-9 expression and function in HT1080 fibrosarcoma cells. Gene silencing was performed with either siScrambled (open circles) or siMT1-MMP (closed circles) in HT1080 fibrosarcoma cells. Cells were then serum-starved in the presence or absence of up to 30 nM PMA for 24 hours. (A) Total RNA isolation and qRT-PCR were performed as described in the “Materials and Methods” section to assess the expression of MMP-9. (B) On treatment, conditioned media were harvested and gelatin zymography performed as described in the “Materials and Methods” section to assess the extent of pro-MMP-9, pro-MMP-2, and MMP-2 hydrolytic activity. (C) Scanning densitometry was performed of a representative zymogram. PMA indicates phorbol-12-myristate-13-acetate; qRT-PCR, quantitative real-time polymerase chain reaction.
Discussion
Membrane type-1 matrix metalloproteinase is a membrane-embedded protein which exhibits functions at both sides of the plasma membrane, allowing coordination between the extracellular and intracellular milieus in response to environmental cues such as inflammatory stimuli. It is therefore well positioned to sense and to modify the extracellular environment by processing matrix components, transmembrane proteins, and soluble factors, as well as to regulate, through its intracellular membrane-anchored domain, cell motility, metabolism, and gene transcription. In support of its contribution toward gene regulation, our results provide evidence that MT1-MMP exerts transcriptional control of inflammasome components and positions MT1-MMP as an important signal-transducing intermediate relaying extracellular pro-inflammatory cues.
Noncatalytic MT1-MMP regulation of gene transcription in endothelial cells recently highlighted genes involved in cell proliferation, vasculature development, and blood vessel morphogenesis.43 Furthermore, indirect evidence of MT1-MMP–mediated transcriptional regulation of target genes has also been reported to require a functional MT1-MMP intracellular domain. These results were, in part, obtained using concanavalin-A–treated cells and involved growth factors and cytokines,8,44 inflammation,23,45 and autophagy15,46 biomarkers. Here, we not only confirm the signal-transducing intermediate role for MT1-MMP, as its absence reduced the PMA-induced phosphorylation status of NF-κB and Erk, but we also extend our observations to the transcriptional modulation of downstream transcription factors such as ELK1, EGR1, ETS1, and ETS2. Interestingly, ETS1/2 was found to be involved in the inflammatory immune response to viral infections of the heart.47 ETS1 particularly is a common mediator of the renal pro-inflammatory and profibrotic effects of Ang II.48 Cross talk between ELK3/ETS1/MT1-MMP related to angiogenesis has also recently been reported.49 High levels of ELK1 mRNA were found in clinical specimens of serous epithelial ovarian cancer chemoresistant tissues50 and in relaxin’s induction of MMP-9.51 Interestingly, EGR1 abrogation reduced collagen-induced transcriptional regulation of MT1-MMP52 and Si0(2)-driven transcription of MT1-MMP.53 E26 oncogene homolog 1 (ETS1) transcriptional activity on the MT1-MMP promoter was also reported in angiogenesis.54 Finally, overexpression of erythroblastosis virus ETS1 gene is correlated with both tumor progression and poor response to chemotherapy in cancer treatment.55
Activation of NF-κB has been reported to be triggered by MT1-MMP45 and requires its release from inhibitor of NF-κB (IκB) proteins in the cytoplasm. Although much work has focused on the identification of pathways regulating this cytosolic rate-limiting step of NF-κB activation, recent data reveal the existence of IκB-independent control of NF-κB activity by modulatory phosphorylations from IκB kinases (IKKα, β, and ε).56 As such, IKKs are believed to phosphorylate p105, p65, and IκB. Whether and how any hierarchical IκB and p105 phosphorylation occurs on PMA stimulation remains unknown. Differential interaction affinities of IKKs toward p105 and IκB were reported57 and may, in part, explain the lag in maximal phosphorylation status as observed in Figure 1B.
