Abstract
Mod5 is a multifunctional protein that modifies a subset of tRNAs in the cytoplasm and is also required for an RNA-mediated form of transcriptional silencing. Previous in vivo studies have shown that the nuclear silencing function of Mod5 does not require that the causative tRNA gene encode a Mod5 substrate, though Mod5 is still required. However, previous data have not directly tested whether Mod5 can directly bind substrate and nonsubstrate RNAs. We herein demonstrate that Mod5 directly binds to both substrate and nonsubstrate RNAs, including a highly structured, non-tRNA sequence (5S-rRNA), consistent with previous in vivo data. Furthermore, we show that some RNAs drastically change the aggregation behavior of Mod5 with implications for tRNA-gene mediated silencing.
Keywords: Mod5, tRNA modification, protein aggregation, amyloid fiber, RNA binding, isopentenyl transferase
Introduction
tRNA isopentenyl transferases are highly conserved enzymes that transfer an isopentenyl group from dimethylallylpyrophosphate (DMAPP) to the N6 position of A37, adjacent to the anticodon [1–9]. The S. cerevisiae enzyme, Mod5, resides primarily in the cytoplasm where it modifies mitochondrial and cytoplasmic tRNAs[10–12]. Mod5 modifies a subset of tRNAs with specific sequence requirements (AAA) surrounding the target nucleotide at A37 [13, 14]. Substrates in yeast and humans [15] include Tyr-, Ser-, Trp-, and Cys-isoacceptor tRNAs. This modification on a tRNA increases tRNA stability [16] and the fidelity and efficiency of translation—an effect related to the tRNA’s affinity for the ribosome [2, 5, 17, 18].
A small fraction of the enzyme also resides in the nucleus[19, 20], where it is required for a form of transcriptional silencing, termed tRNA gene-mediated (tgm) silencing [11, 21–23]. Tgm silencing was originally observed in budding yeast as antagonizing pol II gene promoters adjacent to tRNA genes [24]. The silencing does not involve steric interference, was shown to occur upstream or downstream of the tRNA gene [24], and is dependent on histone modifications and chromatin remodeling [25]. This phenomenon also requires the clustering of the tRNA genes to the nucleolus, demonstrating a role for nuclear architecture and localization [23, 26, 27]. Deletion of the MOD5 gene relieves silencing of a pol II promoter adjacent to a tRNA gene on a reporter plasmid [21, 23]. Recently, a form of tgm silencing has been proposed to exist in human cells [28].
Mod5 involvement in tgm silencing mechanisms was further confirmed by the following observations [22]: (1) Mod5 protein is physically associated with tRNA gene loci, (2) Mod physically associates with pol III transcription complex proteins, (3) Mod5 copurifies with multiple proteins of a chromatin remodeling complex (RSC) and histones that affect tgm silencing, and (4) Mod5 co-immunoprecipitates with precursor-tRNAs (pre-tRNA) that are thought to be found exclusively in the nucleus. Interestingly, Mod5 was found to co-immunoprecipitate with both substrate and nonsubstrate tRNAs for isopentenylation, but preferentially with corresponding nuclear pre-tRNAs in both types of RNA. This was consistent with the observation that tRNA genes encoding substrate or nonsubstrate pre-tRNAs are both capable of conferring tgm-silencing [22], suggesting that the specificity of Mod5 for enzymatic modification of tRNA in the cytoplasm is distinct from that of its nuclear role in transcriptional silencing. Lastly, truncation of the pre-tRNA transcripts by early termination compromised tgm silencing, suggesting the mechanism required a tRNA-like transcript for efficiency.
Based on these data, we hypothesize that complexes between Mod5 and nascent pre-tRNAs at the site of transcription might recruit chromatin modification and remodeling proteins that affect local chromatin structure to impart silencing of nearby transcription by RNA polymerase II. This mechanism would require that Mod5 binds pre-tRNAs whether or not they are substrates for modification at A37, which has not previously been shown. Testing this was complicated by the prion-like tendency of Mod5 to aggregate in solution and in vivo, in agreement with recent studies showing that Mod5 forms heritable amyloid-like aggregates [29]. It is possible to select for this heritable aggregation in vivo by growth in certain fungicides that require loss of Mod5 modifications of tRNAs (e.g. fluconazole). However, this aggregation of Mod5 affects only cytoplasmic tRNA modification and not the nuclear silencing function [11, 22].
In this study, we show for the first time that Mod5 directly binds to both substrate and non-substrate RNAs in vitro, as well as the corresponding primary transcript pre-tRNAs. Further, binding to tRNA-like molecules facilitates aggregation of Mod5. We propose a model for tgm silencing that includes binding of pre-tRNAs by Mod5.
