Abstract
The genotypic diversity of antibiotic-producing Pseudomonas spp. provides an enormous resource for identifying strains that are highly rhizosphere competent and superior for biological control of plant diseases. In this study, a simple and rapid method was developed to determine the presence and genotypic diversity of 2,4-diacetylphloroglucinol (DAPG)-producing Pseudomonas strains in rhizosphere samples. Denaturing gradient gel electrophoresis (DGGE) of 350-bp fragments of phlD, a key gene involved in DAPG biosynthesis, allowed discrimination between genotypically different phlD+ reference strains and indigenous isolates. DGGE analysis of the phlD fragments provided a level of discrimination between phlD+ genotypes that was higher than the level obtained by currently used techniques and enabled detection of specific phlD+ genotypes directly in rhizosphere samples with a detection limit of approximately 5 × 103 CFU/g of root. DGGE also allowed simultaneous detection of multiple phlD+ genotypes present in mixtures in rhizosphere samples. DGGE analysis of 184 indigenous phlD+ isolates obtained from the rhizospheres of wheat, sugar beet, and potato plants resulted in the identification of seven phlD+ genotypes, five of which were not described previously based on sequence and phylogenetic analyses. Subsequent bioassays demonstrated that eight genotypically different phlD+ genotypes differed substantially in the ability to colonize the rhizosphere of sugar beet seedlings. Collectively, these results demonstrated that DGGE analysis of the phlD gene allows identification of new genotypic groups of specific antibiotic-producing Pseudomonas with different abilities to colonize the rhizosphere of sugar beet seedlings.
Antibiotic compounds produced by fluorescent Pseudomonas strains play key roles in the suppression of various soilborne plant pathogens (41, 47, 52). 2,4-Diacetylphloroglucinol (DAPG) produced by Pseudomonas fluorescens has activity against a range of plant pathogens, including bacteria, fungi, and nematodes (reviewed in reference 41). Recently, the broad-spectrum activity of DAPG also has drawn attention in the medical area because of the bacteriolytic activity of DAPG against multidrug-resistant Staphylococcus aureus (18). DAPG-producing Pseudomonas spp. have been isolated from the rhizospheres of different crops grown in soils from diverse geographic regions (19), and they are predominant constituents of the rhizosphere of wheat plants grown in soils that naturally suppress take-all disease (4, 30, 39, 58). They also have been isolated from soils that naturally suppress black root rot of tobacco (19, 43) or Fusarium wilt disease (23).
Multiple genes are involved in biosynthesis and regulation of DAPG production in P. fluorescens (reviewed in reference 12). One of these genes, the polyketide synthase gene phlD, is essential for synthesis of the DAPG precursor monoacetylphloroglucinol (1). It has been well documented that the phlD gene is conserved among DAPG-producing Pseudomonas strains found worldwide (19, 39) but displays a certain degree of polymorphism (23, 28, 42). Given that the genotypic diversity among DAPG-producing Pseudomonas strains provides an enormous resource for identifying strains that are highly rhizosphere competent and superior for biological control of plant diseases (40, 47), the sequence heterogeneity of the phlD gene is now routinely used to assess the diversity of this group of antagonistic bacteria (24, 28, 31, 42, 56). A range of other methods have been used to determine the genotypic diversity of DAPG-producing Pseudomonas strains; these methods include amplified ribosomal DNA restriction analysis (19, 37), random amplified polymorphic DNA (RAPD) analysis (28, 40), and BOX-PCR (31). A notable difficulty with all of these methods is the requirement for isolation and cultivation of phlD+ pseudomonads from soil and rhizosphere environments prior to genotypic characterization of the organisms. Isolation of phlD+ pseudomonads can be achieved by plating on semiselective media, followed by colony hybridization (39), a time-consuming method. Alternatively, direct characterization of phlD+ Pseudomonas isolates in rhizosphere samples can be performed by a rapid PCR assay (32). However, this method also requires cultivation of a rhizosphere sample in semiselective nutrient broth prior to characterization of the phlD+ genotype and may be biased toward detecting the most predominant genotype.
The aim of this work was to develop a simple and rapid method to study the presence and genotypic diversity of phlD+ Pseudomonas strains directly in rhizosphere samples without prior isolation or enrichment on nutrient media. New phlD-specific primers were developed, and their specificity was tested with a range of different phlD+ genotypes, alone and in mixtures. Polymorphisms within the amplified 350-bp phlD fragments were assessed by denaturing gradient gel electrophoresis (DGGE) analysis, sequencing, and phylogenetic analysis. The specificity and resolving capacity of the PCR-DGGE system were compared to the specificity and resolving capacity of currently used techniques, including phlD restriction fragment length polymorphism (RFLP) (31), RAPD analysis (19), and the rapid PCR assay (32). Finally, the biological significance of the newly developed PCR-DGGE classification was tested in root colonization assays with sugar beet seedlings treated with eight genotypically different phlD+ genotypes of Pseudomonas.
MATERIALS AND METHODS
Pseudomonas strains and culture conditions.
All Pseudomonas strains used in this study were cultured on King's medium B (KMB) agar (20) at 25°C for 48 h. To determine the specificity and resolving capacity of the PCR-DGGE method developed in this study, we tested multiple phlD+ reference strains (Table 1), as well as 184 indigenous phlD+ Pseudomonas isolates obtained from the rhizospheres of three plant species (wheat, sugar beet, and potato) by colony hybridization with a phlD-specific probe.
TABLE 1.
phlD+ Pseudomonas strains used in this study
| Strain or isolate | Code | Accession no. for phlD | Reference |
|---|---|---|---|
| Q2-87 | PfQ287 | U41818 | 1 |
| F113 | PfF113 | AJ278811 | 8 |
| CHA0 | PfCHA0 | AJ278806 | 49 |
| Pf-5 | PfPf5 | AF214457 | 16 |
| Q8R1-96 | PfQ8R196 | AF207693 | 40 |
| Q65C-80 | PfQ65c80 | AJ278807 | 13 |
| CMIA2 | PfCMIA2 | AJ278808 | 9 |
| MI-96 | PfMI96 | AF207692 | 40 |
| PILH1 | PfPILH1 | AJ278810 | 19 |
| PITR2 | PfPITR2 | AJ278809 | 19 |
| HR3-A13 | PfY | AY391780 | 29 |
| PR3-A52 | PfZ | AY391779 | 29 |
| 42-36 | Pf4236 | AF396857 | 38 |
| 42-27 | Pf4227 | AF396856 | 38 |
| 39-8 | Pf398 | AF396855 | 38 |
| 37-27 | Pf3727 | AF396854 | 38 |
| 22-27 | Pf2227 | AF396853 | 38 |
| 19-41 | Pf1941 | AF396852 | 38 |
| 19-30 | Pf1930 | AF396851 | 38 |
| 19-7 | Pf197 | AF396850 | 38 |
| 18-33 | Pf1833 | AF396849 | 38 |
| 11-18 | Pf1118 | AF396848 | 38 |
| 7-37 | Pf737 | AF396847 | 38 |
| 6-28 | Pf628 | AF396846 | 38 |
| 3-1 | Pf31 | AF396845 | 38 |
| D27B1 | D27B1 | NAa | 23 |
| PSB459 | Psp A1 | AY486314 | This study |
| PWB152 | Psp A2 | AY486317 | This study |
| PWB134 | Psp B | AY486316 | This study |
| PSB516 | Psp C | AY486315 | This study |
| PSB211 | Psp D | AY486313 | This study |
| PSB113 | Psp E1 | AY486312 | This study |
| PPB433 | Psp E2 | AY486310 | This study |
| PWB522 | Psp E3 | AY486319 | This study |
| PPB239 | Psp E4 | AY486309 | This study |
| PWB257 | Psp E5 | AY486318 | This study |
| PPB617 | Psp F | AY486311 | This study |
| PWB5516 | Psp Z | AY486320 | This study |
NA, not available.