One interesting aspect from our study relates to the MT1-MMP/MMP-9 interrelationship in PMA-stimulated cells. The deregulation of mmp-9 gene expression is, in part through epigenetic mechanisms, implicated in several pathophysiological processes, including inflammation, chronic obstructive pulmonary disease, autoimmune diseases, and cancer.58 Here, we found that PMA-induced MMP-9 expression was abrogated in cells where MT1-MMP was silenced, suggesting that MT1-MMP acts as an important signaling intermediate in MMP-9 gene and protein expression. Given that the foremost mechanism of action of MMP-9 in brain disorders appears to be its involvement in immune/inflammation responses through processing and activation of various cytokines and chemokines,59 the involvement of a MT1-MMP–dependent event may be envisioned as a secondary process that could regulate the response to a given pro-inflammatory stimuli. More recently, the MT1-MMP expression level status was found to dictate the in vitro action of lupeol on PMA-induced inflammatory biomarkers MMP-9 and COX-2 in a pediatric brain tumor cell model.60
Therapies which can directly or indirectly target the inflammasome and its downstream cytokines to quiet inflammation are currently envisioned.61,62 Because we have shown that the induction of inflammasome activation by carcinogens partially requires MT1-MMP, one may envision that any strategy that alters MT1-MMP’s noncatalytic functions may also alter the expression of inflammasome components and downstream cytokines. To this end, the green tea polyphenol epigallocatechin-3-gallate (EGCG) is among the most potent natural inhibitors of the catalytic and noncatalytic functions of MT1-MMP.63–66 Epigallocatechin-3-gallate was found to suppress melanoma growth by inhibiting inflammasome and IL-1β secretion67 and to inhibit NLRP3 inflammasome activation.68 More recently, EGCG targeting of MT1-MMP–mediated Src and JAK/STAT3 signaling was found to inhibit transcription of colony-stimulating factors 2 and 3 in mesenchymal stromal cells.8 Finally, the non-antibiotic cellular effects exerted by the anti-inflammatory tetracycline derivative minocycline was recently assessed in a concanavalin-A–activated human HepG2 hepatoma cell model, a condition known to increase the expression of MT1-MMP and to trigger inflammatory and autophagy processes.69 Parts of minocycline’s effects were found to be exerted through the inhibition of MT1-MMP signaling functions.70 In line with that observation, minocycline exerted protective properties against NLRP3 inflammasome-induced inflammation and P53-associated apoptosis in an early brain injury model.71
Among the downstream pro-inflammatory cytokines screened in our study, we found that the transcription of IL-33 and IL-12A was MT1-MMP dependent in carcinogen-stimulated HT1080 cells. Newly discovered roles for IL-33 in obesity, intestinal inflammation, and tumorigenesis have recently been reported,72,73 whereas low IL-12 production or a high IL-33/IL-12 ratio in patients is believed to contribute to tumor development.74 Whether MT1-MMP is involved in IL-12 expression as an antitumor cytokine or in IL-33 expression, a cytokine which bears both pro-tumor and antitumor activities, remains to be addressed. Interestingly, selective inhibition of MT1-MMP abrogated the progression of experimental inflammatory arthritis and upregulation of IL-12.75 The connection between inflammation and tumor initiation is not a 1-way street, and there is also evidence that DNA damage can lead to inflammation and thereby promote tumorigenesis. The first critical genetic evidence for inflammatory cells as a source of tumor-promoting cytokines was obtained in a mouse model of Colitis-Associated Cancer (CAC), where inactivation of NF-κB in myeloid cells reduced tumor growth and blocked production of IL-6 and other cytokines in response to colitis.76 Subsequent work demonstrated the effect mediated through IL-6, IL-11, and TNF-α in gastric cancer,77 in which IL-1β is also a tumor promoter.78 Both IL-6 and IL-1β have been linked to chronic inflammation which might mediate the association between obesity and endometrial cancer79 and found in high-risk colorectal neoplasia.80
Conclusions
Altogether, our data show a significant involvement of MT1-MMP in the transcriptional regulation of inflammatory biomarkers, consolidating its contribution to signal-transducing functions, in addition to its classical hydrolytic activity (Figure 8). In this study, we investigated MT1-MMP transcriptional effects on genes which received positive stimulus from MT1-MMP following pro-inflammatory cues (PMA treatment, Table 1). As the silencing of MT1-MMP led to increased expression of several genes in PMA-stimulated cells (Figures 3A and 4A), such transcriptional control needs to be further investigated to determine its impact on inflammatory diseases. Although of importance to eventually validate the data presented here in other cell lines, we definitely do acknowledge, however, that different cell lines would present different base levels of inflammatory cytokines and possible differential expression of inflammatory signaling depending on basal MT1-MMP expression levels. Development and assessment of MT1-MMP catalytic and noncatalytic inhibitors67–69 may therefore be envisioned as possibly also contributing to the reduction in cancer-related inflammation processes.