Materials and Methods
Recombinant Mod5 expression and purification
The endogenous Mod5 ORF was amplified by PCR from a genomic DNA preparation of yeast BY4741 cells using primers with Nde1 and Xho1 sequences. PCR products were digested with Nde1 and Xho1 and ligated into the pet15B vector, a bacterial expression plasmid with a 6× histidine C-terminal tag. The plasmid was transformed into BL21 E. coli by electroporation using a BioRad Gene Pulser at at 2.5 kV, 25 uF, 200 Ohms (http://springerlab.tch.harvard.edu/springer/uploads/Equipment/Genepulsermanual.pdf). 8 L of E. coli were grown to OD (600 nm) 0.55 in LB + 100 ug/mL Ampicillin (37°C), then induced with 1.5 mM IPTG (5 hours, 17°C). Cells were pelleted at 2,620 RCF, 15 minutes, 4°C, and frozen at −80°C. Pellets were resuspended in 120 mL buffer (50 mM Tris-HCl, 50 mM NaCl, pH 7.5) on ice. Cells were lysed using a Divitech Microfluidizer (model 110Y) using two passes with 200 μm orifice cartridge, then 8 passes with 100 μm cartridge. Lysate was centrifuged at 5,470 RCF, 15 minutes, 4°C, and the supernatant combined with 60 mL packed, pre-equilibrated BioRex 70 cation exchange resin (50 mM Tris-HCL, 50 mM NaCl, pH 7.5). The lysate was allowed to bind for 1 hour (4°C), spun for 5 minutes at 152 RCF, and the supernatant was discarded. Resin was washed twice with one volume buffer (200 mM NaCl, 50 mM Tris-HCl, pH 7.5). Protein was eluted with two resuspensions using 500 mM NaCl, 50 mM Tris-HCl, pH 7.5 using one packed resin volume. Elutions were loaded onto a 5 mL packed column of TALON nickel affinity resin at 4°C. The column was washed with 50 mL buffer (300 mM NaCl, 20 mM NaPO4, pH 7.0) and eluted with 5.5 mL 450 mM imidazole, 50 mM NaPO4, 300 mM NaCl, pH 8.0. Eluent was run on an 80 mL Sephacryl S-100 column (2.7 cm diameter × 19.1 cm height) using 50 mM Tris-HCl, 50 mM NaCl, pH 7.5 buffer at a rate of 1 mL flow-through per minute at 4°C. 750 μL fractions were collected, examined by SDS-PAGE, then the 10 highest concentration fractions were combined, aliquoted, and stored at −80°C.
DNA Template Synthesis
Template DNA fragments for transcription (Supplemental Figure 2) were ordered through Integrated DNA Technologies. Lyophilized DNA pellets were dissolved in 10mM Tris-HCl containing 1mM EDTA, to a concentration of 100 μM. Equal volumes of overlapping, complementary strand pairs were combined and hybridized at 37°C overnight. Ten μL of the 50 μM solutions of hybridized DNA were then added to a solution with a final concentration of 0.4 μM DNTP’s, 10 mM Tris-HCl, pH 7.4, 50 mM KCl, 1.5 mM MgCl2, and up to 5 units Taq DNA polymerase [30] in a final volume of 50 μL. The solutions were then incubated at 35°C for 5 min, 45°C for 5 min, 55°C for 5 min, 65°C for 30 min and 37°C for 15 min, ethanol precipitated, resuspended in 15 μL of water, and stored at −20 °C.
In vitro transcription and RNA purification
For each transcription reaction, 200 μL solutions of 50 mM Tris-HCl pH 7.5, 15 mM MgCl2, 5 mM DTT, 2 mM spermidine, 2 mM ATP, UTP, GTP, CTP, 2/25 of volume template (see above), empirically titrated His6-tagged T7 RNA polymerase (He et al. 1997) were incubated 12 hours at 42°C. Solutions were centrifuged at 15,400 RCF for 5 minutes to remove pyrophosphate precipitate and the supernatants were ethanol precipitated, gel-purified through a 12% polyacrylamide gel, visualized by shadowing with longwave UV light, and passively eluted out of gel slices. RNA was ethanol precipitated, resuspended in 1X TE (10 mM Tris-HCl pH 7.5, .1 mM EDTA) and stored at −80°C. Radioactive transcripts were prepared as above, except 0.5 mM ATP containing 10 μCi radioactive α-ATP (Perkin-Elmer) was substituted in the above reaction and MgCl2 was reduced to 12 mM.
End-labeling of 5S-rRNA
5S-rRNA was prepared as described above. RNA was incubated with calf intestinal alkaline phosphatase (New England Biolabs) (20 μL reaction, 1 unit CIP, 17 pmol 5S-rRNA, 1× final (New England Biolabs) cutSmart buffer) for 30 minutes at 37°C. RNA was diluted to 50 μL using H2O, then extracted twice, first with 50 μL of 25:24:1 phenol:chloroform:isoamyl alcohol, then once with 50 μL of chloroform. RNA was ethanol precipitated then resuspended for radioactive end-labeling (50 μL volume, 1× (New England Biolabs) polynucleotide kinase buffer, 20 units PNK (New England Biolabs), 10 μCi radioactive gamma 32P-ATP). The reaction was incubated for 30 minutes at 37°C, extracted and ethanol precipitated, then resuspended in 50 μL of 200 mM NaCl.
