Abstract
A general procedure for efficient generation of gene knockouts in gram-negative bacteria by the adaptation of the Saccharomyces cerevisiae URA3 selection system is described. A Pseudomonas putida strain lacking the URA3 homolog pyrF (encoding orotidine-5′-phosphate decarboxylase) was constructed, allowing the use of a plasmid-borne copy of the gene as the target of selection. The delivery vector pTEC contains the pyrF gene and promoter, a conditional origin of replication (oriR6K), an origin of transfer (mobRK2), and an antibiotic selection marker flanked by multiple sites for cloning appropriate DNA segments. The versatility of pyrF as a selection system, allowing both positive and negative selection of the marker, and the robustness of the selection, where pyrF is associated with uracil prototrophy and fluoroorotic acid sensitivity, make this setup a powerful tool for efficient homologous gene replacement in gram-negative bacteria. The system has been instrumental for complete deletion of the P. putida choline-O-sulfate utilization operon betCDE, a mutant which could not be produced by any of the other genetic strategies available.
Homologous gene replacement is a basic tool for the study of biological systems, and as such the efficiency of methods for gene knockout generation contributes to the ease with which a system can be characterized by permitting genetic manipulation. The diversity of bacterial species under study has harbored the development of a plethora of cloning, expression, and mutagenesis tools, but those for the generation of gene knockouts are still in need of improvement (39). PCR-based one-step techniques for gene replacement and disruption (6, 46, 47) greatly facilitate gene knockout construction but thus far have only been shown to work in a limited number of species (i.e., Escherichia coli and Salmonella spp.). For other bacteria, including a large number of genera of considerable environmental and biotechnological interest, exchange of DNA segments to and from the chromosome depends on the use of counterselection markers, but these are few and of limited efficiency.
The dearth of suitable tools to effect such exchanges limits the possibilities of genetic analysis and chromosomal engineering of most non-E. coli strains. The most widely used counterselection marker in gram-negative bacteria is the sacB gene of Bacillus subtilis, whose product, levan sucrase, confers sucrose sensitivity on a large number of gram-negative species (11, 40). Also, in Streptomyces spp., the glkA gene product, glucose kinase, confers sensitivity to 2-deoxyglucose (44). Use of the glkA system in other bacteria has not been reported. In Bacillus subtilis, uracil phosphoribosyltransferase has been used successfully as a counterselection marker in one-step generation of chromosomal point mutations, in-frame deletions, and gene knockouts (9). Uracil phosphoribosyltransferase converts 5-fluorouracil into a compound, 5-fluoro-UMP, that is metabolized into 5-fluoro-dUMP, which strongly inhibits thymidylate synthetase (28).
Except for sacB-based vectors, none of the counterselection markers have been employed in Pseudomonas putida to any significant extent. Yet, although useful in many cases, sacB is by no means an optimal gene for this end, as its function depends on the transport of sucrose. This often has to be forced by adding high extracellular concentrations (5% or more) of the sugar, causing undue stress and favoring the rise of mutations that bypass the stringency of the selection. Having a reliable system for allelic exchange and chromosomal engineering of P. putida is a prerequisite to realize the full potential of this species in biocatalysis and bioremediation. The same applies to many other gram-negative bacteria which are not yet amenable to genetic analysis (34).
In our pursuit of improved genetic tools for P. putida, we came across the uracil biosynthesis gene URA3, which has been used widely in positive and negative selection in Saccharomyces cerevisiae. URA3 encodes an enzyme, orotidine-5′-phosphate decarboxylase (ODCase), that can catalyze the transformation of 5-fluoroorotic acid (FOA, a uracil analogue) into a highly toxic compound (3). Thus, counterselection works on the basis that the presence of URA3 confers sensitivity to FOA, while URA3-negative cells are FOA resistant. Alternatively, URA3 as a positive selection marker works based on the ability of an exogenous or plasmid-borne URA3 gene complementing uracil auxotrophy of a URA3- negative strain.
Assuming that URA3/FOA counterselection could work so long as an ODCase activity exists in an organism, successful adaptation of the URA counterselection system has allowed homologous gene replacement in the archaea Halobacterium salinarum (33), Thermococcus kodakaraensis KOD1 (38), and Halorerax volcanii (2). Also, URA3-based counterselection has been used in Mycobacterium smegmatis for obtaining unmarked gene integration into the chromosome (20) and in shuttle vectors for the hyperthermophilic archaeon Pyrococcus abyssi (21). The pyr genes are URA homologs, such that adaptation of the URA3 system to bacteria is based on identification of the ODCase-encoding pyr gene and isolation of mutants that are FOA resistant and uracil auxotrophs, followed by the use of allelic exchange to isolate the gene replacement.
Using the complete genome sequence of P. putida KT2440 (27), we identified a single pyrF/URA3 ortholog in the chromosome. This opened the possibility of recreating in P. putida the many advantages and utilities of the URA3 system of S. cerevisiae. In this work, we present a complete genetic system for generation of knockouts in gram-negative bacteria that uses the pyrF gene of Pseudomonas putida as a positive and negative marker. We prove that pyrF-based selection is highly efficient by creating a P. putida strain entirely deleted of the osmotic stress-related operon betCDE, which could not have been generated by any other method.
MATERIALS AND METHODS
Biological materials, culture conditions, and general procedures.
