Abstract
Hypoxia is an important environmental stressor leading to endocrine disruption and reproductive impairment in fish. Although the hypoxia-inducible factor 1 (HIF-1) is known to regulate the transcription of various genes mediating oxygen homeostasis, its role in modulating steroidogenesis-related gene expression remains poorly understood. In this study, the regulatory effect of HIF-1 on the expression of 9 steroidogenic enzyme genes was investigated in zebrafish embryos using a “gain-of-function and loss-of-function” approach. Eight of the genes, CYP11a, CYP11b2, 3β-HSD, HMGCR, CYP17a1, 17β-HSD2, CYP19a, and CYP19b, were found to be differentially upregulated at 24 and 48 hpf following zHIF-1α-ΔODD overexpression (a mutant zebrafish HIF-1α protein with proline-414 and proline-557 deleted). Knockdown of zHIF-1α also affected the expression pattern of the steroidogenic enzyme genes. Overexpression of zHIF-1α and hypoxia exposure resulted in downregulated StAR expression but upregulated CYP11a and 3β-HSD expression in zebrafish embryos. Conversely, the expression patterns of these 3 genes were reversed in embryos in which zHIF-1α was knocked down under normoxia, suggesting that these 3 genes are regulated by HIF-1. Overall, the findings from this study indicate that HIF-1–mediated mechanisms are likely involved in the regulation of specific steroidogenic genes.
Keywords: HIF-1, hypoxia, steroidogenic enzyme genes, zebrafish, steroid hormones
Introduction
Hypoxia caused by aquatic ecosystem enrichment with nutrients and organic matter is a major global threat that is predicted to worsen as climate change progresses. The adverse impacts of aquatic hypoxia include significant reductions in fisheries production, fish growth, alteration of species composition, and mass fish mortality.1,2 Hypoxia impairs fish reproduction by inhibiting testicular and ovarian development, reducing sperm and egg quality, affecting fertilization, and hatching and influencing the survival of larvae and juvenile fitness.3 Ovarian and testicular growth, gametogenesis, and oocyte and sperm production were significantly reduced in Atlantic croakers in hypoxic regions of the Mississippi River4,5 and in the northern Gulf of Mexico.6
In fish, the inhibition of key reproductive processes by hypoxia is associated not only with reduced metabolism but also with the repression of specific hormones and hormone receptors along the hypothalamus-pituitary-gonad (HPG) axis.3,7 Numerous in vivo studies have shown that fish reproduction is affected by hypoxia via alteration of the steroidogenesis pathway.7–11 Steroid hormones biosynthesis is a fundamental process for reproduction in vertebrates, and all steroid hormones are derived from cholesterol. The rate-limiting step in steroid production is the delivery of cholesterol from the outer to the inner mitochondrial membrane, which is mediated first by the steroidogenic acute regulatory (StAR) protein12 followed by the conversion of cholesterol to pregnenolone by the cytochrome P450 cholesterol side-chain cleavage (P450scc) enzyme encoded by CYP11a1.13 Pregnenolone is then metabolized to progesterone by 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase (encoded by 3β-HSD)14 or hydroxylated to 17α-hydroxyprogesterone by cytochrome P450 17α-hydroxylase (encoded by CYP17a1). Progesterone and 17α-hydroxyprogesterone are then converted through a series of steps to androgen and estrogen by enzymes such as 17β-hydroxysteroid dehydrogenase (17β-HSD) and P450 aromatase (CYP19).15 Estradiol and testosterone are steroid hormones that are important for sexual and reproductive development in animals.16,17 Molecular oxygen is essential for the synthesis of sex steroid hormones,18 and recent studies revealed that the expression of certain steroidogenic enzyme genes, such as StAR, CYP19, and CYP17A1 (which specifies 17α-monooxygenase), may be regulated by hypoxia.19
Hypoxia-inducible factor 1 is a heterodimeric protein consisting of an oxygen-labile HIF-1α subunit and a constitutively expressed HIF-1β subunit.20 Due to the oxygen-sensitive oxygen-dependent degradation domain (ODD) in the HIF-1α subunit, HIF-1 is rapidly degraded under normoxia.21 HIF-1α is stabilized under hypoxia and is transported into the nucleus where it dimerizes with HIF-1β22 to form a heterodimeric complex that binds to the hypoxia-responsive elements (HREs) in the promoters of HIF-target genes.23 HIF-1 has been shown to negatively regulate StAR expression and steroidogenesis in granulosa cells under hypoxia.24
Although more than 100 different genes are now known to be controlled by HIF-1, whether steroidogenic enzyme genes are regulated under hypoxia through HIF-1 remains poorly understood. Zebrafish (Danio rerio) have 2 zHIF-1α paralogs: zHIF-1aA and zHIF-1aB. Recent studies found that zHIF-1aB was more sensitive to oxygen than zHIF-1aA in zebrafish.25,26 Using a gain-of-function and loss-of-function approach, we describe here the effects of zHIF-1α (zHIF-1aB) on the expression of 9 selected steroidogenic genes and embryonic development in zebrafish (D. rerio).
Materials and Methods
Zebrafish maintenance
Wild-type adult zebrafish were maintained under a constant 14 hours:10 hours light:dark cycle at 28°C. Flow through systems were set up in the laboratory to provide a constant temperature and normoxic (7.0 ± 0.2 mg O2 L−1) or hypoxic (1.0 ± 0.2 mg O2 L−1) environments during experimental periods as described previously by Shang et al.10 Embryos were obtained by natural spawning and cultured in zebrafish E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, and 5% methylene blue) at 28.5°C and staged according to Kimmel et al.27
Plasmid construction
The full-length zHIF-1α complementary DNA (cDNA) was amplified by reverse transcription polymerase chain reaction (RT-PCR) from zebrafish embryos and cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA) to form pGEMT-zHIF-1α. Due to the short half-life of the wild-type HIF-1α protein under normoxia,28 a zHIF-1α mutant construct (pGEMT-zHIF-1α-ΔODD) was synthesized to increase the stability of the zHIF-1α protein during its overexpression in normoxic embryos. To form pGEMT-zHIF-1α-ΔODD, 2 sites containing proline residues (proline-414 and proline-557) in wild-type ODD (at nucleotide positions 1234–1251: TTG GCA CCT GCA GGC and at nucleotide positions 1659–1680: ATG CTG GCT CCT TAC ATC CCA) were deleted from pGEMT-zHIF-1α using the Gene Tailor Site-Directed Mutagenesis Kit (Invitrogen, Carlsbad, CA USA).
To monitor the overexpression of zHIF-1α in developing zebrafish embryos, a zHIF-1α-eGFP fusion expression vector was constructed. The open reading frame (ORF) of enhanced green fluorescent protein (eGFP) was amplified from the pEGFP-N1 (Clontech Laboratories, Mountain View, CA, USA) plasmid using the gene-specific primers EGFP-F-Sma: 5′-TCC CCC GGG GGA ATG GTG AGC AAG GGC GA-3′ and EGFP-R-Not: 5′-TTG CGG CCG CAA TTA CTT GTA CAG CTC-3′, which contained built-in SmaI and NotI sites (underlined), respectively. SmaI-digested and NotI-digested eGFP PCR products and the pCMV-TNT vector (Promega) were purified and ligated overnight at 16°C. The plasmid for which the sequence was verified, pCMV-eGFP, was subsequently used in the subcloning experiment. The zHIF-1α-ΔODD ORF was amplified from pGEMT-zHIF-1α-ΔODD with the specific primers HIF-1α-F-Kpn: 5′-CGG GGT ACC CCG ATG GAT ACT GGA GTT GTC AC-3′ and HIF-1α-R-Sal: 5′-ACG CGT CGA CGT CGG TTG ACT TGG TCC AGA GCA C-3′, which contained built-in KpnI and SalI sites, respectively. The reverse primer was designed to eliminate the stop codon by converting TGA (stop codon) to CGA (arginine). The KpnI-digested and SalI-digested pCMV-eGFP plasmid and zHIF-1α-ΔODD PCR products were ligated with T4 ligase (Invitrogen) to produce pCMV-zHIF-1α-ΔODD-eGFP. The sequence of this construct was verified by sequencing the DNA.
