Abstract
It is known that an ineffective antibiotic treatment can induce morphological shifts in uropathogenic Escherichia coli (UPEC) but the virulence properties during these shifts remain to be studied. The present study examines changes in global gene expression patterns and in virulence factor-associated genes in an extended spectrum beta-lactamase (ESBL)-producing UPEC (ESBL019) during the morphologic transitions induced by an ineffective antibiotic and in the presence of human primary bladder epithelial cells. Microarray results showed that the different morphological states of ESBL019 had significant transcriptional alterations of a large number of genes (Transition; 7%, Filamentation; 32%, and Reverted 19% of the entities on the array). All three morphological states of ESBL019 were associated with a decreased energy metabolism, altered iron acquisition systems and altered adhesion expression. In addition, genes associated with LPS synthesis and bacterial motility was also altered in all the morphological states. Furthermore, the transition state induced a significantly higher release of TNF-α from bladder epithelial cells compared to all other morphologies, while the reverted state was unable to induce TNF-α release. Our findings show that the morphological shifts induced by ineffective antibiotics are associated with significant transcriptional virulence alterations in ESBL-producing UPEC, which may affect survival and persistence in the urinary tract.
Keywords: filamentation, extended-spectrum β-lactamase, uropathogenic Escherichia coli, ineffective antibiotics, morphological plasticity
Introduction
Urinary tract infections (UTIs) are one of the most common bacterial infections. UTI is especially common in women as nearly half of all women will be infected during their lifetime (Foxman, 2003). The vast majority of all urinary tract infections are caused by uropathogenic Escherichia coli (UPEC), accounting for approximately 80% of community-acquired symptomatic UTI and approximately 25% of the nosocomial infections (Ronald, 2003). UPEC isolates have been shown to express an array of virulence factors like adhesins (such as P- and type-1 fimbriae), toxins (α-hemolysin), lipopolysaccharide (LPS), capsular, siderophores (iron scavenger system) and TcpC to evade the host defenses and colonize the urinary tract (Bower et al., 2005; Yadav et al., 2010).
During UTI, UPEC may invade the urothelial cells where they can replicate and form biofilm-like intracellular bacterial communities (IBCs) (Anderson et al., 2003). Likewise, after the resolution of an active infection non-replicating bacteria can remain within vesicles in deeper layers of the urothelium as quiescent intracellular reservoirs (QIRs) (Mysorekar and Hultgren, 2006). These QIRs can remain quiescent for several months without eliciting an inflammatory response. The intracellular niche not only protects the bacteria from many of the innate immune defenses, such as phagocytosis by neutrophils (Justice et al., 2004), but also from antibiotics that often are ineffective in eliminating intracellular bacteria (Blango and Mulvey, 2010). The subsequent resurgence of these intracellular reservoirs can be the cause of recurrent or chronic UTI (Rosen et al., 2007). UPEC exhibit morphological plasticity within the IBCs (Justice et al., 2004, 2006; Horvath et al., 2011). In the initial phase, bacteria transform from a rod shape to coccoid morphology that may follow by an additional transition into a filamentous form (Justice et al., 2004; Horvath et al., 2011). As bacteria emerge from the IBC they may do so in a filamentous form (Justice et al., 2004). Filamentous E. coli have been observed in urine from women with UTI and the filamentation per se may serve as a strategy to resist host immune responses, such as phagocytosis (Horvath et al., 2011), but may also be seen as a more general stress response. Filamentation can be induced by components of an activated host defense system (Justice et al., 2006), shear stress (Khandige et al., 2016) and also in response to treatment with different beta-lactam antibiotics, in particular, aminothiazolyl cephalosporins like ceftibuten. Beta-lactam antibiotics induce E. coli filamentation by inhibiting penicillin-binding protein-3 (PBP-3) that catalyzes septa formation (Gould and MacKenzie, 1997). SulA, a member of the bacterial SOS DNA damage repair system, has also been shown to induce bacterial filamentation. It was recently shown that cell division gene damX is also a mediator of reversible filamentation of UPEC (Khandige et al., 2016). We have previously shown that filamentation of extended spectrum β-lactamase (ESBL)-producing UPEC isolates can alter the ability of UPEC to evoke pro-inflammatory responses in renal epithelial cells and neutrophils (Demirel et al., 2015). Therefore, knowledge of the virulence factors expressed by different morphological forms of UPEC may be used to devise novel therapeutic strategies to obstruct these virulence factors in order to inhibit pathogenesis.
A major concern and emerging health problem is the increasing incidence of antibiotic resistance among uropathogenic strains (Coque et al., 2008; Brolund, 2014). A majority of the ESBL-producing bacteria are isolated from urine samples (Pitout and Laupland, 2008; Khanfar et al., 2009). In addition, mortality has been shown to increase in ESBL-induced infections proportionally with the duration of the delay in effective treatment, making the choice of correct antibiotics critical for patients (Tumbarello et al., 2007).
The virulence capacity of ESBL-producing UPECs during their morphological shifts remains to be properly studied. The aim of the present study was to investigate changes in global gene expression patterns and in virulence factor-associated genes in ESBL-producing UPEC during the morphologic transitions induced by ineffective antibiotics in the presence of human primary bladder epithelial cells.
Material and methods
Cell and bacterial culture
Primary human bladder epithelium progenitor cells (HBEP, CELLnTEC Advanced Cell Systems AG, Bern, Switzerland) were cultured in wells with CnT-58 cell culture medium (CELLnTEC) supplemented with 100 U/mL penicillin and 100 μL/mg streptomycin (both from Invitrogen Ltd, Paisley, UK) in a humidified atmosphere with 5% CO2 at 37°C. At confluency the culture was differentiated during 4 days using CnT-21 differentiation medium (CELLnTEC) supplemented with 1 mM CaCl2. The ESBL-producing UPEC was originally isolated from a patient at Örebro University hospital, Sweden. The isolate, designated ESBL019 (previously named ESBL7, Demirel et al., 2015), has previously been characterized and determined to be multidrug resistant (CTX-M-15) and resistant to ceftibuten (MIC 7,680 ng/ml) and belongs to the ST131 clone (Onnberg et al., 2011). ESBL019 was grown in Luria broth (Difco Laboratories, Detroit, MI, USA) overnight on shake at 200 rpm 37°C. The bacteria were resuspended in sterile phosphate buffered saline prior to inoculation of CnT-21 cell culture medium (CCM) with or without ceftibuten (480 ng/mL). The antibiotic concentration used is a reference minimal inhibitory concentration (MIC) of ceftibuten that was determined on two antibiotic susceptible UPEC isolates as previously defined (Demirel et al., 2015).
Morphologic transitions and evaluation
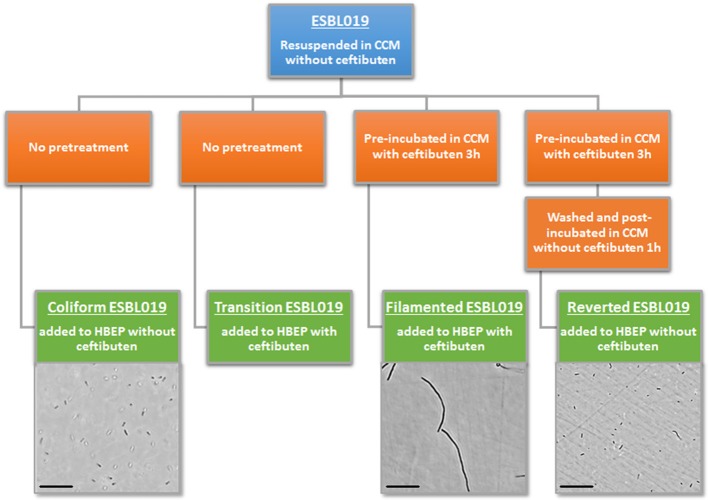
Four morphologic states of ESBL019 were used during the experiments (Figure 1). A first ESBL019 morphological state was prepared by resuspension of ESBL019 in CCM without ceftibuten supplementation prior to its inoculation of HBEP cells without ceftibuten (designated; ESBL019 Coliform). A second ESBL019 morphological state was prepared by resuspension in CCM and then used to inoculate HBEP cells with ceftibuten supplementation (480 ng/mL) (designated; ESBL019 Transition). This transition from coliform into filamented occurs in the presence of HBEP cells. A third ESBL019 morphological state was prepared by resuspension and pre-incubation for 3 h in CCM with ceftibuten in a tube, in order to induce complete filamentation of all bacteria. Filamentation was confirmed by microscopy. The third pre-filamented ESBL019 fraction was used to inoculate HBEP cells in the presence of ceftibuten to keep the bacteria filamented (designated; ESBL019 Filamented). The CCM of the pre-filamented ESBL019 fraction was replaced with CCM without ceftibuten and the filamented ESBL019 were allowed to revert completely back to its coliform during 1 h in a tube prior to the inoculation of HBEP cells without ceftibuten (designated; ESBL019 Reverted) (Figure 1). The bacterial morphologies were evaluated with an Olympus CKX41 inverted light microscope (Olympus, Segrate, Italy). Prior to inoculation the bacterial concentrations of all fractions were adjusted in order to inoculate HBEP cells with a multiplicity of infection (MOI) of 10. All inoculated cells were incubated for 4 h in 5% CO2 at 37°C after which supernatants were collected and centrifuged 5 min at 5,000 × g to collect the extracellular bacteria. The viability of the HBEP cells was >90% after the infection as determined by the neutral red assay. The HBEP cells were then lysed with sterile MQ water for 3 min and the intracellular bacteria were collected and centrifuged for 5 min at 5,000 × g followed by a washing step with PBS.
