Abstract
Fluorescent microspheres were applied in a novel fashion during subsurface drilling of permafrost and ground ice in the Canadian High Arctic to monitor the exogenous microbiological contamination of core samples obtained during the drilling process. Prior to each drill run, a concentrated fluorescent microsphere (0.5-μm diameter) solution was applied to the interior surfaces of the drill bit, core catcher, and core tube and allowed to dry. Macroscopic examination in the field demonstrated reliable transfer of the microspheres to core samples, while detailed microscopic examination revealed penetration levels of less than 1 cm from the core exterior. To monitor for microbial contamination during downstream processing of the permafrost and ground ice cores, a Pseudomonas strain expressing the green fluorescent protein (GFP) was painted on the core exterior prior to processing. Contamination of the processed core interiors with the GFP-expressing strain was not detected by culturing the samples or by PCR to detect the gfp marker gene. These methodologies were quick, were easy to apply, and should help to monitor the exogenous microbiological contamination of pristine permafrost and ground ice samples for downstream culture-dependent and culture-independent microbial analyses.
Cold-adapted microorganisms have recently been discovered in some of the harshest environments on Earth, including Antarctic subglacial and permanently ice-covered lakes, cloud droplets, ice cores, and snow (for an overview, see references 11 and 23), Siberian permafrost and tundra soil (14, 28, 31), and an Arctic glacier (30). Significantly, evidence of microbial activity and growth at below-freezing temperatures in Antarctic snow (6), ice (9, 19), and Siberian permafrost (2, 14, 25) has been demonstrated recently. Microbial habitats may exist in thin films of liquid water present in permafrost or in permafrost brine lenses (cryopegs) at below-freezing temperatures (2, 14). Such habitats allow for microbial metabolic activity at temperatures as low as −20°C and even microbial reproduction at −10°C (2). However, relatively little is known about the microbial biodiversity present or the traits that enable such microorganisms to survive in permafrost, and virtually nothing is known of microbial populations and activity in ground ice. Studying microbial communities in permafrost and ground ice will increase our basic knowledge of these microbial communities and allow for exploitation of the potential biotechnological applications (cold-adapted enzymes and compounds) of indigenous microorganisms (7). These unique environments also represent extraterrestrial analogs in the nascent field of exobiology, especially in light of the recent evidence of massive amounts of shallow ground ice near the surface of Mars (4, 14, 18). This has necessitated the development of novel research techniques, including sample collection and processing, for obtaining pristine subsurface permafrost and ground ice samples.
Subsurface sampling techniques have been refined for various matrices ranging from unconsolidated sands to solid bedrock (for a review, see reference 22). In all cases, assessment of exogenous microbial contamination is imperative in ensuring that the samples being examined are representative of the indigenous microbial population. The primary sources of microbial contamination during subsurface drilling include the penetration of drilling fluids, which may transfer exogenous microorganisms from the core exterior to the interior, and improper handling of the sample during postdrilling processing. The use of drilling equipment that requires no drilling fluid or that utilizes filtered compressed air to remove borehole cuttings reduces but does not eliminate the potential for microbial contamination. Techniques used to eliminate (26) and detect possible microbial contamination in subsurface samples include the comparison of community level physiological and membrane lipid profiles (20), addition of exogenous microbial species (28), and addition of fluorescent latex microspheres (10, 13, 17).
Fluorescent latex microspheres act as easily detectable microbial surrogates and permit the determination of contaminant penetration into the core sample. Fluorescent latex microspheres have been applied directly in drilling fluids (8), by mixing them with a dry carrier molecule and placing the mixture at the bottom of the hole before drilling starts (13), or by puncturing a Whirlpak bag containing a liquid mixture of microspheres when the core sample enters into the core tube (10). These methods of fluorescent-microsphere application are not appropriate for use in the coring of frozen subsurface matrices for several reasons. The use of liquid microsphere carriers (either in drilling fluids or in specific tracer solutions released during drilling) risks melting the frozen cores and compromising the core integrity or potentially freezing the drill string in the hole if any stoppage is experienced. The application of fluorescent microspheres as a dry powder to the bottom of the hole is also not effective when compressed air is used to remove cuttings, as the powder is immediately blown out of the hole when drilling is resumed.
Previous studies examining the microbiology of permafrost have employed fluidless drilling techniques combined with an exogenous bacterial tracer such as Serratia marcescens (15, 24, 28). In this study, we developed a novel method for the application of fluorescent microspheres as a microbial surrogate tracer in the drilling of permafrost and ground ice in the Canadian High Arctic. Unlike other methods previously described, the fluorescent microspheres were applied directly to the drill bit, core catcher, and core tube as a concentrated solution and allowed to dry before drilling commenced. This resulted in significant transfer of the fluorescent microspheres directly to the cores and minimized loss of the microspheres when compressed air was used to remove cuttings. A psychrotolerant bacterial strain (Pseudomonas sp. strain Cam1-gfp2) expressing the green fluorescent protein (GFP) was used to monitor for possible contamination during downstream core processing in the laboratory. These innovations provided advantages over other techniques in the sampling of permafrost and ground ice for subsequent culture-dependent and culture-independent analyses.