The bacteria and plasmids used in this work are listed in Table 1. Pseudomonas putida KT2442 (15) and Escherichia coli strains were grown at 30°C and 37°C, respectively, in LB or 2YT broth (37). P. putida strains were also grown in M9 minimal medium (37) with citrate (2 g/liter) as the carbon source. Antibiotics were used at the following concentrations: rifampin, 50 μg/ml; ampicillin, 150 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml (E. coli) or 100 μg/ml (P. putida); gentamicin, 10 μg/ml; and chloramphenicol, 30 μg/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| P. putida KT2442 | Rifr; derived from P. putida KT2440 | 15 |
| P. putida TEC1 | Rifr; P. putida KT2442 with a nearly complete internal deletion of pyrF | This work |
| P. putida TEC1 ΔbetCDE | Rifr Gmr; TEC1 whose betCDE putative operon was deleted by use of pTGLB | This work |
| E. coli CC118λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE recA1 λpir lysogen | 15 |
| E. coli HB101 | Smr; rpsL recA thi pro leu hsdR−M+ (E. coli K-12/E. coli B. hybrid) | 37 |
| E. coli X82 | lacZ53(Am) λ−trpC50 pyrF287 hisG1(Fs) rpsL8; uracil auxotroph | E. coli Stock Center CGSC 5889 ID 6952 |
| Plasmids | ||
| pUC19NotI | Apr; pUC-type plasmid with MCS of pUC19 flanked by NotI sites | 15 |
| pUP | Apr; pUC19NotI inserted with a 2-kb EcoRI-HindIII PCR fragment containing the P. putida pyrF chromosome region | This work |
| pUPX | Apr; pUP with an xylE SmaI fragment from pXYLE10 cloned into the Eco47III and SmaI sites of pUP | This work |
| pKNG101 | Smr; sacRB+, broad-host-range plasmid of conditional replication | 17 |
| pKPX | Smr; pKNG1 with the pUPX ΔpyrF::xyIE NotI fragment | This work |
| pKT231 | Kmr, Smr, broad-host-range plasmid derived from RSF1010 | 10 |
| pKP | Kmr; pKT231 with the P. putida pyrF gene and its promoter region | This work |
| pGAPN | Apr; pGEM-T Easy carrying a PCR fragment of pyrF (with endogenous promoter) | This work |
| pGP704 | Apr; oriR6K, mobRK2, MCS of M13 tg131 | 25 |
| pGEM-T Easy | Apr; PCR fragment cloning plasmid | Promega |
| pGGLB | Apr; pGEM-T Easy with the betCDE deletion cassette cloned into the NotI site | Galväo et al., unpublished |
| pTEC | Kmr FOAs Ura+; ligation of MCS-Km-MCS, oriR6K/origin of transfer mobRK2, and pyrF PCR fragments | This work |
| pTGLB | Kmr Gmr FOAs Ura+; pTEC with the betCDE deletion cassette cloned into the NotI site | This work |
| pRK600 | Cmr; oriColE1, mobRK2, traRK2 | 19 |
For the P. putida ΔpyrF TEC1 strain, uracil (Sigma Aldrich, Madrid, Spain) was used at 20 μg/ml. 5-Fluoroorotic acid (FOA; Zymo Research) was used at 250 μg/ml. Plasmids were transferred from E. coli to P. putida by tripartite conjugation on membrane filters (0.45 μm, Millipore), including helper strain E. coli HB101(pRK600). After 6 to 8 h of incubation at 30°C on LB agar, the conjugation mixture was plated on minimal selective medium as indicated in each case. All oligonucleotides were synthesized by Sigma-Genosys. DNA constructs were sequenced with the dideoxy method (ABI-Prism automated DNA sequencer, Perkin Elmer). Endurance to osmotic stress was assayed by plating serial dilutions of cultures onto solid M9 minimal medium supplemented with citrate and sodium chloride to 0.6 or 0.7 M.
Plasmid and strain construction.
The P. putida ΔpyrF TEC1 strain was created as follows. A 1.8-kb region of the P. putida genome spanning the pyrF gene (identified with sequence information from the Pseudomonas putida genome, http://www.tigr.org) was amplified by PCR with primers 37pyr5 (5′-CCGTAAAGCTTGGATGCACTTAGCCTGCAAAAGGAGC-3′) and 37pyr3 (5′-TGCAGAATTCCTGAAGCGCGAATGCCCCAAAGGCGTG-3′), which added EcoRI and HindIII sites at the ends of the targeted DNA segment. The resulting product was then cloned into pUC19Not (Table 1), yielding plasmid pUP. This plasmid was cut with Eco47III and SmaI, releasing an internal 404-bp segment of the 702-bp-long pyrF gene, and treated with T4 DNA polymerase (Roche Applied Science) to produce blunt ends at the sequences left attached to the plasmid. This was ligated with the SmaI fragment from pXYLE10 (42) containing the xylE gene (for catechol 2,3-dioxygenase), yielding pUPX. Plasmid pKPX, used for chromosomal delivery of the resulting assembly ΔpyrF::xylE, was generated by excising the 2,418-bp NotI fragment of pUPX (containing the deleted pyrF genomic region inserted with xylE) into the NotI site of the sacB+ suicide vector pKNG101 (17).
