Abstract
Uveal melanoma (UM) is the most common primary intraocular malignant tumor in adults and arises from the transformation of melanocytes in the uveal tract. Even after treatment of the primary tumor, up to 50% of patients succumb to metastatic disease. The liver is the predominant organ of metastasis. There is an important need to provide effective treatment options for advanced stage UM. In order to provide the preclinical basis for new treatments, it is important to understand the molecular underpinnings of the disease. Recent genomic studies have shown that mutations within components of G protein-coupled receptor (GPCR) signaling are early events associated with ~98% of UMs.
Implications
This review discusses the alterations in GPCR signaling components (GNAQ and GNA11), dysregulated GPCR signaling cascades, and viable targeted therapies with the intent to provide insight into new therapeutic strategies in UM.
Introduction
UM is a rare cancer with ~2,500 new patient diagnoses being reported per year in the U.S. Primary UM tumors are treated effectively by radiation plaque therapy or enucleation; however, approximately 50% of patients develop metastatic disease, frequently in the liver (1, 2). In some cases, metastases are found decades after successful treatment of the primary tumors and one likely explanation is dissemination of tumor cells from the primary site followed by cellular dormancy (3). Metastatic UM responds poorly to clinically available therapies and patients often succumb within 1 year of diagnosis of the metastases; hence, there is an urgent unmet need for effective therapeutic strategies for advanced UM (4). There is a low mutational burden in UM tumors, unlike cutaneous melanoma, but identification of mutations in components of GPCR signaling in UM tumors may uncover new therapeutic targets in UM. These components include GNAQ, GNA11, PLCB4 and CYSLTR2, mutations of which are found in ~98% of UM. Here, we review the molecular alterations in GPCR pathway components and discuss the therapeutic possibilities directed at targeting GPCR signaling.
GNAQ and GNA11 mutations
GNAQ and GNA11 encode the alpha subunits of guanine nucleotide-binding proteins (G proteins), Gαq and Gα11, respectively. They form a heterotrimeric complex with β and γ subunits and are important intermediates between membrane-bound GPCRs and intracellular signaling cascades. G proteins are normally inactive when bound by guanosine diphosphate (GDP) but agonist activation of GPCRs triggers recruitment of the G proteins to the receptors where a switch from GDP to guanosine triphosphate (GTP) occurs, rendering the G proteins active to bind/stimulate proteins associated with downstream pathways. Normally, the GTPase activity of G proteins hydrolyzes GTP to GDP (5), an inactivation process that is catalyzed by the regulator of G protein signaling (RGS) proteins. GNAQ and GNA11 mutations occur in a mutually exclusive manner in ~93% of UM tumors (The Cancer Genome Atlas (TCGA): GNAQ and GNA11 mutations detected in ~50% and ~43% of UM tumors, respectively) (6). They are almost exclusively found in exon 5 codon 209 (Q209) although mutations in exon 4 codon 183 (R183) have been determined in a minority of cases (7–9). Common substitutions in GNAQ are glutamine-to-leucine (Q209L) and glutamine-to-proline (Q209P) whereas in GNA11, the most frequent substitution is Q209L. Q209 is crucial for the GTPase activity of G proteins; thus, hydrolysis of GTP is abolished in GNAQ and GNA11 mutants, leading to constitutive activation of the Gαq and Gα11 proteins in UM. Interestingly, the levels of Gαq Q209L mutant proteins may be regulated by Ric-8A, a molecular chaperone that contributes to folding of the G protein (10). Deletion of Ric-8A in GNAQ Q209L mutant melanocytes grafted into NSG mice led to a marked reduction in levels of membrane-associated mutant Gαq proteins and inhibition of GNAQ Q209L driven tumor progression.