Figure 8.
Summarizing scheme of MT1-MMP–mediated regulation of pro-inflammatory events. The classical role of MT1-MMP is in the promotion of cell invasion through ECM degradation and activation of latent pro-MMPs. The combined pro-inflammatory signaling from PMA and MT1-MMP–mediated signaling (through documented Src, NF-κB, Jak/Stat. RhoA/ROK, Erk) leads to increased cytokine transcription. Increased cytokine expression then promotes an angiogenic and inflammatory phenotype which contributes to tumor development. ECM indicates extracellular matrix; NF-κB, nuclear factor κB; PMA, phorbol-12-myristate-13-acetate; qRT-PCR, quantitative real-time polymerase chain reaction.
Footnotes
PEER REVIEW: Eight peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1479 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Institutional Research Chair in Cancer Prevention and Treatment held by Dr Borhane Annabi at UQAM.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
SS performed all the experiments, analyzed the data, and drafted the manuscript. BA designed the study, analyzed the data, and drafted the manuscript. All authors read and approved the final manuscript.
REFERENCES
- 1.Snyman C, Niesler CU. MMP-14 in skeletal muscle repair. J Muscle Res Cell Motil. 2015;36:215–225. doi: 10.1007/s10974-015-9414-4. [DOI] [PubMed] [Google Scholar]
- 2.Itoh Y. Membrane-type matrix metalloproteinases: their functions and regulations. Matrix Biol. 2015;44–46:207–223. doi: 10.1016/j.matbio.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Chen Q, Jin M, Yang F, Zhu J, Xiao Q, Zhang L. Matrix metalloproteinases: inflammatory regulators of cell behaviors in vascular formation and remodeling. Mediators Inflamm. 2013;2013:14. doi: 10.1155/2013/928315. Article 928315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parks WC, Wilson CL, López-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 5.Al-Raawi D, Abu-El-Zahab H, El-Shinawi M, Mohamed MM. Membrane type-1 matrix metalloproteinase (MT1-MMP) correlates with the expression and activation of matrix metalloproteinase-2 (MMP-2) in inflammatory breast cancer. Int J Clin Exp Med. 2011;4:265–275. [PMC free article] [PubMed] [Google Scholar]
- 6.Löffek S, Schilling O, Franzke CW. Series “matrix metalloproteinases in lung health and disease”: biological role of matrix metalloproteinases: a critical balance. Eur Respir J. 2011;38:191–208. doi: 10.1183/09031936.00146510. [DOI] [PubMed] [Google Scholar]
- 7.Sun J. Matrix metalloproteinases and tissue inhibitor of metalloproteinases are essential for the inflammatory response in cancer cells. J Signal Transduct. 2010;2010:7. doi: 10.1155/2010/985132. Article 985132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zgheib A, Lamy S, Annabi B. Epigallocatechin gallate targeting of membrane type 1 matrix metalloproteinase-mediated Src and Janus kinase/signal transducers and activators of transcription 3 signaling inhibits transcription of colony-stimulating factors 2 and 3 in mesenchymal stromal cells. J Biol Chem. 2013;288:13378–13386. doi: 10.1074/jbc.M113.456533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annabi B, Laflamme C, Sina A, Lachambre MP, Béliveau R. A MT1-MMP/NF-kappaB signaling axis as a checkpoint controller of COX-2 expression in CD133+ U87 glioblastoma cells. J Neuroinflammation. 2009;6:8. doi: 10.1186/1742-2094-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porta C, Larghi P, Rimoldi M, et al. Cellular and molecular pathways linking inflammation and cancer. Immunobiology. 