Figure 1.
Summary of the experimental design, morphological transition and visual evaluation by light microscope of the different forms of ESBL019. Scale bar: 100 μm.
RNA preparation and microarray
Total RNA was isolated from the combined intra-and extracellular bacteria with the RNeasy Mini Kit (Qiagen Technologies, Hilden, Germany) according to manufacturer instructions. Contaminating DNA was removed by DNase digestion (TURBO DNase, Life technologies, MA, USA) according to manufacturer instructions. RNA concentration and purity were determined using a Nano-Drop ND-1000 Spectrophotometer (Nano-Drop Technology Inc., Wilmington, DE, USA). All samples had OD260/280 and OD260/230 ratios above 1.9. RNA quality was further evaluated using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) according to manufacturer instructions. All samples had RNA integrity number (RIN) values above 9. High-quality total RNA from four biological replicates was used to prepare labeled cRNA with Low Input Quick Amp WT Labeling Kits (Agilent) according to manufacturer instructions. Labeled cRNA samples were hybridized in a G2545A hybridization oven (Agilent) onto Agilent SurePrint G3 E. coli Gene Expression 8x15k (Agilent Technologies) glass arrays according to manufacturer instructions and subsequently scanned with a G2565 CA array laser scanner (Agilent Technologies). Image analysis and data extraction was performed with Feature Extraction Software (version 10.7.3.1, Agilent Technologies). The virulence factor list for E. coli was generated through the use of virulence association in the PATRIC database (www.patricbrc.org) with the addition of MESH virulence search term associated genes and literature mining. Gene expression data is available in the GEO database with the accession number GSE99661.
Quantitative real-time PCR (qPCR)
cDNA synthesis (200 ng of total RNA) was performed by using High Capacity cDNA Reverse Transcription Kit for single-stranded cDNA synthesis (Applied Biosystems, CA, USA) according to manufacturer‘s protocol. Maxima SYBR Green qPCR Master Mix (ThermoFisher Scientific, MA, USA) was used for the qPCR according to manufacturer's instructions. Two hundred and fifty nanometer of primer and 10 ng template cDNA was added to each reaction. Primers were ordered from Eurofins MWG Synthesis GmbH (Ebersberg, Munich, Germany) (Table S1). The amplification was performed in a CFX96 Touch™ Real-Time PCR Detection System (Biorad, CA, USA) using the following protocol: initial denaturation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s followed by annealing at 60°C for 30 s and extension at 72°C for 30 s. The PCR was followed by a dissociation curve analysis between 60 and 95°C. The Ct values were analyzed by the comparative Ct (ΔΔCt) method and normalized to the endogenous control gapA (encoding glyceraldehyde 3-phosphate dehydrogenase A). Fold difference was calculated as 2−ΔΔCt.
Transmission electron microscopy
ESBL019 was prepared for electron microscopy with or without ceftibuten as previously mentioned, but without the presence of HBEP cells. ESBL019 was washed in PBS before fixation with 5% glutaraldehyde in 0.1 M phosphate buffer. The bacteria were absorbed on carbon-coated copper grids and contrasted with 1% uranyl acetate solution. A JEOL JEM 1230 transmission electron microscope at 80-kV accelerating voltage was used for examination and images were recorded with a Gatan Multiscan 791 CCD camera.
Agglutination assays
ESBL019 in its four different morphological states were collected after HBEP inoculation to measure Type-1-and P-fimbriae phenotype expression. The expression of Type-1 fimbriae was evaluated using 5% yeast diluted in PBS and the expression of P-fimbriae was evaluated using 2% of P-positive human erythrocytes diluted in PBS. All the bacteria from the well were mixed 1:1 (v/v) with yeast or erythrocytes on a glass slide and agglutination was evaluated with an Olympus CKX41 inverted light microscope.
Measurement of TNF-α release from primary bladder epithelial cells
An enzyme-linked immunosorbent assay (ELISA) was performed to measure the TNF-α release from HBEP cells in response to the different morphological states of ESBL019. TNF-α was measured using the human TNF-α kit (ELISA MAX™ Standard, BioLegend, San Diego, CA, USA) according to the manufacturer's protocol and measured on a spectrophotometer (Multiskan Ascent, Thermo Labsystems, Helsingfors, Finland).
Statistical analysis and microarray data processing
All microarray data analysis was performed using Gene Spring GX version 13.0 (Agilent Technologies) after per chip and gene 75th percentile shift normalization of samples. Different expression between groups was analyzed with one-way analysis of variance (ANOVA) parametric test. Significantly expressed entities (p < 0.05) was obtained by Tukey HSD post hoc test followed by Bonferroni multiple testing correction and a fold change set at ≥2. Significance for GO term enrichment and single experiment pathway analysis were set at a p-value < 0.05 and p-value < 0.1 respectively. The virulence factor list for E. coli was generated through the use of virulence association in the PATRIC database (www.patricbrc.org) with the addition of MESH virulence search term associated genes and literature mining. In total the list was composed of 305 genes classified as associated with or designated to virulence. The differences between groups in the ELISA assay were evaluated with one-way ANOVA followed by Bonferroni test. The differences compared to ESBL019 Coliform in the qPCR were evaluated with Student's unpaired t-test. Differences were considered statistically significant when p < 0.05. Data are presented as mean ± standard error of the mean (SEM), n = number of independent experiments.
Results
Morphological plasticity of E. coli in response to ceftibuten exposure
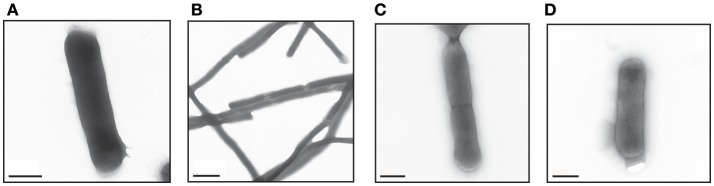
ESBL019 is a strain resistant to ceftibuten. It changed morphology from its normal coliform into a filamentous form (>100 μm) after exposure to ceftibuten in the presence of human primary bladder epithelial cells (HBEP). This transition in the presence of HBEP is defined as ESBL019 Transition. The filamentation process started as early as 1 h after the addition of ceftibuten and the bacteria reached full filamentation approximately 3 h after exposure to ceftibuten. After removing ceftibuten from the growth medium, the filamentous ESBL019 reverted completely back to its coliform within 1 h (Figure 1). Transmission electron microscopy showed a normal Coliformed bacteria (Figure 2A) and that the septa between the bacteria was not fully formed in the filamented phase (Figure 2B). The reversion from the filamentous form back to the coliform was associated with formation of the bacterial septa (Figures 2C,D).
Figure 2.
Visual evaluation of the different forms of ESBL019 by transmission electron microscope. ESBL019 Coliform (A) was grown for 3 h without ceftibuten, ESBL019 Filamented (B) was grown for 3 h in the presence of ceftibuten and ESBL019 Reverted (C,D) was grown in the presence of ceftibuten for the first 3 h and then without for 1 h to allowed the bacteria to revert completely back to its coliform. Septa formation during reversion can also be observed (C). Scale bar: 0.5 μm (A,C,D) 2 μm (B).