Hypoxia exposure experiment
Zebrafish embryos were divided into 2 groups: 1 group was reared in a hypoxic system and the other in a normoxic system. Tanks used for hypoxia exposure were set up as previously described.10 Dissolved O2 concentrations were monitored continuously using dissolved oxygen meters (Cole-Parmer 01972-00, Vernon Hills, IL, USA) and polarographic probes (Cole-Parmer 5643-00), and adjustment was made using a dissolved oxygen controller (Cole-Parmer 01972-00). Fertilized embryos (4 hours postfertilization [hpf ]) were placed in net cages and allowed to develop to desired stages of development (24, 48, or 72 hpf). Each treatment consisted of 5 replicates of 60 embryos each. At the end of the experiments, the embryos were immersed in 200 µL of TRI Reagent (Molecular Research Center, Cincinnati, OH, USA) and stored at −80°C until RNA was extracted. The embryos used for hormone analysis were snap-frozen in liquid nitrogen and stored at −80°C. Animal care and experiments were conducted in accordance with the City University of Hong Kong animal care guidelines.
RNA isolation and first-strand cDNA synthesis
Total RNA was extracted using TRI Reagent (Molecular Research Center) according to the manufacturer’s instructions. First-strand cDNA was synthesized in a 25-µL reaction that contained 0.25 µg (0.5 µg/µL) of random primer, 5 µL of 5 × reaction buffer, 1.25 µL of 10 mM deoxynucleotide triphosphates, 0.65 µL of 40 U ribonuclease (RNase inhibitor) (Invitrogen), 1 µL of 200 U M-MLV Reverse Transcriptase (Promega), and 1 µg of total RNA, which was pretreated with RNase-free deoxyribonuclease I (Invitrogen) to eliminate genomic DNA contamination. cDNA synthesis was performed at 37°C for 1 hour. The cDNAs were kept at −20°C until they were analyzed by real-time PCR.
Real-time PCR
Real-time PCR, which was used to quantify gene expression, was performed using a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) as described previously.11 PCR assays were conducted using the SYBR Green–based detection method (#4367659; Applied Biosystems, Warrington, UK) according to the manufacturer’s instructions. Sequences of the primers used for real-time PCR are shown in Table 1. Melting curve analysis was performed after the PCR was completed to assess the amplification specificity. The identity of the PCR amplicons was confirmed by DNA sequencing. The cycling conditions were as follows: initial denaturation at 95°C for 10 minutes, followed by 40 cycles performed at 95°C for 15 seconds, and 0°C for 30 seconds. β-Actin was employed as an endogenous control for normalization. All PCR reactions were performed in triplicate. Fold change was calculated according to the following formula: fold , where ΔCT = CT(target) − CT(β-actin) and Δ(ΔCT) = ΔCT(stimulated) − ΔCT(control). All data were expressed as the mean ± SD and P ⩽ .05 was considered statistically significant.
Table 1.
Sequences of real-time PCR primers.
Estradiol (E2) and testosterone (T) measurement
In total, 60 zebrafish embryos from each replicate were collected and immediately stored at −80°C for hormone extraction (E2 and T) as described by Yu et al.11 E2 and T levels were measured using a commercial competitive enzyme-linked immunosorbent assay (ELISA) kit (Cayman Chemical, Ann Arbor, MI, USA).
Overexpression of zHIF-1α
Capped eGFP and zHIF1α-ΔODD-eGFP messenger RNAs (mRNAs) were synthesized from linearized pCMV-eGFP and pCMV-zHIF-1α-ΔODD-eGFP, respectively, using the T7 mMESSAGE mMACHINE Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. mRNAs were microinjected at a concentration of 0.5 µg/µL in Hanks buffer containing 0.15% phenol red, and each embryo was injected with 2 nL mRNA at a 1 to 2 cell stage using a FemtoJet microinjection system (Eppendorf AG, Hamburg, Germany). Five replicates of 60 fertilized embryos each were used for both treatments. Injected embryos were incubated under normoxic conditions and sampled at various developmental stages for analyses.
zHIF-1α knockdown
Antisense morpholino oligonucleotides (MOs) were obtained from Gene Tools (Philomath, OR, USA). An MO targeted against the translation initiation region of zHIF-1α (zHIF-1α MO: 5′-CAG TGA CAA CTC CAG TAT CCA TTC C-3′) was used to knockdown the zHIF-1α protein. The standard control MO (5′-CCT CTT ACC TCA GTT ACA ATT TAT A-3′) was used as an injection control. The optimized morpholino amount for zHIF-1α knockdown experiments was 8 ng/embryo. Injected embryos were reared to the desired developmental stages under hypoxic or normoxic conditions as described above.
Statistical analysis
The Student t test was used to test the null hypothesis that there is no significant difference in mean gene expression between the treatment and concurrent control groups. Significance was set at P ⩽ .05. All statistical analyses were conducted using Prism 3.02 (GraphPad, San Diego, CA, USA).