Gel Mobility Shift Assays
16 μM Mod5 was serially diluted 3-fold into buffer (50 mM Tris-HCl pH 7.2, 50 mM NaCl), in 10 μL aliquots. Next, for all tRNA 5 μL of 32P-labeled RNA (~100 CPM/μL) diluted in 3× binding buffer (24 mM Tris-HCl pH 7.2, 150 mM NaCl, 15 mM MgCl2) was added to each tube and incubated at 37 °C for 30 min. For the 5S binding, ~36,000 CPM/μL was used, approximately a 6-fold higher RNA concentration than for tRNA binding. After incubation, 3 μL of 6× loading buffer (60% glycerol, 0.1% xylene cyanol) was added and 10 μL was run on an 8% native polyacrylamide gel in TBE buffer at 30 mA (1 mm thick gel, 7 cm length, Aquebogue Machine Shop Model 200). Gels were dried overnight, exposed on Phosphorimager screens overnight and imaged on a Typhoon 9210 Imager (GE Healthcare). Band intensities were analyzed using ImageQuant software and statistical analysis was performed with GraphPad Prism.
RNA competition Assays
10 μL of 20 μM unlabeled competitor RNAs were titrated using 2-fold serial dilutions in 1X TE (10 mM Tris-HCl pH 7.5, .1 mM EDTA) to a final volume of 5 μL each. 5 μL of 32P-labeled pre-Tyr RNA (~100 CPM/μL) diluted in 3× binding buffer (24 mM Tris-HCl pH 7.2, 150 mM NaCl, 15 mM MgCl2) was added to unlabeled competitor RNA tubes. 5 μL of 16 μM purified Mod5 was added to the above mixture for a final Mod5 concentration of 5.3 μM, 15 μL final reaction volume. Samples were incubated at 37 °C for 30 min.; then 3 μL of 6× loading buffer (60% glycerol, 0.1% xylene cyanol) were added, and samples were run on an 8% native polyacrylamide gel in TBE buffer at low power (30 mA, 1 mm thick gel, 7 cm length, Aquebogue Machine Shop Model 200)). Gels were analyzed as above. IC50 values and confidence intervals were calculated using GraphPad Prism. Data was fit to:
“Top” was given a constraint of ≤ 100% bound, while “Bottom” was constrained to ≥ 0%.
Isopentenylation assay
Reactions were performed in 58 mM Tris-HCl pH 7.2, 1.2 mM ATP, 5.8 mM MgCl2, +/− 0.2 mM DMAPP, 10 U SuperRNase-In (Ambion), 40,000 CPM of internally 32P-labeled RNA, 5.3 μM Mod5, 1.2 mM β-mercaptoethanol in a 17 μL final reaction volume. Reactions were incubated at 37°C for 1 hour and RNAs were ethanol precipitated. RNA pellets were washed one time with 70% ethanol, air dried, and resuspended in 10 μl of 8 M urea and 150 U of RNase T1 (Roche) was added to each. Samples were incubated overnight at 37 °C. Next, 2 μL of 6× loading buffer (60% glycerol, 0.1% xylene cyanol) was added to each and 10 μL was loaded onto a 20% polyacrylamide, 7M urea denaturing gel (pre-run at 20 mA, for 2 hrs, 1 mm thick gel, 70 cm length, Aquebogue Machine Shop Model 200). Samples were run at 25 mA, 1 mm thick gel, 7 cm length, Aquebogue Machine Shop Model 200, 2 hrs. The gel was exposed on a phosphorscreen for 3 hrs and imaged on Typhoon 9210 (GE Healthcare).
Light Scattering Assay
Samples (7.5 μM Mod5, +/− 7.5 μM RNA, in 125 mM NaCl, 5 mM potassium phosphate, 12.5 mM Tris-HCl, pH 7.4), 100 μL final volume, were loaded into a 96-well Costar plate. A405 was recorded using a Tecan Safire II at 37 °C. Readings were performed in duplicate and normalized to buffer-only A405.
Fluorescent microscopy
Imaging used an Olympus IX70 inverted microscope with a CoolSNAP HQ2 monochrome camera, 100× oil immersion lens. Mod5 was incubated with a fluorescein-conjugated RNA samples (1 μM Fl-pre-Ser-tRNA, 7.5 μM Mod5, 125 mM NaCl, 5 mM potassium phosphate, 12.5 mM Tris-HCl, pH 7.4, 20 μL volume) for 1.5 hours in humid conditions at 37 °C on No. 1.5 glass coverslips. Imaging used 492 nm excitation, 535 nm emission.