2009;214:761–777. doi: 10.1016/j.imbio.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Biddlestone J, Bandarra D, Rocha S. The role of hypoxia in inflammatory disease. Int J Mol Med. 2015;35:859–869. doi: 10.3892/ijmm.2015.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iurlaro R, León-Annicchiarico CL, Muñoz-Pinedo C. Regulation of cancer metabolism by oncogenes and tumor suppressors. Methods Enzymol. 2014;542:59–80. doi: 10.1016/B978-0-12-416618-9.00003-0. [DOI] [PubMed] [Google Scholar]
- 13.Proulx-Bonneau S, Guezguez A, Annabi B. A concerted HIF-1α/MT1-MMP signalling axis regulates the expression of the 3BP2 adaptor protein in hypoxic mesenchymal stromal cells. PLoS ONE. 2011;6:e21511. doi: 10.1371/journal.pone.0021511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annabi B, Lee YT, Turcotte S, et al. Hypoxia promotes murine bone-marrow-derived stromal cell migration and tube formation. Stem Cells. 2003;21:337–347. doi: 10.1634/stemcells.21-3-337. [DOI] [PubMed] [Google Scholar]
- 15.Pratt J, Annabi B. Induction of autophagy biomarker BNIP3 requires a JAK2/STAT3 and MT1-MMP signaling interplay in concanavalin-A-activated U87 glioblastoma cells. Cell Signal. 2014;26:917–924. doi: 10.1016/j.cellsig.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell S, Vargas J, Hoffmann A. Signaling via the NF-κB system. Wiley Interdiscip Rev Syst Biol Med. 2016;8:227–441. doi: 10.1002/wsbm.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato H, Takino T. Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances pericellular proteolysis and invasion. Cancer Sci. 2010;101:843–847. doi: 10.1111/j.1349-7006.2010.01498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zucker S, Pei D, Cao J, Lopez-Otin C. Membrane type-matrix metalloproteinases (MT-MMP) Curr Top Dev Biol. 2003;54:1–74. doi: 10.1016/s0070-2153(03)54004-2. [DOI] [PubMed] [Google Scholar]
- 19.Ando K, Ishibashi T, Ohkawara H, et al. Crucial role of membrane type 1 matrix metalloproteinase (MT1-MMP) in RhoA/Rac1-dependent signaling pathways in thrombin- stimulated endothelial cells. J Atheroscler Thromb. 2011;18:762–773. doi: 10.5551/jat.6783. [DOI] [PubMed] [Google Scholar]
- 20.Annabi B, Bouzeghrane M, Moumdjian R, Moghrabi A, Béliveau R. Probing the infiltrating character of brain tumors: inhibition of RhoA/ROK-mediated CD44 cell surface shedding from glioma cells by the green tea catechin EGCg. J Neurochem. 2005;94:906–916. doi: 10.1111/j.1471-4159.2005.03256.x. [DOI] [PubMed] [Google Scholar]
- 21.Gingras D, Bousquet-Gagnon N, Langlois S, Lachambre MP, Annabi B, Béliveau R. Activation of the extracellular signal-regulated protein kinase (ERK) cascade by membrane-type-1 matrix metalloproteinase (MT1-MMP) FEBS Lett. 2001;507:231–236. doi: 10.1016/s0014-5793(01)02985-4. [DOI] [PubMed] [Google Scholar]
- 22.Ohkawara H, Ishibashi T, Sugimoto K, Ikeda K, Ogawa K, Takeishi Y. Membrane type 1-matrix metalloproteinase/Akt signaling axis modulates TNF-α-induced procoagulant activity and apoptosis in endothelial cells. PLoS ONE. 2014;9:e105697. doi: 10.1371/journal.pone.0105697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akla N, Pratt J, Annabi B. Concanavalin-A triggers inflammatory response through JAK/STAT3 signalling and modulates MT1-MMP regulation of COX-2 in mesenchymal stromal cells. Exp Cell Res. 2012;318:2498–2506. doi: 10.1016/j.yexcr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Zgheib A, Pelletier-Bonnier É, Levros LC, Jr, Annabi B. Selective JAK/STAT3 signalling regulates transcription of colony stimulating factor-2 and -3 in concanavalin-A-activated mesenchymal stromal cells. Cytokine. 