Results
Hypoxia alters the expression pattern of steroidogenic enzyme genes
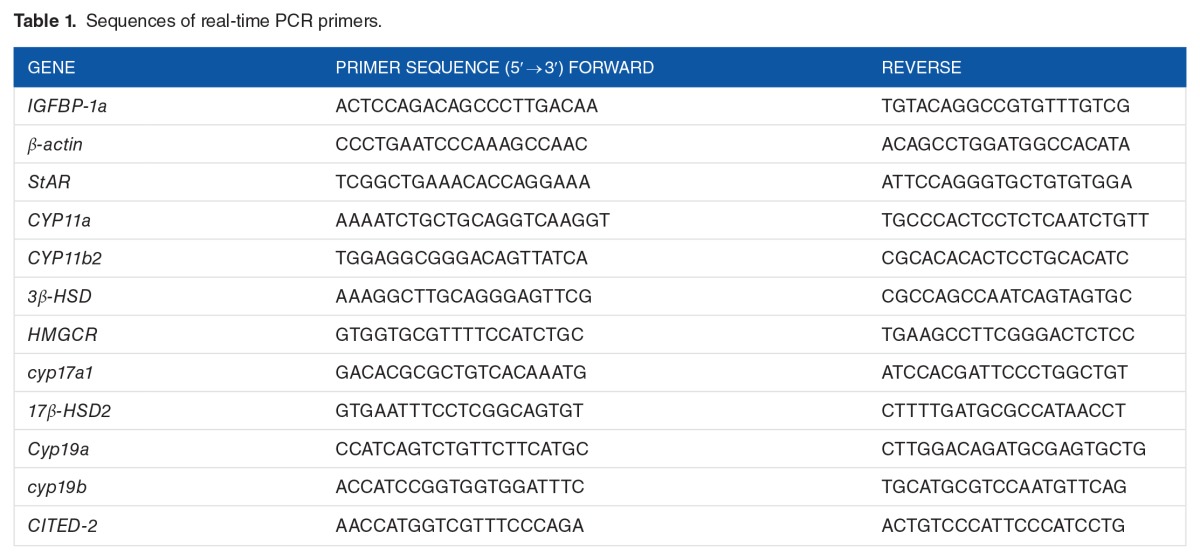
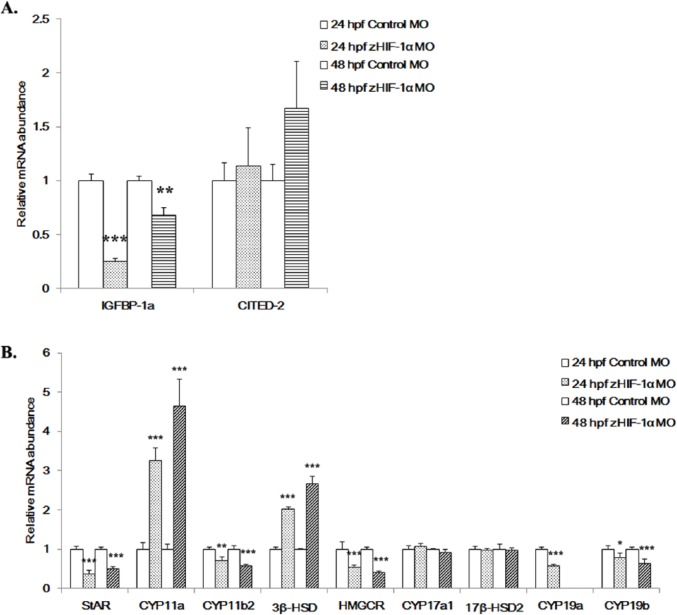
To confirm that HIF-1 signaling was activated in the zebrafish embryos under the experimental hypoxic conditions employed in this study, real-time PCR was performed to measure the expression of 2 HIF-1 target genes, IGFBP-1a29 and CITED-2,30 in embryos at 24, 48, and 72 hpf following exposure to hypoxia. Both the IGFBP-1a and CITED-2 mRNAs were significantly upregulated in embryos under hypoxia at all detected stages (Figure 1A), which indicated that the hypoxic conditions used in the study were appropriate.
Figure 1.
Effects of hypoxia on the expression of hypoxia markers and steroidogenic genes. Effect of hypoxia on the expression of (A) the HIF-1 target genes, IGFBP-1a and CITED-2 and (B) 9 steroidogenic enzyme genes. Fertilized zebrafish embryos were exposed to normoxic (7.0 ± 0.2 mgO2L−1) or hypoxic (1.0 ± 0.2 mg O2L−1) conditions for 24, 48, or 72 hours. Gene expression was quantified using real-time PCR and normalized against β-actin mRNA. Data are presented as the mean relative fold change ± SD with respect to the normoxia control (its expression level was arbitrarily set to 1). Expression levels significantly different from the control are indicated by asterisks (t test, n ⩾ 4, *P ⩽ .05, **P ⩽ .01, ***P ⩽ .001). HIF-1 indicates hypoxia-inducible factor 1; mRNA, messenger RNA.
The mRNA expression levels for 9 selected steroidogenic genes in embryos exposed to hypoxia were differentially affected (Figure 1B). Five steroidogenic genes were significantly downregulated. StAR expression was downregulated by 2.1 ± 0.05 (P ⩽ .01) and 2.6 ± 0.2 (P ⩽ .001) fold at 24 and 72 hpf, respectively; HMGCR was reduced by 1.4 ± 0.02 (P ⩽ .01) and 7.1 ± 2.78 (P ⩽ .001) fold at 24 and 72 hpf, respectively; 17β-HSD2 was reduced by 1.8 ± 0.19 (P ⩽ .01) and 1.9 ± 0.2 (P ⩽ .01) fold at 48 and 72 hpf, respectively; CYP19a was downregulated by 1.5 ± 0.01 (P ⩽ .01) and 2.1 ± 0.02 (P ⩽ .05) fold at 48 and 72 hpf, respectively; and CYP19b was downregulated by 0.5 ± 0.01 (P ⩽ .001) fold at 72 hpf. In contrast, 3 genes were significantly upregulated in response to hypoxia. CYP11a was upregulated by 3.5 ± 0.62 (P ⩽ .01), 3.6 ± 0.8 (P ⩽ .01), and 1.3 ± 0.19 (P ⩽ .05) fold at 24, 48, and 72 hpf, respectively; CYP11b2 was upregulated by 1.6 ± 0.11 (P ⩽ .05) and 1.4 ± 0.09 (P ⩽ .01) fold at 24 and 48 hpf, respectively; and 3β-HSD was increased by 1.6 ± 0.11 (P ⩽ .05) and 5.4 ± 1.04 (P ⩽ .001) fold at 48 and 72 hpf, respectively. CYP17a1 mRNA expression showed no significant changes in hypoxic embryos in any of the 3 developmental stages.
Overexpression of zHIF-1α mRNA delays embryonic development and the expression pattern of steroidogenic genes
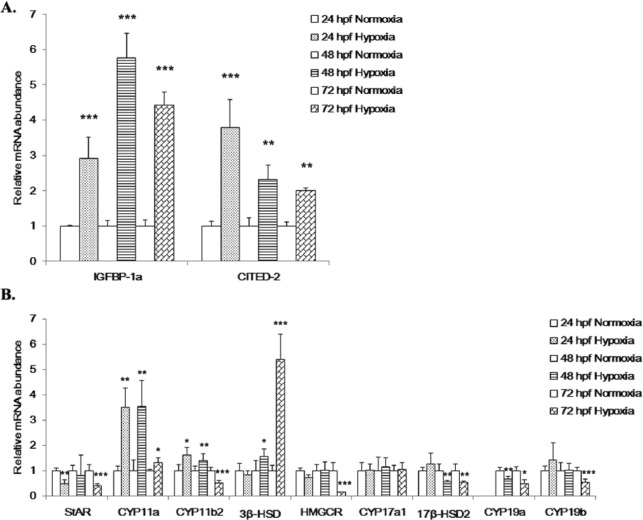
To investigate the relationship between zHIF-1α expression and steroidogenesis in zebrafish, overexpression and knockdown of zHIF-1α were conducted by microinjection of the in vitro–transcribed zHIF-1α-ΔODD-eGFP mRNAs and zHIF-1α translation-blocking morpholinos (MO) into embryos, respectively. Five treatment replicates were used in each experiment. The total numbers of embryos injected with the eGFP mRNA (control) and zHIF-1α-ΔODD-eGFP mRNA were 719 and 1081, respectively. Based on the embryo staging criteria described by Westerfield,31 the development of embryos microinjected with zHIF-1α-ΔODD-eGFP mRNA was found to be delayed by at least 6 hours when compared with that of the eGFP-injected and uninjected controls at 24 hpf (Figure 2A).
Figure 2.
