ABSTRACT
Identification of biomarkers for early detection of lung cancer (LC) is important, in turn leading to more effective treatment and reduction of mortality. Serological proteome analysis (SERPA) was used to identify proteins around 34 kD as ECH1 and HNRNPA2B1, which had been recognized by serum autoantibody from 25 LC patients. In the validation study, including 90 sera from LC patients and 89 sera from normal individuals, autoantibody to ECH1 achieved an area under the curve (AUC) of 0.799 with sensitivity of 62.2% and specificity of 95.5% in discriminating LC from normal individuals, and showed negative correlation with tumor size (rs = −0.256, p = 0.023). Autoantibody to HNRNPA2B1 performed an AUC of 0.874 with sensitivity of 72.2% and specificity of 95.5%, and showed negative correlation with lymph node metastasis (rs = −0.279, p = 0.012). By using longitudinal preclinical samples, autoantibody to ECH1 showed an AUC of 0.763 with sensitivity of 60.0% and specificity of 89.3% in distinguishing early stage LC from matched normal controls, and elevated autoantibody levels could be detected greater than 2 y before LC diagnosis. ECH1 and HNRNPA2B1 are autoantigens that elicit autoimmune responses in LC and their autoantibody can be the potential biomarkers for the early detection of LC.
KEYWORDS: Autoantibody, ECH1, HNRNPA2B1, lung cancer, serological proteome analysis (SERPA)
Introduction
Lung cancer (LC) is the leading cause of cancer death worldwide. In 2015, LC is estimated to account for more than 220,000 new cancer cases and 158,000 cancer deaths in the United States.1 Presently, early detection of LC is limited to low-dose spiral computed tomography (LDCT) to screen LC in at-risk individuals.2-4 While LDCT offer mortality benefit in high-risk individuals,5 its application as a screening procedure have been restricted by the plentiful costs6 and high false positives (50%), requiring individuals to have unnecessary follow-up examinations and unnecessary surgery therapy.7 Hence, the search for improving biomarkers that can complement with imagining for LC screening remains an important goal.
Serum biomarkers would enhance diagnostic capabilities to complement the currently available diagnostic tests, since they are noninvasive and reproducible screening tools. Many studies have demonstrated that cancer sera contain autoantibodies that react with a unique group of autologous cellular antigens called tumor-associated antigens (TAAs).8-10 The immune response appear months to years before the clinical diagnosis of a tumor, which is triggered by the release of TAAs from tumors in cancer patients, suggesting serum autoantibody detection is highly suitable for early cancer diagnosis.11 Early detection is essential for the optimal management of cancer. Thus, extensive studies are being conducted to identify and validate new biomarkers that would add to current markers and increase the sensitivity and specificity of cancer detection. Some studies have been developing autoantibody assays that could complement CT scanning in LC diagnosis and management.12-18 The only one commercial test currently is a blood test that measures autoantibodies to LC-associated antigens called EarlyCDT-Lung. It was developed to assist physicians in the early detection of LC in a high-risk population with pulmonary nodules detected by CT.16 Due to the low sensitivity of this test, additional antigens need to be explored, and alternative ways to improve the assay performance must also be pursued.
In the present study, we identified ECH1 and HNRNPA2B1 as new autoantigens for LC using immunoproteomic approach (serological proteome analysis, SERPA), and found their autoantibodies could improve the performance characteristics for distinguish those with early-stage LC patients without clinical symptoms from the high-risk population, who are heavy smokers.
Materials and methods
Serum sample collection
Five groups in two independent sample sets (discovery and validation) were used for this study. All of the serum samples were collected from the New York University (NYU) Lung Cancer Biomarker Center, a member of the National Cancer Institute-sponsored Early Detection Research Network (NCI-EDRN). The discovery set (set 1) included two groups (group 1 and group 2). Group 1 consisted of 28 LC patients with 219 serial serum samples, including samples from prior, at-diagnosis and post-treatment, in which, 94 serum samples from 25 patients included at least 2 to 11 serial samples, which have been collected on average 32 mo (range, 0.5–120 mo) before the diagnosis of LC, and 125 serum samples from 19 patients included at least 2 to 13 serial samples collected on average 29.9 mo (range, 3–110 mo) post-treatment of LC. All of the patients are heavy smokers with average 54.4 pack years of smoking. When every patient in group 1 was diagnosed as LC, two normal individuals were collected by matching for age, gender and smoking pack-year in the same hospital. Therefore, total 56 normal individuals were included in group 2.
A separate set of samples (set 2: validation set), including three groups (group 3, group 4 and group 5) were used for validation of the autoantibody. Group 3 (n = 90) consisted of LC serum samples collected within eight weeks of biopsy-proven LC diagnosis. LC cases were staged as pathological stage I–III by the thoracic surgeons. Group 4 was the controls to group 3, in which there were 89 normal serum samples from healthy people who are never smokers or light smokers (<20 pack year). Group 5 consisted of 90 COPD (Chronic obstructive pulmonary disease) patients who were high-risk smokers and ex-smokers and participated in CT scan LC screening.
The detailed characterizes of five groups are shown in Table 1. The utilization of human samples was approved by the Institutional Review Board (IRB) at the Scripps Research Institute and at NYU. All clinical information were collected under Health Insurance Portability and Accountability Act (HIPAA) compliance from study participants with written informed consent under clinical research protocols approved by appropriate IRBs.
Table 1.
Characterization of lung cancer patients and controls.
| Set 1 (Discovery) |
Set 2 (Validation) |
||||
|---|---|---|---|---|---|
| Groups | Group 1 | Group 2 | Group 3 | Group 4 | Group 5 |
| n | 28 | 56 | 90 | 89 | 90 |
| Age | |||||
| Mean ± SD | 64.0 ± 5.5 | 59.3 ± 7.3 | 67.5 ± 10.7 | 58.0 ± 9.7 | 61.0 ± 8.2 |
| Range | 56–78 | 40–79 | 41–87 | 30–84 | 36–81 |
| Gender | |||||
| Male (%) | 11 (39.3) | 22(39.3) | 35 (38.9) | 53 (59.6) | 35 (38.9) |
| Female (%) | 17 (60.7) | 34(60.7) | 55 (61.1) | 36 (40.4) | 55 (61.1) |
| Smoking | |||||
| Average pack years | 54.4 | 53.0 | 35.0 | 8.4 | 35.5 |
| 0 pack years (%) | 0 (0.0) | 0 (0.0) | 15 (16.7) | 18 (20.2) | 8 (8.9) |
| <20 pack years (%) | 1 (4.0) | 2 (3.6) | 38 (42.2) | 71 (79.8) | 20 (22.2) |
| ≥20 pack years (%) | 24 (96.0) | 54 (96.4) | 31 (34.4) | 0 | 62 (68.9) |
| Unknown (%) | 0 | 0 | 6 (6.7) | 0 | 0 |
| Clinical stage | |||||
| Stage I (%) | 22 (78.6) | 30 (33.3) | |||
| Stage II (%) | 1 (3.6) | 30 (33.3) | |||
| Stage III (%) | 3 (10.7) | 30 (33.3) | |||
| Unknown (%) | 2 (7.1) | 0 | |||
| Histologic type | |||||
| NSCLC | 26 (92.9) | 90 (100.0) | |||
| Adenocarcinoma (%) | 21 (75.0) | 81 (90.0) | |||
| Squamous (%) | 3 (10.7) | 6 (6.7) | |||
| Unknown (%) | 2 (7.1) | 3 (3.3) | |||
| SCLC | 2 (7.1) | 0 | |||
| Tumor size | |||||
| <3 cm (%) | 51 (56.7) | ||||
| ≥3 cm (%) | 28 (31.1) | ||||
| Unknown (%) | 11 (12.2) | ||||
| Lymph node number | |||||
| No (%) | 16 (64.0) | 32 (35.6) | |||
| Yes (%) | 4 (16.0) | 49 (54.4) | |||
| 1–3 (%) | 0 | 39 (43.3) | |||
| 4–9 (%) | 0 | 8 (8.9) | |||
| ≥10 (%) | 0 | 2 (2.2) | |||
| Unknown (%) | 5 (20.0) | 9 (10.0) | |||
Group 1: 28 LC patients with 129 serial serum samples.