PCR-DGGE analysis.
PCR amplification was carried out in a 25-μl reaction mixture which contained 3 μl of a 40-fold-diluted heat-lysed cell suspension (39), 1× GeneAmp PCR buffer (Perkin-Elmer Corp., Norwalk, Conn.), each deoxynucleoside triphosphate (Promega) at a concentration of 500 μM, 40 pmol of the reverse primer, 40 pmol of the forward primer (Amersham Pharmacia Biotech), 1.5 mM MgCl2, and 1.0 U of AmpliTaq DNA polymerase (Perkin-Elmer). The PCR program consisted of an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 67°C for 30 s, and 72°C for 60 s. The reactions were performed by using a Peltier Thermal Cycler-200 (Biozym, Landgraaf, The Netherlands). Fifteen microliters of the PCR product was used for analysis by DGGE with the Dcode universal mutation detection system (Bio-Rad Laboratories, Hercules, Calif.). The DGGE analysis protocol was based on the initial protocol of Muyzer et al. (35) and was performed by using an 8% (wt/vol) acrylamide gel with a linear denaturing gradient (100% denaturant contained 7 M urea plus 40% [vol/vol] deionized formamide). In almost all cases a gradient from 32% denaturant at the top to 41% denaturant at the bottom gave optimal separation of the amplified products, and this gradient was routinely used. Gels were run for 10 min at 200 V and subsequently for 16 h at 85 V (60°C), stained with ethidium bromide (0.5 μg/ml in 1× Tris-acetate-EDTA [TAE] [pH 8.3]) for 30 min, and visualized on an UV transilluminator.
DNA sequence analysis.
phlD fragments from multiple representative strains were amplified by using the Expand High Fidelity Taq polymerase (Roche, Almere, The Netherlands) and were subsequently sequenced by BaseClear (Leiden, The Netherlands). Alignment of phlD sequences obtained in this study and phlD sequences present in the databases was performed with Clustal W (53). Distance matrices were computed with MEGA, and phylogenetic trees were constructed by using the neighbor-joining method (44); the topology was checked by bootstrap analysis (1,000 data sets).
RFLP and RAPD analyses.
To determine the resolving capacity of the classifications assessed by PCR-DGGE analysis, multiple phlD+ strains were also subjected to phlD RFLP and RAPD analyses, two techniques that are currently used to determine the genotypic diversity of DAPG-producing Pseudomonas spp. For the phlD RFLP analysis, 629-bp fragments of the phlD gene were amplified with primers B2BF and BPR4, and this was followed by restriction with HaeIII, MspI, or TaqI (32). Restriction fragments were separated on a 2% agarose gel in 1× TAE for 2 to 3 h at 120 V. RAPD analysis with the 10-mer primers M12, M13 and D7 was performed according to protocols described previously (19, 40). The amplification products were separated on a 2% agarose gel in 1× TAE at 120 V for 3 h. phlD RFLP patterns and RAPD markers were visualized with a UV transilluminator and were photographed by using a digital camera. All PCR-RAPD amplifications were repeated at least two times, and only the consistent RAPD markers were included in the evaluation. The sizes of the restriction fragments obtained in the phlD RFLP analysis and the RAPD markers were determined with the Phoretix 1D software (version 3.0; Phoretix International, Newcastle, England). Band positions were converted to Rf values (0 and 1), and profile similarities were calculated by determining the pairwise coefficients of similarity (Nei-Li distances) for the total number of lane patterns. Cluster analysis with neighbor joining (44) and the corresponding bootstrap analysis (1,000 data sets) were performed with the Treecon software (version 1.3b) for Windows (54).
Plant cultivation and DNA extraction from the rhizosphere.
Wheat plants (Triticum aestivum L. cv. Bussard) were grown in a soil consisting of agricultural CB soil (4) mixed at a 1:1 ratio (wt/wt) with quartz sand. Sixteen wheat seeds were sown in square polyvinyl chloride pots containing 250 g (dry weight) of soil. A spontaneous rifampin-resistant derivative of phlD+ Pseudomonas isolate PWB532, representing DGGE group E, was introduced into soil at densities of 0, 10, 102, 103, 104, and 106 cells/g with an initial water content of 20% (vol/wt). One additional control treatment consisted of soil that was autoclaved twice (with 24 h between the two autoclave runs) to eliminate putative indigenous phlD+ isolates. Each treatment consisted of three replicates. After 10 to 12 days of cultivation in a growth chamber at 20°C with a 16-h photoperiod, the wheat plants were harvested, and rhizosphere samples were prepared for (i) enumeration of the introduced strain on selective agar plates and (ii) direct DNA extraction, followed by PCR-DGGE analysis. For enumeration of the introduced strain, 0.5 g of roots with associated rhizosphere soil was suspended in 5.0 ml of sterile distilled water and shaken vigorously for 1 min on a Vortex mixer; samples were subsequently sonicated in an ultrasonic cleaner for 1 min and dilution plated onto KMB agar supplemented with delvocid (100 mg/liter), chloramphenicol (13 mg/liter), ampicillin (40 mg/liter), and rifampin (100 mg/liter) (48). The plates were incubated for 3 days at 25°C, and colonies were enumerated. For direct DNA extraction from the rhizosphere, 0.5 g of roots with associated rhizosphere soil was suspended in 1.0 ml of saline phosphate buffer, shaken vigorously for 1 min on a Vortex mixer, and sonicated in an ultrasonic cleaner for 1 min. The roots were discarded, and the suspension was centrifuged for 1 min at 10,000 rpm (19,000 × g). An additional 0.5 g of rhizosphere soil that had been subjected to the same treatment was added, and the sample was subsequently processed by bead beating (three times for 90 s each). Cells were lysed by using the protocol of a FastDNA SPIN kit for soil (Bio 101). The DNA pellet was dissolved in 50 μl of Tris-EDTA (10 mM Tris, 0.1 mM EDTA; pH 8). PCR amplification of extracted DNA was performed in 50-μl reaction mixtures containing approximately 10 to 50 ng of DNA. In most cases, this amount of DNA was acquired after 100-fold dilution of the DNA obtained with the FastDNA SPIN kit. To enhance the specificity of the PCR, a ramping PCR was carried out as follows: the annealing temperature was initially 60°C, and it was increased to 72°C in steps of 0.1°C. The PCR program consisted of an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, with an increase at a rate of 0.1°C/s up to 72°C, and 72°C for 1 min. PCR amplification was carried out as described above. The primer concentration used was 20 pmol per reaction mixture instead of 40 pmol per reaction mixture.