MATERIALS AND METHODS
Permafrost and ground ice sampling.
Subsurface permafrost and ground ice located at Eureka, Ellesmere Island, Nunavut, Canada, were sampled using a modified H/PP 150 portable drilling rig (12) (Webster Drilling Limited, Porirua, New Zealand) equipped with two different drill bits. The first bit, used primarily for permafrost, was a tungsten carbide cutter bit (62-mm inside diameter, 79-mm outside diameter [o.d.]) attached to a single core catcher tube, which in turn was protected by a core barrel. Chilled, compressed air was used to remove cuttings. The second drill bit, used primarily for ground ice drilling, was a Sipre auger with tungsten carbide cutters (73-mm inside diameter, 110-mm o.d.) which did not require compressed air for cuttings removal.
Three sites, each possessing a distinct subsurface vertical profile, were sampled. Site Eu1 (79°59.900′N, 85°53.755′W), drilled to 12.61 m, consisted of heterogeneous silty-clay and clayey-silt permafrost laminations interspersed with thin ice veins. Site Eu2 (79°59.893′N, 85°52.614′W) was situated above an ice wedge (consisting of vertically stratified ice) which was located approximately 0.8 m below the surface. The transition from ice wedge to permafrost in Eu2 was a clearly defined interface at 6.03 m. Drilling in Eu2 was stopped when the ice wedge-permafrost interface was exited. At site Eu3 (80°0.029′N, 85°50.367′W), ground ice (primarily horizontally stratified ice) was encountered at 1.82 m and was still present at 16.05 m, when the drilling was stopped.
Fluorescent microspheres.
Fluorescent latex microspheres (Fluoresbrite yellow-green carboxylated microspheres; Polyscience, Inc., Warrington, Pa.) of 0.5-μm (3.64 × 1011 microspheres/ml) and 0.05-μm (3.64 × 1014 microspheres/ml) diameters were used as a microbial surrogate and particulate tracer to monitor potential contamination during drilling. Three methods for microsphere application were employed. Method A consisted of mixing a microsphere solution (1 ml of each stock microsphere solution in 40 ml of sterile deionized water) with approximately 400 g of snow (9.11 × 1011 microspheres/g of snow) to make seven equally sized snowballs (approximately 5 cm in diameter), which were frozen overnight at −20°C. The snow was dropped down the hole, crushed and packed with a weight, and allowed to set for 10 to 15 min before drilling was reinitiated. Method B involved delivering a mixture of microspheres (1 ml of each stock microsphere solution in 200 ml of sterile deionized water) to the bottom of the drill hole by using a thin plastic bag that was punctured, releasing the microsphere solution (1.82 × 1012 microspheres/ml). The pooled microsphere solution was allowed to freeze for 2 to 16 h in the hole (average temperature of −15°C), creating a fluorescent-microsphere plug, after which drilling was reinitiated. Method C consisted of painting, with a 1-ml pipette tip, the inside of the bit, core catcher, and first inch of the core tube with 333 μl each of a concentrated solution of microspheres (250 μl of each stock microsphere solution in 500 μl of 100% ethanol). The microsphere solution, delivering a total of 9.11 × 1013 microspheres, was allowed to dry before drilling was resumed. The drill string was reintroduced into the borehole, and drilling was resumed with a penetration depth of up to 30 cm. Upon retraction of the drill string from the borehole and disassembly of the drill bit and core catcher, the core samples were immediately placed in sterile Whirlpak bags and kept at below-freezing temperatures during transportation and storage. On-site visualization of the fluorescent microspheres was performed using both ambient light and a portable UV light source (320 nm).
Core processing.
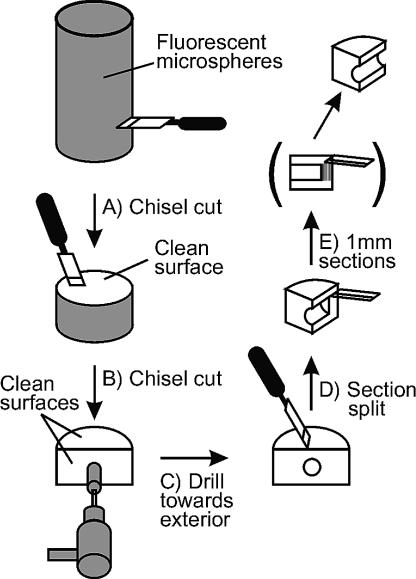
Processing of the cores for microscopic visualization of fluorescent microspheres was performed using a strategy that avoided transfer of microspheres toward the interior of the core (Fig. 1). The first lateral break in the core produced a hockey puck-shaped sample (Fig. 1A). The sample was then placed clean side up and cut vertically once again from the clean side outward, producing a second clean face (Fig. 1B). The center of the second clean face served as the starting point from which drilling was initiated using a sterile 19-mm-o.d. drill bit (Fig. 1C). The sample was securely placed in a vise and drilled to a distance approximately 10 mm from the microsphere-covered exterior. A final vertical cut through the drilled hole split the sample (Fig. 1D), exposing a third clean face, from which 10 consecutive 1-mm-wide sections were sampled for microscopic analysis (Fig. 1E). The 1-mm-wide sections, carefully measured with a ruler, were parallel to the external core surface and were obtained by sectioning the prepared area by using sterile razor blades, starting from the interior surface and working outwards.