Effects of zHIF-1α overexpression on embryonic development and the expression of hypoxia markers and steroidogenic genes. (A) Development of zebrafish embryos at 24 hpf. a, uninjected control embryo; b, embryo microinjected with eGFP mRNA shows normal development (Prim-6 stage); c, embryo microinjected with zHIF-1α-ΔODD-eGFP mRNA shows developmental retardation (at approximately 18-somite stage). (B) Percentage of mortality and morphological abnormality in zebrafish embryos (24 hpf) microinjected with zHIF-1α mRNA. The effect of zHIF-1α overexpression on steroidogenic gene expression was examined by microinjecting zHIF-1α mRNA into 1- to 2-cell stage zebrafish embryos and maintained under normoxic conditions. Zebrafish embryos microinjected with eGFP mRNA and maintained under normoxia were used as the control. Because no statistically significant difference was observed between the data sets of the microinjected control and the no-injection control (One-way analysis of variance with a P < .05 threshold), the latter data set is presented here. Control: uninjected embryos (n = 373, dead = 31, abnormal = 4); eGFP: embryos microinjected with eGFP mRNA (n = 719, dead = 48, abnormal = 16); and zHIF-1α: embryos microinjected with zHIF-1α-ΔODD-eGFP mRNA (n = 1081, dead = 193, abnormal = 297). (C) Effects of zHIF-1α overexpression on IGFBP-1a and CITED-2 expression. (D) Effects of zHIF-1α overexpression on steroidogenic gene expression. Gene expression was quantified using real-time PCR and normalized with β-actin mRNA. Data are presented as the mean relative fold change ± SD with respect to the gene expression level in the control (its expression level was arbitrarily set to 1) for each experiment. Expression levels significantly different from the control are indicated by asterisks (t test, n ⩾ 4, *P ⩽ .05, **P ⩽ .01, ***P ⩽ .001). eGFP indicates enhanced green fluorescent protein; mRNA, messenger RNA.
At 24 hpf, 17.8% of the embryos microinjected with zHIF-1α-ΔODD-eGFP mRNA were dead compared with the 6.1% dead embryos observed with eGFP mRNA microinjection. The percentage of mortality of the embryos microinjected with zHIF-1α mRNAs was approximately 3-fold greater than that of the eGFP control (Figure 2B). In addition, the percentage of abnormal embryos observed in the zHIF-1α microinjection experiments was 27.5% (297/1081), whereas only 2.2% (16/719) of abnormal embryos were observed in the eGFP control group. Following microinjection with the zHIF-1α-ΔODD-eGFP mRNAs, IGFBP-1a and CITED-2 were significantly upregulated in embryos at 24 and 48 hpf (Figure 2C), which suggested successful overexpression of zHIF-1α in the embryos up until 48 hpf.
Following microinjection of the zHIF-1α mRNAs, the expression patterns of all 9 steroidogenic genes were found to be affected in zebrafish embryos at 24 and 48 hpf (Figure 2D). With the exception of StAR which was downregulated by 1.82 ± 0.12 (P ⩽ .01) and 1.43 ± 0.12 (P ⩽ .01) fold at 24 and 48 hpf, respectively, all other steroidogenic genes were upregulated by the overexpression of zHIF-1α. CYP11a was upregulated by 2.4 ± 0.04 (P ⩽ .01) and 7.0 ± 1.2 (P ⩽ .01) fold at 24 and 48 hpf, respectively; CYP11b2 was upregulated by 2.1 ± 0.6 (P ⩽ .01) and 3.4 ± 0.13 (P ⩽ .001) fold at 24 and 48 hpf, respectively; CYP17a1 was upregulated by 2.0 ± 0.3 (P ⩽ .01) and 3.2 ± 0.06 (P ⩽ .001) fold at 24 and 48 hpf, respectively; 17β-HSD2 was upregulated by 2.98 ± 0.1 (P ⩽ .05) and 1.6 ± 0.39 (P ⩽ .05) fold at 24 and 48 hpf, respectively; CYP19a was upregulated by 1.34 ± 0.05 (P ⩽ .05) and 2.8 ± 0.27 (P ⩽ .01) fold at 24 and 48 hpf, respectively; CYP19b was upregulated by 1.6 ± 0.11 (P ⩽ .01) and 4.9 ± 0.21 (P ⩽ .001) fold at 24 and 48 hpf, respectively; and 3β-HSD and HMGCR were upregulated by 1.9 ± 0.4 (P ⩽ .05) and 3.5 ± 0.4 (P ⩽ .001) fold, respectively, at 48 hpf.
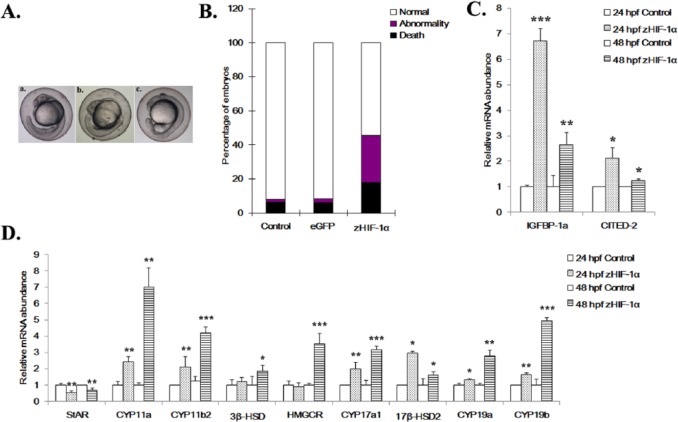
Knockdown of zHIF-1α alters expression of steroidogenic genes
The experiment in which zHIF-1α was knocked down with a morpholino was conducted to complement the study in which zHIF-1α was overexpressed. Following zHIF-1α knockdown, morphological abnormalities were observed in morphant embryos at 48 hpf under normoxia (Figure 3A). The observed deformities included small head circumference, bent notochords, deformed hearts, distorted abdomens, and short curved tails. Knocking down zHIF-1α also resulted in significantly shorter zebrafish embryo body length at the same developmental stage (Figure 3A). In morphant embryos at 24 hpf, the expression levels of IGFBP-1a and CITED-2 were significantly reduced by 2.7 ± 0.48 (P ⩽ .001) and 1.9 ± 0.14 (P ⩽ .01) fold, respectively, compared with those of the control. However, IGFBP-1a was elevated by 1.6 ± 0.18 (P ⩽ .001) fold in morphants at 48 hpf, whereas CITED-2 expression was unaffected.
Figure 3.
Effects of zHIF-1α knockdown on zebrafish embryonic development and the expression of hypoxia markers and steroidogenic genes under normoxia. (A) Morphological abnormalities of the zHIF-1α knockdown embryos at 48 hpf under normoxia. The effect of zHIF-1α knockdown on steroidogenic gene expression was examined by microinjecting zHIF-1α MOs into 1- to 2-cell stage zebrafish embryos and maintained under either normoxia or hypoxia. Zebrafish embryos microinjected with a standard control MO were used as controls. All morphants were imaged at the left lateral view. a, Control: embryo injected with control MO; b-j, embryos microinjected with zHIF-1α MO; b, c, and d, morphants with decreased body length, bended notochords (black arrows), and distorted abdomens (blue arrows); e-j, morphants with bended notochords, small head circles (red circles), and short curved tail (yellow arrows). (B) Effects of zHIF-1α knockdown on IGFBP-1a and CITED-2 expression under normoxia. (C) Effects of zHIF-1α knockdown on steroidogenic gene expression under normoxia. Gene expression was quantified by real-time PCR and normalized against β-actin mRNA. Data are presented as the mean relative fold change ± SD with respect to the gene expression level in the control (its expression level was arbitrarily set to 1) for each experiment. Expression levels significantly different from the control are indicated by asterisks (t test, n ⩾ 4, *P ⩽ .05, **P ⩽ .01, ***P ⩽ .001). mRNA indicates messenger RNA; MO, morpholino oligonucleotide.