To create a pyrF+ broad-host-range plasmid, the P. putida pyrF gene and its promoter (890 bp) was amplified with primers NotIpyrFprom (5′-GATATACAGCGGC CGCATCGGGCCGTGGGCAACGGT-3′) and Asc pyrF (5′-GAATGGCGCGCCCCTTACCCACGGATCTC-3′), introducing NotI and AscI sites to the 5′ and 3′ ends of the amplified segment, respectively. The resulting 882-bp product was ligated to pGEM-T Easy (Promega), resulting in pGAPN. Digestion of pGAPN with EcoRI (vector sites flanking the pyrF insert) released a 920-bp fragment containing pyrF, which was ligated with EcoRI-digested plasmid pKT231 (10) and electroporated into E. coli X82, a uracil auxotroph (from S. Brenner, through the E. coli Stock Center, CGSC 5889 ID 6952), and plated on M9 minimal medium (37) supplemented with glucose (2 g/liter), tryptophan (0.1 mM), histidine (1 mg/liter), thymidine (0.5 mg/liter), and kanamycin. The plasmid borne by transformants growing without uracil was named pKP and consisted of pKT231 with a functional pyrF insertion.
pTEC, the pyrF+ vector for allelic exchange, was generated as follows. The same primers (NotIpyrFprom and Asc pyrF) employed for construction of pKP were used to amplify the 870-bp region of the P. putida pyrF gene and its promoter. A 1.0-kb kanamycin resistance gene was amplified separately from pUT mini-Tn5 Km1 (7) with primers 5′MCS Km (5′-ATAAGAATGCGGCCGCTCTAGAAGTACTAGGCCTATCGATGAGCTCCGCCACGTTGTGTCTCAAAATCTCTGATGTTACATTGCACAAGATAA-3′) and 3′MCS Km (5′-TGCAGTTTCATTTGATGCTCGATGAGTTTTTCTAAG GTACCAGATCTGATATCACGCGTGAATTCAGAC-3′) which flanked the kanamycin resistance determinant with multiple restriction sites. Finally, a 2.0-kbp fragment from plasmid pGP704 (25) containing the origin of transfer (oriT) of RK2 (12, 45) and the vegetative origin of replication (oriV) of R6K (which functions only in concert with the replication protein ∏ encoded by gene pir) (16) was amplified with primers 5′ORI EcoNotAvr (5′-TACGGAATTCGCGGCCGCCCTAGGCCATGTCAGCCGTTAAGTGTTCCTGT-3′) and 3′ORI EagApaAsc (5′-CATGTCGGCGCGCCGGGCCCCGGCCGCCGTGGCTCCGGCGTCTT-3′), which add EcoRI and AscI sites to the corresponding DNA. The three PCR products (one bearing pyrF, other kanamycin resistance, and the last the oriV/oriT origins) were digested with the cognate enzymes required to produce cohesive ends, ligated, and transformed into E. coli CC118λpir (15). Transformants growing on LB-kanamycin agar plates were found to bear the plasmid shown in Fig. 3, which was subsequently named pTEC (EMBL accession number BN000674).
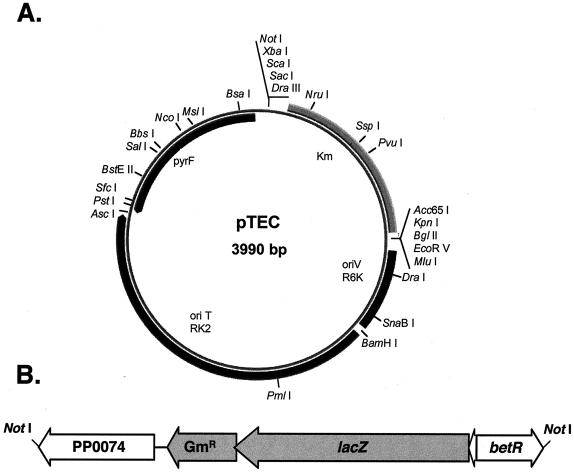
FIG. 3.
Map of pTEC. (A) The plasmid contains the P. putida pyrF gene and its promoter, a kanamycin resistance cassette flanked by multiple cloning sites, the broad-host-range origin of replication oriR6K, and the origin of transfer mobRK2. Only unique restriction sites are shown. The diagram was drawn with PlasMapper (http://wishart.biology.ualberta.ca/PlasMapper/index.html) (8). (B) Schematic diagram of the cassette cloned into the pTEC NotI site for generating a betCDE deletion mutant. In the cassette, the betCDE genes have been replaced by the gentamicin resistance cassette and lacZ genes while maintaining the flanking sequences, ORF PP0074 and betR.
ΔpyrF P. putida strain TEC1 was constructed by mobilizing pKPX from E. coli CC118λpir into P. putida KT2442 by tripartite mating with E. coli HB101(pRK600) as the helper and selecting exconjugants in minimal medium plates with citrate and streptomycin. Putative cointegrates were grown in LB without selection, and resolution of the cointegrate resulting in chromosomal acquisition of the deleted pyrF gene was selected by plating on minimal medium plates with citrate, uracil, and FOA (counterselection of sacB with sucrose never worked in this case).
The plasmid used for deletion of the P. putida putative betCDE operon, pTGLB, was constructed as follows. A PCR product with a gentamicin resistance cassette and a β-galactosidase gene flanked by betCDE upstream (ORF PP0074) and downstream (betR) sequences, forming a transcriptional fusion of the betCD promoter with the β-galactosidase gene (Galvão et al., unpublished data) was cloned into plasmid pGEM-T Easy (Promega) to generate pGGLB. pGGLB was cut with NotI, and the deletion cassette was cloned into the NotI site of pTEC to generate pTGLB.