FIG. 1.
Schematic diagram of sample processing for fluorescent-microsphere penetration analysis. (A) Initial chisel cut exposing the first clean core surface is shown. (B) The second chisel cut in the center of the clean core surface created a second clean surface. (C) A clean, sterile 19-mm-o.d. drill bit was used to drill a hole in the center of the second clean surface. The hole was drilled until it was approximately 10 mm from the exterior surface. (D) A third chisel cut was made on the first clean surface, positioned to cleave the drill hole in half. (E) At this point, 1-mm-wide sections, starting from the core interior, were sampled by using clean, sterile razor blades and using the drill hole as a guide. The 1-mm-wide sections were placed in clean, sterile tubes and kept at −20°C until processing for microscopic analysis.
Quantitative analysis of fluorescent microspheres.
The 10 consecutive 1-mm-wide sections (Fig. 1E) from each selected ground ice (n = 4) and permafrost (n = 3) sample (see Table 1) were examined for the presence or absence of microspheres. For each core sample, all 10 sections (10 by 1 mm; approximate volume, 142 μl) were prepared and analyzed for the presence of the fluorescent microspheres. The ground ice sample sections were prepared for microscopic analysis by melting, pooling, and then drying the slurry onto microscope slides (25 by 75 by 1 mm; Fisher Scientific, Inc., Nepean, Ontario, Canada) previously prepared by the application of nail polish to create a rectangular dam that was 22 by 40 mm. The permafrost sample sections were liquefied by the addition of water, homogenized, and dried onto the prepared microscope slides. Before microscopy was performed, the sample sections were hydrated with 20 μl and 40 to 60 μl of H2O for ground ice and permafrost samples, respectively. The samples were examined quantitatively with a Leitz Laborlux S epifluorescence microscope (excitation filter wavelength of 360 nm and emission filter wavelength of 460 nm) equipped with a 40× Fluotar Fluoreszenz oil immersion objective (type B; R. P. Cargille Laboratories, Inc., Cedar Grove, N.J.). For each 1-mm-wide section, 25 random fields were counted.
TABLE 1.
Summary of samples, drill bit types, fluorescent-microsphere application methods, and fluorescent-microsphere penetration
| Sample site | Depth (m) | Sample type | Application methoda | Drill bitb | Depth of penetration (mm)c |
|---|---|---|---|---|---|
| Eu1d | 11.58 | Permafrost | C | TC | 5 |
| 11.58 | Permafrost | C | TC | 2 | |
| 11.58 | Permafrost | C | TC | 2 | |
| Eu2 | 2.06 | Ground ice | C | TC | 5 |
| Eu3 | 6.48 | Ground ice | C | SA | 6 |
| 6.61 | Ground ice | C | SA | 2 | |
| 7.15 | Ground ice | B | SA | 4 |
Method of fluorescent-microsphere application: B, fluorescent-microsphere plug; C, fluorescent-microsphere painting.
Drill bits used: TC, tungsten carbide; SA, Sipre auger.
Depth of fluorescent-microsphere penetration as measured from the exterior of the core sample.
Site Eu1 at 11.58 m was sampled three separate times for fluorescent-microsphere analysis.
Application of a GFP-marked Pseudomonas strain to detect contamination during microbial sampling.