Gene expression alterations in the different morphological phases
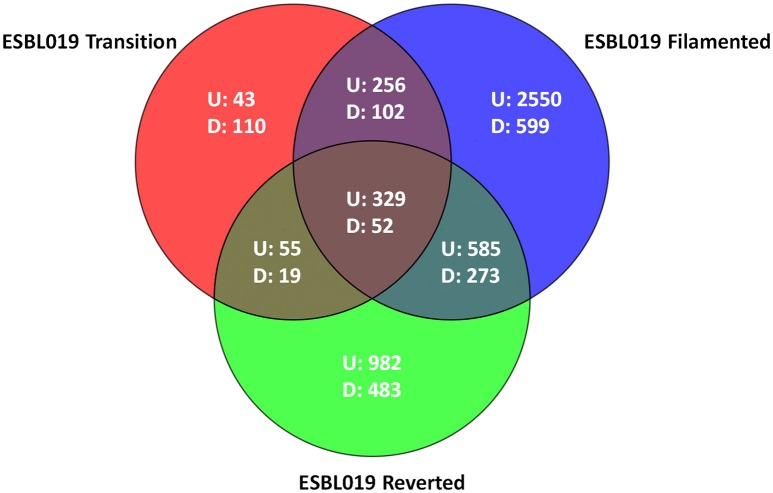
Microarray analysis was performed on total RNA isolated from (intra and extracellular) bacteria prior to and following their morphological transition from coliform to filamentous form and then reverted to their original coliform. In total 6438 entities were differentially expressed (p < 0.05) with at least a ≥2 fold change compared to the ESBL019 Coliform. All three morphological states, ESBL019 Transition, ESBL019 Filamented and ESBL019 Reverted, induced changes in gene expressions with 683, 3,720, and 1,951 up-regulated and 283, 1,026, and 827 down-regulated gene expressions, respectively (Figure 3). A total of 739 altered gene entities are shared between ESBL019 Transition and ESBL019 Filamented with 585 up-regulated and 157 down-regulated entities, and 1,239 gene entities were shared between ESBL019 Filamented and ESBL019 Reverted with 914 up-regulated and 325 down-regulated entities. In addition, 381 shared gene entities were differentially expressed in all three groups (Figure 3).
Figure 3.
Venn diagram of differentially expresses entities. Red represents ESBL019 Transition compared to ESBL019 Coliform, blue represents E7 Filamented compared to ESBL019 Coliform and green represents ESBL019 Reverted compared to E7 Coliform. Overlapping regions represents shared entities. Up- and down-regulated entities are designated U and D respectively. n = 4 per group.
Gene ontology and pathway analysis
Gene ontology (GO) analysis was performed on significantly altered gene entities compared to ESBL019 Coliform expression. In total, 14 (ESBL019 Transition), 9 (ESBL019 Filamented) and 4 (ESBL019 Reverted) gene ontologies were uniquely enriched (Table S2). Single experiment pathway analysis (SEA) (p < 0.05) was performed in order to determine significantly affected cellular pathways. In total, 33 pathways were altered, of which 30 pathways were related to metabolism and the rest to enterobactin biosynthesis and polymyxin resistance (Table S3).
Virulence factors in the different morphological states
Next, a more detailed analysis of the alterations in expression patterns of virulence associated genes of ESBL019 was conducted. The compiled list of virulence factor associated genes, 305 entities in total, was used to filter out significantly altered entities associated with virulence in the different groups. In total, 199 altered gene entities were identified with association to virulence. These 199 altered gene entities were grouped into 10 different functional groups; Adhesion, Growth, LPS synthesis, Metabolism, Motility, Protection, Siderophore, Toxins, Secretion and Transcription regulation. Of the 199 genes, 19 genes had significantly altered expressions in all three morphological states compared to the original ESBL019 Coliform (Table 1A). The ESBL019 Transition state showed altered expression of 33 virulence associated genes (Tables 1A,B). The ESBL019 Filamented state showed the single largest alteration in virulence associated gene expression. A total of 149 altered expression in genes was observed in the ESBL019 Filamented state (Tables 1A,B). The ESBL019 Reverted state showed altered expression of 92 virulence associated genes (Tables 1A,B).
Table 1A.
Shared virulence factors associated genes present among significantly altered entities in ESBL019 Transition, ESBL019 Filamented and ESBL019 Reverted compared to ESBL019 Coliform.
| Function | Gene | ESBL019 Coliform | Fold change vs. ESBL019 coliform | Description | ||
|---|---|---|---|---|---|---|
| Normalized entity fluorescence intensity signals | ESBL019 transition | ESBL019 filamentous | ESBL019 reverted | |||
| Adhesin | papG | 5 | 2.4 | 3.0 | 2.7 | PapG protein |
| papF | 5 | 2.8 | 4.2 | 2.2 | PapF protein | |
| Growth | carB | 7,750 | −2.5 | −6.0 | −2.7 | Carbamoyl-phosphate synthase large subunit |
| LPS synthesis | waaV | 4 | 3.0 | 4.3 | 2.3 | Putative beta1.3-glucosyltransferase |
| rfbC | 4 | 2.9 | 6.5 | 2.7 | dTDP-6-deoxy-D-glucose-3.5 epimerase | |
| Metabolism | mglB | 2,552 | −3.7 | −10.0 | 3.5 | D-galactose-binding periplasmic protein precursor |
| Motility | fliC | 5 | 2.4 | 3.8 | 2.2 | Flagellar biosynthesis flagellin. filament structural protein |
| Protection | tcpC | 6 | −4.2 | −4.1 | −2.1 | TCP pilus biosynthesis protein tcpC |
| ompT | 5,577 | 2.5 | −2.7 | −5.2 | Protease VII precursor | |
| wcaE | 13 | 3.6 | 31.7 | 3.2 | Putative colanic acid biosynthesis glycosyl transferase | |
| ibpA | 155 | 7.1 | 37.4 | 4.3 | Heat shock protein | |
| ibpB | 124 | 6.0 | 56.6 | 12.0 | Heat shock protein | |
| Siderophore | sitB | 1,841 | −4.5 | −10.9 | −2.8 | SitB protein |
| chuT | 1,255 | −7.0 | −9.1 | −2.3 | Putative Periplasmic binding protein | |
| fepE | 5,165 | −2.4 | −5.4 | −2.9 | Ferric enterobactin transport protein fepE | |
| fepA | 2,007 | −3.0 | −3.9 | −2.6 | Ferrienterobactin receptor precursor | |
| iroN | 5 | 2.6 | 3.5 | 2.9 | Siderophore receptor IroN | |
| entF | 6 | 2.4 | 3.7 | 2.0 | ATP-dependent serine activating enzyme | |
| Toxins | hlyC | 7 | 2.8 | 3.7 | 2.1 | Hemolysin C |
Table 1B.
Virulence factors associated genes present among significantly altered entities in ESBL019 Transition, ESBL019 Filamented and ESBL019 Reverted compared to ESBL019 Coliform.