GNAQ and GNA11 are the most frequently mutated genes in UM; however, their mutations do not correlate with UM patient outcome, patient survival or factors that would indicate high risk of metastasis (9, 11). GNAQ and GNA11 mutations occur at similar frequencies in metastasizing and non-metastasizing tumors. Similarly, these mutations are not associated with class 1 (low metastatic potential) or class 2 (high metastatic potential) of UM tumors (11, 12). It has been shown that the Q209 mutation in GNAQ and GNA11 are found in benign nevi such as blue nevi in addition to primary and metastatic UM tumors (7, 8). Particularly, GNAQ is frequently mutated in blue nevi (~83%). This suggests that mutations in the G proteins are early events in UM development. Despite this notion, the GNA11 Q209 mutation is more commonly identified in UM metastases (~57%) and found only in ~7% of benign blue nevi, indicating that in comparison to GNAQ, alteration in GNA11 is associated with higher risk of metastasis of UM (7, 8, 13).
Effector and receptor mutations: PLCB4 and CYSLTR2
In addition to the GNAQ and GNA11 mutations, other, less common, driver mutations in UM have been identified recently by next-generation sequencing. Mutations in phospholipase C β4 (PLCB4) and cysteinyl leukotriene receptor 2 (CYSLTR2) have been identified in 1% and 4 UM tumors, respectively (14, 15) and are mutually exclusive with GNAQ and GNA11 mutations. The alteration in PLCβ4, D630Y, affects the Y-domain of the highly conserved catalytic core of PLCβ4 which controls signal transduction (14). Consistent with this finding, PLCβ4 interacts directly with Gαq (16) (Fig 1). PLCβ4 mediates signal transduction by catalyzing the conversion of phosphatidylinositol 4,5-bisphosphate (PIP2) in the plasma membrane into inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). Subsequently DAG and IP3 activate downstream signaling components such as protein kinase C (PKC). IP3, particularly, translocates into the cytosol where it induces the release of calcium (Ca2+) from the endoplasmic reticulum to activate PKC.
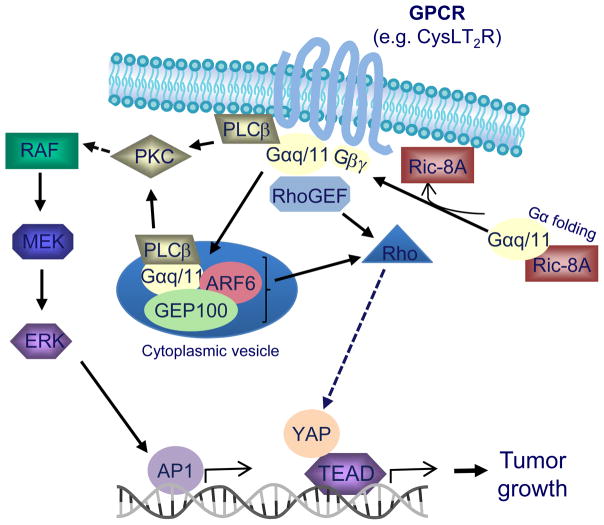
Fig 1.
Alterations in GPCR signaling in UM. GPCRs (e.g. CysLT2R) activate the Gα subunit proteins, Gαq and Gα11, which stimulate downstream effector pathways including ERK1/2 and YAP. Ric-8A chaperones participate in folding of G proteins which is important for membrane localization of the G proteins. ARF6 promotes localization of Gαq/11 to cytoplasmic vesicles where oncogenic Gαq/11 signaling can be active. In UM, mutations in GNAQ, GNA11, CYSLTR2 or PLCB4 lead to constitutive activation of pathways downstream of Gαq and Gα11 proteins.
The other alteration in the GPCR pathway in UM is at the level of GPCRs. Out of 136 UM patient specimens analyzed by Moore and colleagues, 4 samples harbored a leucine to glutamine substitution at codon 129 (Leu129Gln) in the CYSLTR2 gene, and all four samples lacked mutations in GNAQ, GNA11 or PLCB4 (15). CYSLTR2 encodes a seven transmembrane GPCR that activates Gαq, a finding that is consistent with the mutual exclusive profile of the mutations (17) (Fig 1). Activation of Gαq by CysLT2R promotes binding of Gαq/Gα11 to PLCβ4. The Leu129Gln mutation is located in the third transmembrane helix of the receptor and promotes ligand independent activation of the GPCR. Expression of Leu129Gln CYSLTR2 in HEK293 cells increased basal levels of calcium and promoted the growth of melanocyte cell lines in vitro and in vivo (15). Collectively, these findings raise the possibility of targeting mutant forms of GNAQ/GNA11, PLCB4 and CYSLTR2 in UMs as well as pathways downstream of mutant GNAQ/GNA11.