To detect possible microbial contamination during sampling of permafrost and ground ice core interiors, Pseudomonas sp. strain Cam1-gfp2 was used as a tracer for exogenous microbial contamination. Pseudomonas sp. strain Cam1-gfp2 is a psychrotolerant strain originally isolated from Arctic soil (21) and was genetically modified to carry a stable chromosomal insertion expressing GFP and kanamycin resistance (1). Pseudomonas sp. strain Cam1-gfp2 was grown overnight at 30°C in tryptic soy broth (Difco Laboratories, Sparks, Md.) to a density of ∼107 cells/ml. Cells were applied to the entire exterior surface of the core with a sterile cotton swab. The culture density in the original culture was determined by serial dilution in sterile saline (0.85% NaCl) and the spread plate technique on tryptic soy agar (TSA) after 2 days' growth at 30°C. The cores were processed for molecular and microbiological analyses by laterally cutting the core to expose a clean surface (Fig. 1A). A sample subcore was then excised from the center of the clean surface by using sterile 38- and 51-mm-o.d. drill bits for permafrost and ice cores, respectively. The subcore samples were immediately transferred to sterile Whirlpak bags and stored at −20°C until further analyses. All permafrost samples were rapidly processed in a biological safety cabinet, while ground ice samples were processed in a −20°C walk-in freezer to minimize thawing. All sampling equipment was sterilized before use and kept in a freezer (−20°C) or on dry ice prior to sample processing to minimize thawing of the samples.
Culture analysis of subcore samples.
To detect possible contamination of the subcores with Pseudomonas sp. strain Cam1-gfp2, samples were taken from the subcore and the remaining outer material with a sterile scalpel and forceps. For permafrost, 1.0-g samples were taken and placed in sterile 20- by 150-mm sterile screw-top test tubes containing 2.5 g of 3-mm-diameter glass beads (Fisher Scientific). Permafrost samples were diluted in 3.0 ml of ice-cold 0.1% Na4P2O7 and vortexed for 1 min. Serial dilutions were plated on TSA containing 6 mg of kanamycin/ml. Plates were incubated for 48 h at 30°C. For ground ice samples, subcore samples and the remaining exterior ground ice material were melted at 4°C for 3 h. Samples of the melted ice (1 and 0.1 ml) were collected and plated as described above for permafrost samples. Enumeration and screening for Pseudomonas sp. strain Cam1-gfp2 fluorescence, due to the presence of GFP, were performed using an Imager Fx (Bio-Rad Laboratories, Hercules, Calif.) at an excitation wavelength of 488 nm.
To determine the survivability of Pseudomonas sp. strain Cam1-gfp2 on the frozen core samples, 100 μl of an overnight culture of Pseudomonas sp. strain Cam1-gfp2 (1.4 × 109 CFU/ml) was applied to 0.9 g of either a permafrost (n = 2) or a ground ice (n = 2) sample and placed at −20°C for 20 min. The samples were thawed at 4°C, and viable cell counts were determined by serial dilution and spread plating on TSA containing 6 mg of kanamycin/ml. The plates were incubated at 30°C for 48 h, and to verify that the enumerated colonies were Pseudomonas sp. strain Cam1-gfp2, the colonies were assayed for the presence of GFP production as described above.
Molecular analysis of pristine samples.
Total community DNA was extracted from permafrost and ground ice samples by using an UltraClean soil DNA isolation kit (Mo Bio Laboratories, Solona Beach, Calif.), modified by vortexing the samples for only 1 min. DNA extracts were purified with polyvinylpolypyrrolidone spin columns as described by Berthelet et al. (3). To test for the presence of the Cam1-gfp2 strain, PCR was performed to detect the chromosomal gfp insertion with the primers gfpF (5′-TGTGCTCTCTCTTTTCGTTGGG-3′) and gfpR (5′-TGGTGTTCAATGCTTTGCCAG-3′), which produced a 455-bp PCR amplicon. To detect the presence of bacterial DNA in the extracts as a positive PCR amplification control, bacterial 16S rRNA genes were amplified by PCR using the bacterial universal primers 341F (5′-CCTACGGGAGGCAGCAG-3′) and 926R (5′-CCGTCAATTCITTTGAGTTT-3′), which yields a 585-bp PCR amplicon (29). PCR mixtures were prepared with reagents and procedures from Invitrogen Canada, Inc. (Burlington, Ontario, Canada), with the exception that 0.625 μl of a 10-mg/ml concentration of bovine serum albumin was added to the reaction mixture. Thermal cycling was performed in a Touchgene gradient system (Techne, Inc., Princeton, N.J.) using the following program: initial denaturation at 95°C for 3 min; 30 cycles of 94°C for 1 min, 53°C for 1 min, and 72°C for 55 s; and a final extension at 72°C for 7 min. PCR fragments were visualized by agarose gel electrophoresis (0.8% agarose) and ethidium bromide staining essentially as described by Sambrook et al. (27).
RESULTS AND DISCUSSION
Fluorescent-microsphere detection.
The ability to monitor exogenous microbial contamination during subsurface sampling and downstream core processing is critical in ensuring that subsequent analyses are truly representative of the indigenous microbial community (16, 22). The use of fluorescent microspheres as a surrogate bacterial tracer is generally the preferred approach (10, 13, 17). In addition to exogenous microbial contamination, challenges arise during drilling when the subsurface matrix is permafrost and freezing temperatures need to be maintained. Two possible solutions include the use of chilled, sterile compressed air to cool the drill bit cutting face (12) and dry drilling (without drilling mud) at reduced speeds (15).