2013;63:187–193. doi: 10.1016/j.cyto.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 25.de Zoete MR, Palm NW, Zhu S, Flavell RA. Inflammasomes. Cold Spring Harb Perspect Biol. 2014;6:a016287. doi: 10.1101/cshperspect.a016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martins JD, Liberal J, Silva A, Ferreira I, Neves BM, Cruz MT. Autophagy and inflammasome interplay. DNA Cell Biol. 2015;34:274–281. doi: 10.1089/dna.2014.2752. [DOI] [PubMed] [Google Scholar]
- 27.Li TH, Huang CC, Yang YY, et al. Thalidomide improves the intestinal mucosal injury and suppresses mesenteric angiogenesis and vasodilatation by down-regulating inflammasomes-related cascades in cirrhotic rats. PLoS ONE. 2016;11:e0147212. doi: 10.1371/journal.pone.0147212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey S, Singh S, Anang V, Bhatt AN, Natarajan K, Dwarakanath BS. Pattern recognition receptors in cancer progression and metastasis. Cancer Growth Metastasis. 2015;8:25–34. doi: 10.4137/CGM.S24314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu A, Wu H. Structural mechanisms of inflammasome assembly. FEBS J. 2015;282:435–444. doi: 10.1111/febs.13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulasov I, Borovjagin AV, Kaverina N, et al. MT1-MMP silencing by an shR-NA-armed glioma-targeted conditionally replicative adenovirus (CRAd) improves its anti-glioma efficacy in vitro and in vivo. Cancer Lett. 2015;365:240–250. doi: 10.1016/j.canlet.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Langlois S, Di Tomasso G, Boivin D, et al. Membrane type 1-matrix metallo-proteinase induces endothelial cell morphogenic differentiation by a caspase-dependent mechanism. Exp Cell Res. 2005;307:452–464. doi: 10.1016/j.yexcr.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Genís L, Gálvez BG, Gonzalo P, Arroyo AG. MT1-MMP: universal or particular player in angiogenesis? Cancer Metastasis Rev. 2006;25:77–86. doi: 10.1007/s10555-006-7891-z. [DOI] [PubMed] [Google Scholar]
- 34.Seiki M, Mori H, Kajita M, Uekita T, Itoh Y. Membrane-type 1 matrix metal-loproteinase and cell migration. Biochem Soc Symp. 2003;70:253–262. doi: 10.1042/bss0700253. [DOI] [PubMed] [Google Scholar]
- 35.Zitvogel L, Kepp O, Galluzzi L, Kroemer G. Inflammasomes in carcinogenesis and anticancer immune responses. Nat Immunol. 2012;13:343–351. doi: 10.1038/ni.2224. [DOI] [PubMed] [Google Scholar]
- 36.Gutiérrez-Fernández A, Soria-Valles C, Osorio FG, et al. Loss of MT1-MMP causes cell senescence and nuclear defects which can be reversed by retinoic acid. EMBO J. 2015;34:1875–1888. doi: 10.15252/embj.201490594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fortier S, Touaibia M, Lord-Dufour S, Galipeau J, Roy R, Annabi B. Tetra- and hexavalent mannosides inhibit the pro-apoptotic, antiproliferative and cell surface clustering effects of concanavalin-A: impact on MT1-MMP functions in marrow-derived mesenchymal stromal cells. Glycobiology. 2008;18:195–204. doi: 10.1093/glycob/cwm133. [DOI] [PubMed] [Google Scholar]
- 38.Toufaily C, Charfi C, Annabi B, Annabi B. A role for the Cavin-3/Matrix metalloproteinase-9 signaling axis in the regulation of PMA-activated human HT1080 fibrosarcoma cell neoplastic phenotype. Cancer Growth Metastasis. 2014;7:43–51. doi: 10.4137/CGM.S18581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 40.Karin M, Delhase M. The I kappa B kinase (IKK) and NF-kappa B: key elements of proinflammatory signalling. Semin Immunol. 2000;12:85–98. doi: 10.1006/smim.2000.0210. [DOI] [PubMed] [Google Scholar]