In addition to wheat, experiments were performed with sugar beet (Beta vulgaris cv. Auris). Twenty-eight sugar beet seeds were sown in small square pots containing 250 g (dry weight) of soil and cultivated in a climate room with a controlled environment at 20°C and a 16-h photoperiod. Genotypically different phlD+ isolates were introduced separately into soil at a density of approximately 104 CFU/g of soil. The phlD+ isolates were spontaneous rifampin-resistant derivatives of isolates PWB233 (DGGE group A), PSC2218 (DGGE group B), PPB2310 (DGGE group C), PSB211 (DGGE group D), PWB532 (DGGE group E), PPB3512 (DGGE group F), PSC415 (DGGE group Z), and Q8R1-96 (DGGE group G). For short-term colonization experiments, the sugar beet plants were harvested after 10 to 12 days of cultivation. For long-term colonization studies, sugar beet plants were grown in the same pots for six successive cycles consisting of 10 to 12 days each. Twice a week, the plants were treated with one-third-strength Hoaglund's solution (macroelements only). After 10 to 12 days of growth, plants were harvested, and their root systems with rhizosphere soil were collected. Excess root material was mixed with the cultivated soil and represented approximately 0.125% (wt/wt) of the soil dry weight. The cultivated soil was subsequently returned to the same pot, and sugar beet seeds were replanted. This process of plant growth and harvesting was repeated for six successive cycles. Four replicates were included per treatment. For both short-term and long-term colonization experiments, rhizosphere samples were plated onto selective media and subjected to direct DNA extraction as described above for the experiments with wheat.
Statistical analysis.
Population densities of the introduced phlD+ fluorescent Pseudomonas strains were log10 transformed prior to statistical analysis. For the colonization assays with sugar beet seedlings, differences in population densities between the introduced strains were analyzed for each successive growth cycle by analysis of variance, followed by Tukey's Studentized range test (SAS Institute Inc., Cary, N.C.).
Nucleotide sequence accession numbers.
phlD sequences obtained in this study have been deposited in the GenBank database under the accession numbers shown in Table 1.
RESULTS AND DISCUSSION
Primer design and specificity.
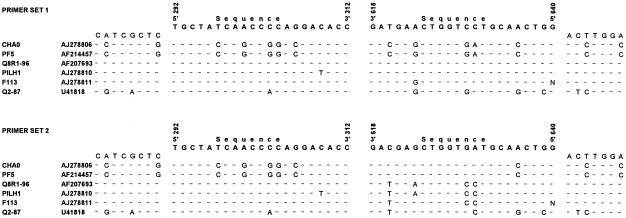
For detection of DAPG-producing Pseudomonas spp., a number of primers directed against sequences in the phlD gene have been developed previously (32, 39). The sizes of the amplification products of these primers range from approximately 600 to 750 bp, which is relatively large for further analysis by DGGE. Therefore, two new sets of oligonucleotide primers were developed for conserved sequences in the phlD gene of multiple reference strains (Fig. 1). In both primer sets, the forward primer is identical (DGGE292for), whereas there is a four-nucleotide difference between the two different reverse primers (DGGE618rev and 6DGGE618rev). Additionally, a 40-bp GC clamp is attached at the 5′ end of the forward primer (Table 2). The first primer set resulted in amplification of fragments of the predicted size (approximately 350 bp) from DNA of four genotypically different phlD+ reference strains (PILH1, F113, Q8R1-96, and Q2-87) and from DNA of each of 184 phlD+ isolates obtained previously from the rhizospheres of three different plant species. No amplification product was obtained from DNA of phlD mutant R1SS101 or from DNA of phlD+ reference strains CHA0 and Pf-5. At this stage, several attempts were made to further optimize the first primer set, including adjustment of the primer concentrations, the annealing temperature (the temperatures tested ranged from 48 to 70°C), and the Mg2+ concentration. However, none of these attempts were successful. The second primer set resulted in amplification of the predicted 350-bp fragment from DNA of all six phlD+ reference strains, including strains CHA0 and Pf-5, and from DNA of each of the 184 phlD+ isolates (Fig. 2). For successful amplification with the second primer set, a two-step PCR approach was required. In the first PCR step, no GC clamp was present at the 5′ end of the forward primer. After the first step, the PCR products were diluted 100- to 1,000-fold, after which the second PCR step was performed with the forward primer containing the GC clamp. The PCR programs for the one-step and two-step PCR amplifications were the same as the program described above.
FIG. 1.
Comparison of partial phlD sequences from several Pseudomonas strains. Bases that are identical in all sequences are indicated by dashes. The sequences and positions of the two primer sets used in PCR-DGGE analysis are shown above each of the two alignments. The positions of the 5′ and 3′ ends of each of the primers correspond to the positions in the phlD sequence of Pf-5 (accession number AF214457).
TABLE 2.
Properties of the oligonucleotide primers used in the PCR-DGGE analysis
| Primer | Sequence (5′-3′) | G+C content (%) | Melting temp (°C) |
|---|---|---|---|
| DGGE618rev | CCAGTTGCAGGACCAGTTCATC | 55 | 67.9 |
| 6DGGE618rev | CCAGTTGCATCACCAGCTCGTC | 59 | 67.9 |
| DGGE292for | TGCTATCAACCCCAGGACACC | 57 | 68.1 |
| DGGE292forCG | CGCCGGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGTGCTATCAACCCCAGGACACC | 84 | 57.9 |
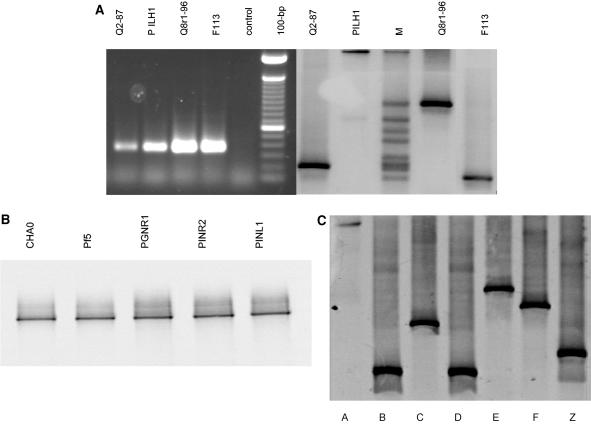
FIG. 2.
(A) Agarose gel (left gel) and 32 to 41% denaturant DGGE gel (right gel) of phlD fragments amplified from phlD+ reference strains Q2-87, PILH1, Q8r1-96, and F113. Lane M contained the DGGE marker, which included independently amplified phlD fragments from nine reference and indigenous strains. (B) A 40 to 51% denaturant DGGE gel of phlD fragments amplified from phlD+ reference strains CHA0, Pf-5, PGNR1, PINR2, and PINL1. (C) A 32 to 41% denaturant DGGE gel of phlD fragments amplified from phlD+ indigenous isolates obtained from the rhizosphere of multiple plant species. DGGE groups A (PWB152), B (PWB134), C (PSB516), D (PSB211), E (PSB113), F (PPB617), and Z (PWB5516) were included.
DGGE analysis.