To ensure the retrieval of uncontaminated samples, two methodologies were developed and validated: application of fluorescent microspheres during the permafrost and ground ice drilling and painting of permafrost-ground ice core samples with a GFP-marked bacterial strain for detecting microbial and nucleic acid contamination during downstream core processing. Several aspects of microsphere application were examined, including the type of drill bit, the method used to apply the microspheres, and the sample type (Table 1). Initial field experiments were evaluated based on visual inspection of the core samples for the presence and distribution of fluorescent microspheres by using a handheld portable UV light.
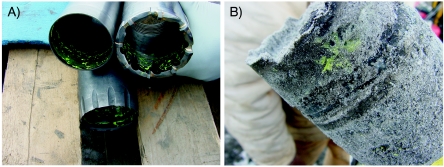
Method A (fluorescent snowball) proved to be unsatisfactory and was tested only once. During drilling, most of the microsphere-snow mixture was forced into the center of the core tube, which resulted in little transfer of the microspheres to the core sample. Additionally, when the tungsten carbide bit and compressed air were used, a large amount of the microsphere-snow mixture was blown out of the hole before the drill bit reached the bottom of the hole. With method B (fluorescent-microsphere plug), the primary criticism was that once the microsphere plug was frozen and drilling was resumed, a large portion of the material recovered was not core sample but rather cuttings originating from the hole's sidewalls. This greatly reduced the recovery of core material available for analyses. Method C (painting of the microspheres onto the drill bits) was a novel approach in that the fluorescent microspheres were applied directly to the drill bit, core catcher, and core tube and demonstrated significant transfer of the microspheres from the painted surfaces to the exterior of the core sample (Fig. 2). Method C was also relatively quick and could be executed without a stoppage in drilling, while methods A and B required a minimum of 30 min between microsphere application and resumption of drilling. In method C, the drill bit could be cleaned, painted with the fluorescent-microsphere solution, dried, and be ready for the next run while the previous samples were removed from the core tube and packaged for transport. An additional advantage to method C was that, when compressed air was used to cool the cutting face of the drill bit and remove cuttings from the hole, the microspheres were not blown out of the hole, as was the case with method A and, to a limited extent, with method B. Based on field examination of the core samples with a UV light, method C was chosen as the optimal procedure for fluorescent-microsphere application and was used for all subsequent sampling for two reasons: (i) method C had the highest level of fluorescent microsphere transfer to the core samples and (ii) the overall integrity of the recovered core was equal to or significantly better than that obtained by the other methods.
FIG. 2.
Field demonstration of fluorescent microsphere application. (A) Drill bit, core catcher, and core tube, painted with fluorescent microspheres before drilling; (B) visualization of fluorescent microspheres on the exterior of a ground ice core sample with ambient light.
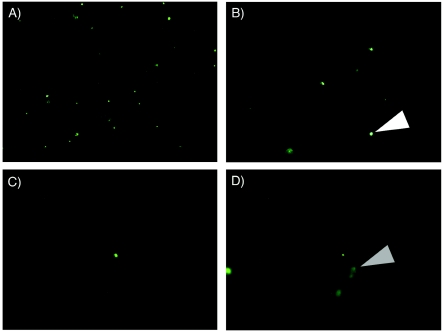
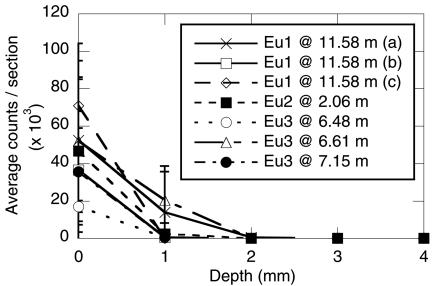
Microscopic examination of core samples to determine the extent of microsphere penetration was performed in the laboratory on the uppermost portion of each sample to maximize the presence of the fluorescent microspheres. Only 0.5-μm-diameter microspheres could be readily visualized microscopically (results not shown). The fluorescent microspheres were easily distinguished from other fluorescent particles (Fig. 3) by microscopic comparison under white light. There was no significant penetration of the microspheres in any of the intact cores examined, with microsphere counts per section approaching 0 after only 2 to 3 mm (Fig. 4). In the case of one sample taken from site Eu2 (at 6.82 m) by method B, microspheres were observed in all 10 of the 1-mm-wide sections (i.e., microsphere penetration of ≥10 mm). Closer examination revealed that this sample was composed almost entirely of cuttings originating from the sidewall of the hole. This was typical of method B, with only a single usable sample recovered (Eu3 at 7.15 m). When the sections were exhaustively examined for the presence of the microspheres (i.e., scanning of the entire section for the presence of single fluorescent microspheres), the maximum depth of microsphere penetration ranged from 2 to 6 mm (Table 1). In permafrost samples, the maximum microsphere penetration averaged 3.0 mm (±1.7 mm) whereas maximum penetration in the ground ice samples averaged 4.3 mm (±1.7 mm). The type of drill bit employed also did not affect maximum microsphere penetration; the microsphere penetration in samples obtained with the tungsten carbide cutters averaged 3.5 mm (±1.7 mm), and that in samples obtained with the Sipre auger averaged 4.0 mm (±2.0 mm). Based on these results, a minimum of 10 mm from the core exterior should be sacrificed to ensure an uncontaminated core sample.