Group 2: 56 serum samples from normal individuals.
Group 3: 90 serum samples from LC patients.
Group 4: 89 serum samples from normal individuals.
Group 5: 90 serum samples from COPD patients.
Cell culture and subcellular fractionation
The LC cell line (H1299) was purchased from American Type Culture Collection (ATCC, Manassas, VA), and cultured in DMEM (Dulbecco's modified Eagle's medium, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin and 100 units/mL streptomycin. Cells grown in 75-cm2 Falcon tissue culture flasks were allowed to reach 95% confluence. Then, cells were rinsed once with DMEM without FBS and removed from the flask by incubating them with a solution containing trypsin–EDTA (Gibco, Carlsbad, CA), and harvested in a 15 mL centrifuge tube. Subcellular fractionation of H1299 was performed using ProteoExtract® Subcellular Proteome Extraction Kit (Millipore, Billerica, MA) according to the manufacturer's recommendation. Briefly, differential extraction of proteins according to their localization (including cytosolic, membrane/organelle and nucleic fractions) were obtained from H1299 cells using fraction-specific buffers provided in the kit. Then, the protein fractions were purified and concentrated by ice-cold acetone for further study.
Two-dimensional gel electrophoresis (2DE) analysis
Subcellular fractions of H1299 cells were directly lysed in rehydration sample buffer (8 M Urea, 50 mM dithiothreitol (DTT), 4% 3-[(3-cholamidopropyl) dimethylammonio] −1-propanesulfonate (CHAPS), 0.2% carrier ampholytes) as provided by Bio-Rad Laboratories (Hercules, CA) and were vortexed vigorously for 1 h at room temperature (RT). Insoluble substances were removed by centrifuge at 16,000g for 30 min at 4°C. Supernatant was collected and protein concentration was measured by the Bradford assay (Bio-Rad, Hercules, CA). For the first dimensional gel electrophoresis (1-DE) analysis, a total of 200 μg of protein was mixed with rehydration buffer, a trace bromophenol blue prepared in proteomics-grade water and applied on a pH 3–10, 11-cm isoelectric focusing (IEF) strip (Bio-Rad, Hercules, CA). IEF was performed at a current of 50 mA per gel, 300 V for 30 min, followed by 3,500 V for 2.5 h, and additional 8,000 V for 5 h. Strips were immediately stored at −80°C for the second dimensional gel electrophoresis (2-DE) analysis. For 2-DE, 10% SDS-polyacrylamide gels (SDS-PAGE) were used. Proteins were transferred onto nitrocellulose membrane (Osmonics Inc., MA) for Western blotting analysis or stained with 0.1% Coomassie blue R-250 prepared in 40% methanol/10% acetic acid. The spots were visualized using PDQuest 2-DE analysis software as described in the manufacturer's manual (Bio-Rad, Hercules, CA).
One- and two-dimensional Western blotting and proteomic analysis
To screen the autoantibody-positive sera, H1299 cells were lysed directly in Laemmli's sample buffer (Bio-Rad, Hercules, CA) and loaded onto 10% SDS-PAGE gel followed by running the gel at 80 V for 2 h, which is then transferred onto nitrocellulose membrane (Osmonics Inc., MA) for Western blotting. The membrane was then cut into 0.5-cm wide stripes. After blocking with 5% nonfat milk prepared in Tris-buffered saline (TBS), containing 0.05% Tween-20 (TBST), for 1 h at RT, the nitrocellulose membrane strips were incubated with sera at a dilution of 1:200. Horseradish peroxidase-conjugated goat anti-human IgG (Caltag Laboratories, San Francisco, CA) was used as secondary antibody with a dilution of 1:10,000 for 1 h at RT. The positive bands were detected with Enhanced Chemiluminescence (ECL) kit (Amersham, Arlington Heights, IL). For 2-DE Western blotting, the proteins on 2-DE gel are directly transferred onto nitrocellulose membrane and incubated with two pools of five sera from patients with LC in group 1 and from normal individuals in group 2 at a dilution of 1:500. All pools of sera were assayed in duplicated.
After identifying the interesting protein spots, gel spots in 2-DE gel were excised and digested to perform liquid chromatography tandem mass spectrometry (LC–MS/MS) analysis. MS/MS spectra derived from peptides were submitted for database search using TurboSequest (available in Bioworks version 3.3.1) against the human IPI database (v3.48), in both correct and reverse orientations to enable false-discovery rate (FDR) calculation. The following filters were applied in Bioworks: DCn ≥0.85; consensus score ≥10.0; protein probability ≤1 × 10−3 and Xcorr ≥ 1.5, 2.0 and 2.5, for singly, doubly and triply charged peptides, respectively.
The commercial recombinant proteins of ECH1, GAPDH and HNRNPA2B1 were electrophoresed by SDS-PAGE and transferred onto a nitrocellulose membrane that was then cut into strips. The strips were incubated with sera diluted 1:200 and subsequently with HRP-conjugated goat anti-human IgG diluted 1:10,000.
Enzyme-linked immunosorbent assay (ELISA)
Enoyl Coenzyme A hydratase 1 (ECH1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) recombinant proteins were purchased from Abcam (Cambridge, MA, USA) and heterogeneous nuclear ribonucleoproteins A2/B1 isoform B1(HNRNPA2B1) from Abnova (Walnut, CA, USA). ECH1, GAPDH and HNRNPA2B1 autoantibodies in human sera were detected by ELISA. Briefly, proteins were diluted in PBS to a final concentration of 0.5 μg/mL for coating 96-well microtiter plates (Fisher Scientific, Pittsburgh, PA). Human serum samples at 1:200 dilutions were added to the antigen-coated wells and incubated for 2 h at RT after washed twice by PBS with 0.5% Tween 20 (PBST). Horseradish peroxidase-conjugated goat antihuman IgG (Santa Cruz Biotechnology Inc., Dallas, TX, USA) at 1:4,000 dilution and the substrate 2,2′-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid) (Sigma, Ronkonkoma, NY, USA) were used as detecting reagents. Each sample was tested in duplicate, and the average OD at 405 nm was used for data analysis.