DGGE and temperature gradient gel electrophoresis are widely used to study the microbial diversity in environmental samples and to monitor changes in specific microbial groups or communities (7, 10, 11, 22, 35, 45, 55). DGGE allows analysis of a large number of samples, which is essential for studying spatial and temporal variations in microbial populations (14, 33, 34, 35, 36, 51). To date, most primers used in DGGE analysis target rRNA genes of different microbial genera. In this context, group-specific primers have been developed for a number of bacterial genera, including Pseudomonas and Bacillus (10, 11), as well as for Burkholderia (45), Actinomycetes (15), and ammonium oxidizers and methanotrophs (2, 22). Recently, primers targeting specific biosynthetic genes have been developed, and, when combined with DGGE fingerprinting, these primers have led to a better level of discrimination within specific bacterial groups. For example, primers directed against the fliC gene allowed specific detection of the bacterial wilt pathogen Ralstonia solanacearum in soil and subsequent discrimination between strains obtained from various places (46). Rapid assessment of the diversity of methanogens was performed by DGGE analysis of the nifH gene (25, 57). In the same line of research, terminal RFLP analysis of PCR-amplified nifH fragments was shown to be a rapid technique for profiling diazotrophic microbial communities (50). Additionally, the community structures of ammonia-oxidizing bacteria (17) and bacteria from the marine environment (3) were explored by DGGE analysis of the amoA and rpoB genes, respectively.
In the present study, DGGE analysis was performed with the phlD fragments amplified with the first and second primer sets described above. A linear 32 to 41% denaturant gradient allowed detection of the amplified fragments and gave optimal discrimination between genotypically different phlD+ reference strains and isolates, except for strains CHA0 and Pf-5 (Fig. 2A and C). For CHA0 and Pf-5, a 40 to 51% denaturant gradient was required to detect the PCR products amplified with the second primer set (Fig. 2B). Attempts to design a DGGE gradient (32 to 51% denaturant) for all phlD+ strains, including strains CHA0 and Pf-5, resulted in a loss of discrimination between phlD+ genotypes other than those of strains CHA0 and Pf-5. The difference in the behavior of strains CHA0 and Pf-5 compared to other phlD+ strains in both PCR and DGGE is supported by phlD sequence data (see below) and has been described previously for PCR with other phlD primers (32). For strains other than CHA0 and Pf-5, the migration positions of PCR fragments obtained with the first and second primer sets were different, but the DGGE groups of the phlD+ genotypes were the same for both primer sets (data not shown). DGGE analysis of PGNR1, PINR2, and PINL1, three other strains that are also very closely related to CHA0 and Pf-5 (19), showed that their amplified phlD fragments migrated to the same positions as the phlD fragments of strains CHA0 and Pf-5 (Fig. 2B).
DGGE analysis of the 184 phlD+ isolates obtained from the rhizospheres of wheat, sugar beet, and potato resulted in seven DGGE groups, designated DGGE groups A, B, C, D, E, F, and Z (Fig. 2C and Table 3). Several of the 184 phlD+ isolates were assigned to the same DGGE group as reference strains PILH1 (DGGE group A) and Q2-87 (DGGE group B). None of the 184 phlD+ isolates were assigned to DGGE groups containing the reference strains Q8R1-96 (DGGE group G), F113 (DGGE group I), and CHA0 (DGGE group M). Therefore, the isolates assigned to DGGE groups C, D, E, F, and Z may represent phlD+ genotypes not described previously. DGGE group E was the dominant phlD+ genotype found among the collection of 184 indigenous phlD+ isolates, representing approximately 52% of the diversity (Table 3).
TABLE 3.
Genotypic classification of 184 indigenous phlD+ Pseudomonas isolates originating from the rhizospheres of multiple plant species by DGGE and RAPD analyses
| DGGE group | RAPD group | na | Frequency (% of all isolates)
|
|
|---|---|---|---|---|
| DGGE | RAPD | |||
| A | A1 | 28 | 16.85 | 15.22 |
| A2 | 3 | 1.63 | ||
| B | B1 | 19 | 10.33 | 10.33 |
| C | C1 | 11 | 5.98 | 5.98 |
| D | D1 | 2 | 1.09 | 1.09 |
| E | E1 | 68 | 52.16 | 36.96 |
| E2 | 18 | 9.78 | ||
| E3 | 3 | 1.62 | ||
| E4 | 6 | 3.26 | ||
| E5 | 1 | 0.54 | ||
| F | F1 | 5 | 2.72 | 2.72 |
| Z | Z1 | 20 | 10.87 | 10.87 |
| Total | 184 | 100 | 100 | |
Total number of isolates belonging to each RAPD group.
Comparison of PCR-DGGE, RAPD, and phlD RFLP analyses.
To determine the resolving capacity of the classifications obtained by PCR-DGGE analysis of the phlD gene, RAPD analysis with three 10-mer primers and phlD RFLP analysis were performed. The results of RAPD analysis of the 184 indigenous phlD+ isolates correlated well with the results obtained by PCR-DGGE, but there was a higher degree of discrimination than there was in the PCR-DGGE analysis (Table 3). RAPD analyses resulted in 12 different RAPD groups, whereas PCR-DGGE resulted in seven different groups. Isolates belonging to DGGE group E were assigned to five different RAPD groups (RAPD groups E1 thru E5), and isolates belonging to DGGE group A were assigned to two RAPD groups (RAPD groups A1 and A2). A subset of five phlD+ reference strains and 12 indigenous phlD+ isolates, representing the 12 different RAPD groups, were analyzed by RFLP analysis of a 629-bp phlD fragment with three restriction enzymes, a technique routinely used to determine the genotypic diversity of DAPG-producing Pseudomonas spp. (32). Based on the RFLP analysis, the two isolates (isolates A1 and A2) belonging to DGGE group A were identical to D27B1, a reference strain representing DGGE genotype A (Table 4). Representative isolates belonging to DGGE groups D, F, and Z could be distinguished on the basis of the RFLP analysis. In contrast, however, the RFLP profiles of reference strain Q8r1-96 (DGGE group G) were identical to those of all five isolates belonging to DGGE group E (RAPD groups E1 thru E5). Similarly, the RFLP profiles of isolates belonging to DGGE groups B and C were identical. These results indicated that PCR-DGGE provides a higher level of discrimination between phlD+ genotypes than the currently used phlD RFLP analysis.
TABLE 4.
Genotypic classification of 12 indigenous phlD+ Pseudomonas isolates originating from the rhizospheres of multiple plant species and five phlD+ reference Pseudomonas strains by DGGE and phlD RFLP analysesa
| DGGE genotype | RFLP genotype | Representative strain |
|---|---|---|
| A | M | D27B1 |
| A | M | PWB152 |
| A | M | PSB459 |
| B | B | Q2-87 |
| B | NEW1 | PWB134 |
| C | NEW1 | PSB516 |
| D | NEW2 | PSB211 |
| E | D | PSB113 |
| E | D | PPB433 |
| E | D | PWB522 |
| E | D | PPB239 |
| E | D | PWB257 |
| F | NEW3 | PPB617 |
| G | D | Q8r1-96 |
| I | K | F113 |
| M | A | CHA0 |
| Z | NEW4 | PWB5516 |
For the phlD RFLP analysis, 629-bp fragments of the phlD gene were amplified with primers B2BF and BPR4 and this was followed by restriction with HaeIII, MspI, and TaqI (32). NEW indicates groups based on the RFLP analysis which do not correspond with any of the previously described 17 genotypes (23).
Sequence and phylogenetic analyses.