| Function | Gene | ESBL019 coliform | Fold change vs. ESBL019 Coliform | Description | ||
|---|---|---|---|---|---|---|
| Normalized entity fluorescence intensity signals | ESBL019 transition | ESBL019 filamentous | ESBL019 reverted | |||
| Protection | degP | 6,718 | 2.1 | Periplasmic serine protease Do; heat shock protein HtrA | ||
| Siderophore | shiF | 344 | −3.0 | ShiF protein | ||
| fepC | 119 | −2.1 | ATP-binding component of ferric enterobactin transport | |||
| Toxins | hlyA | 6 | −2.6 | Hemolysin A | ||
| Growth | carA | 3,789 | −2.2 | −2.3 | Carbamoyl-phosphate synthetase. glutamine | |
| Metabolism | mglA | 801 | −4.5 | −16.2 | ATP-bind. comp. of methyl-galactoside transp. and galactose taxis | |
| dctA | 3,224 | −5.0 | −8.0 | Uptake of C4-dicarboxylic acids | ||
| Protection | ivy | 1,292 | 3.3 | 4.4 | orf. hypothetical protein | |
| Siderophore | entE | 763 | −2.3 | −5.7 | 2.3-dihydroxybenzoate-AMP ligase | |
| entC | 708 | −2.9 | −4.3 | isochorismate hydroxymutase 2. enterochelin biosynthesis | ||
| iucA | 641 | −2.2 | −2.9 | IucA protein | ||
| fepB | 269 | −2.2 | −2.6 | Ferric enterobactin | ||
| iroB | 10 | 2.0 | 2.7 | Putative glucosyltransferase | ||
| chuA | 595 | −2.0 | −2.5 | Outer membrane heme/hemoglobin receptor | ||
| Adhesin | papX | 3,768 | −5.5 | PapX protein | ||
| focX | 6,009 | −5.4 | Putative Regulatory protein | |||
| csgE | 234 | −2.5 | Curli production assembly/transport component. 2nd curli operon | |||
| flu | 796 | −2.4 | Outer membrane fluffing protein. similar to adhesin | |||
| ompA | 145,147 | −2.2 | Outer membrane protein 3a | |||
| sfaB | 40 | 2.4 | Putative F1C and S fimbrial switch Regulatory | |||
| papI | 20 | 2.6 | PapI protein | |||
| phoU | 1,346 | 2.7 | neg. reg. for pho regulon and putative enzyme in phosphate metab. | |||
| focG | 18 | 2.7 | F1C minor fimbrial subunit protein G presursor | |||
| focD | 11 | 2.9 | F1C fimbrial usher | |||
| papH | 12 | 3.1 | PapH protein | |||
| focC | 14 | 3.1 | F1C periplasmic chaperone | |||
| focH | 23 | 3.1 | F1C Putative fimbrial adhesin precursor | |||
| csgA | 5 | 3.3 | Curlin major subunit. coiled surface structures; cryptic | |||
| focF | 14 | 3.4 | F1C minor fimbrial subunit F precursor | |||
| papA | 21 | 3.6 | PapA protein | |||
| papK | 10 | 3.7 | PapK protein | |||
| papD | 1,267 | 3.9 | PapD protein | |||
| papC | 9 | 3.9 | PapC protein | |||
| papE | 13 | 4.0 | PapE protein | |||
| fimE | 351 | 5.5 | Recombinase involved in phase variation; regulator for fimA | |||
| fimB | 40 | 6.8 | Recombinase involved in phase variation; regulator for fimA | |||
| LPS synthesis | waaI | 14 | 2.2 | Putative LPS biosynthesis enzyme | ||
| waaJ | 3 | 2.2 | Putative LPS biosynthesis enzyme | |||
| waaL | 39 | 2.5 | Lipid A-core. surface polymer ligase | |||
| waaA | 105 | 2.7 | 3-deoxy-D-manno-octulosonic-acid transferase | |||
| yibD | 93 | 3.2 | Putative regulator | |||
| rfbA | 23 | 3.2 | Glucose-1-phosphate thymidylyltransferase | |||
| waaY | 11 | 3.8 | Putative LPS biosynthesis protein | |||
| Metabolism | dppA | 8,306 | −8.6 | Dipeptide transport protein | ||
| narH | 262 | −4.1 | Nitrate reductase 1. beta subunit | |||
| narG | 214 | −3.9 | Nitrate reductase 1. alpha subunit | |||
| fruR | 3,876 | −3.3 | Transcriptional repressor of fru operon and others | |||
| deoC | 1,595 | −2.6 | 2-deoxyribose-5-phosphate aldolase | |||
| malX | 201 | −2.3 | PTS system. maltose and glucose-specific II ABC | |||
| lysR | 81 | 2.1 | Positive regulator for lys | |||
| ycjV | 10 | 2.5 | Putative ATP-binding component of a transport system | |||
| gatC | 88 | 2.6 | PTS system galactitol-specific enzyme IIC | |||
| btuB | 147 | 2.8 | Outer memb. Rec. for transp. of vit B12. E colicins and bact.phage | |||
| wbdP | 3 | 3.0 | Glycosyl transferase | |||
| ycfQ | 354 | 3.2 | orf. hypothetical protein | |||
| yedL | 5 | 3.6 | orf. hypothetical protein | |||
| ycjM | 69 | 8.6 | Putative sucrose phosphorylase | |||
| Motility | flhD | 638 | −5.6 | reg. of flag. biosynth. acting on class 2 operons; transcrip. Init. factor | ||
| flhC | 365 | −3.5 | reg. of flag. Biosynth. acting on class 2 operons; transcrip. Init. factor | |||
| fliY | 3,253 | −2.5 | Putative periplasmic binding transport protein | |||
| flgM | 281 | −2.1 | Anti-FliA | |||
| fliG | 22 | 2.1 | flag. biosynth. component of motor switching and energizing | |||
| fliZ | 156 | 2.2 | orf. hypothetical protein | |||
| flhE | 8 | 2.2 | Flagellar protein | |||
| flgA | 134 | 2.4 | Flagellar biosynthesis; assembly of basal-body periplasmic P ring | |||
| fliD | 17 | 2.5 | Flag. biosynth; filament capping protein; enables filament assembly | |||
| fliI | 39 | 2.5 | Flagellum-specific ATP synthase | |||
| fliK | 168 | 3.4 | Flagellar hook-length control protein | |||
| Protection | ompF | 45,769 | −7.3 | Outer membrane protein 1a | ||
| kdpE | 132 | 2.1 | Regulator of kdp operon | |||
| sbmA | 231 | 2.1 | Sensitivity to microcin B17. possibly envelop protein | |||
| gfcC | 9 | 2.3 | orf. hypothetical protein | |||
| sdiA | 295 | 2.5 | Regulatory protein sdiA | |||
| etk | 23 | 2.7 | orf. hypothetical protein | |||
| etp | 26 | 2.7 | Putative phosphatase | |||
| yadK | 10 | 2.7 | Protein yadK | |||
| ybcL | 84 | 3.0 | Protein ybcL precursor | |||
| gfcE | 10 | 3.1 | Putative function in exopolysaccharide production | |||
| gfcD | 553 | 3.9 | orf. hypothetical protein | |||
| ycbQ | 8 | 4.3 | Putative fimbrial-like protein | |||
| gfcA | 278 | 4.6 | orf. hypothetical protein | |||
| sulA | 2,130 | 5.4 | Suppressor of lon; inhibits cell division and ftsZ ring formation | |||
| recA | 1,662 | 6.0 | DNA strand exchange and renaturation. DNA-dep ATPase | |||
| lexA | 489 | 6.3 | Regulator for SOS | |||
| bhsA | 241 | 8.8 | orf. hypothetical protein | |||
| Secretion | tir | 39 | 2.2 | Putative translocated intimin receptor protein | ||
| escJ | 15 | 2.5 | escJ | |||
| escN | 15 | 2.8 | escN | |||
| espB | 8 | 2.8 | Secreted protein EspB | |||
| sepQ | 3 | 3.1 | sepQ | |||
| sepZ | 11 | 3.3 | sepZ | |||
| eae | 20 | 3.4 | Intimin adherence protein | |||
| escU | 49 | 3.6 | escU | |||
| Siderophore | entB | 345 | −3.6 | 2.3-dihydro-2.3-dihydroxybenzoate synthetase. isochroismatase | ||
| entF | 6 | −3.5 | Enterobactin synthetase component F | |||
| iucC | 867 | −3.5 | IucC protein | |||
| entA | 262 | −3.4 | 2.3-dihydro-2.3-dihydroxybenzoate dehydrogenase | |||
| chuS | 655 | −2.8 | Putative heme/hemoglobin transport protein | |||
| fepG | 967 | −2.7 | Ferric enterobactin transport protein | |||
| iucB | 365 | −2.6 | IucB protein | |||
| feoB | 937 | −2.5 | Ferrous iron transport protein B | |||
| iroE | 10 | 3.7 | IroE protein | |||
| Toxins | stx1A | 22 | 2.3 | Shiga-like toxin 1 subunit A encoded within | ||
| hlyE | 135 | 2.8 | Hemolysin E | |||
| stx2B | 6 | 3.6 | Shiga-like toxin II B subunit | |||
| stx1B | 12 | 4.3 | Shiga-like toxin 1 subunit B | |||