RESULTS AND DISCUSSION
P. putida pyrF gene and FOA sensitivity.
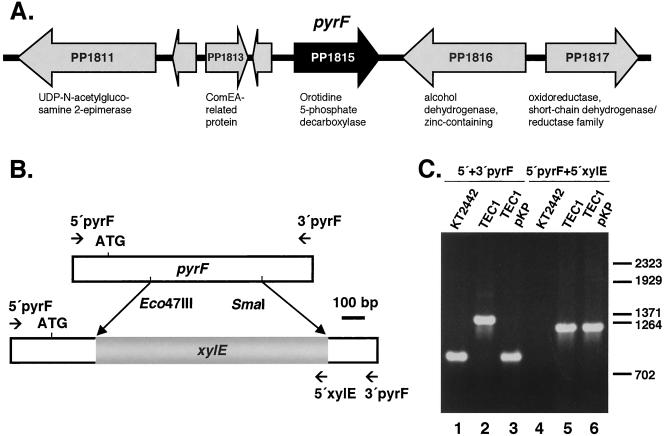
A BLAST search on the P. putida genome with the P. aeruginosa pyrF gene sequence yielded an open reading frame (ORF) with 90% identity (locus PP1815 in the P. putida genome). The P. aeruginosa pyrF gene has been shown to encode orotidine-5′-monophosphate decarboxylase (ODCase) by enzymatic assays of the gene product (43). In P. putida, pyrF is not part of an operon (Fig. 1A), being located between a conserved hypothetical protein and a predicted alcohol dehydrogenase.
FIG. 1.
Generation of ΔpyrF P. putida TEC1. (A) Schematic diagram of the P. putida pyrF gene (ORF PP1815) chromosomal context. The locus name of each ORF with assigned or predicted function is shown; hypothetical ORFs are unmarked. (B) Schematic diagram of the P. putida pyrF gene and of the ΔpyrF in P. putida TEC1. The xylE gene was cloned in the Eco47III and SmaI sites of pyrF. Primers are shown as arrows, indicating primer combinations used to amplify the pyrF and xylE sequences. (C) PCR analysis of wild-type and ΔpyrF strains. Lanes 1 to 3: primers annealing to the pyrF promoter and to the 3′ end of the gene were used to amplify the pyrF gene of P. putida KT2442, of the ΔpyrF strain P. putida TEC1, and of TEC1 with the pyrF-containing pKP plasmid. Lanes 4 to 6: the chromosomal copy of ΔpyrF in P. putida TEC1 carrying pKP was amplified with primers annealing at the 5′ end of pyrF and xylE.
In a first step to develop a counterselection system based on pyrF, we tested the sensitivity of P. putida KT2442 to FOA. Plating of up to 109 CFU on solid minimal medium containing uracil and FOA revealed sensitivity to the compound, the spontaneous mutation frequency to FOA resistance being 0.8 × 10−6. In T. kodakaraensis and H. salinarum, the frequency of FOA resistance arising is 1 × 10−5 to 5 × 10−5 and 6.9 × 10−6, respectively (33, 38). Growth of FOA-resistant colonies on solid minimal medium showed that, contrary to what happens in these two microorganisms, mutation to FOA resistance was not associated with uracil auxotrophy. Instead, of 50 FOA-resistant P. putida colonies tested, all grew in the absence of uracil, such that resistance is likely to be associated either with FOA uptake or with the specific reaction catalyzed by the ODCase on FOA but not on uracil. The particularity in FOA resistance of P. putida is further noted by the fact that the plating of 109 CFU on solid medium with FOA but lacking uracil did not yield any resistant colonies. In any case, these figures were well within the range of what is useful for selection and counterselection of chromosome segments.
As a prerequisite to set up a counterselection system based on pyrF, we pursued the generation of a ΔpyrF P. putida strain so that pyrF in a plasmid could serve as a marker. To this end, a 2.0-kb region containing the pyrF gene was PCR cloned as described in the Materials and Methods section, and the subsequent cloning of an xylE cassette was carried out to allow pyrF deletion (Fig. 1B). The deletion construct (Fig. 1B) was cloned in pKNG101 (17), yielding pKPX, and introduced into P. putida KT2442. Chromosomal integration of the plasmid was selected by streptomycin resistance. Following growth without selection, a second recombination event that maintained the ΔpyrF copy was pursued. Efforts to use the sacB gene of pKPX as a counterselection marker in the presence of 5% or 8% sucrose to force resolution of the cointegrates repeatedly generated sucrose-resistant spontaneous mutants. On the contrary, plating the streptomycin-resistant exconjugants on minimal medium containing uracil and FOA resulted in the rise of FOA-resistant colonies, of which two of eight tested were uracil auxotrophs. These ura FOA-resistant colonies were also sensitive to streptomycin and turned yellow when sprayed with catechol (10 g/liter), the substrate of the xylE gene product, catechol-2,3-dioxygenase (not shown). These properties were indicative of the successful double recombination of the ΔpyrF::xylE segment into the chromosome of P. putida.
To confirm this, one of the ura clones that was FOA resistant, streptomycin-sensitive, and xylE+ (hereafter referred to as P. putida TEC1) was subjected to PCR analysis of the relevant chromosomal sequences. Figure 1C shows that the uracil auxotrophy and FOA resistance of P. putida TEC1 are associated with a band that corresponds in size to the deletion cassette with xylE (lanes 2 and 5). Note that when P. putida TEC1 carried a plasmid with the wild-type pyrF gene, the pyrF::xylE deletion cassette was not amplified with the pyrF primers, possibly due to preferential annealing of the primers to the wild-type gene. Indeed, whenever a wild-type copy of pyrF was present, amplification of the pyrF::xylE deletion cassette was not observed (see also Fig. 5, lane 6). P. putida TEC1 was thus chosen as the pyrF mutant of reference. That pyrF turned out to be far more powerful than sacB to select resolution of the cointegrate of pKPX with the chromosome of P. putida provided a good indication of the potential of the URA3 homologue for vector development.
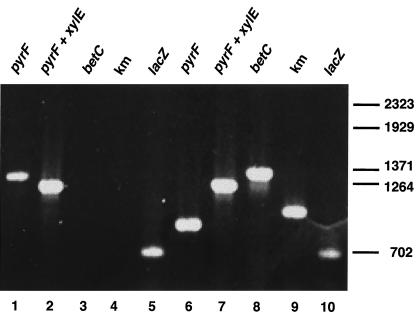
FIG. 5.