Dysregulated pathways downstream of mutant GNAQ/GNA11
Since the identification of mutations in GPCR signaling components in a high proportion of UM tumors, molecular understanding of pathways downstream of Gαq and Gα11 has become crucial for development or discovery of effective treatment options for metastatic UM. These pathways include the ERK1/2, Rho/Rac/YAP and PI3K/AKT pathways.
ERK1/2 Pathway
The ERK1/2 pathway usually involves binding of ligands to tyrosine kinase receptors on the cell membrane and activation of downstream intermediates, RAS, RAF and MEK. MEK phosphorylates and activates ERK1/2, which subsequently undergoes dimerization and translocates into the nucleus to regulate cellular processes including proliferation, survival, differentiation and apoptosis (18). The ERK1/2 pathway is frequently activated in UM with 86% of primary UM tumors reported to have elevated ERK1/2 phosphorylation (19). However, unlike cutaneous melanoma, in which BRAF is commonly mutated, mutations in RAS and BRAF are rare in UM tumors (20). Only one patient with choroidal melanoma has been shown to harbor the BRAF V600E mutation (21). MEK-ERK1/2 activation in UM is expected to be induced rather by mutant Gαq/Gα11 proteins. The G proteins activate PLCβ and PKC, which then stimulates MEK/ERK1/2 (18) (Fig 1). Transfection of GNAQ Q209L in human melanocytes enhanced phosphorylated ERK1/2 protein levels and was associated with increased anchorage-independent growth (7). Consistently, these results were reversed by siRNA-mediated knockdown of GNAQ which decreased phospho-ERK1/2 levels (7). However, it is noteworthy that in some UM cases, ERK1/2 may not be activated by G proteins as a study of 22 UM patient tumors did not observe a correlation between GNAQ mutation and ERK1/2 activation although samples with GNAQ mutations had a higher average of the total ERK1/2 expression level compared to GNAQ wild-type tumors (22).
Rho/Rac/YAP Pathway
A major role of Gαq/11 is to directly bind and activate PLCβ, but additional effectors and signaling pathways may also play an important role. Particularly relevant to UM, Gαq/11 stimulates Rho and Rac small GTPase-mediated signaling through direct binding and activation of several members of the large Rho guanine-nucleotide exchange factor (RhoGEF) family, including p63RhoGEF and Trio (23, 24) (Fig 1). Trio appears to be a key player in mediating mitogenic signals in UM since knockdown of Trio inhibited tumor growth and DNA synthesis in two UM cell line models (25). Interestingly, Trio is a very large multi-domain protein that has two GEF domains, one that selectively activates RhoA and one that activates Rac1/RhoG. Notably, activation of both RhoA and Rac1 by Trio are required for Gαq/11 mitogenic signaling (25, 26).
Multiple pathways downstream of Rho/Rac are likely to mediate Trio-dependent cell proliferation in mutant Gαq/11 UM cells. Depletion of Trio by siRNA did not inhibit ERK1/2 activation, but instead blocked activation of the MAPKs JNK and p38; JNK and p38 can both regulate the transcription factor AP-1 which controls the transcription of a number of growth-promoting genes (25). In addition, two reports strongly implicate two transcriptional co-activators in the Hippo pathway, Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ), as being critical mediators of oncogenic Gαq/11 downstream of Rho/Rac (27, 28). These reports showed that mutationally activated Gαq/11 induced the cytoplasm to nucleus translocation of YAP/TAZ and transcription of several YAP/TAZ-dependent genes that promote cell proliferation (27, 28). Moreover, siRNA depletion of YAP or inhibition of YAP with the pharmacological inhibitor verteporfin suppressed the growth of UM cells in culture and in a mouse model (27, 28). Robust nuclear localization of YAP was also observed in uveal melanocytes in a recently described zebrafish model for UM (29). The mechanisms underlying Rho and Rac activation of YAP appear to involve both inhibition of upstream kinases, large tumor suppressor homolog 1 and 2 (LATS1/2), as well as increased sequestration to F-actin of the YAP-binding protein angiomotin (AMOT), resulting in both cases in increased levels of YAP available to translocate to the nucleus (27, 28, 30). The exciting implication of the above studies is that the Trio/Rho/Rac/YAP pathway of oncogenic Gαq/11 signaling suggests numerous new therapeutic targets in UM.