| Transcription regulation | rfaH | 384 | 2.1 | Transcription act. of lipopolysaccharide core. F pilin. and haemolysin | ||
| Adhesin | fimC | 26 | 2.1 | 3.5 | Periplasmic chaperone. required for type 1 fimbriae | |
| fimI | 39 | 2.1 | 3.4 | Fimbrial protein | ||
| fimH | 36 | 2.1 | 3.6 | Minor fimbrial subunit. D-mannose specific adhesin | ||
| fimD | 25 | 2.2 | 2.9 | Outer membrane protein; export and assembly of type 1 fimbriae | ||
| fimA | 126 | 2.0 | 2.3 | Major type 1 subunit fimbrin | ||
| csgC | 19 | 2.6 | 2.0 | Putative curli production protein | ||
| fimG | 27 | 2.8 | 2.5 | Fimbrial morphology | ||
| fimF | 8 | 3.6 | 3.3 | FimF protein precursor | ||
| LPS synthesis | yijP | 4,354 | −2.3 | −3.8 | Protein yijP | |
| Metabolism | glnA | 8,617 | −86.0 | −15.1 | Glutamine synthetase | |
| ilvD | 3,921 | −11.3 | −8.2 | Dihydroxy-acid dehydratase | ||
| serA | 2,494 | −9.6 | −3.7 | D-3-phosphoglycerate dehydrogenase | ||
| tdcE | 123 | −3.6 | 3.7 | Keto-acid formate acetyltransferase | ||
| trpB | 573 | −3.0 | −2.5 | Tryptophan synthase. beta protein | ||
| kbaZ | 9 | 2.3 | 16.0 | Putative tagatose 6-phosphate kinase 2 | ||
| pitB | 26 | 2.8 | 3.2 | Low-affinity phosphate transport | ||
| aer | 29 | 2.8 | 3.3 | Aerotaxis sensor receptor. flavoprotein | ||
| trxC | 276 | 3.5 | 3.3 | Thioredoxin 2 | ||
| Motility | fliJ | 13 | 3.4 | 2.3 | Flagellar fliJ protein | |
| Protection | ompW | 4,714 | −9.1 | 2.6 | Outer membrane protein W precursor | |
| matB | 614 | −2.5 | 2.2 | orf. hypothetical protein | ||
| evgS | 526 | −2.2 | −2.9 | Putative sensor for regulator EvgA | ||
| ECs0850 | 5 | 4.6 | 2.2 | Hypothetical protein | ||
| manB | 11 | 8.5 | 2.3 | Phosphomannomutase | ||
| Siderophore | yjjQ | 68 | 2.1 | 2.4 | putative regulator | |
| Transcription regulation | rpoB | 12,927 | −2.3 | −4.5 | RNA polymerase. beta subunit | |
| rpoA | 68,440 | −2.3 | −3.7 | RNA polymerase. alpha subunit | ||
| Adhesin | ppdD | 21 | 2.3 | Prelipin peptidase dependent protein | ||
| LPS synthesis | rfaS | 12,88 | −4.3 | Lipopolysaccharide core biosynthesis | ||
| rfaP | 3,909 | −3.8 | LPS core biosynthesis; phosphorylation of core heptose | |||
| rfaZ | 2,091 | −3.0 | Lipopolysaccharide core biosynthesis | |||
| rfaY | 1,690 | −2.5 | Lipopolysaccharide core biosynthesis | |||
| waaU | 999 | −2.1 | Probably hexose transferase; lipopolysaccharide core biosynthesis | |||
| rfaG | 1,009 | −2.0 | Lipopolysaccharide core biosynthesis protein rfaG | |||
| Metabolism | eda | 3,246 | −3.1 | 2-keto-3-deoxygluc 6-phos aldo and 2-keto-4-hydroxyglut aldo | ||
| tktA | 2,823 | −2.2 | Transketolase 1 isozyme | |||
| dsbA | 27,226 | −2.1 | Protein disulfide isomerase I. essential for cytochrome c synthesis | |||
| prfB | 3,054 | −2.0 | Peptide chain release factor 2 | |||
| gnsA | 3,461 | −2.0 | Predicted regulator of phosphatidylethanolamine synthesis | |||
| yjhS | 37 | 2.1 | orf. hypothetical protein | |||
| agaA | 113 | 2.5 | Putative N-acetylgalactosamine-6-phosphate deacetylase | |||
| agaC | 42 | 2.7 | PTS system N-acetylgalactosamine-specific IIC component 1 | |||
| dctA | 3,224 | 3.2 | Uptake of C4-dicarboxylic acids | |||
| astA | 93 | 3.6 | orf. hypothetical protein | |||
| lacZ | 20 | 3.7 | Beta-galactosidase | |||
| gatZ | 18 | 4.1 | Putative tagatose 6-phosphate kinase 1 | |||
| gatB | 15 | 5.0 | Galactitol-specific enzyme IIB of phosphotransferase system | |||
| agaS | 72 | 5.3 | Putative tagatose-6-phosphate aldose/ketose isomerase | |||
| agaB | 32 | 7.0 | PTS system. cytopl. N-acetylgalactosamine-spec IIB component 1 | |||
| ulaB | 27 | 7.2 | Putative transport protein | |||
| ulaC | 86 | 8.6 | Putative PTS system enzyme II A component | |||
| ulaA | 76 | 11.2 | orf. hypothetical protein | |||
| agaW | 40 | 18.1 | PTS system N-acetylgalactosameine-specific IIC component 2 | |||
| agaV | 19 | 18.9 | PTS system. cytopl. N-acetylgalactosamine-spec IIB component 2 | |||
| kbaY | 19 | 28.0 | Tagatose-bisphosphate aldolase 2 | |||
| Motility | fliT | 31 | 2.1 | Flagellar biosynthesis; repressor of class 3a and 3b operons | ||
| flgJ | 29 | 2.1 | Flagellar biosynthesis | |||
| fliO | 11 | 2.1 | Flagellar protein fliO | |||
| fliL | 23 | 2.2 | Flagellar biosynthesis | |||
| flgH | 20 | 2.2 | Flagellar biosynthesis. basal-body outer-membrane L | |||
| flhB | 38 | 2.3 | Putative part of export apparatus for flagellar proteins | |||
| flgB | 24 | 2.3 | Flagellar biosynthesis. cell-proximal portion of basal-body rod | |||
| fliE | 37 | 2.7 | Flagellar biosynthesis; basal-body component | |||
| fliF | 27 | 2.8 | Flagellar biosynthesis; basal-body MS | |||
| lfhA | 8 | 3.1 | Flagellar biosynthesis | |||
| fliP | 11 | 3.3 | Flagellar biosynthetic protein fliP precursor | |||
| fliQ | 21 | 4.9 | Flagellar biosynthesis | |||
| Protection | kpsC | 986 | −3.4 | KpsC protein | ||
| bamA | 6,961 | −3.3 | orf. hypothetical protein | |||
| evgS | 526 | −2.9 | putative sensor for regulator EvgA | |||
| degP | 6,718 | −2.6 | Periplasmic serine protease Do; heat shock protein HtrA | |||
| kpsM | 3,290 | −2.2 | KpsM protein | |||
| Transcription regulation | fis | 61,722 | −3.9 | Site-specific DNA inversion stimulation factor; DNA-binding protein | ||
In order to confirm the microarray results, real-time qPCR was carried out on seven genes belonging to the functional categories Adhesion (FimAH), Protection (ibpAB), and Siderophore (chuAT, sitB) that were significantly altered compared to ESBL019 Coliform. In agreement with the microarray, a significant upregulation of FimAH was observed in ESBL019 Filamented and ESBL019 Reverted state compared to ESBL019 Coliform (Table 2). In addition, ibpAB was observed to be significant upregulated in all the three different states compared to ESBL019 Coliform, as shown by the microarray data. We also found that chuAT, sitB were significantly downregulated in ESBL019 Transition and ESBL019 Filamented state compared to ESBL019 Coliform (Table 2). However, this was not observed to be true for the ESBL019 Reverted state.
Table 2.
Quantitative real-time PCR data for ESBL019 Transition, ESBL019 Filamented and ESBL019 Reverted compared to ESBL019 Coliform.
| Gene | Fold change vs. ESBL019 coliform | Description | ||
|---|---|---|---|---|
| ESBL019 transition | ESBL019 filamentous | ESBL019 reverted | ||
| FimA | −1.2 ± 0.86a | 15 ± 3.1a | 8.5 ± 2.1a | Major type 1 subunit fimbrin |
| FimH | −1.1 ± 0.74a | 10 ± 1.8a | 11 ± 2.3a | Minor fimbrial subunit. D-mannose specific adhesin |
| ibpA | 5.3 ± 0.90a | 230 ± 49a | 22 ± 3.7a | Heat shock protein |
| ibpB | 7.2 ± 1.5a | 330 ± 80a | 130 ± 33a | Heat shock protein |
| chuA | −6.3 ± 1.2a | −1.6 ± 0.1a | 0.90 ± 1.0 | Putative Periplasmic binding protein |
| chuT | −15 ± 3.2a | −4.4 ± 1.5a | 1.1 ± 1.0 | Outer membrane heme/hemoglobin receptor |
| sitB | −11 ± 1.6a | −5.4 ± 1.6a | 0.71 ± 0.82 | SitB protein |
n = 3–4.