PCR analysis of P. putida TEC1 ΔbetCDE and FOA-resistant cointegrate colonies. Lanes 1 to 5, P. putida TEC1 ΔbetCDE colonies. Lanes 6 to 10, FOA-resistant colonies carrying a chromosomal insertion of pTGLB. Primers: lanes 1 and 6, anneal at pyrF gene promoter and 3′ end (as shown in Fig. 1B); lanes 2 and 7, anneal at pyrF gene 5′ end and xylE gene 5′ end (as shown in Fig. 1B); lanes 3 and 8, betC internal primers; lanes 4 and 9, kanamycin internal primers; lanes 5 and 10, lacZ internal primers.
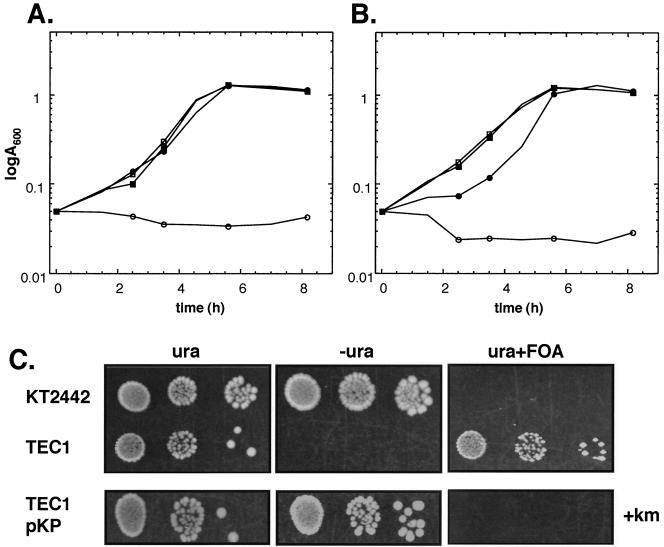
To confirm that pyrF encodes the P. putida orotidine-5′-monophosphate decarboxylase, growth of the wild-type and mutant strains was compared. Clearly, while the ΔpyrF P. putida strain TEC1 failed to grow in standard minimal medium, uracil addition sufficed to make growth of P. putida TEC1 and its wild-type counterpart P. putida KT2442 indistinguishable in liquid medium (Fig. 2A). This suggests that so long as an adequate amount of uracil is present, deletion of pyrF does not alter the growth rate. The uracil auxotrophy of P. putida TEC1 can be complemented by the pyrF+ plasmid pKP, carrying pyrF under its own promoter (Fig. 2B). That pyrF deletion confers FOA resistance, as should the deletion of ODCase, was shown by a growth assay on solid medium (Fig. 2C). While P. putida TEC1 is FOA resistant, P. putida KT2442 is sensitive. Uracil prototrophy and FOA sensitivity in P. putida TEC1 is regained upon complementation with pKP. Thus, the P. putida pyrF gene is a bona fide orotidine-5′-monophosphate decarboxylase, and this activity is associated with FOA sensitivity. Reversion of the ΔpyrF strains back to the wild-type pyrF genotype is straightforward, as P. putida TEC1 and its derivatives can be electroporated with the pyrF+ pUP plasmid (which does not replicate in P. putida) (Table 1), and following a round of antibiotic selection and growth without selection, plated on minimal medium without uracil. Spraying colonies with catechol allows visualization of xylE marker loss (not shown).
FIG. 2.
Uracil auxotrophy and FOA resistance conferred by pyrF deletion is complemented by plasmid-borne pyrF. (A) Strains P. putida KT2442 (squares) and P. putida TEC1 (circles) were grown at 30°C in liquid M9 minimal medium supplemented with citrate in the presence (solid symbols) or absence (open symbols) of uracil at 20 μg/ml. Growth was monitored at 600 nm after inoculation to 0.05 at 0 h. (B) Strains P. putida TEC1 carrying pKT231 (circles) or pKP (pKT231 carrying pyrF under its own promoter) (squares) were grown as in A. (C) Upper panel: Deletion of pyrF confers uracil auxotrophy and FOA resistance. Serial dilutions of P. putida KT2442 or TEC1 were plated on solid M9 minimal medium in the presence or absence of uracil (20 μg/ml) or with uracil and 5-fluoroorotic acid (250 μg/ml). Lower panel: Serial dilutions of P. putida TEC1/pKP were plated as above but with kanamycin, the resistance marker of pKP.
Structure and qualities of pTEC, a suicide plasmid for homologous gene replacement.
On the basis of the data above, we anticipated that the ΔpyrF background should permit the use of pyrF as a counterselection marker in a suicide vector. Thus, the essential elements of such a plasmid were put together by PCR cloning to make pTEC (Fig. 3A). These elements were the pyrF gene under its own promoter, a broad-host-range kanamycin resistance cassette flanked by multiple cloning sites, facilitating cloning of the desired genomic regions through which recombination is to take place, a segment of plasmid pGP704 (25) carrying a conditional origin of replication (oriV R6K), and an origin of transfer (oriT) for RK2-mediated mobilization (see Materials and Methods for details). This design allows, in a vector of small size, the insert of any DNA segment of interest and its propagation in specialized E. coli hosts (such as E. coli CC118λpir) engineered to express in trans the replication protein ∏ of plasmid R6K. Finally, pTEC derivatives can be passed to P. putida ΔpyrF by bipartite or tripartite conjugation mediated by the tra genes of plasmid RK2 (which can be provided by a helper strain, such as HB101 carrying pRK600; see Table 1). Formation of a cointegrate between pTEC derivatives and the chromosome of P. putida TEC1 can be selected by plating in minimal medium without uracil (and, if desired, with kanamycin), while its resolution can be forced by replating in medium containing FOA.