ADP-ribosylation factor 6 (ARF6)
Another monomeric small GTPase, ARF6, has been proposed as a key mediator of most pathways activated by oncogenic Gαq/11 (31). Inhibition or depletion of ARF6 in UM cells inhibited cell proliferation and the downstream signaling targets PLCβ, ERK1/2, Rho, Rac and YAP. Activation of ARF6 by Gαq/11 appears to be mediated by direct binding of Gαq/11 to the ARF-GEF, GEP100. A provocative mechanism for the role of ARF6 in controlling Gαq/11 signaling was proposed, in which ARF6 promoted localization of mutationally activated Gαq/11 to cytoplasmic vesicles, rather than the plasma membrane, and these cytoplasmic vesicles are the site of oncogenic signaling by Gαq/11 (31) (Fig 1). Regardless of the exact mechanism of how ARF6 regulates Gαq/11 signaling, ARF6 provides another potential therapeutic target in UM.
PI3K/AKT Pathway
Phosphatidylinositol (4,5)-bisphosphate 3-kinase (PI3K) catalyzes the formation of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) from PIP2, and PIP3 induces membrane translocation of AKT where it becomes active and signals to promote cell proliferation and survival (18). PI3K signaling is negatively regulated by phosphatase and tensin homolog (PTEN), which reverses PIP2 conversion to PIP3. Complete loss of PTEN is identified in 16% of UM tumors (n=75) and weaker immunostaining of PTEN is found in 42.7% of UM tumors (32). Patients with PTEN-null tumors are associated with a shorter disease-free survival compared to patients with UM tumors expressing normal levels of PTEN (32).
Targeted therapies in UM
Patients with metastatic UM typically die within one year of diagnosis; hence, there is an urgent unmet need to identify novel approaches for treatment which includes targeted therapies as either monotherapy or in combinational strategies.
GPCR Inhibitors
Since activating mutations in either CYSLTR2, GNAQ, GNA11 or PLCB4 are found in ~98% of UMs, this signaling pathway represents a viable therapeutic target for the treatment of UM. One approach to targeting this pathway is the development of specific inhibitors to individual proteins in the pathway. For example, constitutively active GPCRs such as the CysLT2R-L129Q mutant found in UM (15) could potentially be targeted using a receptor specific inverse agonist, which would bind to and stabilize an inactive conformation of the receptor that should no longer activate Gαq/11. Unfortunately, CysLT2R-L129Q is only a viable target in ~4% of UM patients. A more compelling target is the activated G protein, either Gαq or Gα11, which together are mutated in ~93% of UM patients. In this regard, there are two potent and specific inhibitors that have been reported for Gαq, YM-254890 and FR900359 or UBO-QIC. YM-254890 is a cyclic depsipeptide isolated from Chromobacerium sp. broth and showed inhibition of Gαq signaling mediated processes such as ADP-induced platelet aggregation and Ca2+ mobilization through a Gαq-coupled receptor (33). A crystal structure of the Gαq protein, loaded with GDP, in complex with YM-254890 suggests the inhibitor works by allosterically stabilizing the protein in the GDP bound form, preventing GDP dissociation, and locking it in the inactive state (34). More recently, FR900359, a slightly different cyclic depsipeptide isolated from the Ardisia crenatasims plant, has shown similar pharmacological activity and specificity against Gαq (35). Docking models and molecular dynamics studies suggest FR900359 inhibits Gαq in an identical manner to YM-254890 (36). While these compounds appear to be potent and selective inhibitors of Gαq (they have not been tested on Gα11), it’s unclear whether they would be able to inhibit the constitutively active forms of Gαq typically found in UM. For example, while YM-254890 inhibited a Gαq-R183C mutant, it was unable to inhibit Gαq-Q209L (33). Moreover, one also needs to consider Gαq vs Gα11 selectivity when inhibiting these proteins since a non-selective Gαq/11 inhibitor would likely be toxic, as knockout of both Gαq and Gα11 in mice leads to death in utero (37). Finally, one could consider targeting PLCβ4 with a selective inhibitor since this is mutated in ~1% of UM and is a downstream effector of Gαq/11. However, PLCβ4 is unlikely to be the only downstream target of activated Gαq/11 in UM so such an inhibitor might only prove useful in the small number of patients that have a PLCβ4 mutation.