The expression patterns of the steroidogenic genes in the zHIF-1α morphants were different from those of the control embryos (microinjected with control MO) under normoxia (Figure 3C). Three steroidogenic genes were specifically upregulated in the zHIF-1α morphants. StAR was significantly increased by 1.7 ± 0.24 (P ⩽ .01) fold, CYP11b2 was upregulated by 2.1 ± 0.5 (P ⩽ .05) fold, and CYP17a1 was upregulated by 1.3 ± 0.04 (P ⩽ .05) fold at 24 hpf. In contrast, CYP11a was downregulated by 4.8 ± 0.01 (P ⩽ .01) at 24 hpf, whereas 3β-HSD and 17β-HSD2 were downregulated by 1.7 ± 0.02 (P ⩽ .01) and 1.8 ± 0.1 (P ⩽ .05) fold, respectively, at 24 hpf. No significant changes in HMGCR or CYP19b expression were observed in embryos in the zHIF-1α morphants at 24 hpf compared with embryos microinjected with control MO. Among the 9 steroidogenic genes examined, only StAR, CYP11a, 3β-HSD, and 17β-HSD2 in the zHIF-1α morphants exhibited opposite patterns of expression to those of the embryos overexpressing zHIF-1α. These observations suggest that expressions of these genes might be directly and/or indirectly regulated by HIF-1.
To confirm whether changes in the expression of steroidogenic genes under hypoxia (Figure 1B) were due to regulation by zHIF-1α, embryos in which zHIF-1α had been knocked down were exposed to hypoxia. Comparing embryos in which zHIF-1α had been knocked down with the control MO-injected embryos under hypoxia, IGFBP-1a was downregulated by 4 ± 0.04 (P ⩽ .001) and 1.47 ± 0.02 (P ⩽ .01) fold at 24 and 48 hpf, respectively, whereas CITED-2 was unaffected at both stages (Figure 4A). These observations suggest that zHIF-1α was successfully downregulated in the knockdown treatment.
Figure 4.
Effects of zHIF-1α knockdown on the expression of hypoxia markers and steroidogenic genes under hypoxia. (A) Effects of zHIF-1α knockdown on the hypoxia marker genes IGFBP-1a and CITED-2 under hypoxia. Methods for zHIF-1α knockdown are detailed in Figure 3. (B) Effects of zHIF-1α knockdown on steroidogenic gene expression under hypoxia. Gene expression was quantified by real-time PCR and normalized against β-actin mRNA. Data are presented as the mean relative fold change ± SD with respect to the gene expression level in the hypoxia control (its expression level was arbitrarily set to 1) for each experiment. Expression levels significantly different from the control are indicated by asterisks (t test, n ⩾ 4, *P ⩽ .05, **P ⩽ .01, ***P ⩽ .001). mRNA indicates messenger RNA.
In the zHIF-1α morphant embryos under hypoxia, StAR was significantly downregulated by 2.8 ± 0.08 (P ⩽ .001) and 2.0 ± 0.01 (P ⩽ .001) fold at 24 and 48 hpf, respectively; CYP11a was upregulated by 3.2 ± 0.26 (P ⩽ .001) and 4.7 ± 0.86 (P ⩽ .001) fold at 24 and 48 hpf, respectively; CYP11b2 was downregulated by 1.4 ± 0.13 (P ⩽ .01) and 1.7 ± 0.09 (P ⩽ .001) fold at 24 and 48 hpf, respectively; 3β-HSD was upregulated by 2.0 ± 0.05 (P ⩽ .001) and 2.7 ± 0.34 (P ⩽ .001) fold at 24 and 48 hpf, respectively; HMGCR was downregulated by 1.8 ± 0.29 (P ⩽ .001) and 2.5 ± 0.05 (P ⩽ .001) fold at 24 and 48 hpf, respectively; and CYP19a was downregulated by 1.8 ± 0.01 (P ⩽ .001) fold at 24 hpf but was undetectable at 48 hpf; CYP19b was downregulated by 1.3 ± 0.11 (P ⩽ .05) and 1.6 ± 0.17 (P ⩽ .001) fold at 24 and 48 hpf, respectively; and CYP17a1 and 17β-HSD2 were not affected in the zHIF-1α morphants at any of the time points.
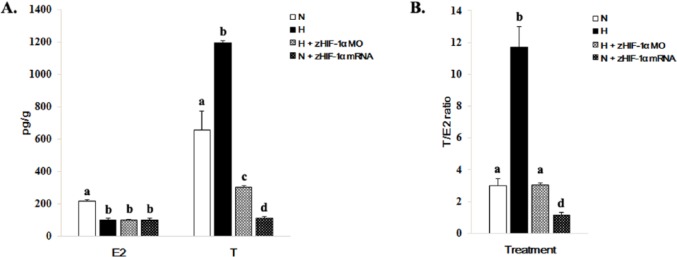
E2 and T levels
The concentrations of E2 and T in whole embryos varied under normoxic and hypoxic conditions and with or without overexpression and knockdown of zHIF-1α (Figure 5). The concentrations of E2 and T in whole embryos under hypoxia were 2.2-fold less and 1.8-fold greater than those of the normoxic controls (Figure 5A), which resulted in a 3.9-fold greater T/E2 ratio (from 3.0 ± 0.42 (normoxia) to 11.73 ± 1.28 (hypoxia) (Figure 5B). The concentrations of both E2 and T were lower in embryos overexpressing zHIF-1α under normoxic conditions and in zHIF-1α knockdown embryos under hypoxic conditions (Figure 5A). The T/E2 ratio in the zHIF-1α morphants under hypoxia was similar to that of the control embryos under normoxia. However, the T/E2 ratio in embryos overexpressing zHIF-1α under normoxia was 10.3-fold lower than those maintained under hypoxia (Figure 5B).
Figure 5.
Effects of hypoxia and overexpression of zHIF-1α on E2 and T levels in zebrafish embryos. (A) Effects of hypoxia and zHIF-1α expression on E2 and T levels. Methods for hypoxic exposure and zHIF-1α overexpression and knockdown are detailed in Figures 1 to 3, respectively. Hormone levels in whole zebrafish embryos (5 replicates of 60 pooled embryos each) were measured by the ELISA method. The data are presented as the mean concentration of hormone (in picograms per gram of embryo wet weight) ± SD. The different letters above the bars indicate statistically significant differences between the indicated groups for the same hormone (P < .05, Student-Newman-Keuls). (B) Effects of hypoxia and zHIF-1α expression on the T/E2 ratios. Data are presented as the mean ± SD (P < .05, Student-Newman-Keuls). The different letters above the bars indicate statistically significant differences between the indicated groups.