Generation and characterization of a P. putida ΔbetCDE strain.
The performance and survival of microorganisms, including P. putida, in soil and the rhizosphere of plants relies to a large extent on their ability to endure desiccation and osmotic stress (18, 24). Choline-O-sulfate has been shown to be an efficient osmoprotectant in several plants (14, 36) and microbial species (4, 13, 14, 23, 26, 32). It is thought that choline-O-sulfate is found in soil, as plants (30), fungi (1, 22), and bacteria have transport systems for this ester (4, 26, 29). In bacteria, choline-O-sulfate can be transformed into choline (31, 35), which is subsequently transformed into glycine betaine (41), a potent osmoprotectant (5).
We have found that the genome of P. putida bears a putative choline-O-sulfate utilization operon, named betCDE, for choline-O-sulfate uptake (ORFs PP0075 and PP0076) and conversion to choline (ORF PP0077; Galvão et al., unpublished data). To examine the role of choline-O-sulfate in osmoprotection of P. putida in a rhizosphere microcosm system, we set out to generate a lacZ+-traceable ΔbetCDE strain, the expectation being that such a deletion would affect P. putida's response to desiccation and osmotic stress. Production of such a strain with a standard sacB-based vector system such as pKNG101 (17) failed repeatedly. Besides the problems inherent in the use of sacB mentioned above, mutations involved in osmotic stress may be particularly difficult to produce with a counterselection system which involves the addition of ≥5% solute, which causes considerable osmotic pressure. This scenario provided the most appropriate case to test the performance of the pyrF-based allelic exchange system presented above.
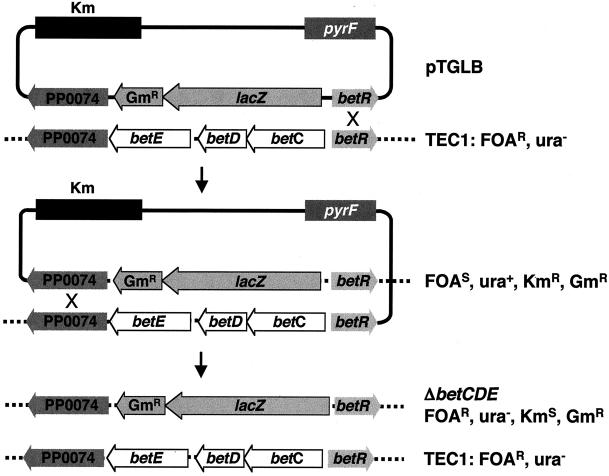
To this end, a DNA segment carrying a gentamicin resistance cassette and the β-galactosidase (lacZ) gene flanked by the genomic sequences upstream and downstream of betCDE (ORF PP0074 and betR) was assembled (Fig. 3B). The β-galactosidase gene is cloned as a transcriptional fusion to the betCD promoter, as indicated by a small arrow ahead of the lacZ gene (Galvão et al., unpublished data). This segment was cloned into the NotI site of pTEC to yield plasmid pTGLB. This construct was transferred to P. putida TEC1 by tripartite conjugation. Recombination of pTGLB into the chromosome was selected on minimal medium with kanamycin and gentamicin but lacking uracil, thus using positive selection for uracil prototrophy upon plasmid integration. Following growth in liquid medium supplemented with uracil but without antibiotic selection, cells in which a second recombination event had taken place (resulting in the betCDE deletion) were selected by plating on minimal medium with gentamicin, uracil, and FOA (Fig. 4).
FIG. 4.
Schematic map for pyrF-based counterselection for generation of homologous gene recombination with pTEC. Chromosomal integration events by homologous recombination between PP0074 or betR chromosomal and plasmid (pTGLB) sequences are selected by plating on medium with kanamycin (Km) or gentamicin (Gm) or both but lacking uracil. Cells with the plasmid integrated in the chromosome are grown without selection to allow a second recombination event that maintains either the wild-type or the deleted betCDE operon. The ΔbetCDE mutant is isolated by selecting cells that grow in the presence of FOA, gentamicin, and uracil but are kanamycin sensitive and uracil auxotrophs.
A total of 810 colonies of two morphologies grew when 108 CFU were plated in these conditions. The major morphotype (760, or 88%) represented ΔbetCDE deletion mutants, as judged by the uracil auxotrophy and kanamycin sensitivity of 25 colonies analyzed. This represents a respectable double-recombination frequency of nearly 1.3 × 10−5. PCR analysis showed that the FOA-resistant, kanamycin-sensitive colonies no longer had pTGLB in the chromosome, as judged by loss of the wild-type pyrF copy, the betC gene, and the kanamycin cassette while maintaining the β-galactosidase gene in the deletion cassette (Fig. 5, lanes 1 to 5). The second morphotype consisted of faster-growing colonies, which were found to be FOA resistant uracil prototrophs as well as resistant to kanamycin. That these colonies carry mutation to FOA resistance containing integrated pTGLB was confirmed by PCR analysis, which showed the presence of both wild-type and ΔpyrF copies as well as of the betC gene and the kanamycin cassette (Fig. 5, lanes 6 to 10). The frequency at which these FOA-resistant colonies arose is 2 × 10−6 (50 FOA-resistant colonies per 108 CFU plated), near that of P. putida KT2442 spontaneous mutation to FOA resistance (see above). That efficient homologous gene replacement is possible through use of pyrF-based selection is made clear by the fact that the frequency of double-recombination events generating knockout strains (1.3 × 10−5) is at least 6.5 times higher than that of spontaneous FOA resistance (2 × 10−6).