Electron microscopy
Eighteen μM Mod5, +/− 7.5 μM pLeu/mLeu/pTyr/mTyr/5S-rRNA, 75 mM NaCl, 3 mM NaPO4, 7.5 mM Tris-HCl (pH 7.4) was incubated for 24 hours at 37°C and sonicated briefly [31]. Samples were prepared using conventional methods for negative stain preparation [32]. Imaging was at room temperature using a Morgagni 268 at 100kV. Images were acquired at 22,000× with an Orius 2000 CCD camera with a pixel size of 2.4 A.
RESULTS
Mod5 directly binds to substrate and nonsubstrate RNAs
In our in vitro assays we worked with three tRNAs that are predicted substrates for Mod5-modification: mat-Tyr-tRNA (mature tRNA with 5′leader, intron and 3′ trailer removed), pre-Ser-tRNA (with 5′ leader and 3′ trailer), and mat-Ser-tRNA (with leader and trailer sequences removed). We also used nonsubstrate mature tRNAs (mat-Leu-tRNA), nonsubstrate pre-tRNAs (pre-Tyr-tRNA, pre-Leu-tRNA, and pre-Ala-tRNA), and nonsubstrate 5S rRNA (Supplementary Table 1). Mat-Tyr, pre-Ser, and mat-Ser-tRNAs contain the AAA36–38 sequence requirement for modification, while the intron in the pre-Tyr-tRNA disrupts the AAA and thus is not predicted to be modified by the enzyme at A37. The mat-Leu, pre-Leu, mat-Ala, and pre-Ala-tRNA sequences do not contain the required AAA at the anticodon loop, nor an A at position 37, and are therefore predicted to be nonsubstrates for isopentenylation (Supplementary Table 1).
To confirm these predictions and the activity of the isolated Mod5 enzyme we performed isopentenyl-transferase assays in the presence or absence of DMAPP substrate and each RNA. The modification reaction was followed by the digestion of the products with RNase T1 (which cleaves on the 3′ side of guanosine residues leaving a 3′GMP), and analysis of oligonucleotide fragments on denaturing polyacrylamide gels. The transfer of a dimethylallyl/isopentenyl group from DMAPP to the RNA would result in a DMAPP-dependent shift of the indicated RNase T1-fragment containing the modified base (Supplemental Figure 1). As predicted, mat-Tyr, mat-Ser, and pre-Ser tRNAs are modified at the oligo containing the A37 position (Figure 1 A), while pre-Ala, pre-Tyr-tRNA, pre-Leu-tRNA, and Leu-tRNA are not modified (Figure 1 B) (summarized in Supplementary Table 1). These data confirm the Mod5 substrate specificity in our recombinant enzyme [1, 3, 5] and show that the enzyme is capable of recognizing the folded substrates used for in vitro binding assays. We note that we see an additional 7mer oligonucleotide that becomes modified in the substrate mat-Ser and pre-Ser-tRNAs in the presence of Mod5 and DMAPP (Figure 1A). A possible explanation for this is that there is high sequence similarity in this 7 nt fragment compared to the 10 nt A37-containing fragment (GAAUCC compared to AAAUCC in the A37-containing fragment). Furthermore, although these two sequences reside in different arms of the tRNA, they are at similar relative positions within their respective arms (Supplemental Figure 3). The lack of a third A in the 7nt fragment as well as differences in positioning within the tRNA might explain the differences in efficiency of modification between the two, in which only ~30% of the 7 nt fragment becomes modified under the conditions used compared to nearly 100% in the A37-containing 10 nt fragment.
Figure 1.

Mod5 isopentenylation assays. (A) Substrate or (B) nonsubstrate tRNA genes were transcribed in the presence of 32P-adenosine creating internally labeled RNAs. RNAs were then incubated with Mod5 in the presence or absence of DMAPP. The samples were subsequently digested with RNase T1 and fragments separated with 20% denaturing PAGE. Fragments containing the modifiable adenosine in the absence of DMAPP are indicated with an asterisk*. A shifted band in the presence of DMAPP indicates the presence of a modified-adenosine containing an isopentenyl group (See Supplemental Figure 1).
Gel mobility shift assay (GMSA) analyses demonstrate that Mod5 binds to both substrate and nonsubstrate tRNAs and pre-tRNAs, as well as 5S RNA, within a similar range of protein concentrations (Figure 2 A–H). At low concentrations of Mod5, in some cases there are discrete shifted RNA bands that likely represent monomer, dimer, or higher complexes of RNAs with Mod5, though there is some smearing of signal in lanes consistent with either semi-stable complexes or variably structured complexes under these conditions. At higher concentrations of protein, these give way for all RNAs to supershift at the gel interface, suggesting a much larger or insoluble complex (Figure 2 A–H, discussed below). We will address the implications of these slow-migrating complexes below, but at a minimum these data suggest that Mod5 directly binds to RNAs regardless of whether the RNA is a substrate for modification.