Statistical analysis
Data regarding the different seroreactivity of the autoantibodies were summarized by mean of OD value. Due to the sera antibodies against three TAAs were not normally distributed (Shapiro Wilk's test), nonparametric Mann–Whitney U-tests were used to compare differences of antibody levels between two groups, and nonparametric Kruscal-Wallis test were used to compare differences of antibody levels among multiple groups. χ2 tests were used to compare the differences of frequency between two groups and among multiple groups. Spearman's test was used to evaluate the correlation between autoantibody level and tumor size or lymph nodes number. The receiver operating characteristic (ROC) analysis of single-variable was conducted for each autoantibody for the distinguishing of LC from controls, leading to estimates of area under the curve (AUC) with 95% confidence interval (CI). The optimal cutoff thresholds for designating positive reaction were determined at the point on the ROC curve at which Youden's index (sensitivity + specificity −1) was maximal. Differences were considered statistically significant when p < 0.05. Statistical analyses were performed using SPSS software (version 18.0).
Results
A novel autoantibody against 34 kD autoantigen was found in serial serum samples from patients with early stage of lung cancer (discovery set 1)
Ninety-four serial sera collected before the diagnosis from 25 patients with early stage of LC (group 1) and 56 sera from normal human individuals (group 2) matched to group 1 were examined for autoantibodies using Western blotting with LC cell line H1299. Interestingly, there were two patients who appeared to have an increase in autoantibody for antigen/antigens with molecular weight 34 kD before the diagnosis of LC (Fig. 1A and B).
Figure 1.
Western blotting analysis. (1A and 1B): Reactivity of serial serum samples from two representative LC patients in group 1 to LC cell line H1299. Patient 1 was a 69-y-old Caucasian male who was a current heavy smoker (more than 50 pack years) and started to smoke at the age of 16-y-old. He was drawn sequential serum samples between year of 2003 and 2011 and diagnosed as lung adenocarcinoma of stage IA in Oct. 2009. As shown in 1A, in the initial serum sample collected in May 2003, autoantibodies were barely observed, but appeared one year later, 5 y before the diagnosis of LC. The 34 kD band (arrow) arose at the time point of 1.5 y before the lung cancer diagnosis (lane 5 in 1A) and reached at the highest titer at the time of 3 mo before the diagnosis (lane 6 in 1A). Fig. 1(B) Shows a 64-y-old Hispanic male, who has been smoking for 44 y with average 32 pack years, since he was 20 y old and diagnosed as small cell LC in stage IA when he was 64-y-old. A 34 kD reactive band (arrow) was observed in the initial serum sample collected in Sept. 2003 (lane 1 in 1B), but the titer was sharply increased 4 y later that was the time point of 3 y (lane 4 in 1B) before LC diagnosis. (1C) Reactivity of serum samples from five lung cancer patients in group 1, which drawn at the diagnosis of LC and five normal individuals in group 2 to H1299 cell line. 1D-1I: Immunoreaction of recombination protein ECH1 (1D–1F) and HNRNPA2B1 (1G–1I) to the serial serum samples from two representative patients the same with 1A and 1B, and to 10 representative individual sera (1F and 1I) from LC patients in group 3. *Time of lung cancer diagnosis (patient 1 was diagnosed in Oct. 2009 and patient 2 in April 2001).
Of 25 patients who had sequential serum samples in group 1, 10/25 patients (40.0%) were identified by Western blotting analysis to contain antibodies against 34 kD cellular protein, while no reactivity with the 34 kD protein was detected in 56 matched normal human sera. Western blotting analysis of five representative sera from LC (group 1) and normal individuals (group 2) are shown in Fig. 1C.
34kD proteins were identified to be ECH1, GAPDH and HNRNPA2B1 by SERPA approach
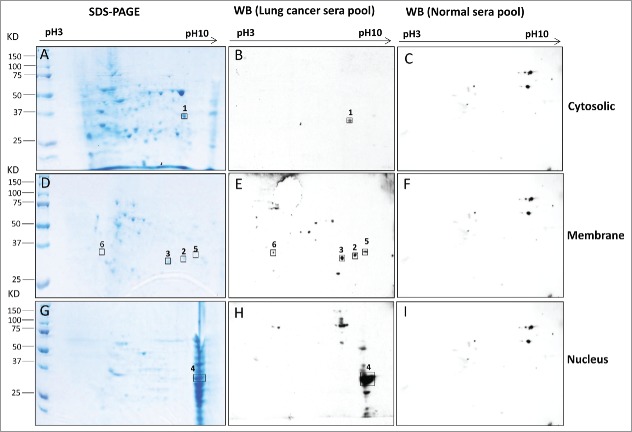
Cytosolic, membrane and nuclear fractions of H1299 cell line were obtained using differential extraction of proteins. Proteins were separated by 2-DE and TAAs were screened with two pools of sera, which consisted of patients with early-stage LC in group 1 (Fig. 1C: lane 1–5) and matched healthy individuals in group 2 (Fig. 1C: lane 6–10). A total of six spots with 34 kD molecular weight exhibited reactivity with early-stage LC sera pool but not with controls (Fig. 2), in which one spot was from cytosolic (Fig. 2A and B), four from membrane (Fig. 2D and E) and one from nucleus (Fig. 2G and H). These spots were excised from the gel, trypsin-digested and subsequently analyzed by LC–MS/MS. Ultimately, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), ECH1 (Enoyl Coenzyme A hydratase 1) and HNRNPA2B1 (heterogeneous nuclear ribonucleoproteins A2/B1 isoform B1) was successfully identified by MS in the cytosolic, membrane and nuclear fractions, respectively (Table 2).
Figure 2.
Immunoreactive proteins were detected by autoantibodies in serum samples from lung cancer patients. 2-DE gel electrophoresis of H1299 subcellular fractions (cytosolic, membrane and nucleus) were stained by Coomassie blue (2A, 2D and 2G). 2-DE Western blotting of H1299 subcellular fractions with the pool of sera from LC patients (n = 5 from group 1: line 1–5 in Fig. 1C) (2B, 2E and 2H) and with the pool of sera form normal individuals (n = 5 from group 2: line 6–10 in Fig. 1C) (2C, 2F and 2I).
Table 2.
