Abstract
The pioneering contributions of Ondrej Krivanek to the development of electron energy loss spectrometers, energy filters, and detectors for transmission and scanning transmission electron microscopes have provided researchers with indispensible tools across a wide range of disciplines in the physical sciences, ranging from condensed matter physics, to chemistry, mineralogy, materials science, and nanotechnology. In addition, the same instrumentation has extended its reach into the life sciences, and it is this aspect of Ondrej Krivanek’s influential contributions that will be surveyed here, together with some personal recollections. Traditionally, electron microscopy has given a purely morphological view of the biological structures that compose cells and tissues. However, the availability of high-performance electron energy loss spectrometers and energy filters offers complementary information about the elemental and chemical composition at the subcellular scale. Such information has proven to be valuable for applications in cell and structural biology, microbiology, histology, pathology, and more generally in the biomedical sciences.
Keywords: Electron energy loss spectroscopy, energy-filtered transmission electron microscopy, spectroscopic imaging, STEM-EELS, CCD cameras, detective quantum efficiency, detection limits
Introduction
From the time of its origin, electron energy loss spectroscopy (EELS) was recognized as being a particularly sensitive technique for the detection of lighter elements because those atoms have favorable inelastic scattering cross sections for inner shell excitations [1–3]. It was therefore understood at the outset that EELS had potential for analyzing the composition of biological specimens, which are predominantly composed of lighter atoms.
In addition, early studies in Albert Crewe’s laboratory at the university of Chicago showed that EELS could distinguish between biological molecules, such as different amino acids, and different nucleic acid bases by analyzing details of the EELS fine structure caused by excitation of valence or core shell electrons [4, 5]. However, in the 1970s electron spectrometers were not readily available commercially. Instead, the few existing EELS systems in laboratories around the world were home-built and often unsuitable for users who were non-physicists. For example, my own work on EELS began at the Department of Physics, Cambridge University, UK, where I used a 45° magnetic prism spectrometer on an AEI EM6G electron microscope operating at an accelerating voltage of 60 kV with the task of obtaining a better characterization of EELS core-edges and investigating the potential of EELS as a quantitative microanalytical tool. The spectrometer that I used had been built by David Wittry a few years earlier, when he was on sabbatical leave from the University of Southern California [6]. It was also in Cambridge where I met Ondrej Krivanek in 1975 since we were both graduate students at the Cavendish Laboratory. In fact, Ondrej’s Ph.D. research did not involve EELS, but was concerned with analyzing high-resolution TEM images of amorphous materials to discriminate nanometer-sized ordered domains from random fluctuations in the structure [7]. I completed my studies at the Cavendish one year after Ondrej, and moved to Mike Whelan’s group in the Department of Metallurgy and Materials in Oxford where I continued work on EELS with an in-column energy filter built by Egerton et al. [8] based on the original design by Castaing and Henry [9]. Then the following year, I moved to the Department of Applied and Engineering Physics at Cornell University in Ithaca, NY, where I worked in John Silcox’s group using a third type of EELS, a Wien-Filter spectrometer, which had been constructed by Curtis and Silcox [10], and which was attached to a somewhat ancient Hitachi HU11A TEM. Experience with these three instruments illustrates the point that in the 1970s most EELS and EFTEM systems were home-built and often quite difficult to use.
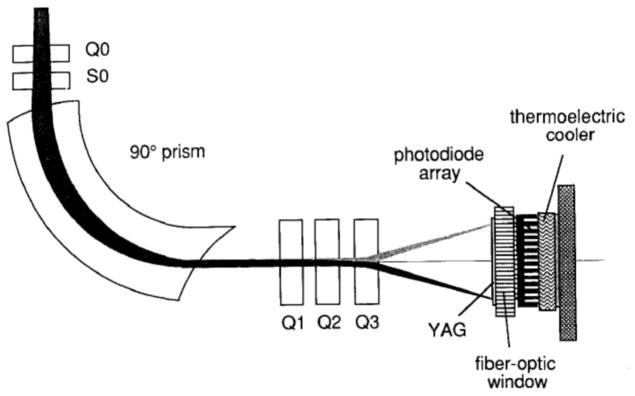
In the early 1980s this situation changed radically due to two events: first, Ondrej Krivanek designed a compact electron energy loss spectrometer offering relatively high performance in terms of collection efficiency and energy resolution; and second, Peter Swann, an ex-Professor of Materials Science from the University of London, who was an accomplished electron microscopist and inventor, and co-founder of Gatan Inc., entered into a partnership with Ondrej, who oversaw the launch of the first commercial post-column EELS system. Initially, the spectrometer collected spectra serially using a photomultiplier tube coupled to a scintillator [11], but soon afterwards Ondrej and his team developed an electron energy loss spectrometer with a parallel detection based on a photodiode array coupled to a YAG scintillator, as indicated in Figure 1 which is taken from Ondrej’s Ultramicroscopy paper from 1987 [12]. Shuman working on muscle physiology at the University of Pennsylvania with a home-built system had also demonstrated in 1981 that electron energy loss spectra could be collected in parallel [13]. The Gatan detector provided an enormous improvement in sensitivity, expanded the scientific community pursuing EELS, and enabled near single atom detectability in STEM with a sufficiently small probe. In particular, exciting results were obtained from a wide range of specimens, often using what was then the state-of-the-art STEM, the VG Microscope HB501 [14–17].