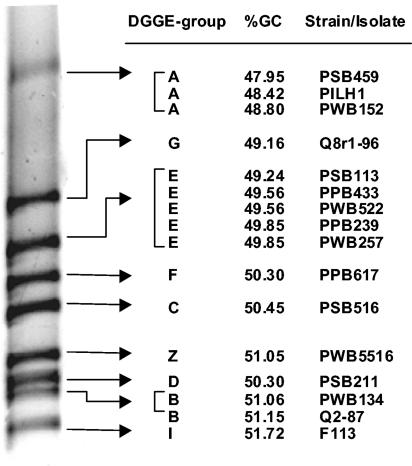
In general, sequence analyses of the 350-bp phlD fragments amplified from DNA of 12 isolates representing the different DGGE genotypes (genotypes A, B, C, D, E, F, and Z) showed a good correlation between the G+C contents of the amplified fragments and the migration patterns in the denaturing gel (Fig. 3). The G+C contents ranged from 47.95% for DGGE genotype A (located in the upper part of the 32 to 41% denaturant gradient) to 51.72% for DGGE genotype I (located at the bottom). The results further showed that not only the G+C content but also the position of the so-called melting domains (34) within the amplified fragment determines the electrophoretic mobility. The relatively high G+C content (59.6%) of the phlD fragments of reference strains CHA0 and Pf-5 supported the requirement for a gradient with a higher percentage of denaturant (40 to 51% denaturant), as described previously.
FIG. 3.
Relationship between the electrophoretic mobilities of phlD fragments and their G+C contents. The electrophoretic mobilities of 350-bp phlD fragments in a denaturing gradient gel (32 to 41% denaturant) are shown for various strains and isolates representing nine different DGGE groups. For three DGGE groups (DGGE groups A, B, and E), multiple isolates were included in the analysis.
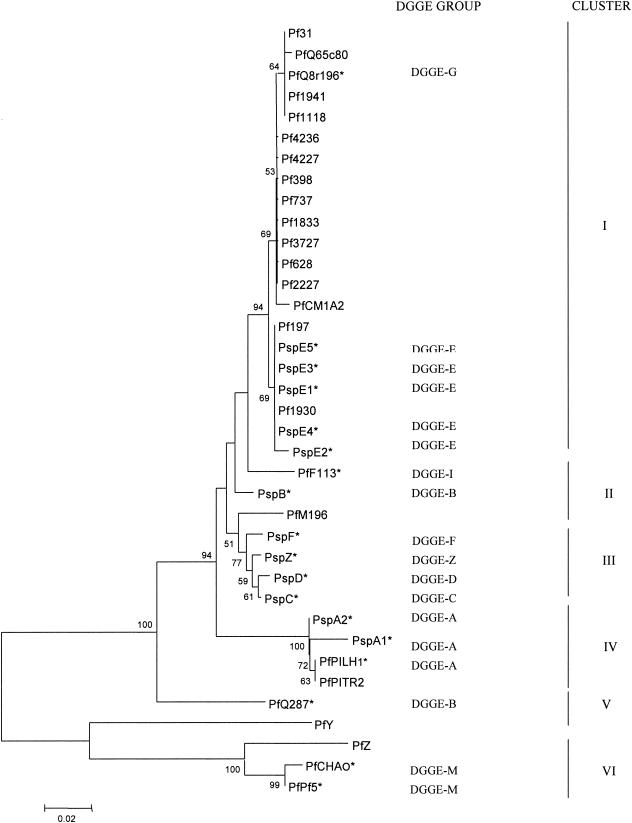
Phylogenetic analyses of the phlD sequences obtained in this study and present in the database (Table 1) revealed a total of six distinct clusters based on bootstrap values higher than 75% (Fig. 4). Cluster I contained the reference strains Q8R1-96 (DGGE genotype G), Q65c80, and CM1A2, as well as isolates E1 through E5, which were five representatives of DGGE genotype E. Representative isolates of DGGE genotype B were clustered together (cluster II) with phlD+ reference strains F113 (DGGE genotype I) and MI-96. Cluster III contained only representative isolates of DGGE genotypes C, D, F, and Z. Isolates A1 and A2, representing DGGE genotype A, were classified in the same cluster (cluster IV) together with reference strains PILH1 and PITR2, both of which were also classified by DGGE analysis as genotype A. Reference strain Q2-87 (DGGE genotype B) formed a unique cluster (cluster V) that included the recently described strain HR3-A13 (PfY) (29). However, Q2-87 was not grouped close to PWB134, a strain also classified in DGGE genotype B. These results are in accordance with results obtained by the RFLP analysis of strain Q2-87 and isolate PWB134. Cluster VI was the most distant cluster obtained in the phylogenetic analyses and contained reference strains CHA0 and Pf-5 (both DGGE genotype M) and the recently described strain PR3-A52 (PfZ) (29). Phylogenetic analyses of the phlD gene sequences by neighbor joining or maximum likelihood yielded similar results and tree topologies.
FIG. 4.
Phylogenetic tree (not rooted) of 325-bp phlD fragments inferred by the neighbor-joining method. Bootstrap values higher than 50% are indicated at the nodes. Asterisks indicate the representative reference phlD+ strains or isolates belonging to the different DGGE groups. Clusters I through VI are defined by bootstrap values of >75%.
PCR-DGGE of rhizosphere samples.
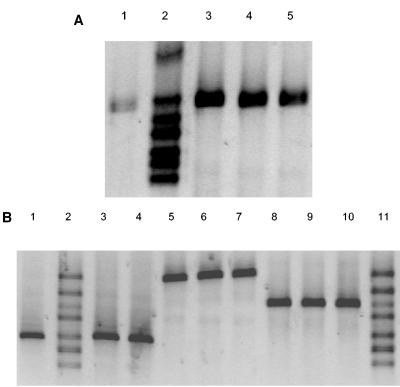
A notable difficulty of phlD RFLP and RAPD analyses is the requirement for isolation and cultivation of phlD+ Pseudomonas isolates from soil and rhizosphere environments prior to genotypic characterization of the organisms. A PCR-DGGE analysis was performed with DNA extracted from rhizosphere samples obtained from roots of wheat plants grown for 10 to 12 days in soil treated with a spontaneous rifampin-resistant derivative of isolate PWB532 (DGGE group E). Isolate PWB532 was introduced into the soil at initial densities ranging from 10 to 106 CFU/g. Rhizosphere samples were subjected to direct DNA extraction, followed by PCR-DGGE, and they were also dilution plated onto KMB supplemented with rifampin for comparison purposes. Based on dilution plating, the rhizosphere population densities of introduced isolate PWB532 ranged from 5 × 102 to 5 × 106 CFU/g of root after 10 to 12 days of cultivation. PCR with DNA directly extracted from the wheat rhizosphere resulted in amplification of the 350-bp phlD fragment when the density of PWB532 was equal to or higher than 5 × 103 CFU/g (fresh weight) of root. Subsequent DGGE analysis of the 350-bp fragments amplified from DNA extracted from the rhizosphere of wheat colonized by isolate PWB532 showed a single band corresponding to DGGE group E (Fig. 5A). No other DGGE genotypes were detected. Additionally, no 350-bp amplification products were detected in the control treatments, which included rhizosphere samples from natural CB soil and from CB soil autoclaved twice prior to wheat cultivation (data not shown). The ramping PCR protocol used for DNA extracted from rhizosphere samples was crucial as it considerably increased the sensitivity of the PCR amplification. Similar results were obtained in short-term experiments with sugar beet plants grown in soils treated with isolates PSC415 (DGGE group Z), Q8R1-96 (DGGE group G), and PPB3512 (DGGE group F) (Fig. 5B). Collectively, these results indicate that PCR-DGGE can be used to detect specific phlD+ genotypes directly in rhizosphere samples with a detection limit of approximately 5 × 103 CFU/g of root when ethidium bromide staining of the gel after electrophoresis is used. For ethidium bromide-stained gels, a detection limit of 105 CFU/g of soil was reported previously for R. solanacearum (46). When the results were combined with Southern hybridization, however, cell densities of R. solanacearum of approximately 103 CFU/g of soil could be detected (46). The results of the latter study suggest that the detection limit for indigenous phlD+ isolates may be increased further when the technique is combined with Southern hybridization or when silver staining is used instead of ethidium bromide staining.