Summary of identified protein spots by Mass Spectrometry.
| Spot no. | Accession no. | Protein name | Official Symbol | Localization | Score | PI | Expected MW | Function |
|---|---|---|---|---|---|---|---|---|
| 1 | gi|378404908 | Glyceraldehyde-3-phosphate dehydrogenase (isoform 2) | GAPDH | Cytosolic | 109 | 8.57 | 31.5 | An important energy-yielding step in carbohydrate metabolism |
| 2 | gi|194374345 | Unnamed protein product | Membrane | 141 | 30.4 | Unknown | ||
| 3 | gi|16924265 | Enoyl Coenzyme A hydratase 1, peroxisomal | ECH1 | Membrane | 277 | 8.47 | 35.7 | Functions in the auxiliary step of the fatty acid β-oxidation pathway |
| 4 | gi|14043072 | Heterogeneous nuclear ribonucleoproteins A2/B1 isoform B1 | HNRNPA2B1 | Nucleus | 169 | 9.97 | 37.4 | Associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport |
PtdIns: isoelectric point; MW: molecular weight.
Potential of autoantibody to ECH1 and HNRNPA2B1 as biomarkers in lung cancer diagnosis: Evaluation in an independent validation set 2
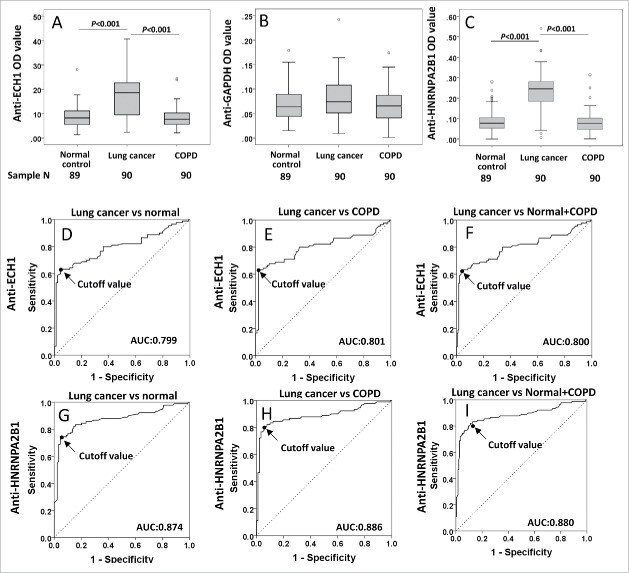
Table 3 and Fig. 3A and B show the individual and average OD values of autoantibodies against GAPDH, ECH1 and HNRNPA2B1 in the three groups. Autoantibodies to ECH1 (Fig. 3A) and HNRNPA2B1 (Fig. 3C) were revealed significant higher OD value in LC (group 3) than that in normal individuals (group 4) and COPD patients (group 5) (p < 0.01), while not for GAPDH (Fig. 3B). To evaluate the value of anti-ECH1 and anti-HNRNPA2B1 autoantibodies in LC diagnosis, we performed ROC analysis further. When the discrimination between the LC patients and normal individuals was performed, the overall accuracy of autoantibodies against ECH1 and HNRNPA2B1 was 78.8% and 83.8% (Table 4), with AUC of 0.799 (Fig. 3D) and 0.874 (Fig. 3G), respectively. The similar results were observed when LC was compared with COPD patients (Fig. 3E and H, Table 4), as well as normal controls combined with COPD patients (Fig. 3F and I, Table 4). Commercial recombinant protein ECH1 and HNRNPA2B1 were further used for Western blotting analysis to verify the sera with positive reactivity in the discovery study (Fig. 1D–F).
Table 3.
Comparison of individual markers in the validation set 2.
| Anti-ECH1 |
Anti-GAPDH |
Anti-HNRNPA2B1 |
||
|---|---|---|---|---|
| Group | N | Median, IQR | Median, IQR | Median, IQR |
| NC | 89 | 0.082, 0.056 | 0.064, 0.047 | 0.077, 0.054 |
| LC | 90 | 0.187, 0.132 | 0.074, 0.057 | 0.245, 0.098 |
| COPD | 90 | 0.077, 0.048 | 0.066, 0.047 | 0.076, 0.056 |
| p value | p value | p value | ||
| Three groups | <0.001a | 0.129a | <0.001a | |
| LC vs. NC | <0.001b | — | <0.001b | |
| LC vs. COPD | <0.001b | — | <0.001b | |
| COPD vs. NC | 0.765b | — | 0.673b |
COPD: Chronic obstructive pulmonary disease; NC: normal control; LC: lung cancer; IQR: inter-quartile range
Kruscal–Wallis test.
Mann–Whitney test.
Figure 3.
Autoantibody levels and receiver operating characteristic (ROC) curves of autoantibody to ECH1, GAPDH and HNRNPA2B1 in validation set 2. 3A–3C: Box and Whisker plots for serum levels of autoantibody to ECH1 (3A), GAPDH (3B) and HNRNPA2B1 (3C) in LC, normal control and COPD patients. The line within the box marks the median, and the 25th and 75th percentiles are presented by the edges of the area, which is known as inter-quartile range (IQR). The bars indicate 1.5 times of the IQR from upper or lower percentiles. 3D–3I: ROC curves of autoantibody to ECH1 (3D–3F) and HNRNPA2B1 (3G–3I). AUC: area under the curve. COPD: chronic obstructive pulmonary disease.
Table 4.
Evaluation of autoantibodies to ECH1 and HNRNPA2B1 in lung cancer diagnosis.
| AUC (95% CI) | p | Sensitivity (%) | Specificity (%) | YI | PPV (%) | NPV (%) | Accuracy (%) | Cutoff | |
|---|---|---|---|---|---|---|---|---|---|
| Anti-ECH1 | |||||||||
| LC vs NC | 0.799 (0.731–0.867) | <0.001 | 62.2 | 95.5 | 0.577 | 93.3 | 71.4 | 78.8 | 0.163 |
| COPD vs NC | 0.482 (0.396–0.567) | 0.672 | — | — | — | — | — | — | — |
| LC vs COPD | 0.801 (0.732–0.870) | <0.001 | 62.2 | 97.8 | 0.600 | 93.3 | 72.1 | 80.0 | 0.163 |
| LC vs NC+COPD | 0.800 (0.735–0.865) | <0.001 | 62.2 | 96.6 | 0.589 | 90.3 | 83.6 | 85.1 | 0.163 |
| Anti-HNRNPA2B1 | |||||||||
| LC vs NC | 0.874 (0.818–0.929) | <0.001 | 72.2 | 95.5 | 0.677 | 81.2 | 77.2 | 83.8 | 0.202 |
| COPD vs NC | 0.479 (0.394–0.564) | 0.627 | — | — | — | — | — | — | — |
| LC vs COPD | 0.886 (0.832–0.940) | <0.001 | 80.0 | 94.4 | 0.744 | 93.5 | 82.5 | 87.2 | 0.144 |
| LC vs NL+COPD | 0.880 (0.828–0.932) | <0.001 | 82.2 | 88.8 | 0.710 | 78.7 | 90.9 | 86.6 | 0.137 |
COPD: Chronic obstructive pulmonary disease; NC: normal control; LC: lung cancer; PPV: positive predictive value; NPV: negative predictive value; YI: Youden's Index
Additionally, we evaluated the correlation of these two autoantibodies with histological and clinical parameters in LC patients (group 3) (Table 5 and Fig. 4A–H). The frequency of anti-ECH1 autoantibody in LC patients is statistically higher than controls who are non-smokers or light smokers (p < 0.001, Table 5), but the frequency does not have relationship with smoking pack years (p = 0.811, Table 5). In addition, the patients with tumor size <3 cm were observed to have significantly higher serum level of autoantibodies against ECH1 (OD median: 0.206) than those with tumor size ≥3 cm (OD median: 0.163) (Fig. 4A), and there is a negative correlation between anti-ECH1 antibody level and tumor size (rs = −0.256, P = 0.023) (Fig. 4E). For the anti-HNRNPA2B1, the patients with stage I have the highest autoantibody level and frequency among all of the stages (p = 0.012 and p = 0.015, Table 5). The patients who have not developed lymph node metastasis showed higher autoantibody level to HNRNPA2B1 than those who have lymph node (OD value: 0.261 vs. 0.213, Fig. 4D), and the negative correlation between autoantibody level and lymph node number was observed (rs = −0.279, p = 0.012, Fig. 4H).