- 41.Tahanian E, Sanchez LA, Shiao TC, Roy R, Annabi B. Flavonoids targeting of IκB phosphorylation abrogates carcinogen-induced MMP-9 and COX-2 expression in human brain endothelial cells. Drug Des Devel Ther. 2011;5:299–309. doi: 10.2147/DDDT.S19931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Ma J, Wang KS, Mi C, Lee JJ, Jin X. Blockade of TNF-α-induced NF-κB signaling pathway and anti-cancer therapeutic response of dihydrotanshinone I. Int Immunopharmacol. 2015;28:764–772. doi: 10.1016/j.intimp.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Koziol A, Martín-Alonso M, Clemente C, Gonzalo P, Arroyo AG. Site-specific cellular functions of MT1-MMP. Eur J Cell Biol. 2012;91:889–895. doi: 10.1016/j.ejcb.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Domoto T, Takino T, Guo L, Sato H. Cleavage of hepatocyte growth factor activator inhibitor-1 by membrane-type MMP-1 activates matriptase. Cancer Sci. 2012;103:448–454. doi: 10.1111/j.1349-7006.2011.02162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sina A, Proulx-Bonneau S, Roy A, Poliquin L, Cao J, Annabi B. The lectin concan-avalin-A signals MT1-MMP catalytic independent induction of COX-2 through an IKKgamma/NF-kappaB-dependent pathway. J Cell Commun Signal. 2010;4:31–38. doi: 10.1007/s12079-009-0084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pratt J, Roy R, Annabi B. Concanavalin-A-induced autophagy biomarkers requires membrane type-1 matrix metalloproteinase intracellular signaling in glioblastoma cells. Glycobiology. 2012;22:1245–1255. doi: 10.1093/glycob/cws093. [DOI] [PubMed] [Google Scholar]
- 47.Corsten M, Heggermont W, Papageorgiou AP, et al. The microRNA-221/-222 cluster balances the antiviral and inflammatory response in viral myocarditis. Eur Heart J. 2015;36:2909–2919. doi: 10.1093/eurheartj/ehv321. [DOI] [PubMed] [Google Scholar]
- 48.Feng W, Chumley P, Hua P, et al. Role of the transcription factor erythroblastosis virus E26 oncogen homolog-1 (ETS-1) as mediator of the renal proinflammatory and profibrotic effects of angiotensin II. Hypertension. 2012;60:1226–1233. doi: 10.1161/HYPERTENSIONAHA.112.197871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heo SH, Cho JY. ELK3 suppresses angiogenesis by inhibiting the transcriptional activity of ETS-1 on MT1-MMP. Int J Biol Sci. 2014;10:438–447. doi: 10.7150/ijbs.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shuang T, Wang M, Zhou Y, Shi C, Wang D. NF-κB1, c-Rel, and ELK1 inhibit miR-134 expression leading to TAB1 upregulation in paclitaxel-resistant human ovarian cancer. Oncotarget. 2017;8:24853–24868. doi: 10.18632/oncotarget.15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmad N, Wang W, Nair R, Kapila S. Relaxin induces matrix-metalloproteinases-9 and -13 via RXFP1: induction of MMP-9 involves the PI3K, ERK, Akt and PKC-ζ pathways. Mol Cell Endocrinol. 2012;363:46–61. doi: 10.1016/j.mce.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbolina MV, Adley BP, Ariztia EV, Liu Y, Stack MS. Microenvironmental regulation of membrane type 1 matrix metalloproteinase activity in ovarian carcinoma cells via collagen-induced EGR1 expression. J Biol Chem. 2007;282:4924–4931. doi: 10.1074/jbc.M608428200. [DOI] [PubMed] [Google Scholar]