MEK Inhibitors
Due to the high incidence of MEK-ERK1/2 activation in UM, targeting of components of the Gαq/Gα11-induced ERK1/2 cascade such as MEK and PKC has been investigated as a therapeutic approach in UM. Inhibition of MEK1/2 either by trametinib, selumetinib or PD0325901, induced growth arrest and apoptosis in GNAQ/GNA11 mutant UM cell lines and xenograft tumors (38–41). Trametinib is FDA-approved and used in combination with the BRAF inhibitor, dabrafenib for patients with unresectable or metastatic melanoma harboring BRAF V600E and V600K mutations (42). However, reports from clinical studies indicated variable outcomes for MEK inhibition in UM. In a phase 1 trial involving 16 metastatic UM patients, trametinib was shown to have limited efficacy (43). A phase II trial which enrolled 120 advanced UM patients showed that selumetinib improved the median progression-free survival (PFS) by ~9 weeks compared to chemotherapy (dacarbazine or temozolomide) but only modestly increased median overall survival (44). In the most recent phase III clinical trial (SUMIT), the combination of selumetinib and dacarbazine failed to improve PFS compared to chemotherapy alone and was associated with a low response rate (3–4%) (45). Thus, while pre-clinical studies support MEK inhibitors as part of a therapeutic approach for metastatic UM, it is important to identify agents that in combination enhance the responses of UM to MEK inhibition.
The poor clinical response of metastatic UM to MEK inhibitors may be in part due to factors produced by the tumor microenvironment. Hepatocyte growth factor (HGF) provided resistance to trametinib growth inhibitory effects in metastatic UM cell lines and targeting of cMET (the receptor for HGF) enhanced the effects of trametinib in metastatic UM (40, 46). HGF-CMET signaling induced downregulation of BH3 pro-apoptotic proteins, BIM and BMF, which correlated with HGF-mediated inhibition of apoptosis in trametinib-treated cultures (46). Importantly, HGF is secreted by quiescent hepatic stellate cells that can be found in the liver microenvironment and phosphorylated/activated cMET is detected in the majority of UM liver metastases (40, 46). HGF promotes activation of the PI3K/AKT pathway as a mechanism of resistance to trametinib and the increase in phosphorylation of AKT is mediated by PI3K isoforms α, δ and γ. Use of a β-sparing PI3K inhibitor, GDC0032, reversed HGF-induced resistance to trametinib (46). These findings support an earlier study which reported the combination of MEK and PI3K inhibitors for UM and these agents induce marked early apoptosis in GNAQ mutant UM cell lines compared to the inhibitors alone which have moderate effects on apoptosis (40, 41).
MEK inhibitors have also been tested in combination with PKC inhibitors for UM. Inhibition of PKC alone such as by AEB071 or sotrastaurin induced cell cycle G1 phase arrest and suppressed PKC and ERK1/2 signaling (39). However, these effects were not durable, whereas in combination with MEK inhibitors, PD0325901 or MEK162, sustained inhibition of the ERK1/2 pathway was observed (39). Furthermore, the inhibitors caused a strong synergistic growth inhibitory effect on UM cell lines and marked tumor regression in UM xenograft models. Recently, screening of drug combinations involving AEB701 in a large panel of UM patient-derived xenografts (PDX) also determined two non MEK inhibitors that could potentially be used in combination with PKC inhibitors as therapeutic strategies for metastatic UM (47). These were the p53-MDM2 inhibitor, CGM097 and the mTORC1 inhibitor, RAD001. AEB701 and CGM097 or AEB701 and RAD001 markedly reduced tumor growth and induced tumor regression (47). These studies provide evidence for a number of targeted therapies that may be evaluated in combination with MEK inhibitors or PKC inhibitors in clinical studies to improve the survival of patients with metastatic UM.