Table 5.
Autoantibodies against ECH1 and HNRNPA2B1 on effect of patients and disease characteristics in validation set 2.
| Anti-ECH1 (cutoff: 0.163) |
Anti-HNRNPA2B1 (cutoff: 0.202) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Median, IQR | p | Frequency (%) | p | Median, IQR | p | Frequency (%) | p | |
| Gender | 0.275a | 0.921b | 0.631a | 0.406b | |||||
| Male | 35 | 0.200, 0.144 | 22 (62.9) | 0.251, 0.103 | 27 (77.1) | ||||
| Female | 55 | 0.180, 0.126 | 33 (60.7) | 0.245, 0.129 | 38 (69.1) | ||||
| Age | 0.341a | 0.586b | 0.891a | 0.537b | |||||
| <65 y | 35 | 0.174, 0.072 | 23 (65.7) | 0.239, 0.078 | 24 (68.6) | ||||
| ≥ 65 y | 55 | 0.204, 0.139 | 33 (60.0) | 0.248, 0.083 | 41 (74.5) | ||||
| Smoke | <0.001a | <0.001b | 0.107a | 0.070b | |||||
| No | 15 | 0.091, 0.075 | 3 (20.0) | 0.201, 0.142 | 8 (53.3) | ||||
| Yes | 69 | 0.197, 0.064 | 53 (76.8) | 0.243, 0.076 | 55 (79.7) | ||||
| <20 pack years | 12 | 0.217, 0.070 | 0.811c | 10 (83.3) | 0.583b | 0.266, 0.121 | 0.924c | 10 (83.3) | 0.921b |
| 20–40 pack years | 30 | 0.200, 0.062 | 24 (80.0) | 0.255, 0.080 | 24 (80.0) | ||||
| >40 pack years | 27 | 0.200, 0.105 | 19 (70.4) | 0.244, 0.076 | 21 (77.8) | ||||
| Stage | 0.411c | 0.407b | 0.012c | 0.015b | |||||
| I | 30 | 0.207, 0.115 | 21 (70.0) | 0.269, 0.064 | 27 (90.0) | ||||
| II | 30 | 0.175, 0.146 | 16 (53.3) | 0.224, 0.142 | 17 (56.7) | ||||
| III | 30 | 0.183, 0.119 | 19 (63.3) | 0.235, 0.099 | 21 (70.0) | ||||
| Histological type | |||||||||
| NSCLC | 0.750a | 1.000b | 0.893a | 1.000b | |||||
| Adenocarcinoma | 81 | 0.184, 0.136 | 50 (61.7) | 0.246, 0.100 | 58 (71.6) | ||||
| Squamous | 6 | 0.207, 0.132 | 4 (66.7) | 0.250, 0.176 | 4 (66.7) | ||||
| LM | 0.134a | 0.293b | 0.013a | 0.006b | |||||
| No | 32 | 0.203, 0.133 | 22 (68.8) | 0.261, 0.076 | 28 (87.5) | ||||
| Yes | 49 | 0.185, 0.139 | 28 (57.1) | 0.213, 0.135 | 29 (59.2) | ||||
LM: lymph node metastasis; NSCLC: non-small cell lung cancer; IQR: inter-quartile range;
Mann–Whitney test;
χ2 test;
Kruscal–Wallis test
Figure 4.
Correlation of autoantibodies to ECH1 and HNRNPA2B1 with tumor size and lymph node number in lung cancer patients in validation set 2 group 3. 4A–4D: Box and Whisker plots for serum levels of anti-ECH1 in different tumor size (4A) and lymph node number (4C), and anti-HNRNPA2B1 levels in different tumor size (4B) and lymph node number (4D). The line within the box marks the median, and the 25th and 75th percentiles are presented by the edges of the area, which is known as inter-quartile (IQR). The bars indicate 1.5 times of the IQR from upper or lower percentiles. Mann–Whitney test was used to compare the difference of antibody levels in different tumor size and lymph node numbers. 4E–4H: Scatter graph for serum levels of anti-ECH1 in tumor size (4E) and lymph node number (4G), and anti-HNRNPA2B1 levels in tumor size (4F) and lymph node number (4H). Spearman's rank-order test was used to evaluate the correlation of autoantibody level with tumor size and lymph node numbers. LM: lymph node metastasis number.
Autoantibodies against ECH1 can be detected in the pre-diagnostic sera of lung cancer patients but not HNRNPA2B1: Evidence from study set 1
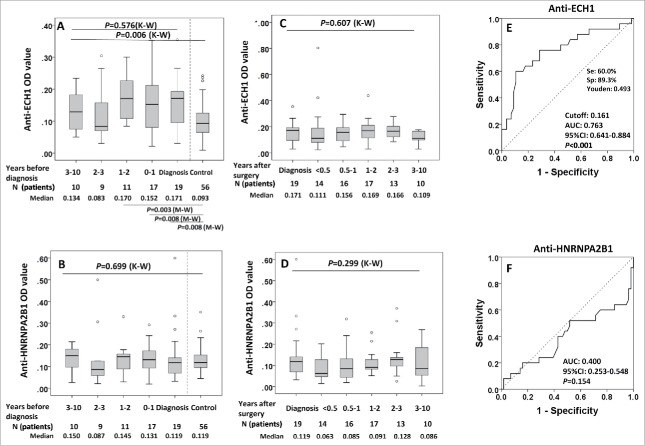
We next questioned whether these two serum autoantibodies can be detected in sera drawn before time of diagnosis, and whether anti-ECH1 and anti-HNRNPA2B1 antibodies might develop over time. Ninety-four pre-diagnostic serum samples drawn at a various range of time (ranging 0.5–120 mo before diagnosis) from 25 patients in group 1 were used to validate these 2 biomarkers discovered. When there was more than one sample in a calendar year in a LC patient, we plotted the average OD for that year. We selected the OD values from the samples with collection date closest to diagnosis for each autoantibody in 25 LC individuals and compared it with the 56 matched-controls. This analysis showed an AUC of 0.763 (95% CI: 0.641–0.884, p < 0.001) with sensitivity of 60.0% and specificity of 89.3% for anti-ECH1 (Fig. 5E), however, AUC for anti-HNRNPA2B1 was only 0.400 with p value of 0.154 (Fig. 5F).