Figure 2. Mod5 binds to substrate and nonsubstrate RNAs.

Phosphorimage of representative nondenaturing gel electrophorotograms of purified recombinant Mod5 and 32P-labeled RNAs (A) pre-Leu-tRNA, (B) mat-Leu-tRNA, (C) pre-Tyr-tRNA, (D) mat-Tyr-tRNA, (E) pre-Ala-tRNA, (F) h-pre-Ser-tRNA, and (G)mat-Ser-tRNA (H) 5S-rRNA with decreasing Mod5 concentrations (5.33 μM, 1.78 μM, 0.593 μM, 0.198 μM, 0.0658 μM, 0.0219 μM, 0.00732 μM, 0.00244 μM, 0.000813 μM, 0.00 μM. In (H) the lane with .00244 uM Mod5 was omitted).
To investigate the relative binding strengths of these RNAs with Mod5 we used a competition assay in which Mod5 was bound to a constant concentration of 32P-labeled pre-Tyr-tRNA, in the presence of increasing amounts of unlabeled-competitor RNAs. We determined apparent IC50 values for the nonsubstrate pre-Leu-tRNA, pre-Ala-tRNA, and pre-Tyr-tRNA to be similar (0.4, 1.0, and 0.8 μM, respectively, Figure 3, A, D, and E.). We further determined the competitive strength of two other nonsubstrates including a highly structured non-tRNA (5S rRNA, Figure 3 F) and Leu-tRNA (Figure 3 C), and one other substrate RNA, Tyr-tRNA (Figure 3 B). Leu-tRNA, Tyr-tRNA, and 5S-rRNA sequences (IC50 values 3.5, 1.5, and 1.6 μM, respectively) competed with similar strengths to other RNAs tested. In general, these data are consistent with our previously published in vivo data showing that Mod5 co-immunoprecipitates with both substrate and nonsubstrate tRNAs [22] and suggests that the capacity to serve as a substrate for modification does not predict which RNAs Mod5 can bind.
Figure 3. Effect of Mod5 binding to 32P-labeled pre-Tyr RNA in the presence of substrate and nonsubstrate RNAs.

Phosphorimages of representative (of three repetitions) nondenaturing gel electrophorotograms with indicated concentrations of unlabeled (A) pre-Leu-tRNA, (B) mat-Tyr-tRNA, (C) mat-Leu-tRNA, (D) h-pre-Ala-tRNA, (E) pre-Tyr-tRNA, and (F) 5S-rRNA, with a graphic representation (to the left of each phosphorimage) of concentration-dependent effect of Mod5 binding to 32P-labeled pre-Tyr-tRNA by unlabeled competitor RNAs. (G) A table of IC50 values and 95% confidence intervals by RNA.
tRNA accelerates aggregation of Mod5
As noted above, at the higher Mod5:RNA ratios the majority of the RNA is bound in a complex with proteins that remains near the well interface of the gel, suggesting that the RNA is bound to a multimerized or aggregated form of Mod5. This suggested that aggregation of Mod5 is not prevented by its interaction with RNA and possibly takes the form of amyloid fibers as observed by others [29]. We thus examined the physical form of these interactions further.
To compare the rates of aggregation we measured solution light scattering using absorbance at 405 nm as a measure of Mod5 precipitation/aggregation over time (Figure 4 A). The sample containing Mod5 without RNA demonstrated a modest increase in A405, while the addition of substrates mat-Tyr-tRNA, pre-Ser-tRNA, or mat-Ser-tRNA led to a 3 to 8 fold increase in A405 by 10 hours compared to the Mod5-only sample. Similarly, addition of nonsubstrate pre-Leu-tRNA or pre-Tyr-tRNA led to 6 to 7 fold increases in A405. In contrast, combining Mod5 with 5S-rRNA did not lead to a significant change, notable given the GMSA and competition results (Figures 2 and 3) showing Mod5 binding to 5S rRNA. These results suggest that tRNA-like binding accelerates the aggregation of Mod5, but that the nature of the bound RNA is important.
Figure 4. tRNAs increase Mod5-aggregation, Mod5 forms amyloid fibers in the presence of RNA, and the fibers bind RNA.