FIG. 3.
Epifluorescent microscopic visualization of fluorescent microspheres at increasing depths from the core surface. A permafrost sample from site Eu1 at 11.58 m was used and was representative of the other samples. Fluorescent microspheres were seen as discrete points (white triangle), whereas background fluorescence from particulates was seen as diffuse smears (gray triangle), which was confirmed by visualization with white light. (A) Section at 1 mm from exterior core surface; (B) section at 2 mm from exterior core surface; (C) section at 4 mm from exterior core surface; (D) section at 6 mm from exterior core surface. No beads were observed in sections more than 6 mm from the exterior core surface.
FIG. 4.
Microscopic examination of fluorescent-microsphere penetration. Average counts per section is the average number of fluorescent microspheres counted in 25 random fields for each 1-mm-wide section of each core sample. The samples are as described in Table 1. Error bars represent the standard deviation for 25 fields/1-mm-wide section.
GFP-tagged Pseudomonas detection.
To detect potential contamination during subsampling of the cores, GFP-expressing Pseudomonas sp. strain Cam1-gfp2 was applied as a microbiological tracer to the exterior of the core. This organism was selected because of its psychrotolerant nature, its ease of detection during routine microbiological analysis, and the presence of a unique molecular marker (i.e., gfp) enabling the detection of contamination at low levels by PCR. In both permafrost and ground ice interior core samples, growth of Pseudomonas sp. strain Cam1-gfp2 was not detected after 48 h of incubation at 30°C, whereas growth was clearly present in corresponding samples from the exterior material after only 24 h (Fig. 5). All bacterial colonies originating from the exterior material displayed green fluorescence, suggesting that kanamycin was effective in selecting for the Cam1-gfp2 strain.
FIG. 5.
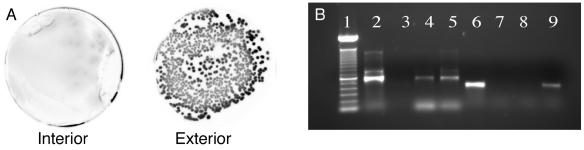
Utilization of Pseudomonas sp. strain Cam1-gfp2 to detect contamination during subsampling of a permafrost sample from site Eu1 at 9.01 m. (A) A sample from the interior core showing the lack of fluorescent Pseudomonas sp. strain Cam1-gfp2 and a sample from the exterior showing the presence of fluorescent Pseudomonas sp. strain Cam1-gfp2 CFU. Samples were spread plated on TSA and incubated for 48 h at 30°C. (B) Detection of gfp and 16S rRNA by PCR analysis and agarose gel electrophoresis of a sample from site Eu1 at 9.01 m. Lanes: 1, 100-bp ladder; 2, Pseudomonas sp. strain Cam1-gfp2 genomic DNA (positive 16S rRNA control); 3, H2O (negative 16S rRNA control); 4, interior core (16S rRNA); 5, exterior core (16S rRNA); 6, Pseudomonas sp. strain Cam1-gfp2 genomic DNA (positive gfp control); 7, H2O (negative gfp control); 8, interior core (gfp); 9, exterior core (gfp).
Survivability assays performed with Pseudomonas sp. strain Cam1-gfp2 on both permafrost and ground ice samples revealed that Pseudomonas sp. strain Cam1-gfp2 is well suited for use as a microbiological tracer with frozen subsurface samples. When applied to permafrost samples, 51.8% (±6.3%; n = 6) of the applied cells remained viable, while with ground ice samples, 24.6% (±6.7%; n = 6) of the cells remained viable.
When we used the more sensitive molecular biology-based method of PCR, the gfp gene was not detected in DNA isolated from the interior core samples, while the 455-bp gfp-amplified fragment was observed in samples taken from the exterior material, indicating the presence of Pseudomonas sp. strain Cam1-gfp2 (Fig. 5). 16S rRNA was detected by PCR in both the interior core and the exterior material, demonstrating that bacterial DNA was successfully extracted and was PCR amplifiable from both samples (Fig. 5). Overall, these results indicate that the interior core permafrost and ground ice were not contaminated during the sampling process. The utilization of a GFP-marked bacterial strain was a quick and readily reproducible technique and represents the first report describing both culture-dependent and -independent methodologies for detecting both bacterial and nucleic acid contamination of permafrost and ground ice drill cores during downstream processing. The ability to detect contaminant nucleic acids in these samples is very important given the widespread utilization of culture-independent molecular techniques for characterizing microbial biodiversity in environmental samples and the high sensitivity of PCR amplification.