Figure 5.
Serial serum analysis of anti-ECH1 and anti-HNRNPA2B1 levels in discovery set 1 study groups. 5A and 5B: Longitudinal analysis of serum anti-ECH1 and anti-HNRNPA2B1 in LC patients at diagnosis and before diagnosis follow-up. Data are presented in quantile box plots. 5C and 5D: Longitudinal analysis of serum anti-ECH1 and anti-HNRNPA2B1 in LC patients at diagnosis, after diagnosis and surgery treatment follow-up. When there was more than one sample in a calendar year in a LC patient, we plotted the average OD for that year. 5E and 5 F: ROC curve of LC patients (n = 28) versus controls (n = 56) for autoantibody to ECH1 (5E) and HNRNPA2B1 (5F). For the patients with serial serum samples before diagnosis, the OD value form the serum samples with collection date closest to diagnosis was used for generating ROC. K–W: Kruscal–Wallis test for multiple groups; M–W: Mann–Whitney test for two groups.
For both of these two autoantibodies, no significant fluctuation of serum autoantibody occurred during the years before LC diagnosis, as well as post treatment (all of p > 0.05) (Fig. 5A–D), however, the difference was observed among the sera groups drown before diagnosis and 56 sera from matched normal controls for anti-ECH1 (p = 0.006) (Fig. 5A). In the further pairwise analysis, it was found that sera from less than 2 y before diagnosis showed an increased level for anti-ECH1 compared with normal controls (all of p < 0.05) (Fig. 5A).
Discussion
Serum autoantibodies detection has been actively explored as a means to provide novel biomarkers to aid in the early clinical diagnosis of cancer, since autoantibodies are typically produced and secreted in the serum before symptoms manifest and highly stable in the serum samples.2,19 Autoantibody signatures might be useful in cancers in which there are high-risk populations and where existing detection methods lack sensitivity and specificity, particularly in LC.2 To identify specific LC biomarker, an immunoproteomic approach combined with MS was used to identify interesting proteins and found that three 2DE-Western blotting-positive spots corresponded to ECH1, HNRNPA2B1 and GAPDH. ECH1 and HNRNPA2B1 were verified in the subsequent validation study.
ECH1 is a member of the hydratase/isomerase superfamily and functions in the auxiliary step of the fatty acid β-oxidation pathway. A variety of studies demonstrated that ECH1 may be associated with tumor progression: abnormal expression has association with hepatocellular carcinoma secondary to hepatitis C virus infection,20 and the pathogenesis of gastric cancer.21 Additionally, the downregulation of ECH1 has been shown to be associated with the DNA damage-induced apoptosis resistance of B cell chronic lymphoid leukemia.22 A higher expression level of ECH1 was confirmed in tissue from patients with gastric carcinoma with lymph node metastases indicating that ECH1 is a critical factor in the development of lymphatic metastasis in gastric cancer.23 However, no study has yet addressed the relevance anti-ECH1 autoantibody to the development of cancers. HNRNPA2B1, one of the most abundant and important nuclear RNA-binding proteins involved in packaging nascent mRNA, alternative splicing,24,25 cytoplasmic RNA trafficking,26 and translation.27 Numerous of researches indicate that HNRNPA2B1was involved in the tumorigenesis of pancreatic cancer,28,29 prostate cancer,30 hepatocellular carcinoma,31 and LC.32 Higher expression level of HNRNPA2B1 in tissues from patients with gastric adenocarcinoma,33 non-small cell LC34 and hepatocellular carcinoma35 has been reported. The autoimmune responses to HNRNPA2B1 have been described previously in patients with hepatitis,36 but not reported for cancer. This study indicated that both autoantibodies could distinguish LC from normal individuals with the accuracy of 78.8% for anti-ECH1 and 83.8% for anti- HRNPNA2B1, as well as COPD patients with accuracy of 80.0% and 87.2%, respectively. The earlyCDT-lung assay test, a currently available commercial kit, included a panel of six autoantibodies against p53, NY-ESO-1, CAGE, GBU4–5, Annexin 1 and SOX2 was reported to have sensitivity of 38.0% and specificity of 88.0% in LC patients, was also shown to have no significant difference based on LC stages.16 In the present study, the frequency of autoantibody to HNRNPA2B1 is 90% in stage I patients, which is significantly higher than that in stage II (56.7%) and stage III (70.0%). The results indicate that anti-HNRNPA2B1 might be potentially taken as a biomarker to detect the early stage of LC. Although the mechanisms leading to autoantibody production in cancer patients are not completely understood, emerging evidence indicates that most TAAs are cellular proteins whose aberrant regulation of function could be linked to malignancy.8 Further analysis of autoantibodies in the clinical characterizations of LC disclosed that anti-ECH1 level showed negative correlation with tumor size, anti-HRNPNA2B1was inversely correlated with lymph node metastasis. It may be suggested that the presences of these two autoantibodies are associated with aggressiveness of LC.
Advanced cancer may have different molecular characteristics compared with preclinical disease; therefore, it is necessary to determine whether biomarkers discovered from patients with established disease can also be applied to samples from earlier time points before diagnosis, or even at a preclinical stage. In our recent study, we demonstrated that autoantibodies discoverable in sera from established LC may also be detectable in pre-diagnostic sera.37 In this study, we found anti-ECH1 level in the serum samples drawn as early as 2 y before diagnosis increase compared with the matched normal individuals (group 2), along with high sensitivity (60.0%) and specificity (89.3%) with an observed AUC of 0.763 (95%CI: 0.641–0.884), while anti-HNRNPA2B1 has the similar antibody level among serial serum samples from LC patients and normal controls. This suggested that autoantibody to ECH1 may also be used as potential biomarkers to help identify those who may be at risk for developing LC later on in their life. However, more works are needed to further evaluate how to apply this clinically as there is a frequent temporal change in positive reactivity through the pre-diagnostic phase.37
In recent decades, the potential utility of autoantibodies as cancer biomarkers to monitor therapeutic outcomes, or as indicators of cancer prognosis post-therapy, has been explored.38-40 It was reported the increased anti-NPM1 autoantibody level in the sera from prostate cancer patients after surgery treatment.41 Using 125 serial serum samples drawn after treatment from 19 LC patients in this study, autoantibody level to both ECH1 and HNRNPA2B1 have not shown variation in the follow-up samples post treatment. It suggested that there was no association between these two antibodies and clinical outcome, and it might not show value as a potential prognostic biomarker after treatment.