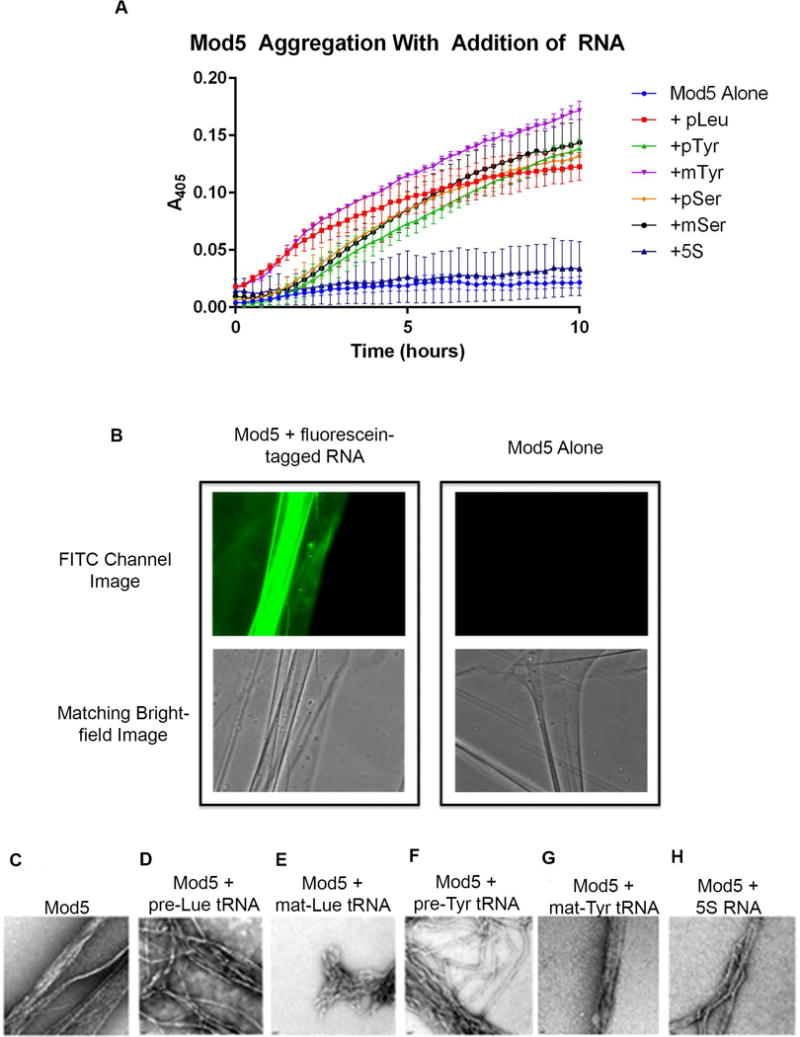
(A) Protein incubated with substrate tRNAs (mat-Tyr-tRNA, pre-Ser-tRNA, mat-Ser-tRNA) or nonsubstrate tRNAs (pre-Tyr-tRNA, pre-Leu-tRNA) demonstrated a ~5–8 fold increase in absorbance after 10 hours compared to the Mod5-only sample. Protein incubated with 5S-rRNA did not increase overall absorbance. (B) Mod5 amyloid fibers bound to a fluorescein-labeled Ser-tRNA. (C-H) Electron micrographs of Mod5 amyloid fibers formed (from left to right) in the (C) absence of RNA, or the presence of (D) pre-Leu-tRNA, (E) mat-Leu-tRNA, (F) pre-Tyr-tRNA, (G) mat-Tyr-tRNA, or (H) 5S-rRNA. 22,000× magnification.
Mod5 forms both punctate aggregates and extended fibers in the presence or absence of RNA
Mod5 was previously shown to form amyloid fibers in vitro and aggregates in vivo under selective pressure [11, 29]. We tested whether RNA affects the ability of Mod5 to form amyloid fibers by examining protein samples +/− RNA using fluorescence and electron microscopy (EM). We also imaged fibers formed in the presence of fluorescently labeled pre-Ser-tRNA to test whether the RNA itself was associated with the protein aggregates. Consistent with our GMSAs (Figure 2 A–G), we observe Mod5 fibers and punctate aggregates bound to the fluorescent RNA (Figure 4 B). For both protein-only samples and those incubated with various RNAs, we observed both punctate particulates and fibers using electron microscopy (Figure 4 C–H) confirming previous observations [29]. The size of the punctate particulates as well as the presence of fibers suggested protein multimers. Individual fibers had an approximate diameter of 5 to 10 nm, and were often seen in bundles with diameters as large as 50 nm. The resolution of this analysis precluded detection of quantitative differences in the dimensions of individual fibers, while spectroscopic assays for changes in the rate of fiber versus particle formation were inconclusive (Supplementary Figure 4): thus, more detailed analysis of differences with or without RNA awaits further investigation.
Discussion
We herein demonstrated that Mod5 directly binds to both substrate and non-substrate RNAs in vitro. This is in agreement with our hypothesis that Mod5-tRNA interactions play a role in the mechanism of tgm-silencing, where Mod5 binds the nascent pre-tRNA transcripts as a prerequisite for subsequent interactions that modify and remodel local chromatin structure. This is consistent with our previous in vivo data showing that Mod5 co-purifies with the precursor and mature forms of both substrate and nonsubstrate tRNAs [22]. The ability of the enzyme to bind with similar affinities to both substrate and nonsubstrate pre-tRNAs suggests that although the presence of an AAA at the A36-A38 position of the active site is predictive of modification activity [14], it is not a major determinant of RNA-binding behavior of Mod5. The observed differences in binding among tRNAs, pre-tRNAs, and 5S-rRNA are relatively small, suggesting substantial tolerance for variations in RNA structure, as well as sequence, for binding to Mod5.