Conclusions.
The work presented here represents a novel method for the application of fluorescent microspheres, used as microbial surrogates, when one drills in permafrost and ground ice matrices with either compressed air or dry drilling. This method of painting a concentrated solution of fluorescent microspheres directly on the drill bit, core catcher, and core tube is also relevant to other situations where dry drilling or maintenance of down-hole freezing temperatures is required, such as in the development of Mars subsurface drilling equipment (5). Painting of the drill bit, core catcher, and core tube is quick, results in reliable transfer to the core samples, is easily visualized in the field under ambient light or with a portable UV light, and is cost effective in that only small amounts of the fluorescent microspheres are required for visualization.
Acknowledgments
We thank Bain Webster and Tony Kingan of Webster Drilling and Alex Pyne and Warren Dickinson of Victoria University of Wellington, New Zealand, for their invaluable drilling expertise. Pseudomonas sp. strain Cam1-gfp2 was kindly provided by J. Trevors of the University of Guelph.
We also acknowledge the logistical and financial support provided by the Polar Continental Shelf Project (project no. 633-03), the Northern Scientific Training Program, and the NASA Astrobiology Technology and Instrument Development Program.
REFERENCES
- 1.Ahn, Y.-B., L. A. Beaudette, H. Lee, and J. T. Trevors. 2001. Survival of a GFP-labeled polychlorinated biphenyl degrading psychrotolerant Pseudomonas spp. in 4 and 22°C soil microcosms. Microb. Ecol. 42:614-623. [DOI] [PubMed] [Google Scholar]
- 2.Bakermans, C., A. I. Tsapin, V. Souza-Egipsy, D. A. Gilichinsky, and K. H. Nealson. 2003. Reproduction and metabolism at −10°C of bacteria isolated from Siberian permafrost. Environ. Microbiol. 5:321-326. [DOI] [PubMed] [Google Scholar]
- 3.Berthelet, M., L. G. Whyte, and C. W. Greer. 1996. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol. Lett. 138:17-22. [DOI] [PubMed] [Google Scholar]
- 4.Boynton, W. V., W. C. Feldman, S. W. Squyres, T. H. Prettyman, J. Bruckner, L. G. Evans, R. C. Reedy, R. Starr, J. R. Arnold, D. M. Drake, P. A. Englert, A. E. Metzger, I. Mitrofanov, J. I. Trombka, C. D'Uston, H. Wanke, O. Gasnault, D. K. Hamara, D. M. Janes, R. L. Marcialis, S. Maurice, I. Mikheeva, G. J. Taylor, R. Tokar, and C. Shinohara. 2002. Distribution of hydrogen in the near surface of Mars: evidence for subsurface ice deposits. Science 297:81-85. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, G. A., H. Cannon, S. Domville, B. Glass, C. McKay, J. George, B. Derkowski, R. Fincher, G. Cooper, K. Zacny, W. Pollard, and S. Clifford. An automated, low mass, low power drill for acquiring subsurface samples of ground ice for astrobiology studies on Earth and on Mars. Third International Conference on Mars Polar Science and Exploration, 13 to 17 October 2003, Banff, Alberta, Canada.
- 6.Carpenter, E. J., S. Lin, and D. G. Capone. 2000. Bacterial activity in South Pole snow. Appl. Environ. Microbiol. 66:4514-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavicchioli, R., K. S. Siddiqui, D. Andrews, and K. R. Sowers. 2002. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 13:253-261. [DOI] [PubMed] [Google Scholar]
- 8.Chapelle, F. H., and P. B. McMahon. 1991. Geochemistry of dissolved inorganic carbon in a Coastal Plain aquifer. 1. Sulfate from confining beds as an oxidant in microbial CO2 production. J. Hydrol. 127:85-108. [Google Scholar]
- 9.Christner, B. C. 2002. Incorporation of DNA and protein precursors into macromolecules by bacteria at −15°C. Appl. Environ. Microbiol. 68:6435-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colwell, F. S., G. J. Stormberg, T. J. Phelps, S. A. Birnbaum, J. McKinley, S. A. Rawson, C. Veverka, S. Goodwin, P. E. Long, B. F. Russell, T. Garland, D. Thompson, P. Skinner, and S. Grover. 1992. Innovative techniques for collection of saturated and unsaturated subsurface basalts and sediments for microbiological characterization. J. Microbiol. Methods 15:279-292. [Google Scholar]