Conclusions
Early detection of LC will allow the patients to receive treatment while still in early stage, even in pre-clinical stage, where it is still possible to prevent the progression to LC. The combinatorial utilization of serum biomarkers with low dose CT examination is one of the promising approaches, which is noninvasive and is of high sensitivity and specificity. This study suggest that ECH1 and HNRNPA2B1 can elicit humoral immune response in LC, and their autoantibodies can be taken as potential biomarkers in the early detection of LC, and even the predictor in the pre-clinical stage of this disease. The subsequent works are needed to furtherly evaluate how to apply these clinically to aid physician to make decision in the early detection of LC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Border Biological Research Center (BBRC) Core Facilities at The University of Texas at El Paso (UTEP) for their support, which were funded by RCMI-NIMHD-NIH grant (8G12MD007592).
Funding
This work was supported by the National Institutes of Health under grant (number: SC1CA166016 and U01CA086137); National Natural Science Foundation of China under grant (number: 81672917, 81372371); and the Major Project of Science and Technology in Henan Province under grant (number: 161100311400).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65:5-29; PMID:25559415; https://doi.org/ 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 2.Field JK, Duffy SW. Lung cancer screening: The way forward. Br J Cancer 2008; 99:557-62; PMID:18665179; https://doi.org/ 10.1038/sj.bjc.6604509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swensen SJ, Jett JR, Hartman TE, Midthun DE, Mandrekar SJ, Hillman SL, Sykes AM, Aughenbaugh GL, Bungum AO, Allen KL. CT screening for lung cancer: Five-year prospective experience. Radiology 2005; 235:259-65; PMID:15695622; https://doi.org/ 10.1148/radiol.2351041662 [DOI] [PubMed] [Google Scholar]
- 4.Sone S, Takashima S, Li F, Yang Z, Honda T, Maruyama Y, Hasegawa M, Yamanda T, Kubo K, Hanamura K et al.. Mass screening for lung cancer with mobile spiral computed tomography scanner. Lancet 1998; 351:1242-5; PMID:9643744; https://doi.org/ 10.1016/S0140-6736(97)08229-9 [DOI] [PubMed] [Google Scholar]
- 5.National Lung Screening Trial Research T, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM et al.. Reduced lung-cancer mortality with low-dose computed tomographic screening. The N Engl J Med 2011; 365:395-409; PMID:21714641; https://doi.org/ 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berrington de Gonzalez A, Mahesh M, Kim KP, Bhargavan M, Lewis R, Mettler F, Land C. Projected cancer risks from computed tomographic scans performed in the United States in 2007. Arch Intern Med 2009; 169:2071-7; PMID:20008689; https://doi.org/ 10.1001/archinternmed.2009.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sone S, Li F, Yang ZG, Honda T, Maruyama Y, Takashima S, Hasegawa M, Kawakami S, Kubo K, Haniuda M et al.. Results of three-year mass screening programme for lung cancer using mobile low-dose spiral computed tomography scanner. Br J Cancer 2001; 84:25-32; PMID:11139308; https://doi.org/ 10.1054/bjoc.2000.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan EM. Autoantibodies as reporters identifying aberrant cellular mechanisms in tumorigenesis. J Clin Invest 2001; 108:1411-5; PMID:11714730; https://doi.org/ 10.1172/JCI14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan EM, Zhang J. Autoantibodies to tumor-associated antigens: Reporters from the immune system. Immunol Rev 2008; 222:328-40; PMID:18364012; https://doi.org/ 10.1111/j.1600-065X.2008.00611.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai L, Lei N, Liu M, Zhang JY. Autoantibodies to tumor-associated antigens as biomarkers in human hepatocellular carcinoma (HCC). Exp Hematol Oncol 2013; 2:15; PMID:23687996; https://doi.org/ 10.1186/2162-3619-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu J, Choi G, Li L, Wang H, Pitteri SJ, Pereira-Faca SR, Krasnoselsky AL, Randolph TW, Omenn GS, Edelstein C et al.. Occurrence of autoantibodies to annexin I, 14-3-3 theta and LAMR1 in prediagnostic lung cancer sera. J Clin Oncol 2008; 26:5060-6; PMID:18794547; https://doi.org/ 10.1200/JCO.2008.16.2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers–blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol 2011; 8:142-50; PMID:21364687; https://doi.org/ 10.1038/nrclinonc.2010.220 [DOI] [PubMed] [Google Scholar]
- 13.Desmetz C, Mange A, Maudelonde T, Solassol J. Autoantibody signatures: Progress and perspectives for early cancer detection. J Cell Mol Med 2011; 15:2013-24; PMID:21651719; https://doi.org/ 10.1111/j.1582-4934.2011.01355.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman CJ, Murray A, McElveen JE, Sahin U, Luxemburger U, Tureci O, Wiewrodt R, Barnes AC, Robertson JF. Autoantibodies in lung cancer: Possibilities for early detection and subsequent cure. Thorax 2008; 63:228-33; PMID:17932110; https://doi.org/ 10.1136/thx.2007.083592 [DOI] [PubMed] [Google Scholar]
- 15.Khattar NH, Coe-Atkinson SP, Stromberg AJ, Jett JR, Hirschowitz EA. Lung cancer-associated auto-antibodies measured using seven amino acid peptides in a diagnostic blood test for lung cancer. Cancer Biol Ther 2010; 10:267-72; PMID:20543565; https://doi.org/ 10.4161/cbt.10.3.12395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam S, Boyle P, Healey GF, Maddison P, Peek L, Murray A, Chapman CJ, Allen J, Wood WC, Sewell HF et al.. EarlyCDT-Lung: An immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res 2011; 4:1126-34; PMID:21733826; https://doi.org/ 10.1158/1940-6207.CAPR-10-0328 [DOI] [PubMed] [Google Scholar]
- 17.Greenberg AK, Lu F, Goldberg JD, Eylers E, Tsay JC, Yie TA, Naidich D, McGuinness G, Pass H, Tchou-Wong KM et al.. CT scan screening for lung cancer: Risk factors for nodules and malignancy in a high-risk urban cohort. PLoS One 2012; 7:e39403; PMID:22768300; https://doi.org/ 10.1371/journal.pone.0039403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rom WN, Goldberg JD, Addrizzo-Harris D, Watson HN, Khilkin M, Greenberg AK, Naidich DP, Crawford B, Eylers E, Liu D et al.. Identification of an autoantibody panel to separate lung cancer from smokers and nonsmokers. BMC Cancer 2010; 10:234; PMID:20504322; https://doi.org/ 10.1186/1471-2407-10-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel K, Farlow EC, Kim AW, Lee BS, Basu S, Coon JS, DeCresce D, Thimothy L, Walters KA, Fhied C et al.. Enhancement of a multianalyte serum biomarker panel to identify lymph node metastases in non-small cell lung cancer with circulating autoantibody biomarkers. Int J Cancer 2011; 129:133-42; PMID:20824709; https://doi.org/ 10.1002/ijc.25644 [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama Y, Kuramitsu Y, Takashima M, Iizuka N, Toda T, Terai S, Sakaida I, Oka M, Nakamura K, Okita K. Proteomic profiling of proteins decreased in hepatocellular carcinoma from patients infected with hepatitis C virus. Proteomics 2004; 4:2111-6; PMID:15221772; https://doi.org/ 10.1002/pmic.200300712 [DOI] [PubMed] [Google Scholar]