Discussion
There is increasing evidence that the HIF-1 signaling system plays essential roles in embryonic development in vertebrates through the activation of genes that regulate energy metabolism and survival. Previous studies have reported the expression of HIF-1α mRNA in the brain, blood vessels, somites, notochord, retina and optic tectum in zebrafish embryos,32 and HIF-1α-deficient mouse embryos (HIF-1α−/−) show defects in the formation of blood vessels and the neural fold as well as malformations in the cardiovascular system.33,34 HIF-1 has also been demonstrated to affect the development of zebrafish embryos through the regulated expression of IGFBP-1.29 In this study, zHIF-1α knockdown using morpholinos resulted in various types of morphological abnormalities in developing zebrafish embryos (Figure 3A) which indicates that the expression of zHIF-1α is essential for the normal development of zebrafish embryos under normoxic conditions and is consistent with the results from previous studies conducted in mice.35 Recently, studies have shown that in addition to its canonical role as a transcription factor, HIF-1α has a nontranscriptional role as a signaling regulator of Notch signaling,36,37 which is involved in many cellular processes during embryonic development including somitogenesis,38 muscle tissue formation,39 blood vessel maturation,40 and heart formation.41 It is reasonable to speculate that the varied morphological abnormalities observed in zebrafish embryos could be attributed to the disruption of Notch signaling as a result of ectopic zHIF-1α expression.
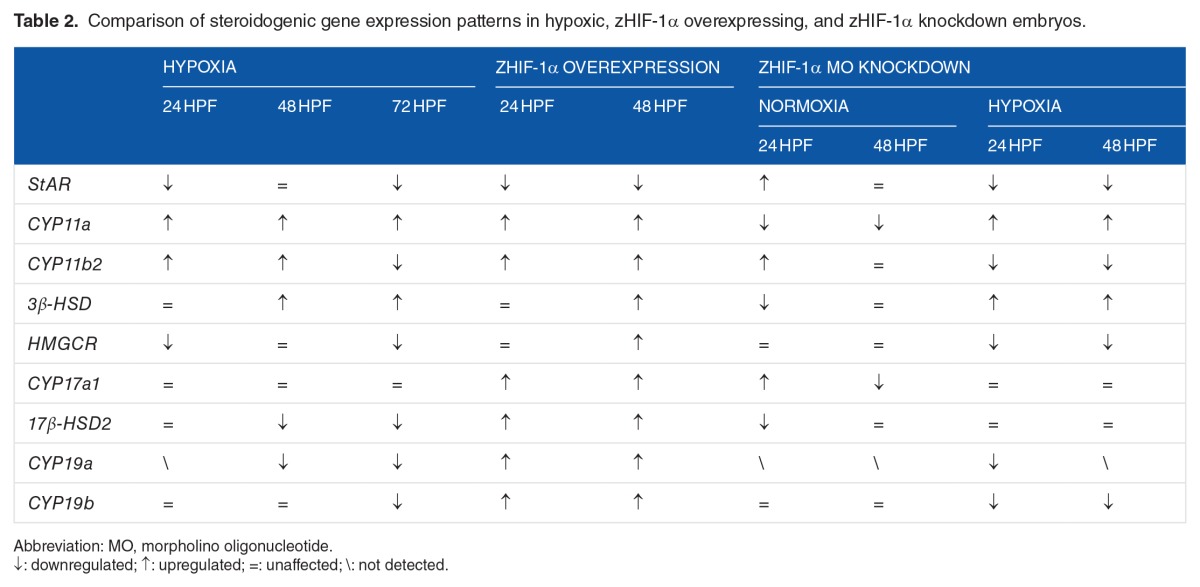
There is ample evidence that hypoxia impairs the sexual and reproductive development of fish through perturbation of the HPG axis at multiple levels, including enzymes that control steroidogenesis.5,10 A qualitative comparison of the results from this study on the expression patterns of steroidogenic genes in zebrafish embryos exposed to hypoxia, embryos overexpressing zHIF-1α mRNA, and zHIF-1α knockdown embryos is provided in Table 2. Changes in the expression of genes encoding steroidogenic enzymes, including StAR, CYP11a, CYP11b2, 3β-HSD, HMGCR, CYP17a1, 17β-HSD2, CYP19a, and CYP19b, would likely affect the production of steroid hormones, such as testosterone, estradiol, aldosterone, and cortisol. Steroid hormones are crucial substances that play a number of important physiological roles. An important function of steroid hormones is coordinating the physiological and behavioral responses for specific biological purposes, such as reproduction. Mutations in StAR are known to cause a complete lack of steroidogenesis in the adrenals and gonads, which, in turn, leads to classical congenital lipoid adrenal hyperplasia in humans.42 Mutations in the CYP11a1 gene causes adrenal insufficiency in humans and rabbits,43,44 whereas CYP11a1-null mice are deficient in steroid synthesis ability.45 In zebrafish embryos in which CYP11a1 was knocked down, the embryos exhibited a shortened axis and defects in cell movement during epiboly.46 CYP11B2 catalyzes 3 sequential reactions that convert 11-deoxycorticosterone to aldosterone.47 Because the biosynthesis of aldosterone is solely catalyzed by CYP11B2,48 downregulation of CYP11b2 might result in the reduced production of aldosterone. 3β-HSD deficiency results in lower amounts of adrenal steroids including mineralocorticoids, glucocorticoids, and sex steroids.14,49 Knockdown studies of CYP17 showed a significant reduction in progesterone formation and de novo synthesis of steroids in mouse tumor Leydig cells.50 Inactivating mutations in CYP19 are rare in humans, but affected individuals are unable to synthesize endogenous estrogens, which results in virilization of both the fetus and the mother.51
Table 2.
Comparison of steroidogenic gene expression patterns in hypoxic, zHIF-1α overexpressing, and zHIF-1α knockdown embryos.
The results of downregulating of StAR and CYP19a under hypoxia were consistent with the results of previous observations reported in mammalian cells52 and in the male and female gonads of zebrafish, which also correlated with significant reductions of estradiol in females and testosterone in males.53 A positive relationship between StAR expression and sex steroid production has also been reported in male rainbow trout, where sex steroid hormone secretion was increased by the elevated expression of StAR.54 Repression of StAR can lead to the decreased production of sex hormones, as StAR is a cholesterol-shuttling protein that facilitates the translocation of cholesterol from the outer to the inner mitochondrial membrane in endocrine tissues, and this represents a rate-limiting step in steroidogenesis.55 In rats under hypoxia, CYP11a expression was previously reported to be stimulated, whereas StAR expression was suppressed.56–58 Similar expression patterns of CYP11a and StAR were found in this study. Concentrations of E2 and T in zebrafish embryos were less under all treatment conditions, with the exception of T, which was upregulated under hypoxia. The findings are consistent with the reduced expression of StAR under all treatment conditions relative to that of the normoxia control. The increased concentration of T under hypoxia presumably reflects the reduced conversion of T to E2 due to the downregulation of CYP19a.
Specifically, we showed that hypoxia and zHIF-1α overexpression significantly downregulated the expression of StAR but upregulated the expression of CYP11a and 3β-HSD. The reversed expression of these 3 genes in the zHIF-1α knockdown embryos under normoxia strongly suggests that these genes are directly regulated by HIF-1. Moreover, computational analysis of the 5′ flanking regions of these 3 genes revealed several putative HREs (5′-A/GCGTG-3′). Within the first 3 kb of the 5′ flanking sequence, 4, 3, and 4 putative HREs (forward/reverse) were found in StAR, CYP11a, and 3β-HSD, respectively. These results indicate that zHIF-1α may directly regulate these genes via HRE binding in the promoter region.
CYP11b2 is the only gene that showed an opposite expression pattern in hypoxia and hypoxic zHIF-1α knockdown embryos. Considering the unaffected expression pattern of CYP11b2 under zHIF-1α overexpression and knockdown treatment under normoxia, these results imply that CYP11b2 is unlikely to be regulated by zHIF-1α directly, or the HREs residing in the CYP11b2 promoter may be occupied by other HIFs under hypoxia.