Significantly altered genes compared to ESBL019 Coliform.
Fimbriae agglutination
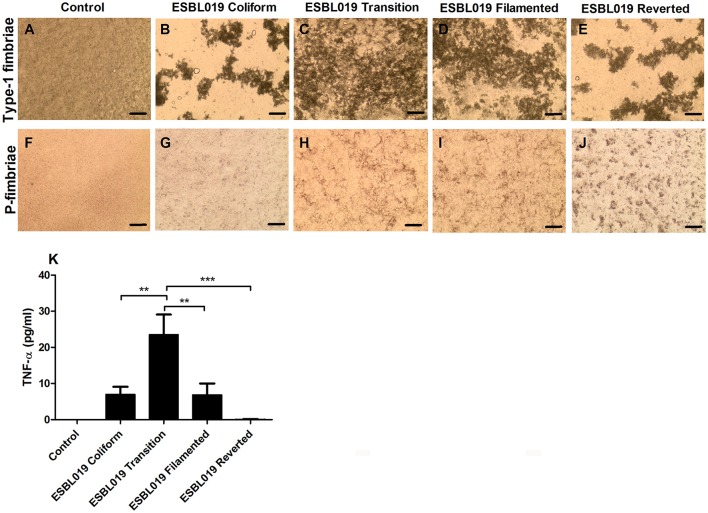
We proceeded with evaluating the phenotypical presence and functionality of the type-1 fimbriae and P-fimbriae in the different morphological states of ESBL019. Yeast agglutination confirmed that all the morphological states of ESBL019 expressed functional type-1 fimbriae (Figures 4B–E) compared to only yeast (Figure 4A). The aggregation pattern between the rod shaped (ESBL019 Coliform and ESBL019 Reverted) and the elongated ESBL019 (ESBL019 Transition and ESBL019 Filamented) morphologies were strikingly different. The yeast was bound along the elongated bacteria and gave a more uniform aggregation (Figures 4C,D), while the rod shaped bacteria formed large isolated yeast aggregates (Figures 4B,E). Human Erythrocytes of blood group P were used to evaluate the P-fimbriae expression. All the morphological states of ESBL019 expressed functional P-fimbriae (Figures 4G–J) compared to only erythrocytes (Figure 4F). We also observed an increased aggregation by the ESBL019 Transition (Figure 4H), ESBL019 Filamented (Figure 4I) and ESBL019 Reverted (Figure 4J) states compared to ESBL019 Coliform (Figure 4G).
Figure 4.
Fimbriae aggregation and TNF-α release from HBEP cells infected with the different morphologies of ESBL019. Type-1 fimbriae functionality was assessed by yeast agglutination without (A) or with ESBL019 Coliform (B), ESBL019 Transition (C), ESBL019 Filamented (D) and ESBL019 Reverted (E) using light microscopy. P-fimbriae functionality was assessed by P-positive human erythrocyte agglutination without (F) or with ESBL019 Coliform (G), ESBL019 Transition (H), ESBL019 Filamented (I), and ESBL019 Reverted (J) using light microscopy. Scale bare: 200 μm. TNF-α release from HBEP cells stimulated with ESBL019 Coliform, ESBL019 Transition, ESBL019 Filamented and ESBL019 Reverted (MOI10) for 4 h in the presence (Transition, Filamented) or absence (Coliform and Reverted) of ceftibuten (K). Data are presented as mean ± SEM of n = 4 independent experiments. Asterisks denote statistical significance **p < 0.01, ***p < 0.001.
Epithelial TNF-α release evoked by the different morphological states
Primary human bladder epithelial cells were stimulated with the different morphological states of ESBL019 to determine their ability to induce the pro-inflammatory cytokine TNF-α. The unstimulated control cells showed no basal release of TNF-α (Figure 4K). ESBL019 Coliform and ESBL Filamented were able to induce an increased release of TNF-α compared to the control. ESBL019 Transition induced a significant higher release of TNF-α compared to control (p < 0.001), ESBL019 Coliform (p < 0.01), ESBL019 Filamented (p < 0.01) and ESBL019 Reverted (p < 0.001). In contrast to the other morphological states of ESBL019, ESBL019 Reverted did not induce any release of TNF-α from the bladder epithelial cells. Taken together, the different morphological states of ESBL019 induce a diverse TNF-α release from bladder epithelial cells.
Discussion
Global gene expression profiling of the different morphological states of ESBL019 demonstrated a significant transcriptional alteration of a large number of genes. In the ESBL019 Transition state, close to 7% of the entities on the array were altered, 32% for ESBL019 Filamented and 19% for ESBL019 Reverted. Thus, it appears that the morphological plasticity of UPEC is associated with an extensive flux in the transcriptome of the bacteria and especially in the filamented state. To the best of our knowledge, this is the first study that investigates changes in global gene expression patterns and in virulence factor associated genes in ESBL-producing UPEC during the morphologic transitions induced by ineffective antibiotics. Importantly, all experiments were conducted in the presence of human primary bladder epithelial cells to mimic the in vivo host-pathogen interactions in a physiologically relevant manner.
The majority of the identified changes in gene ontologies and pathways were associated with metabolic functions. Many metabolic functions are not specific for pathogens and their role in infection fitness remains therefore more or less unexplored (Subashchandrabose and Mobley, 2015). In the ESBL019 Transition state, we observed that genes associated with the tricarboxylic acid cycle (TCA) and glucogenic amino acid (arginine, glutamate, glutamine, and proline) metabolism were in majority downregulated. We also found that the gene dctA, which encodes for the C4-dicarboxylate transporter that is responsible for the uptake of fumarate, succinate and malate was downregulated. Previous studies have shown that gluconeogenesis and the TCA cycle, but not Entner-Doudoroff, glycolysis or pentose-phosphate pathways are important for the fitness of UPEC during in vivo experimental UTI (Alteri et al., 2009). Genes associated with amino acid metabolism, such as dctA, serA (L-serine biosynthesis), ilvD (isoleucine and valine biosynthesis), trpB (tryptophan synthase beta), glnA (catalyzing conversion of glutamate to glutamine) and dppA (dipeptide transporter) were observed to be downregulated in the ESBL019 Filamented state. It has been shown that UPEC grown in urine utilizes primarily peptides and amino acids as carbon sources instead of glucose (Alteri et al., 2009). We also observed that several sugar transporter associated genes such as gatC and ycjV were upregulated in the filamented state, which could lead to an increased acquisition of carbon sources. In the ESBL019 Reverted state, glucose and xylose degradation, glycolysis, Entner-Doudoroff, NAD biosynthesis and gluconeogenesis pathways were downregulated. We also found, in accordance with the filamented state, that glnA, ilvD, and serA were downregulated. In addition, an increase in genes associated with replenishing TCA cycle intermediates (dctA and astA), sugar transporters (agaABCVW), L-ascorbate transport (ulaABC) and degradation of other carbon sources like lactose (lacZ) was observed. It has recently been shown that beta-galactosidase (lacZ) may play an important role in intracellular UPEC growth and IBC maturation (Conover et al., 2016). An interesting finding in the ESBL019 Reverted state was the downregulation of genes associated with transcription, translation, ribosome biosynthesis and tRNA charging. Taken together, these data demonstrates that all three morphological states of UPEC are associated with a decreased energy-yielding cell metabolism, suggesting that survival rather than growth is prioritized. Knowledge of these metabolic shifts could contribute to finding new therapeutic targets to limit the bacteria's ability to acquire alternative carbon sources.