- 53.Xiang F, Bai M, Jin Y, Ma W, Xin J. Egr-1 mediates Si0(2)-driven transcription of membrane type I matrix metalloproteinase in macrophages. J Huazhong Univ Sci Technolog Med Sci. 2007;27:13–16. doi: 10.1007/s11596-007-0104-3. [DOI] [PubMed] [Google Scholar]
- 54.Heo SH, Cho JY. ELK3 suppresses angiogenesis by inhibiting the transcriptional activity of ETS-1 on MT1-MMP. Int J Biol Sci. 2014 Mar 27;10:438–447. doi: 10.7150/ijbs.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu M, Liu X, Jin W, et al. Targeting ETS1 with RNAi-based supramolecular nanoassemblies for multidrug-resistant breast cancer therapy. J Control Release. 2017;253:110–121. doi: 10.1016/j.jconrel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 56.Schmitz ML, Bacher S, Kracht M. I kappa B-independent control of NF-kappa B activity by modulatory phosphorylations. Trends Biochem Sci. 2001;26:186–190. doi: 10.1016/s0968-0004(00)01753-9. [DOI] [PubMed] [Google Scholar]
- 57.Heissmeyer V, Krappmann D, Wulczyn FG, Scheidereit C. NF-kappaB p105 is a target of IkappaB kinases and controls signal induction of Bcl-3-50 complexes. EMBO J. 1999;18:4766–4778. doi: 10.1093/emboj/18.17.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Labrie M, St-Pierre Y. Epigenetic regulation of mmp-9 gene expression. Cell Mol Life Sci. 2013;70:3109–3124. doi: 10.1007/s00018-012-1214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vafadari B, Salamian A, Kaczmarek L. MMP-9 in translation: from molecule to brain physiology, pathology and therapy. J Neurochem. 2015;139:91–114. doi: 10.1111/jnc.13415. [DOI] [PubMed] [Google Scholar]
- 60.Annabi B, Vaillancourt-Jean E, Béliveau R. MT1-MMP expression level status dictates the in vitro action of lupeol on inflammatory biomarkers MMP-9 and COX-2 in medulloblastoma cells. Inflammopharmacology. 2013;21:91–99. doi: 10.1007/s10787-012-0142-8. [DOI] [PubMed] [Google Scholar]
- 61.McCoy SS, Stannard J, Kahlenberg JM. Targeting the inflammasome in rheumatic diseases. Transl Res. 2016;167:125–137. doi: 10.1016/j.trsl.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Rivero Vaccari JP, Dietrich WD, Keane RW. Therapeutics targeting the in-flammasome after central nervous system injury. Transl Res. 2016;167:35–45. doi: 10.1016/j.trsl.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garbisa S, Sartor L, Biggin S, Salvato B, Benelli R, Albini A. Tumor gelatinases and invasion inhibited by the green tea flavanol epigallocatechin-3-gallate. Cancer. 2001;91:822–832. doi: 10.1002/1097-0142(20010215)91:4<822::aid-cncr1070>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 64.Annabi B, Lachambre MP, Bousquet-Gagnon N, Page M, Gingras D, Beliveau R. Green tea polyphenol (−)-epigallocatechin 3-gallate inhibits MMP-2 secretion and MT1-MMP-driven migration in glioblastoma cells. Biochim Biophys Acta. 2002;1542:209–220. doi: 10.1016/s0167-4889(01)00187-2. [DOI] [PubMed] [Google Scholar]
- 65.Maeda-Yamamoto M, Suzuki N, Sawai Y, et al. Association of suppression of extracellular signal-regulated kinase phosphorylation by epigallocatechin gallate with the reduction of matrix metalloproteinase activities in human fibrosarcoma HT1080 cells. J Agric Food Chem. 2003;51:1858–1863. doi: 10.1021/jf021039l. [DOI] [PubMed] [Google Scholar]