Interestingly, upon the addition of RNA to purified Mod5, we observed a marked increase in light scattering with all tRNAs and pre-tRNAs tested, but not 5S-rRNA (Figure 4 A). These observations suggest that tRNA-like molecules can stimulate Mod5 aggregation whether or not they are substrates for isopentenylation (Figure 4 A). The reason for the markedly different aggregation behavior of 5S-rRNA, even though it binds to Mod5 and can compete with tRNA binding, is currently unclear, but could reflect a fundamentally different structure in the ribonucleoprotein. This would not be surprising, since the enzyme evolved specifically to fit a tRNA structure into the active site [14] and the overall effect on the structure of the complex could be quite distinct with a differently shaped RNA.
A second observation of interest comes from the effect of RNAs on aggregation by Mod5. It was previously shown that Mod5 is able to form amyloid-like aggregates, and that this aggregation can be selected in vivo by selecting against functional tRNA-modifying activity in the cytoplasm with an antifungal agent, fluconazole [29]. However, we previously showed that the nuclear silencing function of Mod5 is not ablated when aggregation is selected by this stressor [11], suggesting that the nuclear pool of Mod5 that is associated with RNA polymerase III transcription complexes and pre-tRNAs was not subject to the aggregation behavior of the cytoplasmic pool. Since aggregation of the purified enzyme is not prevented by RNA binding, and if anything is enhanced by it, we infer that the immunity of the nuclear function to fluconazole-selected aggregation might be due to other interactions of Mod5 in the nucleus. We propose an alternative model in which the RNA-bound form of Mod5 could be the “active” conformation of the nuclear enzyme, with the enhanced aggregation a necessary part of its nuclear function. Under this model, tgm-silencing is facilitated by the multimerization of Mod5 which might extend the Mod5-particle to neighboring gene promoters to either attract chromatin modifiers or block access of the transcription machinery to the DNA.
Suzuki and colleagues originally reported that aggregated Mod5 in the cytoplasm conveys resistance to certain fungicides [29], which might be consistent with the property having evolved in fungi to provide a reserve population of yeast that are resistant to anti-fungal attack. Interestingly, the human orthologue, TRIT1, has retained both the cytoplasmic and nuclear functions of Mod5 [22] and we recently demonstrated that it retains the amyloid fibril formation potential of Mod5 [33]. It is not clear why fibril formation would have been retained in an organism as distant from fungi as humans if cytoplasmic aggregation were the selective pressure, though it is possible that this method of removing Mod5 might regulate cholesterol biosynthesis or protein prenylation by modulating the availability of substrates. We suggest an alternative hypothesis, that the fibril formation is a positive attribute in tgm silencing that has been conserved for that reason. The observation that apparent RNA-mediated silencing has been observed near pol III transcription units in human cells [28] would be consistent with such a conservation of function.
Supplementary Material
Acknowledgments
We thank Dr. Alice Telesnitsky for her generous gift of time, lab space, reagents, and guidance. We thank May Tsoi for her technical assistance. We thank Dr. David Barton for helpful discussion. This work was supported by NIH Grant R01 GM082875 to D.R.E. and support from Michigan Undergraduate Research Opportunity Program (UROP) for D.F.R.
Footnotes
DRE and PJS conceived and supervised the study; DRE, PJS, DRS, and DFR designed experiments; PJS, ET, TJW, and DFR performed experiments; PJS, ET, TJW, and DFR analyzed data; PJS, DFR, and DRE wrote the manuscript; PJS, DRE, TJW, and DFR made manuscript revisions.