Conclusions
There is a lack of effective treatment options for advanced UM despite success in the treatment of primary UM tumors. Whilst identification of driver mutations in UM such as GNAQ and GNA11 has shed light to potential therapeutic targets, alterations in GPCR signaling occur early in disease progression. Other mutations and their impact on responses of UM to GPCR targeted therapies will need to be explored (48). For example, inactivating mutations of BAP1 have been found in ~80% of aggressive UM (49) while SF3B1 and EIF1AX were shown to correlate with a favorable prognosis (50). In addition, ongoing investigations will be needed to devise strategies that elicit favorable treatment outcomes using therapies targeting GPCR signaling. As in other forms of advanced-stage melanoma, it is likely that combination approaches will be key to elicit high response rates and the most effective combinations and scheduling of inhibitors will need to be identified.
Acknowledgments
Financial Support: Uveal melanoma research in the Aplin, Benovic and Wedegaertner laboratories is collaboratively funded by a Dr. Ralph and Marian Falk Medical Research Trust Catalyst Award. Additionally, uveal melanoma research in the Aplin lab is supported by a Melanoma Research Alliance team science award, a Cure Ocular Melanoma/Melanoma Research Foundation Established Investigator award and a NIH R01 (CA182635). The Cancer Cell Biology and Signaling (CCBS) program is supported by the Cancer Center Support Grant 5P30CA056036-17. The Wedegaertner lab is also funded by NIH/NCI R03 CA202316-01 and a diversity supplement for NIH R01 GM56444-17 while the Benovic lab is also supported by NIH P01 HL114471-04.
Footnotes
Conflict of Interest: Dr. Andrew Aplin discloses a grant from Pfizer Inc. No other conflicts of interest were disclosed.
References
- 1.Group COMS. Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the Collaborative Ocular Melanoma Study (COMS): COMS report no. 15. Arch Ophthalmol. 2001;119(5):670–6. doi: 10.1001/archopht.119.5.670. [DOI] [PubMed] [Google Scholar]
- 2.Shields CL, Shields JA. Ocular melanoma: relatively rare but requiring respect. Clin Dermatol. 2009;27(1):122–33. doi: 10.1016/j.clindermatol.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Ossowski L, Aguirre-Ghiso JA. Dormancy of metastatic melanoma. Pigment Cell Melanoma Res. 2010;23(1):41–56. doi: 10.1111/j.1755-148X.2009.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diener-West M, Reynolds SM, Agugliaro DJ, Caldwell R, Cumming K, Earle JD, et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch Ophthalmol. 2005;123(12):1639–43. doi: 10.1001/archopht.123.12.1639. [DOI] [PubMed] [Google Scholar]
- 5.Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a G alpha subunit. Science. 1993;262(5141):1895–901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- 6.Decatur CL, Ong E, Garg N, Anbunathan H, Bowcock AM, Field MG, et al. Driver Mutations in Uveal Melanoma: Associations With Gene Expression Profile and Patient Outcomes. JAMA Ophthalmol. 2016;134(7):728–33. doi: 10.1001/jamaophthalmol.2016.0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457(7229):599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koopmans AE, Vaarwater J, Paridaens D, Naus NC, Kilic E, de Klein A. Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br J Cancer. 2013;109(2):493–6. doi: 10.1038/bjc.2013.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel BR, Tall GG. Ric-8A gene deletion or phorbol ester suppresses tumorigenesis in a mouse model of GNAQ(Q209L)-driven melanoma. Oncogenesis. 2016;5(6):e236. doi: 10.1038/oncsis.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onken MD, Worley LA, Long MD, Duan S, Council ML, Bowcock AM, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49(12):5230–4. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64(20):7205–9. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griewank KG, van de Nes J, Schilling B, Moll I, Sucker A, Kakavand H, et al. Genetic and clinico-pathologic analysis of metastatic uveal melanoma. Mod Pathol. 2014;27(2):175–83. doi: 10.1038/modpathol.2013.138. [DOI] [PubMed] [Google Scholar]