Conclusions
In this study, it is demonstrated that zHIF-1 regulates the expression of several steroidogenic enzyme genes in zebrafish embryos. Specifically, the regulated expression of StAR, CYP11a, 3β-HSD, and CYP19a likely affects the production levels of the steroid hormones, E2 and T. This study provides evidence that zHIF-1 has a role in the regulation of embryonic development and steroidogenesis in zebrafish.
Acknowledgments
The authors thank Professor SH Cheng’s group for providing zebrafish embryos and for using microinjection facilities in her lab.
Footnotes
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1240 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a grant from the General Research Fund (PJ9041727) from the Research Grants Council of Hong Kong Special Administrative Region, People’s Republic of China.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
TT conceived and designed the experiments; collected, analyzed, and interpreted the data; wrote the first draft of the manuscript. RMKY performed literature review, contributed to the writing and made critical revisions of the manuscript. RSSW contributed to the writing of the manuscript and approved the final version. RYCK designed the study and performed a critical review of the manuscript. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.Wannink JH, Kashindye JJ, Goudswaard PCK, Witte F. Dwelling at the oxycline: does increased stratification provide a predation refugium for the Lake Victoria sardine Rastrineobola argentea? Freshwater Biol. 2001;46:75–85. [Google Scholar]
- 2.Diaz RJ, Breitburg DL. The hypoxic environment. In: Richards JG, Farrell AP, Brauner CJ, editors. Hypoxia in Fishes. San Diego, CA: Academic Press; 2009. pp. 1–23. [Google Scholar]
- 3.Wu RSS. Effects of hypoxia on fish reproduction and development. In: Richards JG, Farrell AP, Brauner CJ, editors. Fish Physiology. San Diego, CA: Academic Press; 2009. pp. 79–141. [Google Scholar]
- 4.Thomas P, Rahman MS. Region-wide impairment of Atlantic croaker testicular development and sperm production in the northern Gulf of Mexico hypoxic dead zone. Mar Environ Res. 2010;69:S59–S62. doi: 10.1016/j.marenvres.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Thomas P, Rahman MS. Extensive reproductive disruption, ovarian masculinization and aromatase suppression in Atlantic croaker in the northern Gulf of Mexico hypoxic zone. Proc Biol Sci. 2012;B279:28–38. doi: 10.1098/rspb.2011.0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas P, Rahman MS, Picha ME, Tan W. Impaired gamete production and viability in Atlantic croaker collected throughout the 20,000 km(2) hypoxic region in the northern Gulf of Mexico. Mar Pollut Bull. 2015;101:182–192. doi: 10.1016/j.marpolbul.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Wu RSS, Zhou BS, Randall DJ, Woo NYS, Lam PKS. Aquatic hypoxia is an endocrine disruptor and impairs fish reproduction. Environ Sci Technol. 2003;37:1137–1141. doi: 10.1021/es0258327. [DOI] [PubMed] [Google Scholar]
- 8.Hala D, Petersen LH, Martinovic D, Huggett DB. Constraints-based stoichiometric analysis of hypoxic stress on steroidogenesis in fathead minnows, Pimephales promelas. J Exp Biol. 2012;215:1753–1765. doi: 10.1242/jeb.066027. [DOI] [PubMed] [Google Scholar]
- 9.Martinovic D, Villeneuve DL, Kahl MD, Blake LS, Brodin JD, Ankley GT. Hypoxia alters gene expression in the gonads of zebrafish (Danio rerio) Aquat Toxicol. 2009;95:258–272. doi: 10.1016/j.aquatox.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Shang EH, Yu RM, Wu RS. Hypoxia affects sex differentiation and development, leading to a male-dominated population in zebrafish (Danio rerio) Environ Sci Technol. 2006;40:3118–3122. doi: 10.1021/es0522579. [DOI] [PubMed] [Google Scholar]
- 11.Yu RM, Chu DL, Tan TF, et al. Leptin-mediated modulation of steroidogenic gene expression in hypoxic zebrafish embryos: implications for the disruption of sex steroids. Environ Sci Technol. 2012;46:9112–9119. doi: 10.1021/es301758c. [DOI] [PubMed] [Google Scholar]
- 12.Stocco DM, Clark BJ. Regulation of the acute production of steroids in steroidogenic cells. Endocr Rev. 1996;17:221–244. doi: 10.1210/edrv-17-3-221. [DOI] [PubMed] [Google Scholar]
- 13.Simpson ER. Cholesterol side-chain cleavage, cytochrome P450, and the control of steroidogenesis. Mol Cell Endocrinol. 1979;13:213–227. doi: 10.1016/0303-7207(79)90082-0. [DOI] [PubMed] [Google Scholar]
- 14.Simard J, Ricketts ML, Gingras S, Soucy P, Feltus FA, Melner MH. Molecular biology of the 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. Endocr Rev. 2005;26:525–582. doi: 10.1210/er.2002-0050. [DOI] [PubMed] [Google Scholar]
- 15.Nagahama Y. In: Norberg B, Kjesbu OS, Taraanger GL, Andersson E, Stefansson SO, editors. Gonadal steroid hormones: Major regulators of gonadal sex differentiation and gametogenesis in fish; Proceedings of the Sixth International Symposium on the Reproductive Physiology of Fish; Bergen, Norway: University of Bergen; 2000. pp. 211–232. [Google Scholar]
- 16.Hess RA. Estrogen in the adult male reproductive tract: a review. Reprod Biol Endocrinol. 2003;1:52. doi: 10.1186/1477-7827-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stocco C. Aromatase expression in the ovary: hormonal and molecular regulation. Steroids. 2008;73:473–487. doi: 10.1016/j.steroids.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raff H, Bruder ED. Steroidogenesis in human aldosterone-secreting adenomas and adrenal hyperplasias: effects of hypoxia in vitro. Am J Physiol Endocrinol Metab. 2006;290:E199–E203. doi: 10.1152/ajpendo.00337.2005. [DOI] [PubMed] [Google Scholar]
- 19.Lai KP, Li JW, Tse AC, Chan TF, Wu RS. Hypoxia alters steroidogenesis in female marine medaka through miRNAs regulation. Aquat Toxicol. 2016;172:1–8. doi: 10.1016/j.aquatox.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 21.Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 22.Arany Z, Huang LE, Eckner R, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 24.Kowalewski MP, Gram A, Boos A. The role of hypoxia and HIF1α in the regulation of STAR-mediated steroidogenesis in granulosa cells. Mol Cell Endocrinol. 2015;401:35–44. doi: 10.1016/j.mce.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Rytkönen KT, Akbarzadeh A, Miandare HK, et al. Subfunctionalization of cyprinid hypoxia-inducible factors for roles in development and oxygen sensing. Evolution. 2013;67:873–882. doi: 10.1111/j.1558-5646.2012.01820.x. [DOI] [PubMed] [Google Scholar]