Metals such as iron are essential for bacterial life and the low concentration of soluble iron in the urinary tract is regarded as a limiting factor for UPEC growth (Roos et al., 2006). Consequently, it has been shown that iron acquisition is important for UPEC survival and pathogenicity (Subashchandrabose and Mobley, 2015). UPEC can acquire iron through several mechanisms such as siderophores (ferric iron chelators), ferrous iron transporters and heme through outer membrane receptors (Cho et al., 2008). We observed that the iron acquisition systems in all the morphological states of UPEC changed significantly. Genes involved in the enterobactin system (entABF, fepABE), heme system (chuST) and ferrous iron uptake (sitB) were downregulated in all the three morphological states. However, genes in the aerobactin system (iucAC) were downregulated in the ESBL019 Transition and ESBL019 Filamented state but not in the ESBL019 Reverted state. The salmochelin siderophore system (iroEN) was upregulated in all the three morphological states. qPCR analysis confirmed that chuAT and sitB were significantly downregulated in ESBL019 Transition and ESBL019 Filamented state, but not in the ESBL019 Reverted state compared to ESBL019 Coliform. The iron acquisition systems in UPEC have been demonstrated to be redundant, but individual systems have been shown to play key roles in fitness and pathogenicity in different regions of the urinary tract (Garcia et al., 2011). Aerobactin receptors have been shown to significantly contribute to in vivo fitness and they are critical for UPEC virulence (Garcia et al., 2011). It was recently shown that the virulence associated with iron acquisition could be partially due to modulation of amino acid metabolism (Su et al., 2016). Taken together, these data show that the iron acquisition systems in UPEC are altered during morphological shifts, mainly downregulated, but the redundancy in the systems may contribute to preserving in vivo fitness. In addition, the lack of change in the aerobactin system, one of the key siderophores system in uropathogenicity, may indicate that the reverted morphology maintains virulence in the life cycle of UPEC.
During colonization of the urinary tract, UPEC utilizes different adhesion molecules (like fimbriae) to adhere and invade the epithelial cells in order to persist in the urinary tract (Subashchandrabose and Mobley, 2015). The P-fimbriae encoding genes (pap) were upregulated in all the three different morphological states of ESBL019 and we also observed an increased phenotypic expression by agglutination in all the different morphological states compared to ESBL019 Coliform. The P-fimbriae is associated with the pathogenicity of ascending UTI and pyelonephritis (Frendeus et al., 2001). The type-1 fimbriae associated genes (fim), which play an important role in adhesion, invasion, IBC, and biofilm formation during cystitis (Flores-Mireles et al., 2015) were upregulated in ESBL019 Filamented and ESBL019 Reverted, but no changes were observed in ESBL019 Transition state. Consistent with the microarray data, fimAH were markedly upregulated in the Filamented and Reverted state based on qPCR data. We confirmed phenotypically functional type-1 fimbriae in all the different states by yeast agglutination assays. This indicates that the morphological plasticity of UPEC does not compromise on adhesion factors important for colonizing the urinary tract. We hypothesize that the observed upregulation of P-and type-1 fimbriae associated genes is a mechanism to persist in the urinary tract especially after the bacteria emerge from the IBC in a filamented form. Taken together, our results show that all UPEC morphologies maintain fimbrial expression, which could promote persistence and colonization of the urinary tract.
Ascendance from the bladder to the kidneys is an important virulence trait of UPEC associated with motility and flagella expression (Pichon et al., 2009). Studies of morphological plasticity of ESBL019 showed only one significantly upregulated flagellin gene in the ESBL019 Transition state (fliC), but many flagellin genes were both up-and downregulated in the ESBL019 Filamented state. In the ESBL019 Reverted state, all the significantly altered genes were upregulated. These results indicate that morphological plasticity is associated with altered flagellin gene expression, but additional functional experiments are needed to assess the effects on motility.
We and others have shown that filamentation of E. coli is associated with a rapid production and release of endotoxins (Gould and MacKenzie, 1997; Demirel et al., 2015). Genes associated with LPS biosynthesis were almost unchanged in the ESBL019 Transition state, but all LPS genes were upregulated in the ESBL019 Filamented state. However, the majority of the LPS associated genes were downregulated in the ESBL019 Reverted state. The modulation of LPS synthesis during the morphological shifts could be a compensation mechanism to replenish LPS on the surface of ESBL019, as increased shedding of LPS was observed during the transition state (Demirel et al., 2015). This modulation of LPS could play a part in the pathogenicity of UPEC as LPS modulates pro-inflammatory responses (Bussolati et al., 2002).
The morphological plasticity of ESBL019 showed an alteration in many genes associated with bacterial protection. Genes like tcpC, which is associated with inhibition of pro-inflammatory responses (Yadav et al., 2010), was downregulated in all three states. We also observed that the gene encoding for the outer membrane protein ompT, which is involved in the resistance of different antimicrobial peptides (Brannon et al., 2013), was upregulated in the transition state, but downregulated in the two other states. The gene encoding ompF, which is a porin for antibiotics, was downregulated in the filamented state and this has been associated with increased antibiotic resistance (Kishii and Takei, 2009). OmpW, which is required for UPECs resistance against phagocytosis (Wu et al., 2013) was downregulated in the filamented state, but upregulated in the reverted state. We have previously shown that filamented UPEC can be successfully phagocytized by neutrophils (Demirel et al., 2015). Related to neutrophil immunity, the ybcL gene proposed to inhibit neutrophil migration was upregulated in the ESBL019 Filamented state (Lau et al., 2012). The gene ontology SOS response was enriched in the ESBL019 Filamented state and the associated genes recA, lexA, and sulA were upregulated. The SOS system is a response to DNA damage and inhibits cell division and septa formation in order to repair the DNA damage (Justice et al., 2006). sulA has been shown to mediate UPEC filamentation and this filamentation is associated with increased genetic variability and point mutations that can lead to antibiotic resistance (Bos et al., 2015). The filamented form also showed an upregulation of genes (tir, esc, and sep) encoding the Type 3 secretion system (T3SS), which is used by bacteria to inject effectors directly into host cell (Subashchandrabose and Mobley, 2015). T3SS genes are traditionally not found in UPEC, but they have recently been reported to be found in some UPEC isolates (Subashchandrabose et al., 2013), although the role during UTI remains to be determined.
We continued with analyzing the TNF-α production from primary bladder epithelial cells induced by the different morphological states. This was done in order to evaluate the biological relevance of the microarray data. The ESBL019 Transition state induced a significant increase in TNF-α production compared to the control and the other morphologies. We have previously shown that the transition state is associated with increased LPS and ATP release, both known to induce pro-inflammatory cytokines from uroepithelial cells (Demirel et al., 2015). The reverted state did not induce TNF-α production and this may give the reverted UPEC an advantage in colonizing the bladder as host cytokine responses are essential for attracting neutrophils (Smart and Casale, 1994). However, it remains to be investigated if the lack of immune activation by the reverted state is due to an inability to induce an immune response or to an active immune suppression. Our agglutination assays showed no reduced fimbriae function to support that the reverted state is unable to induce an immune response. However, several interesting findings, like the downregulation of LPS associated genes in the reverted state, remains to be evaluated.
Together, these results provide insight into the global gene expression patterns of the different morphological states of ESBL019 induced by an ineffective antibiotic. The decreased energy-yielding metabolism and up-regulated adhesion, suggest that survival and persistence in the bladder is priority rather than growth during the morphological plasticity of ESBL-producing UPEC. These findings are also strengthened by the fact that the reverted state did not induce TNF-α release, which gives the bacteria an advantage in colonizing the bladder. Understanding how ineffective antibiotic treatments affect the virulence and metabolism of ESBL-producing UPEC can promote the identification of new therapeutic targets and new adjuvant combination treatments to reduce the prevalence of multiresistant bacteria.
Author contributions
ID, RK and KP design the study. ID and RK conducted the experiments. ID, RK, IR, UP, and KP analyzed the data. ID, RK, IR, UP, and KP drafted the article. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer JM and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We thank Ola Gustafsson, Department of Biology, Lund University for the assistance with the transmission electron microscopy.