FIG. 5.
(A) DGGE patterns (32 to 41% denaturant) of the 350-bp phlD fragment amplified from DNA extracted from the rhizosphere of wheat colonized by the rifampin-resistant derivative of phlD+ isolate PWB532 representing DGGE group E. Based on dilution plating, isolate PWB532 was present at densities of approximately 5 × 103 CFU/g of root (lane 1), 5 × 104 CFU/g of root (lane 3), 5 × 105 CFU/g of root (lane 4), and 5 × 106 CFU/g of root (lane 5). Lane 2 contained a marker composed of phlD fragments amplified from DNA of seven isolates corresponding to DGGE groups G, E, F, C, Z, B, and I (from top to bottom). (B) DGGE patterns (32 to 41% denaturant) of 350-bp phlD fragments amplified from isolates PSC415 (DGGE group Z), Q8R1-96 (DGGE group G), and PPB3512 (DGGE group F) introduced into the sugar beet rhizosphere. Lane 1, DGGE group Z at a density of 5.3 × 105 CFU/g of root; lane 2, marker; lane 3, DGGE group Z at a density of 2.7 × 105 CFU/g of root; lane 4, DGGE group Z at a density of 1.5 × 105 CFU/g of root; lane 5, DGGE group G at a density of 2.5 × 106 CFU/g of root; lane 6, DGGE group G at a density of 7.6 × 105 CFU/g of root; lane 7, DGGE group G at a density of 1.9 × 106 CFU/g of root; lane 8, DGGE group F at a density of 8.5 × 103 CFU/g of root; lane 9, DGGE group F at a density of 2.8 × 104 CFU/g of root; lane 10, DGGE group F at a density of 2.4 × 104 CFU/g of root; lane 11, marker. The marker consisted of phlD fragments amplified from DNA of seven isolates corresponding to DGGE groups G, E, F, C, Z, B, and I (from top to bottom).
A potential problem for PCR-DGGE analysis of DNA extracted from environmental samples may be that in mixed populations of phlD+ isolates, certain phlD genes are preferentially amplified, leading to incorrect assessment of all the genotypes present. In the present study, a mixture of six genotypically different phlD+ isolates (representatives of DGGE genotypes B, C, E, F, G, and Z) was introduced into wheat rhizosphere samples to a final density of approximately 5 × 105 CFU/g of root for each isolate prior to DNA extraction. PCR-DGGE analysis showed that all six genotypes were detectable in both replicates included. In both mixed samples, one additional band was detected in the DGGE gel; this band may have been a heteroduplex between the different phlD sequence variants, as described previously by Kowalchuk et al. (22), or it may have represented another indigenous phlD+ isolate present in the wheat rhizosphere sample. The latter possibility was not pursued further. Collectively, these results indicated that PCR-DGGE analysis of the phlD gene allows simultaneous detection of multiple genotypes present in a rhizosphere sample.
In the same experiment, the PCR-DGGE methodology was compared with the currently used rapid PCR-based protocol for rhizosphere samples (32). For the latter rapid PCR-based protocol, different dilutions of the rhizosphere samples required incubation in nutrient broth for 48 h prior to PCR and genotypic characterization. PCR and subsequent genotypic characterization showed that the rapid PCR-based protocol resulted in detection of only DGGE group G (strain Q8r1-96), whereas PCR-DGGE analysis resulted in detection of all six genotypes. In conclusion, these results indicated that cultivation of a rhizosphere sample in nutrient broth prior to genotypic characterization introduces a bias toward detecting either the most dominant genotype or the genotypes with higher growth rates or competitive abilities in the nutrient broth relative to the other genotypes present. This bias is circumvented by direct PCR-DGGE analysis of the phlD gene.
Biological significance of PCR-DGGE classification of phlD+ genotypes.
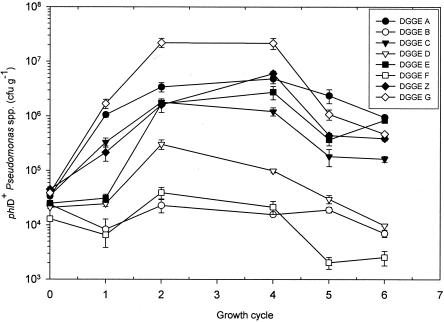
In order to investigate the biological significance of the additional classifications of phlD+ genotypes obtained by the PCR-DGGE methodology described in this study, the population dynamics of eight isolates representing different DGGE groups were monitored during six successive growth cycles of sugar beet seedlings in soil obtained from an agricultural field (Fig. 6). Each of the eight isolates was introduced only once (growth cycle 0) at an initial density of approximately 5 × 104 CFU/g of soil. After the first sugar beet growth cycle, the densities of strains Q8r1-96 (DGGE genotype G) and PWB233 (DGGE genotype A) were the highest densities; the population densities of strain Q8r1-96 (DGGE genotype G) increased further during growth cycles 2 to 4 to values of approximately 3 × 107 CFU/g of root, whereas the population densities of PWB233 (DGGE genotype A) leveled off at values of approximately 5 × 106 CFU/g of root. After the first growth cycle, strains PWB532 (DGGE genotype E), PPB2310 (DGGE genotype C), and PSC415 (DGGE genotype Z) did not exhibit densities as high as those of strains Q8r1-96 (DGGE genotype G) and PWB233 (DGGE genotype A), but they reached densities similar to those of PWB233 (DGGE genotype A) in growth cycles 2 to 4. After six growth cycles of sugar beet seedlings, the population densities of the genotype A, E, and G strains were significantly similar (P < 0.05). Strains PSB211 (DGGE genotype D), PSC2218 (DGGE genotype B), and PPB3512 (DGGE genotype F) colonized the rhizosphere of sugar beet seedlings to a significantly lesser extent than the strains representing the other five DGGE genotypes. The population densities of the three strains (DGGE groups D, B, and F) did not increase above 105 CFU/g of root but instead declined in the last sugar beet growth cycle to values of 104 CFU/g of root (DGGE group D), 8 × 103 CFU/g of root (DGGE group B), and 4 × 103 CFU/g of root (DGGE group F) (Fig. 6).
FIG. 6.
Population dynamics (expressed in CFU per gram of root) of representative isolates for eight different DGGE genotypes in the sugar beet rhizosphere. Isolates were introduced separately into soil only once (cycle 0) at a density of approximately 5 × 104 CFU/g of soil. The population dynamics of each of the isolates was monitored during six successive sugar beet growth cycles consisting of 10 to 12 days each. Cycle 3 was not included. For each growth cycle, mean values for four replicates are shown. The error bars indicate the standard errors.