- 21.Nishigaki R, Osaki M, Hiratsuka M, Toda T, Murakami K, Jeang KT, Ito H, Inoue T, Oshimura M. Proteomic identification of differentially-expressed genes in human gastric carcinomas. Proteomics 2005; 5:3205-13; PMID:16003825; https://doi.org/ 10.1002/pmic.200401307 [DOI] [PubMed] [Google Scholar]
- 22.Vallat L, Magdelenat H, Merle-Beral H, Masdehors P, Potocki de Montalk G, Davi F, Kruhoffer M, Sabatier L, Orntoft TF, Delic J. The resistance of B-CLL cells to DNA damage-induced apoptosis defined by DNA microarrays. Blood 2003; 101:4598-606; PMID:12586635; https://doi.org/ 10.1182/blood-2002-06-1743 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Song M, Wang J, Sun M, Wang B, Li R, Huang Y, Hou L, Jin Y, Wang M et al.. Enoyl coenzyme A hydratase 1 is an important factor in the lymphatic metastasis of tumors. Biomed Pharmacother 2011; 65:157-62; PMID:21616630; https://doi.org/ 10.1016/j.biopha.2011.02.010 [DOI] [PubMed] [Google Scholar]
- 24.Mayeda A, Munroe SH, Caceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J 1994; 13:5483-95; PMID:7957114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdul-Manan N, Williams KR. hnRNP A1 binds promiscuously to oligoribonucleotides: Utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res 1996; 24:4063-70; PMID:8918813; https://doi.org/ 10.1093/nar/24.20.4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem 1999; 274:34389-95; PMID:10567417; https://doi.org/ 10.1074/jbc.274.48.34389 [DOI] [PubMed] [Google Scholar]
- 27.Hamilton BJ, Nagy E, Malter JS, Arrick BA, Rigby WF. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J Biol Chem 1993; 268:8881-7; PMID:8473331 [PubMed] [Google Scholar]
- 28.Siveke JT. The increasing diversity of KRAS signaling in pancreatic cancer. Gastroenterology 2014; 147:736-9; PMID:25167989; https://doi.org/ 10.1053/j.gastro.2014.08.026 [DOI] [PubMed] [Google Scholar]
- 29.Chen ZY, Cai L, Zhu J, Chen M, Chen J, Li ZH, Liu XD, Wang SG, Bie P, Jiang P et al.. Fyn requires HnRNPA2B1 and Sam68 to synergistically regulate apoptosis in pancreatic cancer. Carcinogenesis 2011; 32:1419-26; PMID:21642356; https://doi.org/ 10.1093/carcin/bgr088 [DOI] [PubMed] [Google Scholar]
- 30.Stockley J, Villasevil ME, Nixon C, Ahmad I, Leung HY, Rajan P. The RNA-binding protein hnRNPA2 regulates beta-catenin protein expression and is overexpressed in prostate cancer. RNA Biol 2014; 11:755-65; PMID:24823909; https://doi.org/ 10.4161/rna.28800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shilo A, Ben Hur V, Denichenko P, Stein I, Pikarsky E, Rauch J, Kolch W, Zender L, Karni R. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA 2014; 20:505-15; PMID:24572810; https://doi.org/ 10.1261/rna.042259.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tauler J, Zudaire E, Liu H, Shih J, Mulshine JL. hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res 2010; 70:7137-47; PMID:20807810; https://doi.org/ 10.1158/0008-5472.CAN-10-0860 [DOI] [PubMed] [Google Scholar]
- 33.Jing GJ, Xu DH, Shi SL, Li QF, Wang SY, Wu FY, Kong HY. Aberrant expression and localization of hnRNP-A2/B1 is a common event in human gastric adenocarcinoma. J Gastroenterol Hepatol 2011; 26:108-15; PMID:21175803; https://doi.org/ 10.1111/j.1440-1746.2010.06482.x [DOI] [PubMed] [Google Scholar]
- 34.Boukakis G, Patrinou-Georgoula M, Lekarakou M, Valavanis C, Guialis A. Deregulated expression of hnRNP A/B proteins in human non-small cell lung cancer: Parallel assessment of protein and mRNA levels in paired tumour/non-tumour tissues. BMC Cancer 2010; 10:434; PMID:20716340; https://doi.org/ 10.1186/1471-2407-10-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui H, Wu F, Sun Y, Fan G, Wang Q. Up-regulation and subcellular localization of hnRNP A2/B1 in the development of hepatocellular carcinoma. BMC Cancer 2010; 10:356; PMID:20604928; https://doi.org/ 10.1186/1471-2407-10-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huguet S, Labas V, Duclos-Vallee JC, Bruneel A, Vinh J, Samuel D, Johanet C, Ballot E. Heterogeneous nuclear ribonucleoprotein A2/B1 identified as an autoantigen in autoimmune hepatitis by proteome analysis. Proteomics 2004; 4:1341-5; PMID:15188401; https://doi.org/ 10.1002/pmic.200300757 [DOI] [PubMed] [Google Scholar]
- 37.Dai L, Tsay JC, Li J, Yie TA, Munger JS, Pass H, Rom WN, Zhang Y, Tan EM, Zhang JY. Autoantibodies against tumor-associated antigens in the early detection of lung cancer. Lung Cancer 2016; 99:172-9; PMID:27565936; https://doi.org/ 10.1016/j.lungcan.2016.07.018 [DOI] [PubMed] [Google Scholar]
- 38.Vase MO, Friis S, Bautz A, Bendix K, Sorensen HT, d'Amore F. Breast implants and anaplastic large-cell lymphoma: A danish population-based cohort study. Cancer Epidemiol Biomarkers Prev 2013; 22:2126-9; PMID:23956025; https://doi.org/ 10.1158/1055-9965.EPI-13-0633 [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Werner S, Tao S, Zornig I, Brenner H. Blood autoantibodies against tumor-associated antigens as biomarkers in early detection of colorectal cancer. Cancer Lett 2014; 346:178-87; PMID:24462820; https://doi.org/ 10.1016/j.canlet.2014.01.007 [DOI] [PubMed] [Google Scholar]
- 40.Werner S, Chen H, Tao S, Brenner H. Systematic review: Serum autoantibodies in the early detection of gastric cancer. Int J Cancer 2015; 136:2243-52; PMID:24615018; https://doi.org/ 10.1002/ijc.28807 [DOI] [PubMed] [Google Scholar]
- 41.Dai L, Li J, Xing M, Sanchez TW, Casiano CA, Zhang JY. Using serological proteome analysis to identify serum anti-nucleophosmin 1 autoantibody as a potential biomarker in European-American and African-American patients with prostate cancer. Prostate 2016; 76:1375-86; PMID:27418398; https://doi.org/ 10.1002/pros.23217 [DOI] [PubMed] [Google Scholar]