References
- 1.Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Molecular and cellular biology. 1987;7:177–84. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gefter ML, Bikoff E. Studies on synthesis and modification of transfer RNA. Cancer research. 1971;31:667–70. [PubMed] [Google Scholar]
- 3.Gillman EC, Slusher LB, Martin NC, Hopper AK. MOD5 translation initiation sites determine N6-isopentenyladenosine modification of mitochondrial and cytoplasmic tRNA. Molecular and cellular biology. 1991;11:2382–90. doi: 10.1128/mcb.11.5.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golovko A, Hjalm G, Sitbon F, Nicander B. Cloning of a human tRNA isopentenyl transferase. Gene. 2000;258:85–93. doi: 10.1016/s0378-1119(00)00421-2. [DOI] [PubMed] [Google Scholar]
- 5.Laten H, Gorman J, Bock RM. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic acids research. 1978;5:4329–42. doi: 10.1093/nar/5.11.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemieux J, Lakowski B, Webb A, Meng Y, Ubach A, Bussiere F, Barnes T, Hekimi S. Regulation of physiological rates in Caenorhabditis elegans by a tRNA-modifying enzyme in the mitochondria. Genetics. 2001;159:147–57. doi: 10.1093/genetics/159.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seif E, Hallberg BM. RNA-protein mutually induced fit: structure of Escherichia coli isopentenyl-tRNA transferase in complex with tRNA(Phe) The Journal of biological chemistry. 2009;284:6600–4. doi: 10.1074/jbc.C800235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soderberg T, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: essential elements for recognition of tRNA substrates within the anticodon stem-loop. Biochemistry. 2000;39:6546–53. doi: 10.1021/bi992775u. [DOI] [PubMed] [Google Scholar]
- 9.Warner GJ, Rusconi CP, White IE, Faust JR. Identification and sequencing of two isopentenyladenosine-modified transfer RNAs from Chinese hamster ovary cells. Nucleic acids research. 1998;26:5533–5. doi: 10.1093/nar/26.23.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slusher LB, Gillman EC, Martin NC, Hopper AK. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:9789–93. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smaldino PJ, Read DF, Pratt-Hyatt M, Hopper AK, Engelke DR. The cytoplasmic and nuclear populations of the eukaryote tRNA-isopentenyl transferase have distinct functions with implications in human cancer. Gene. 2015;556:13–8. doi: 10.1016/j.gene.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoladek T, Vaduva G, Hunter LA, Boguta M, Go BD, Martin NC, Hopper AK. Mutations altering the mitochondrial-cytoplasmic distribution of Mod5p implicate the actin cytoskeleton and mRNA 3′ ends and/or protein synthesis in mitochondrial delivery. Molecular and cellular biology. 1995;15:6884–94. doi: 10.1128/mcb.15.12.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benko AL, Vaduva G, Martin NC, Hopper AK. Competition between a sterol biosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense suppression. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:61–6. doi: 10.1073/pnas.97.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou C, Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16142–7. doi: 10.1073/pnas.0805680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamichhane TN, Mattijssen S, Maraia RJ. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Molecular and cellular biology. 2013;33:4900–8. doi: 10.1128/MCB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarham JW, Lamichhane TN, Pyle A, Mattijssen S, Baruffini E, Bruni F, Donnini C, Vassilev A, He L, Blakely EL, Griffin H, Santibanez-Koref M, Bindoff LA, Ferrero I, Chinnery PF, McFarland R, Maraia RJ, Taylor RW. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS genetics. 2014;10:e1004424. doi: 10.1371/journal.pgen.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes & development. 2010;24:1832–60. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. The EMBO journal. 2001;20:4863–73. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boguta M, Hunter LA, Shen WC, Gillman EC, Martin NC, Hopper AK. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Molecular and cellular biology. 1994;14:2298–306. doi: 10.1128/mcb.14.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics. 1999;151:57–75. doi: 10.1093/genetics/151.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13108–13. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pratt-Hyatt M, Pai DA, Haeusler RA, Wozniak GG, Good PD, Miller EL, McLeod IX, Yates JR, 3rd, Hopper AK, Engelke DR. Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3081–9. doi: 10.1073/pnas.1219946110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR. Silencing near tRNA genes requires nucleolar localization. The Journal of biological chemistry. 2005;280:8637–9. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hull MW, Erickson J, Johnston M, Engelke DR. tRNA genes as transcriptional repressor elements. Molecular and cellular biology. 1994;14:1266–77. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Good PD, Kendall A, Ignatz-Hoover J, Miller EL, Pai DA, Rivera SR, Carrick B, Engelke DR. Silencing near tRNA genes is nucleosome-mediated and distinct from boundary element function. Gene. 2013;526:7–15. doi: 10.1016/j.gene.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes & development. 2008;22:2204–14. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science (New York, NY) 2003;302:1399–401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolnough JL, Atwood BL, Giles KE. Argonaute 2 Binds Directly to tRNA Genes and Promotes Gene Repression in cis. Molecular and cellular biology. 2015;35:2278–94. doi: 10.1128/MCB.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science (New York, NY) 2012;336:355–9. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- 30.Engelke DR, Krikos A, Bruck ME, Ginsburg D. Purification of Thermus aquaticus DNA polymerase expressed in Escherichia coli. Analytical biochemistry. 1990;191:396–400. doi: 10.1016/0003-2697(90)90238-5. [DOI] [PubMed] [Google Scholar]
- 31.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. cience (New York, NY) 2000;289:1317–21. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 32.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biological procedures online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waller TJ, Read DF, Engelke DR, Smaldino PJ. The human tRNA-modifying protein, TRIT1, forms amyloid fibers in vitro. Gene. 2016 doi: 10.1016/j.gene.2016.10.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