These results showed that there were considerable differences in the abilities of different phlD+ genotypes to colonize the rhizosphere of sugar beet seedlings, confirming and extending results obtained in previous studies (24, 40). Strain Q8r1-96, representing DGGE genotype G, was shown to be superior for colonization of the sugar beet rhizosphere, especially in the first four growth cycles. Similar observations were described previously for wheat (40) and pea (24). These results indicate that the ability of strain Q8r1-96 to rapidly establish and maintain high population densities in the rhizosphere is not linked to a specific plant species but may be due to specific characteristics that enable this strain to be competitive in different rhizosphere environments. In this context, Mavrodi et al. (27) recently identified possible new traits by subtractive hybridization that may contribute to the superior rhizosphere competence of strain Q8R1-96. These traits include bacteriocin production, a trait that may be advantageous in intraspecific competition with other indigenous pseudomonads. Although strain Q8r1-96 (DGGE genotype G) could not be distinguished from strain PWB532 (DGGE genotype E) on the basis of phlD RFLP analysis, these organisms differed considerably in the ability to colonize the rhizosphere of sugar beet seedlings. Similarly, strains PPB2310 (DGGE genotype C) and PSC2218 (DGGE genotype B), which could not be distinguished by phlD RFLP analysis, differed significantly in the ability to colonize the sugar beet rhizosphere. These results highlight the conclusion that the additional classification of this widely distributed group of antibiotic-producing Pseudomonas spp. by PCR-DGGE analysis of the phlD gene also provides biologically relevant discrimination. Given the level of polymorphism in specific genes involved in the regulation (gacA) (6) or biosynthesis of other antibiotic compounds, including pyrrolnitrin (5, 21) and phenazine antibiotics (26, 39), this technique could easily be used to provide an additional level of discrimination between isolates and strains producing other metabolites involved in rhizosphere competence and biological control of plant pathogens.
Conclusion.
Establishing the presence of individual populations of antagonistic microorganisms in soil and rhizosphere environments is an important first step toward fully understanding the functional roles of the organisms in these natural environments. Additionally, the diversity within such indigenous populations of antagonistic microorganisms with a common biocontrol trait holds promise for further improvement of biological control, especially when this diversity reflects important interactions at the host-antagonist level. The technique described in this paper allows direct detection and assessment of the genotypic diversity of a specific group of bacteria that produce DAPG, a broad-spectrum antibiotic that has been implicated in biological control of multiple plant diseases and in the natural suppressiveness of soils. More specifically, our results indicated that the PCR-DGGE methodology can be used to detect specific phlD+ genotypes directly in rhizosphere samples with a detection limit of approximately 5 × 103 CFU/g of root and that it allows simultaneous detection of multiple genotypes present in a rhizosphere sample. Subsequent bioassays clearly showed that there is differential ability of the genotypic groups with respect to colonization of the sugar beet rhizosphere, confirming the biological significance of this methodology.
Acknowledgments
This research is supported by the Technology Foundation STW (project WBI.4843), the applied science division of NWO, and the technology program of the Ministry of Economic Affairs. The contribution of Jos Raaijmakers was financially supported by the Royal Netherlands Academy of Arts and Science.
We thank Pierre de Wit for critically reading this manuscript and for his valuable suggestions.
REFERENCES
- 1.Bangera, M. G., and L. S. Thomashow. 1999. Identification and characterization of a gene cluster for synthesis of the polyketides antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 181:3155-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boon, N., W. de Windt, W. Verstraete, and E. M. Top. 2002. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101-112. [DOI] [PubMed] [Google Scholar]
- 3.Dahllof, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Souza, J. T., D. M. Weller, and J. M. Raaijmakers. 2003. Frequency, diversity and activity of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in Dutch take-all decline soils. Phytopathology 93:54-63. [DOI] [PubMed] [Google Scholar]
- 5.De Souza, J. T., and J. M. Raaijmakers. 2003. Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiol. Ecol. 43:21-34. [DOI] [PubMed] [Google Scholar]
- 6.De Souza, J. T., M. Mazzola, and J. M. Raaijmakers. 2003. Conservation of the response regulator gene gacA in Pseudomonas species. Environ. Microbiol. 5:1328-1340. [DOI] [PubMed] [Google Scholar]
- 7.Duineveld, B. M., A. S. Rosado, J. D. van Elsas, and J. A. van Veen. 1998. Analysis of the dynamics of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl. Environ. Microbiol. 64:4950-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenton, A. M., P. M. Stephens, J. Crowley, M. O'Callaghan, and F. O'Gara. 1992. Exploitation of genes involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl. Environ. Microbiol. 58:3873-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs, J., and G. Defago. 1991. Protection of cucumber plants against black root rot caused by Phomopsis sclerotioides with rhizobacteria, p. 57-62. In C. Keel, B. Koller, and G. Defago (ed.), Plant growth-promoting rhizobacteria—progress and prospects. IOBC/WPRS bulletin XIV/8. IOBC/WPRS, Interlaken, Switzerland.
- 10.Garbeva, P., J. A. van Veen, and J. D. van Elsas. 2003. Predominant Bacillus sp. in agricultural soil under different management regimes detected via PCR-DGGE. Microb. Ecol. 45:302-316. [DOI] [PubMed] [Google Scholar]
- 11.Garbeva, P., J. A. van Veen, and J. D. van Elsas. 2003. Assessment of the diversity, and antagonism toward Rhizoctonia solani AG3, of Pseudomonas species in soil from different agricultural regimes. FEMS Microbiol. Ecol. 47:51-64. [DOI] [PubMed] [Google Scholar]
- 12.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, L. A., L. Letendre, P. Kovacevich, E. Pierson, and D. M. Weller. 1993. Purification of an antibiotic effective against Gaumannomyces graminis var. tritici produced by a biocontrol agent, Pseudomonas aureofaciens. Soil Biol. Biochem. 25:215-221. [Google Scholar]
- 14.Heuer, H., and K. Smalla. 1997. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities, p. 353-373. In J. D. van Elsas, E. M. H. Wellington, and J. T. Trevors (ed.), Modern soil microbiology. Marcel Dekker, New York, N.Y.