- 26.Köblitz L, Fiechtner B, Baus K, Lussnig R, Pelster B. Developmental expression and hypoxic induction of hypoxia inducible transcription factors in the zebrafish. PLoS ONE. 2015;10:e0128938. doi: 10.1371/journal.pone.0128938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 28.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependant degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajimura S, Aida K, Duan C. Understanding hypoxia-induced gene expression in early development: in vitro and in vivo analysis of hypoxia-inducible factor 1-regulated zebra fish insulin-like growth factor binding protein 1 gene expression. Mol Cell Biol. 2006;26:1142–1155. doi: 10.1128/MCB.26.3.1142-1155.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharya S, Michels CL, Leung MK, Arany ZP, Kung AL, Livingston DM. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- 32.Rojas DA, Perez-Munizaga DA, Centanin L, et al. Cloning of hif-1alpha and hif-2alpha and mRNA expression pattern during development in zebrafish. Gene Expr Patterns. 2007;7:339–345. doi: 10.1016/j.modgep.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotch LE, Iyer NV, Laughner E, Semenza GL. Defective vascularization of HIF-1alpha-null embryos is not associated with VEGF deficiency but with mesenchymal cell death. Dev Biol. 1999;209:254–267. doi: 10.1006/dbio.1999.9253. [DOI] [PubMed] [Google Scholar]
- 36.Hu YY, Fu LA, Li SZ, et al. Hif-1α and Hif-2α differentially regulate Notch signaling through competitive interaction with the intracellular domain of Notch receptors in glioma stem cells. Cancer Lett. 2014;349:67–76. doi: 10.1016/j.canlet.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Villa JC, Chiu D, Brandes AH, et al. Nontranscriptional role of Hif-1 alpha in activation of γ-secretase and notch signaling in breast cancer. Cell Rep. 2014;8:1077–1092. doi: 10.1016/j.celrep.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao J, Zhou Z, Huang L, Li Y, Li J, Zou S. 17β-estradiol regulates the differentiation of cementoblasts via Notch signaling cascade. Biochem Biophys Res Commun. 2016;477:109–114. doi: 10.1016/j.bbrc.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 39.Bi P, Yue F, Sato Y, et al. Stage-specific effects of Notch activation during skeletal myogenesis. Elife. 2016;5:e17355. doi: 10.7554/eLife.17355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krebs LT, Xue Y, Norton CR, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 41.D’Amato G, Luxán G, de la Pompa JL. Notch signalling in ventricular chamber development and cardiomyopathy. FEBS J. 2016;283:4223–4237. doi: 10.1111/febs.13773. [DOI] [PubMed] [Google Scholar]
- 42.Lin D, Sugawara T, Strauss JF, et al. Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis. Science. 1995;267:1828–1831. doi: 10.1126/science.7892608. [DOI] [PubMed] [Google Scholar]
- 43.Pang S, Yang X, Wang M, et al. Inherited congenital adrenal hyperplasia in the rabbit: absent cholesterol side-chain cleavage cytochrome P450 gene expression. Endocrinology. 1992;131:181–186. doi: 10.1210/endo.131.1.1611996. [DOI] [PubMed] [Google Scholar]
- 44.Tajima T, Fujieda K, Kouda N, Nakae J, Miller WL. Heterozygous mutation in the cholesterol side chain cleavage enzyme (p450scc) gene in a patient with 46, XY sex reversal and adrenal insufficiency. J Clin Endocrinol Metab. 2001;86:3820–3825. doi: 10.1210/jcem.86.8.7748. [DOI] [PubMed] [Google Scholar]
- 45.Hu MC, Hsu NC, EI Hadj NB, et al. Steroid deficiency syndromes in mice with targeted disruption of Cyp11a1. Mol Endocrinol. 2002;16:1943–1950. doi: 10.1210/me.2002-0055. [DOI] [PubMed] [Google Scholar]
- 46.Hsu HJ, Hsu NC, Hu MC, Chung BC. Steroidogenesis in zebrafish and mouse models. Mol Cell Endocrinol. 2006;248:160–163. doi: 10.1016/j.mce.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 47.Payne AH, Hales DB. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr Rev. 2004;25:947–970. doi: 10.1210/er.2003-0030. [DOI] [PubMed] [Google Scholar]
- 48.Kawamoto T, Mitsuuchi Y, Toda K, et al. Role of steroid 11 beta-hydroxylase and steroid 18-hydroxylase in the biosynthesis of glucocorticoids and mineralocorticoids in humans. Proc Natl Acad Sci U S A. 1992;89:1458–1462. doi: 10.1073/pnas.89.4.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simard J, Moisan AM, Morel Y. Congenital adrenal hyperplasia due to 3beta-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase deficiency. Semin Reprod Med. 2002;20:255–276. doi: 10.1055/s-2002-35373. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Yao ZX, Papadopoulos V. Cytochrome P450 17alpha hydroxylase/17,20 lyase (CYP17) function in cholesterol biosynthesis: identification of squalene monooxygenase (epoxidase) activity associated with CYP17 in Leydig cells. Mol Endocrinol. 2005;19:1918–1931. doi: 10.1210/me.2004-0271. [DOI] [PubMed] [Google Scholar]
- 51.Jones ME, Boon WC, McInnes K, Maffei L, Carani C, Simpson ER. Recognizing rare disorders: aromatase deficiency. Nat Clin Pract Endocrinol Metab. 2007;3:414–421. doi: 10.1038/ncpendmet0477. [DOI] [PubMed] [Google Scholar]
- 52.Jiang B, Kamat A, Mendelson CR. Hypoxia prevents induction of aromatase expression in human trophoblast cells in culture: potential inhibitory role of the hypoxia-inducible transcription factor Mash-2 (mammalian achaete-scute homologous protein-2) Mol Endocrinol. 2000;14:1661–1673. doi: 10.1210/mend.14.10.0539. [DOI] [PubMed] [Google Scholar]
- 53.Lu X, Yu RM, Murphy MB, Lau K, Wu RS. Hypoxia disrupts gene modulation along the brain-pituitary-gonad (BPG)-liver axis. Ecotoxicol Environ Saf. 2014;102:70–78. doi: 10.1016/j.ecoenv.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 54.Kusakabe M, Nakamura I, Evans J, Swanson P, Young G. Changes in mRNAs encoding steroidogenic acute regulatory protein, steroidogenic enzymes and receptors for gonadotropins during spermatogenesis in rainbow trout testes. J Endocrinol. 2006;189:541–554. doi: 10.1677/joe.1.06684. [DOI] [PubMed] [Google Scholar]
- 55.Stocco DM. StAR protein and the regulation of steroid hormone biosynthesis. Annu Rev Physiol. 2001;63:193–213. doi: 10.1146/annurev.physiol.63.1.193. [DOI] [PubMed] [Google Scholar]
- 56.Bruder ED, Nagler AK, Raff H. Oxygen-dependence of ACTH-stimulated aldosterone and corticosterone synthesis in the rat adrenal cortex: developmental aspects. J Endocrinol. 2002;172:595–604. doi: 10.1677/joe.0.1720595. [DOI] [PubMed] [Google Scholar]
- 57.Bruder ED, Lee PC, Raff H. Metabolomic analysis of adrenal lipids during hypoxia in the neonatal rat: implications in steroidogenesis. Am J Physiol Endocrinol Metab. 2004;286:E697–E703. doi: 10.1152/ajpendo.00502.2003. [DOI] [PubMed] [Google Scholar]
- 58.Bruder ED, Jacobson L, Raff H. Plasma leptin and ghrelin in the neonatal rat: interaction of dexamethasone and hypoxia. J Endocrinol. 2005;185:477–484. doi: 10.1677/joe.1.06159. [DOI] [PMC free article] [PubMed] [Google Scholar]