Footnotes
Funding. This project was financially supported by the Research Committee of Örebro County Council, Nyckelfonden, Swedish Society of Medicine and the Faculty of Medicine and Health at Örebro University.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01058/full#supplementary-material
References
- Alteri C. J., Smith S. N., Mobley H. L. (2009). Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 5:e1000448. 10.1371/journal.ppat.1000448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G. G., Palermo J. J., Schilling J. D., Roth R., Heuser J., Hultgren S. J. (2003). Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301, 105–107. 10.1126/science.1084550 [DOI] [PubMed] [Google Scholar]
- Blango M. G., Mulvey M. A. (2010). Persistence of Uropathogenic Escherichia coli in the Face of Multiple Antibiotics. Antimicrobial. Agents Chemother. 54, 1855–1863. 10.1128/AAC.00014-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J., Zhang Q., Vyawahare S., Rogers E., Rosenberg S. M., Austin R. H. (2015). Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc. Natl. Acad. Sci. U.S.A. 112, 178–183. 10.1073/pnas.1420702111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower J. M., Eto D. S., Mulvey M. A. (2005). Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic 6, 18–31. 10.1111/j.1600-0854.2004.00251.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon J. R., Thomassin J. L., Desloges I., Gruenheid S., Le Moual H. (2013). Role of uropathogenic Escherichia coli OmpT in the resistance against human cathelicidin LL-37. FEMS Microbiol. Lett. 345, 64–71. 10.1111/1574-6968.12185 [DOI] [PubMed] [Google Scholar]
- Brolund A. (2014). Overview of ESBL-producing Enterobacteriaceae from a nordic perspective. Infect. Ecol. Epidemiol. 4:24555. 10.3402/iee.v4.24555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussolati B., David S., Cambi V., Tobias P. S., Camussi G. (2002). Urinary soluble CD14 mediates human proximal tubular epithelial cell injury induced by LPS. Int. J. Mol. Med. 10, 441–449. 10.3892/ijmm.10.4.441 [DOI] [PubMed] [Google Scholar]
- Cho B. K., Knight E. M., Barrett C. L., Palsson B. O. (2008). Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 18, 900–910. 10.1101/gr.070276.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover M. S., Hadjifrangiskou M., Palermo J. J., Hibbing M. E., Dodson K. W., Hultgren S. J. (2016). Metabolic requirements of Escherichia coli in intracellular bacterial communities during urinary tract infection pathogenesis. MBio 7:e00104–16. 10.1128/mBio.00104-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque T. M., Baquero F., Canton R. (2008). Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro. Surveill. 13:pii: 19044. [PubMed] [Google Scholar]
- Demirel I., Kruse R., Onnberg A., Persson K. (2015). Ceftibuten-induced filamentation of extended spectrum beta lactamase (ESBL)-producing uropathogenic Escherichia coli alters host cell responses during an in vitro infection. Microb. Pathog. 78, 52–62. 10.1016/j.micpath.2014.11.015 [DOI] [PubMed] [Google Scholar]
- Flores-Mireles A. L., Walker J. N., Caparon M., Hultgren S. J. (2015). Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13, 269–284. 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman B. (2003). Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Month 49, 53–70. 10.1067/mda.2003.7 [DOI] [PubMed] [Google Scholar]
- Frendeus B., Wachtler C., Hedlund M., Fischer H., Samuelsson P., Svensson M., et al. (2001). Escherichia coli P fimbriae utilize the Toll-like receptor 4 pathway for cell activation. Mol. Microbiol. 40, 37–51. 10.1046/j.1365-2958.2001.02361.x [DOI] [PubMed] [Google Scholar]
- Garcia E. C., Brumbaugh A. R., Mobley H. L. (2011). Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun. 79, 1225–1235. 10.1128/IAI.01222-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould I. M., MacKenzie F. M. (1997). The response of Enterobacteriaceae to beta-lactam antibiotics - ‘round forms, filaments and the root of all evil’. J. Antimicrob. Chemother. 40, 495–499. 10.1093/jac/40.4.495 [DOI] [PubMed] [Google Scholar]
- Horvath D. J., Li B. R., Casper T., Partida-Sanchez S., Hunstad D. A., Hultgren S. J., et al. (2011). Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microb. Infect. 13, 426–437. 10.1016/j.micinf.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice S. S., Hung C., Theriot J. A., Fletcher D. A., Anderson G. G., Footer M. J., et al. (2004). Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 1333–1338. 10.1073/pnas.0308125100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice S. S., Hunstad D. A., Seed P. C., Hultgren S. J. (2006). Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl. Acad. Sci. U.S.A. 103, 19884–19889. 10.1073/pnas.0606329104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandige S., Asferg C. A., Rasmussen K. J., Larsen M. J., Overgaard M., Andersen T. E., et al. (2016). DamX controls reversible cell morphology switching in uropathogenic Escherichia coli. MBio 7:pii: e00642-16. 10.1128/mBio.00642-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanfar H. S., Bindayna K. M., Senok A. C., Botta G. A. (2009). Extended spectrum beta-lactamases (ESBL) in Escherichia coli and Klebsiella pneumoniae: trends in the hospital and community settings. J. Infect. Dev. Ctries. 3, 295–299. 10.3855/jidc.127 [DOI] [PubMed] [Google Scholar]
- Kishii R., Takei M. (2009). Relationship between the expression of ompF and quinolone resistance in Escherichia coli. J. Infect. Chemother. 15, 361–366. 10.1007/s10156-009-0716-6 [DOI] [PubMed] [Google Scholar]
- Lau M. E., Loughman J. A., Hunstad D. A. (2012). YbcL of uropathogenic Escherichia coli suppresses transepithelial neutrophil migration. Infect. Immun. 80, 4123–4132. 10.1128/IAI.00801-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysorekar I. U., Hultgren S. J. (2006). Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl. Acad. Sci. U.S.A. 103, 14170–14175. 10.1073/pnas.0602136103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onnberg A., Molling P., Zimmermann J., Soderquist B. (2011). Molecular and phenotypic characterization of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum beta-lactamases with focus on CTX-M in a low-endemic area in Sweden. APMIS 119, 287–295. 10.1111/j.1600-0463.2011.02730.x [DOI] [PubMed] [Google Scholar]
- Pichon C., Hechard C., Du Merle L., Chaudray C., Bonne I., Guadagnini S., et al. (2009). Uropathogenic Escherichia coli AL511 requires flagellum to enter renal collecting duct cells. Cell Microbiol. 11, 616–628. 10.1111/j.1462-5822.2008.01278.x [DOI] [PubMed] [Google Scholar]
- Pitout J. D., Laupland K. B. (2008). Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- Ronald A. (2003). The etiology of urinary tract infection: traditional and emerging pathogens. Dis. Mon. 49, 71–82. 10.1067/mda.2003.8 [DOI] [PubMed] [Google Scholar]
- Roos V., Ulett G. C., Schembri M. A., Klemm P. (2006). The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74, 615–624. 10.1128/IAI.74.1.615-624.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen D. A., Hooton T. M., Stamm W. E., Humphrey P. A., Hultgren S. J. (2007). Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 4:e329. 10.1371/journal.pmed.0040329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart S. J., Casale T. B. (1994). Tnf-Alpha-Induced transendothelial neutrophil migration is Il-8 dependent. Am. J. Physiol. 266, L238–L245. [DOI] [PubMed] [Google Scholar]
- Su Q., Guan T., He Y., Lv H. (2016). Siderophore biosynthesis governs the virulence of uropathogenic Escherichia coli by coordinately modulating the differential metabolism. J. Proteome Res. 15, 1323–1332. 10.1021/acs.jproteome.6b00061 [DOI] [PubMed] [Google Scholar]
- Subashchandrabose S., Hazen T. H., Rasko D. A., Mobley H. L. (2013). Draft genome sequences of five recent human uropathogenic Escherichia coli isolates. Pathog Dis. 69, 66–70. 10.1111/2049-632X.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose S., Mobley H. L. T. (2015). Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol. Spectr. 3:10.1128/microbiolspec.UTI-0015-2012. 10.1128/microbiolspec.UTI-0015-2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello M., Sanguinetti M., Montuori E., Trecarichi E. M., Posteraro B., Fiori B., et al. (2007). Predictors of mortality in patients with bloodstream infections caused by extended-spectrum-beta-lactamase-producing enterobacteriaceae: Importance of inadequate initial antimicrobial treatment. Antimicrob. Agents Chemother. 51, 1987–1994. 10.1128/AAC.01509-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. B., Tian L. H., Zou H. J., Wang C. Y., Yu Z. Q., Tang C. H., et al. (2013). Outer membrane protein OmpW of Escherichia coli is required for resistance to phagocytosis. Res. Microbiol. 164, 848–855. 10.1016/j.resmic.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Yadav M., Zhang J., Fischer H., Huang W., Lutay N., Cirl C., et al. (2010). Inhibition of TIR domain signaling by TcpC: MyD88-dependent and independent effects on Escherichia coli virulence. PLoS Pathog. 6:e1001120. 10.1371/journal.ppat.1001120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.