- 11.Deming, J. W. 2002. Psychrophiles and polar regions. Curr. Opin. Microbiol. 5:301-309. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson, W., P. Cooper, B. Webster, and J. Ashby. 1999. A portable drilling rig for coring permafrosted sediments. J. Sediment. Res. 69:518-527. [Google Scholar]
- 13.Frederickson, J. K., F. J. Brockman, B. N. Bjornstad, P. E. Long, S. W. Li, J. P. McKinley, J. V. Wright, J. L. Conca, T. L. Kieft, and D. L. Balkwill. 1993. Microbiological characteristics of pristine and contaminated deep vadose sediments from an arid region. Geomicrobiol. J. 11:95-107. [Google Scholar]
- 14.Gilichinsky, D., E. Rivkina, V. Shcherbakova, K. Laurinavichuis, and J. Tiedje. 2003. Supercooled water brines within permafrost—an unknown ecological niche for microorganisms: a model for astrobiology. Astrobiology 3:331-341. [DOI] [PubMed] [Google Scholar]
- 15.Gilichinsky, D. A., G. M. Khlebnikova, D. G. Zvyagintsev, D. G. Fedorov-Davydov, and N. N. Kudryavtseva. 1989. Microbiology of sedimentary materials in the permafrost zone. Int. Geol. Rev. 31:847-858. [Google Scholar]
- 16.Griffin, W. T., T. J. Phelps, F. S. Colwell, and J. K. Frederickson. 1997. Methods for obtaining deep subsurface microbiological samples by drilling, p. 23-44. In P. S. Amy and D. L. Haldeman (ed.), The microbiology of the terrestrial deep subsurface. CRC Press, Boca Raton, Fla.
- 17.Harvey, R. W., L. H. George, R. L. Smith, and D. R. LeBlanc. 1989. Transport of microspheres and indigenous bacteria through a sandy aquifer: results of natural- and forced-gradient tracer experiments. Environ. Sci. Technol. 23:51-56. [Google Scholar]
- 18.Jakosky, B. M., K. H. Nealson, C. Bakermans, R. E. Ley, and M. T. Mellon. 2003. Subfreezing activity of microorganisms and the potential habitability of Mars' polar regions. Astrobiology 3:343-350. [DOI] [PubMed] [Google Scholar]
- 19.Junge, K., H. Eicken, and J. W. Deming. 2004. Bacterial activity at −2 to −20°C in Arctic wintertime sea ice. Appl. Environ. Microbiol. 70:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehman, R. M., F. S. Colwell, D. B. Ringelberg, and D. C. White. 1995. Combined microbial community-level analyses for quality assurance of terrestrial subsurface cores. J. Microbiol. Methods 22:263-281. [Google Scholar]
- 21.Master, E. R., and W. W. Mohn. 1998. Psychrotolerant bacteria isolated from arctic soil that degrade polychlorinated biphenyls at low temperatures. Appl. Environ. Microbiol. 64:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phelps, T. J., and J. K. Frederickson. 2002. Drilling, coring, and sampling subsurface environments, p. 679-695. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 23.Priscu, J. C., C. H. Fritsen, E. E. Adams, S. J. Giovannoni, H. W. Paerl, C. P. McKay, P. T. Doran, D. A. Gordon, B. D. Lanoil, and J. L. Pinckney. 1998. Perennial Antarctic lake ice: an oasis for life in a polar desert. Science 280:2095-2098. [DOI] [PubMed] [Google Scholar]
- 24.Rivkina, E., D. Gilichinsky, S. Wagener, J. Tiedje, and J. McGrath. 1998. Biogeochemical activity of anaerobic microorganisms from buried permafrost sediments. Geomicrobiology 15:187-193. [Google Scholar]
- 25.Rivkina, E. M., E. I. Friedmann, C. P. McKay, and D. A. Gilichinsky. 2000. Metabolic activity of permafrost bacteria below the freezing point. Appl. Environ. Microbiol. 66:3230-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers, S. O., V. Theraisnathan, L. J. Ma, Y. Zhao, G. Zhang, S.-G. Shin, J. D. Castello, and W. T. Starmer. Comparisons of protocols for decontamination of environmental ice samples for biological and molecular examinations. Appl. Environ. Microbiol. 70:2540-2544. [DOI] [PMC free article] [PubMed]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Shi, T., R. H. Reeves, D. A. Gilichinsky, and E. I. Friedmann. 1997. Characterization of viable bacteria from Siberian permafrost by 16S rDNA sequencing. Microb. Ecol. 33:169-179. [DOI] [PubMed] [Google Scholar]
- 29.Sjöling, S., and D. A. Cowan. 2003. High 16S rDNA bacterial diversity in glacial meltwater lake sediment, Bratina Island, Antarctica. Extremophiles 7:275-282. [DOI] [PubMed] [Google Scholar]
- 30.Skidmore, M. L., J. M. Foght, and M. J. Sharp. 2000. Microbial life beneath a high arctic glacier. Appl. Environ. Microbiol. 66:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, J., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]