- 14.Johansson P, Aoude LG, Wadt K, Glasson WJ, Warrier SK, Hewitt AW, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7(4):4624–31. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore AR, Ceraudo E, Sher JJ, Guan Y, Shoushtari AN, Chang MT, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48(6):675–80. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyon AM, Tesmer JJ. Structural insights into phospholipase C-beta function. Mol Pharmacol. 2013;84(4):488–500. doi: 10.1124/mol.113.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans JF. Cysteinyl leukotriene receptors. Prostaglandins Other Lipid Mediat. 2002;68–69:587–97. doi: 10.1016/s0090-6980(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 18.Patel M, Smyth E, Chapman PB, Wolchok JD, Schwartz GK, Abramson DH, et al. Therapeutic implications of the emerging molecular biology of uveal melanoma. Clin Cancer Res. 2011;17(8):2087–100. doi: 10.1158/1078-0432.CCR-10-3169. [DOI] [PubMed] [Google Scholar]
- 19.Weber A, Hengge UR, Urbanik D, Markwart A, Mirmohammadsaegh A, Reichel MB, et al. Absence of mutations of the BRAF gene and constitutive activation of extracellular-regulated kinase in malignant melanomas of the uvea. Lab Invest. 2003;83(12):1771–6. doi: 10.1097/01.lab.0000101732.89463.29. [DOI] [PubMed] [Google Scholar]
- 20.Zuidervaart W, van Nieuwpoort F, Stark M, Dijkman R, Packer L, Borgstein AM, et al. Activation of the MAPK pathway is a common event in uveal melanomas although it rarely occurs through mutation of BRAF or RAS. Br J Cancer. 2005;92(11):2032–8. doi: 10.1038/sj.bjc.6602598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malaponte G, Libra M, Gangemi P, Bevelacqua V, Mangano K, D’Amico F, et al. Detection of BRAF gene mutation in primary choroidal melanoma tissue. Cancer Biol Ther. 2006;5(2):225–7. doi: 10.4161/cbt.5.2.2429. [DOI] [PubMed] [Google Scholar]
- 22.Populo H, Vinagre J, Lopes JM, Soares P. Analysis of GNAQ mutations, proliferation and MAPK pathway activation in uveal melanomas. Br J Ophthalmol. 2011;95(5):715–9. doi: 10.1136/bjo.2009.174417. [DOI] [PubMed] [Google Scholar]
- 23.Lutz S, Freichel-Blomquist A, Yang Y, Rumenapp U, Jakobs KH, Schmidt M, et al. The guanine nucleotide exchange factor p63RhoGEF, a specific link between Gq/11-coupled receptor signaling and RhoA. J Biol Chem. 2005;280(12):11134–9. doi: 10.1074/jbc.M411322200. [DOI] [PubMed] [Google Scholar]
- 24.Rojas RJ, Yohe ME, Gershburg S, Kawano T, Kozasa T, Sondek J. Galphaq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J Biol Chem. 2007;282(40):29201–10. doi: 10.1074/jbc.M703458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaque JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, et al. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell. 2013;49(1):94–108. doi: 10.1016/j.molcel.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt S, Debant A. Function and regulation of the Rho guanine nucleotide exchange factor Trio. Small GTPases. 2014;5:e29769. doi: 10.4161/sgtp.29769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25(6):831–45. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25(6):822–30. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mouti MA, Dee C, Coupland SE, Hurlstone AF. Minimal contribution of ERK1/2-MAPK signalling towards the maintenance of oncogenic GNAQQ209P-driven uveal melanomas in zebrafish. Oncotarget. 2016;7(26):39654–70. doi: 10.18632/oncotarget.9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15(2):73–9. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoo JH, Shi DS, Grossmann AH, Sorensen LK, Tong Z, Mleynek TM, et al. ARF6 Is an Actionable Node that Orchestrates Oncogenic GNAQ Signaling in Uveal Melanoma. Cancer Cell. 2016;29(6):889–904. doi: 10.1016/j.ccell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdel-Rahman MH, Yang Y, Zhou XP, Craig EL, Davidorf FH, Eng C. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J Clin Oncol. 2006;24(2):288–95. doi: 10.1200/JCO.2005.02.2418. [DOI] [PubMed] [Google Scholar]