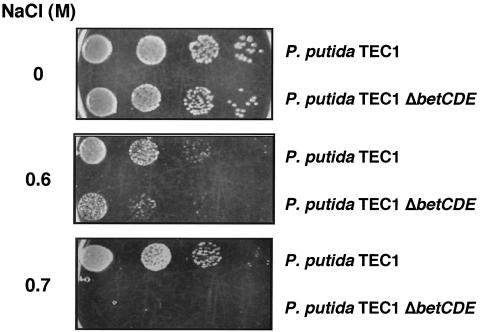
To first characterize the P. putida TEC1 ΔbetCDE strain, its sensitivity to osmotic stress was tested by looking at growth in solid medium containing high salt. Figure 6 shows that the ΔbetCDE mutant was more sensitive to high salt than P. putida TEC1, as in the presence of 0.6 M NaCl the mutant grows much more slowly than P. putida TEC1 and is not viable in 0.7 M NaCl. Thus, deletion of betCDE renders P. putida more sensitive to osmotic stress. Sensitivity to osmotic stress in the absence of protectant compounds is interesting despite the fact that the putative role of the betCDE gene products is related to osmoprotectant uptake and transformation. Because the betCDE mutant lacks a putative transmembrane protein, BetE (Galvão et al., unpublished data), it is possible that the internal membrane is altered, becoming more susceptible to osmotic stress. The betCDE mutant is the basis of further studies under laboratory conditions as well as in the field.
FIG. 6.
Deletion of betCDE renders P. putida TEC1 susceptible to osmotic stress. Serial dilutions of P. putida TEC1 and TEC1 ΔbetCDE were plated on M9 minimal medium supplemented with uracil in the presence or absence of 0.6 or 0.7 M NaCl and grown at 30°C for 24 h (0 M NaCl) or 72 h (0.6 and 0.7 M NaCl).
Use of pTEC in other gram-negative bacteria.
Employing pTEC for homologous gene replacement in other gram-negative bacteria requires generating a pyrF background. This requirement is compensated for by the possibility of obtaining such mutants by plating on FOA, as was done with T. kodakaraensis KOD1 (38) and H. salinarum (33). While full deletion of pyrF has the advantage of avoiding pTEC integration in the pyrF locus, in practice the use of pyrF-negative, FOA-resistant strains generated by spontaneous mutations implies that ≤50% of cointegrates are inadequate and thus require an additional screening step, easily achieved by PCR. Alternatively, pyrF deletion mutants may be generated with pKPX, the pKNG-based suicide plasmid carrying the P. putida gene (see above; Table 1), if there is a high sequence similarity between the pyrF genes of the organism being used and P. putida.
A BLAST search with the P. putida pyrF sequence against the EMBL database shows it to be highly homologous to sequences of several gram-negative bacteria (Pseudomonas syringae, 83%; P. aeruginosa, 82%; Chromobacterium violaceum, 71%; Neisseria meningitidis, 65%; E. coli, 64%; Vibrio vulnificus, 63%; Salmonella enterica serovar Typhimurium, 64%; Shigella flexneri, 63%; Photorhabdus luminescens, 61%; Yersinia pestis, 61%; and Shewanella oneidensis, 60%). Whether these degrees of homology are sufficient to allow homologous recombination will have to be checked for each organism individually with the simple test provided by the xylE+ marker of the ΔpyrF deletion.
Conclusion.
We have developed a vector system that employs the pyrF gene as a selection marker to allow efficient homologous gene replacement. We tested the plasmid in Pseudomonas putida and found that pyrF is a reliable marker in positive and negative selection. At least three strategies can be applied for generating gene knockouts with pTEC. First is the one presented here, where a deletion cassette containing an antibiotic resistance marker is cloned directly into pTEC, yielding a knockout strain in which pTEC can be employed again. Second, PCR products of genomic regions adjacent to the gene of interest may be cloned in the multiple cloning sites flanking the kanamycin resistance cassette in pTEC. Third, a deletion cassette that contains only the genomic regions adjacent to the gene of interest is cloned into pTEC, such that the resulting knockout is antibiotic marker free and additional deletions can be generated with the same vector system. In all cases, the essential features of the system are pyrF-based selection, where the initial recombination event is selected by uracil prototrophy and the double-recombination event is selected by resistance to FOA and by uracil auxotrophy.
Acknowledgments
David Canovas is acknowledged for providing the pGGLB plasmid and for useful discussions. The help of the E. coli Stock Center with strain X82 is greatly appreciated.
This work was supported by the Biocarte and Lindane Contracts of the European Union and by Comunidad Autónoma de Madrid grant 07 M/0075/2002.
REFERENCES
- 1.Bellenger, N., P. Nissen, T. C. Wood, and I. H. Segel. 1968. Specificity and control of choline-O-sulfate transport in filamentous fungi. J. Bacteriol. 96:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 4.Canovas, D., C. Vargas, L. N. Csonka, A. Ventosa, and J. J. Nieto. 1996. Osmoprotectants in Halomonas elongata: high-affinity betaine transport system and choline-betaine pathway. J. Bacteriol. 178:7221-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csonka, L. N., and A. D. Hanson. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569-606. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, X., P. Stothard, I. J. Forsythe, and D. S. Wishart. 2004. PlasMapper: a web server for drawing and auto-annotating plasmid maps. Nucleic Acids Res. 32:660-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabret, C., S. D. Ehrlich, and P. Noirot. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 46:25-36. [DOI] [PubMed] [Google Scholar]
- 10.Franklin, F. 1985. Broad host range cloning vectors for Gram-negative bacteria. DNA cloning: a practical approach. IRL Press, Oxford, England.