- 66.Yamakawa S, Asai T, Uchida T, Matsukawa M, Akizawa T, Oku N. (−)-Epigallocatechin gallate inhibits membrane-type 1 matrix metalloproteinase, MT1-MMP, and tumor angiogenesis. Cancer Lett. 2004;210:47–55. doi: 10.1016/j.canlet.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Ellis LZ, Liu W, Luo Y, et al. Green tea polyphenol epigallocatechin-3-gallate suppresses melanoma growth by inhibiting inflammasome and IL-1β secretion. Biochem Biophys Res Commun. 2011;414:551–556. doi: 10.1016/j.bbrc.2011.09.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsai PY, Ka SM, Chang JM, et al. Epigallocatechin-3-gallate prevents lupus nephritis development in mice via enhancing the Nrf2 antioxidant pathway and inhibiting NLRP3 inflammasome activation. Free Radic Biol Med. 2011;51:744–754. doi: 10.1016/j.freeradbiomed.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 69.Lei HY, Chang CP. Induction of autophagy by concanavalin A and its application in anti-tumor therapy. Autophagy. 2007;3:402–404. doi: 10.4161/auto.4280. [DOI] [PubMed] [Google Scholar]
- 70.Desjarlais M, Pratt J, Lounis A, Mounier C, Haidara K, Annabi B. Tetracycline derivative minocycline inhibits autophagy and inflammation in concanavalin-a-activated human hepatoma cells. Gene Regul Syst Bio. 2014;8:63–73. doi: 10.4137/GRSB.S13946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Chen J, Mo H, et al. Minocycline protects against NLRP3 inflammasome-induced inflammation and P53-associated apoptosis in early brain injury after subarachnoid hemorrhage. Mol Neurobiol. 2016;53:2668–2678. doi: 10.1007/s12035-015-9318-8. [DOI] [PubMed] [Google Scholar]
- 72.Schwartz C, O’Grady K, Lavelle EC, Fallon PG. Interleukin 33: an innate alarm for adaptive responses beyond Th2 immunity—emerging roles in obesity, intestinal inflammation and cancer. Eur J Immunol. 2016;46:1091–1100. doi: 10.1002/eji.201545780. [DOI] [PubMed] [Google Scholar]
- 73.Lu B, Yang M, Wang Q. Interleukin-33 in tumorigenesis, tumor immune evasion, and cancer immunotherapy. J Mol Med (Berl) 2016;94:535–543. doi: 10.1007/s00109-016-1397-0. [DOI] [PubMed] [Google Scholar]
- 74.Jafarzadeh A, Minaee K, Farsinejad AR, et al. Evaluation of the circulating levels of IL-12 and IL-33 in patients with breast cancer: influences of the tumor stages and cytokine gene polymorphisms. Iran J Basic Med Sci. 2015;18:1189–1198. [PMC free article] [PubMed] [Google Scholar]
- 75.Kaneko K, Williams RO, Dransfield DT, Nixon AE, Sandison A, Itoh Y. Selective inhibition of membrane type 1 matrix metalloproteinase abrogates progression of experimental inflammatory arthritis: synergy with tumor necrosis factor blockade. Arthritis Rheumatol. 2016;68:521–531. doi: 10.1002/art.39414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Greten FR, Eckmann L, Greten TF, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Ernst M, Najdovska M, Grail D, et al. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest. 2008;118:1727–1738. doi: 10.1172/JCI34944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tu S, Bhagat G, Cui G, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14:408–419. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Erratum. Cancer Cell. 2008;14:494. [Google Scholar]; Cancer Cell. 2011;19:154. [Google Scholar]
- 79.Dossus L, Rinaldi S, Becker S, et al. Obesity, inflammatory markers, and endometrial cancer risk: a prospective case-control study. Endocr Relat Cancer. 2010;17:1007–1019. doi: 10.1677/ERC-10-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Basavaraju U, Shebl FM, Palmer AJ, et al. Cytokine gene polymorphisms, cytokine levels and the risk of colorectal neoplasia in a screened population of Northeast Scotland. Eur J Cancer Prev. 2015;24:296–304. doi: 10.1097/CEJ.0000000000000087. [DOI] [PMC free article] [PubMed] [Google Scholar]