- 15.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16 rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howell, C. R., and R. D. Stipanovic. 1979. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69:480-482. [Google Scholar]
- 17.Ibekwe, A. M., A. C. Kennedy, P. S. Frohne, S. K. Papiernik, C. H. Yang, and D. E. Crowley. 2002. Microbial diversity along a transect of agronomic zones. FEMS Microbiol. Ecol. 39:183-191. [DOI] [PubMed] [Google Scholar]
- 18.Isnansetyo, A., and Y. Kamei. 2003. MC21-A, a bactericidal antibiotic produced by a new marine bacterium, Pseudoalteromonas phenolica sp. nov. O-BC30T, against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:480-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keel, C., D. M. Weller, A. Natsch, G. Défago, R. J. Cook, and L. S. Thomashow. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 21.Kirner, S., P. E. Hammer, D. S. Hill, A. Altmann, I. Fisher, L. J. Weislo, M. Lanahan, K. H. van Pee, and J. M. Ligon. 1998. Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalchuk, G. A., J. R. Stephen, W. de Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landa, B. B., O. V. Mavrodi, J. M. Raaijmakers, B. B. McSpadden-Gardener, L. S. Thomashow, and D. M. Weller. 2002. Differential ability of genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Appl. Environ. Microbiol. 68:3226-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landa, B. B., D. M. Mavrodi, L. S. Thomashow, and D. M. Weller. 2003. Interactions between strains of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens in the rhizosphere of wheat. Phytopathology 93:982-994. [DOI] [PubMed] [Google Scholar]
- 25.Lovell, C. R., Y. M. Piceno, J. M. Quattro, and C. E. Bagwell. 2000. Molecular analysis of diazotroph diversity in the rhizosphere of smooth cordgrass, Spartina alterniflora. Appl. Environ. Microbiol. 66:3814-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavrodi, D. V., V. N. Ksenzenko, R. F. Bonsall, R. J. Cook, A. M. Boronin, and L. S. Thomashow. 1998. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavrodi, D. V., O. V. Mavrodi, B. B. McSpadden-Gardener, B. B. Landa, D. M. Weller, and L. S. Thomashow. 2002. Identification of differences in genome content among phlD-positive Pseudomonas fluorescens strains by using PCR-based subtractive hybridization. Appl. Environ. Microbiol. 68:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mavrodi, O. V., B. B. McSpadden Gardener, D. V. Mavrodi, R. F. Bonsall, D. M. Weller, and L. S. Thomashow. 2001. Genetic diversity of phlD from 2,4-diacetylphloroglucinol producing fluorescent Pseudomonas spp. Phytopathology 91:35-43. [DOI] [PubMed] [Google Scholar]
- 29.Mazzola, M., D. L. Funnell, and J. M. Raaijmakers. Wheat cultivar-specific selection of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas species from resident soil population. Microb. Ecol., in press. [DOI] [PubMed]
- 30.McSpadden-Gardener, B. B., K. L. Schroeder, S. E. Kalloger, J. M. Raaijmakers, L. S. Thomashow, and D. M. Weller. 2000. Genotypic and phenotypic diversity of phlD-containing Pseudomonas isolated from the rhizosphere of wheat. Appl. Environ. Microbiol. 66:1939-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McSpadden-Gardener, B. B., and D. M. Weller. 2001. Changes in populations of rhizosphere bacteria associated with take-all of wheat. Appl. Environ. Microbiol. 67:4414-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McSpadden-Gardener, B. B., D. V. Mavrodi, L. S. Thomashow, and D. M. Weller. 2001. A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-diacetylphloroglucinol-producing bacteria. Phytopathology 91:44-54. [DOI] [PubMed] [Google Scholar]
- 33.Muyzer, G., T. Brinkhoff, U. Nubel, C. Santegoeds, H. Schafer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruin (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 34.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 35.Muyzer, G. E., C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of PCR-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen, A. T., W. T. Liu, C. Filipe, L. Grady, S. Molin, and D. A. Stahl. 1999. Identification of a novel group of bacteria in sludge from a deteriorated biological phosphorus removal reactor. Appl. Environ. Microbiol. 65:1251-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picard, C., F. Di Cello, M. Ventura, R. Fani, and A. Guckert. 2000. Frequency and diversity of 2,4-diacetylphloroglucinol-producing bacteria isolated from the maize rhizosphere at different stages of growth. Appl. Environ. Microbiol. 66:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picard, C., and M. Bosco. 2003. Genetic diversity of phlD gene from 2,4-diacetylphloroglucinol-producing Pseudomonas spp. strains from the maize rhizosphere. FEMS Microbiol. Lett. 219:167-172. [DOI] [PubMed] [Google Scholar]
- 39.Raaijmakers, J. M., D. M. Weller, and L. S. Thomashow. 1997. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Applied Environmental Microbiol. 63:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raaijmakers, J. M., and D. M. Weller. 2001. Exploiting genotypic diversity of 2,4-diacetylphloroglucinol-producing Pseudomonas spp.: characterization of superior root-colonizing P. fluorescens strain Q8r1-96. Appl. Environ. Microbiol. 67:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raaijmakers, J. M., M. Vlami, and J. T. de Souza. 2002. Antibiotic production by bacterial biocontrol agents. Antonie Leeuwenhoek 81:537-547. [DOI] [PubMed] [Google Scholar]
- 42.Ramette, A., Y. Moënne-Loccoz, and G. Défago. 2001. Polymorphism of the polyketide synthase gene phlD in biocontrol fluorescent pseudomonads producing 2,4-diacetylphloroglucinol and comparison of phlD with plant polyketide synthases. Mol. Plant-Microbe Interact. 14:639-652. [DOI] [PubMed] [Google Scholar]
- 43.Ramette, A., Y. Moënne-Loccoz, and G. Défago. 2003. Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol. Ecol. 44:35-43. [DOI] [PubMed] [Google Scholar]
- 44.Saito, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 45.Salles, J. F., F. A. De Souza, and J. D. van Elsas. 2002. Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl. Environ. Microbiol. 68:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schonfeld, J., H. Heuer, J. D. van Elsas, and K. Smalla. 2003. Specific and sensitive detection of Ralstonia solanacearum in soil on the basis of PCR amplification of fliC fragments. Appl. Environ. Microbiol. 69:7248-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharifi-Tehrani, A., M. Zala, A. Natsch, Y. Moënne-Loccoz, and G. Défago. 1998. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur. J. Plant Pathol. 104:631-643. [Google Scholar]
- 48.Simon, A., and E. H. Ridge. 1974. The use of ampicillin in a simple selective medium for the isolation of fluorescent pseudomonads. J. Appl. Bacteriol. 37:459-460. [DOI] [PubMed] [Google Scholar]
- 49.Stutz, E., G. Defago, and H. Kern. 1986. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76:181-185. [Google Scholar]
- 50.Tan, Z., T. Hurek, and B. Reinhold-Hurek. 2003. Effect of N-fertilization, plant genotype and environmental conditions on nifH gene pools in roots of rice. Environ. Microbiol. 5:1009-1015. [DOI] [PubMed] [Google Scholar]
- 51.Theron, J., and T. E. Cloete. 2000. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 26:37-57. [DOI] [PubMed] [Google Scholar]
- 52.Thomashow, L. S., and D. M. Weller. 1996. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites, p. 187-236. In G. Stacey and N. T. Keen (ed.), Plant-microbe interactions, vol. 1. Chapman & Hall, London, United Kingdom.
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van de Peer, Y., and R. de Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 55.Van Elsas, J. D., G. F. Duarte, A. S. Rosado, and K. Smalla. 1998. Microbiological and molecular biological methods for monitoring microbial inoculants and their effects in the soil environment. J. Microbiol. Methods 32:133-154. [Google Scholar]
- 56.Wang, C., A. Ramette, P. Punjasamarnwong, M. Zala, A. Natsch, Y. Moënne-Loccoz, and G. Défago. 2001. Cosmopolitan distribution of phlD-containing dicotyledonous crop-associated biocontrol pseudomonads of worldwide origin. FEMS Microbiol. Ecol. 37:105-116. [Google Scholar]
- 57.Wawer, C., and G. Muyzer. 1995. Genetic diversity of Desulfovibrio spp. in environmental samples analyzed by denaturing gradient gel electrophoresis of (nifE) hydrogenase gene fragments. Appl. Environ. Microbiol. 61:2203-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weller, D. M., J. M. Raaijmakers, B. B. Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]