- 11.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guiney, D. G., and E. Yakobson. 1983. Location and nucleotide sequence of the transfer origin of the broad host range plasmid RK2. Proc. Natl. Acad. Sci. USA 80:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez, J. A., and L. N. Csonka. 1995. Isolation and characterization of adenylate kinase (adk) mutations in Salmonella typhimurium which block the ability of glycine betaine to function as an osmoprotectant. J. Bacteriol. 177:390-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson, A. D., B. Rathinasabapathi, B. Chamberlin, and D. A. Gage. 1991. Comparative physiological evidence that β-alanine betaine and choline-O-sulfate act as compatible osmolytes in halophytic Limonium species. Plant Physiol. 97:1199-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inuzuka, M., and D. R. Helinski. 1978. Requirement of a plasmid-encoded protein for replication in vitro of plasmid R6K. Proc. Natl. Acad. Sci. USA 75:5381-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 18.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 19.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 20.Knipfer, N., A. Seth, and T. E. Shrader. 1997. Unmarked gene integration into the chromosome of Mycobacterium smegmatis via precise replacement of the pyrF gene. Plasmid 37:129-140. [DOI] [PubMed] [Google Scholar]
- 21.Lucas, S., L. Toffin, Y. Zivanovic, D. Charlier, H. Moussard, P. Forterre, D. Prieur, and G. Erauso. 2002. Construction of a shuttle vector for, and spheroplast transformation of, the hyperthermophilic archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 68:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marzluf, G. A. 1972. Genetic and metabolic control of sulfate metabolism in Neurospora crassa: a specific permease for choline-O-sulfate. Biochem. Genet. 7:219-233. [DOI] [PubMed] [Google Scholar]
- 23.Mason, T., and G. Blunden. 1989. Quaternary ammonium and tertiary sulphonium compounds of algal origin as alleviators of osmotic stress. Bot. Mar. 32:313-316. [Google Scholar]
- 24.Miller, K. J., and J. M. Wood. 1996. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50:101-136. [DOI] [PubMed] [Google Scholar]
- 25.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nau-Wagner, G., J. Boch, J. A. Le Good, and E. Bremer. 1999. High-affinity transport of choline-O-sulfate and its use as a compatible solute in Bacillus subtilis. Appl. Environ. Microbiol. 65:560-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 28.Neuhard, J. 1983. Utilization of preformed pyrimidine bases and nucleosides. p. 95-148. In O. Munch-Petersen (ed.), Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Academic Press, New York, N.Y.
- 29.Nissen, P. 1968. Choline sulfate permease: transfer of information from bacteria to higher plants? Biochem. Biophys. Res. Commun. 32:696-703. [DOI] [PubMed] [Google Scholar]
- 30.Nissen, P., and A. A. Benson. 1964. Active transport of choline sulfate by barley roots. Plant Physiol. 39:586-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osteras, M., E. Boncompagni, N. Vincent, M. C. Poggi, and D. Le Rudulier. 1998. Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: choline-O-sulfate is metabolized into glycine betaine. Proc. Natl. Acad. Sci. USA 95:11394-11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, Y., and J. E. Gander. 1998. Choline derivatives involved in osmotolerance of Penicillium fellutanum. Appl. Environ. Microbiol. 64: 273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peck, R. F., S. Dassarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 34.Pieper, D. H., V. A. Martins dos Santos, and P. N. Golyshin. 2004. Genomic and mechanistic insights into the biodegradation of organic pollutants. Curr. Opin. Biotechnol. 15:215-224. [DOI] [PubMed] [Google Scholar]
- 35.Rathinasabapathi, B., M. Burnet, B. L. Russell, D. A. Gage, P. C. Liao, G. J. Nye, P. Scott, J. H. Golbeck, and A. D. Hanson. 1997. Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: prosthetic group characterization and cDNA cloning. Proc. Natl. Acad. Sci. USA 94:3454-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivoal, J., and A. D. Hanson. 1994. Choline-O-Sulfate Biosynthesis in Plants (Identification and Partial Characterization of a Salinity-Inducible Choline Sulfotransferase from Species of Limonium (Plumbaginaceae). Plant Physiol. 106:1187-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., T. Maniatis, and T. Fritsch. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schweizer, H. P., and V. de Lorenzo. 2004. Molecular Tools for Genetic Analysis of Pseudomonads, p. 317-350. In J. L. Ramos (ed.), Pseudomonas, 1st ed., vol. 1. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 40.Simon, R., B. Hotte, B. Klauke, and B. Kosier. 1991. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J. Bacteriol. 173:1502-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, L. T., J. A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein, D. C. 1992. Plasmids with easily excisable xylE cassettes. Gene 117:157-158. [DOI] [PubMed] [Google Scholar]
- 43.Strych, U., S. Wohlfarth, and U. K. Winkler. 1994. Orotidine-5′-monophosphate decarboxylase from Pseudomonas aeruginosa PAO1: cloning, overexpression, and enzyme characterization. Curr. Microbiol. 29:353-359. [DOI] [PubMed] [Google Scholar]
- 44.van Wezel, G. P., and M. J. Bibb. 1996. A novel plasmid vector that uses the glucose kinase gene (glkA) for the positive selection of stable gene disruptants in Streptomyces. Gene 182:229-230. [DOI] [PubMed] [Google Scholar]
- 45.Yakobson, E. A., and D. G. Guiney, Jr. 1984. Conjugal transfer of bacterial chromosomes mediated by the RK2 plasmid transfer origin cloned into transposon Tn5. J. Bacteriol. 160:451-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]