- 33.Takasaki J, Saito T, Taniguchi M, Kawasaki T, Moritani Y, Hayashi K, et al. A novel Galphaq/11-selective inhibitor. J Biol Chem. 2004;279(46):47438–45. doi: 10.1074/jbc.M408846200. [DOI] [PubMed] [Google Scholar]
- 34.Nishimura A, Kitano K, Takasaki J, Taniguchi M, Mizuno N, Tago K, et al. Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proc Natl Acad Sci U S A. 2010;107(31):13666–71. doi: 10.1073/pnas.1003553107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaima K, Deguchi J, Matsuno Y, Kaneda T, Hirasawa Y, Morita H. Vasorelaxant effect of FR900359 from Ardisia crenata on rat aortic artery. J Nat Med. 2013;67(1):196–201. doi: 10.1007/s11418-012-0644-0. [DOI] [PubMed] [Google Scholar]
- 36.Schrage R, Schmitz AL, Gaffal E, Annala S, Kehraus S, Wenzel D, et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat Commun. 2015;6:10156. doi: 10.1038/ncomms10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Offermanns S, Zhao LP, Gohla A, Sarosi I, Simon MI, Wilkie TM. Embryonic cardiomyocyte hypoplasia and craniofacial defects in G alpha q/G alpha 11-mutant mice. EMBO J. 1998;17(15):4304–12. doi: 10.1093/emboj/17.15.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambrosini G, Musi E, Ho AL, de Stanchina E, Schwartz GK. Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK-dependent autophagic cell death. Mol Cancer Ther. 2013;12(5):768–76. doi: 10.1158/1535-7163.MCT-12-1020. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Wu Q, Tan L, Porter D, Jager MJ, Emery C, et al. Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene. 2014;33(39):4724–34. doi: 10.1038/onc.2013.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng H, Terai M, Kageyama K, Ozaki S, McCue PA, Sato T, et al. Paracrine Effect of NRG1 and HGF Drives Resistance to MEK Inhibitors in Metastatic Uveal Melanoma. Cancer Res. 2015;75(13):2737–48. doi: 10.1158/0008-5472.CAN-15-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khalili JS, Yu X, Wang J, Hayes BC, Davies MA, Lizee G, et al. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner. Clin Cancer Res. 2012;18(16):4345–55. doi: 10.1158/1078-0432.CCR-11-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falchook GS, Lewis KD, Infante JR, Gordon MS, Vogelzang NJ, DeMarini DJ, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13(8):782–9. doi: 10.1016/S1470-2045(12)70269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carvajal RD, Sosman JA, Quevedo JF, Milhem MM, Joshua AM, Kudchadkar RR, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. JAMA. 2014;311(23):2397–405. doi: 10.1001/jama.2014.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvajal RD, Schwartz GK, Mann H, Smith I, Nathan PD. Study design and rationale for a randomised, placebo-controlled, double-blind study to assess the efficacy of selumetinib (AZD6244; ARRY-142886) in combination with dacarbazine in patients with metastatic uveal melanoma (SUMIT) BMC Cancer. 2015;15:467. doi: 10.1186/s12885-015-1470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng H, Chua V, Liao C, Purwin TJ, Terai M, Kageyama K, et al. Co-targeting HGF-cMET signaling with MEK inhibitors in metastatic uveal melanoma. Molecular Cancer Therapeutics. 2017 doi: 10.1158/1535-7163.MCT-16-0552. Article in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carita G, Frisch-Dit-Leitz E, Dahmani A, Raymondie C, Cassoux N, Piperno-Neumann S, et al. Dual inhibition of protein kinase C and p53-MDM2 or PKC and mTORC1 are novel efficient therapeutic approaches for uveal melanoma. Oncotarget. 2016;7(23):33542–56. doi: 10.18632/oncotarget.9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harbour JW, Chao DL. A molecular revolution in uveal melanoma: implications for patient care and targeted therapy. Ophthalmology. 2014;121(6):1281–8. doi: 10.1016/j.ophtha.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45(8):933–6. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]