Figure 1.
Schematic diagram of Gatan parallel EELS showing the magnetic sector, the pre-sector focusing coils, post-spectrometer lens assembly, and the photodiode detection system. From O.L. Krivanek et al. [12].
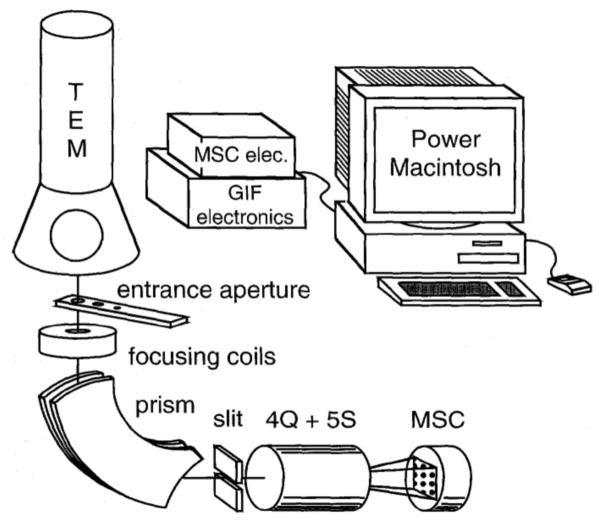
With the advent of CCD detectors, Ondrej realized that it was possible to modify the PEELS system by incorporating a slit at the dispersion plane and a series of other lenses (quadrupoles and sextupoles) after the slit, thus converting the TEM/PEELS into an energy filtering transmission electron microscope (EFTEM). The system could also easily revert to PEELS operation by retracting the slit and changing the lens strengths to image the dispersion plane onto the CCD detector. This provided high-end TEMs with great analytical flexibility since these instruments were equipped with field-emission guns that produced a small probe in STEM. For the first time TEMs could be operated in EFTEM or STEM-EELS mode, which attracted considerable interest from researchers in the life sciences, as indicated in Figure 2, which is taken from Ondrej’s 1995 Ultramicroscopy paper [18]. Over the past two decades there have been many further refinements in energy loss spectrometers and filters, including increased sophistication in the software used to acquire STEM-EELS hyperspectral images, as well as correction of higher order spectrometer aberrations, which give improved collection efficiency with larger spectrometer entrance apertures. However, Ondrej and his colleagues at Gatan had established the essential principles of today’s post-column imaging filters some twenty years ago, i.e., before Ondrej’s embarked upon the next stage of his career: the quest and ultimate realization of aberration-corrected STEMs, and the founding, together with Niklas Dellby, of Nion Inc.
Figure 2.
Principal components of the Gatan imaging filter system: (Q) quadrupole lens, (S) sextupole lens, (MSC) multi-scan CCD camera, (GIF) Gatan imaging filter.
From O.L. Krivanek et al. [18].
It seems clear that Ondrej’s deep understanding of the optics of electron spectrometers and energy filters, and how to correct spectrometer aberrations based on multipole magnetic lenses, helped Ondrej’s path towards correcting the spherical aberration of the probe-forming lens in the STEM. Also, Ondrej’s hands-on experience in interfacing spectrometers to the VG Microscopes HB501 instrument provided him with useful experience on how design features of dedicated STEMs could be improved. Therefore much of Ondrej’s and his colleagues’ experience with energy loss spectrometers helped lead to Nion’s development of the aberration-corrected UltraSTEM [19, 20].
It is should be pointed out that development of the energy filters with high-performance cooled CCD detectors, in turn, led to improvements in detectors for dedicated EELS systems, first the Enfina spectrometer, and more recently the Enfinium spectrometer which incorporates features of the Quantum GIF. In particular, the new generation of EELS enables 1,000 spectra to be read out in 1 second, and the Dual-EELS capability making it possible to acquire two regions of the energy loss spectrum almost simultaneously.
The account below includes examples of the application of electron energy loss spectrometers and post-column imaging filters, which have been developed by Ondrej and his colleagues, and which we have used in our laboratory to study biological specimens. Many other laboratories have been working in this area too, and we do not attempt to present a comprehensive review of EELS and EFTEM in the life sciences. For example, researchers have been using various in-column imaging filters to address important biological questions [21–24], and this is also outside the scope of this article.
Applications of the GIF
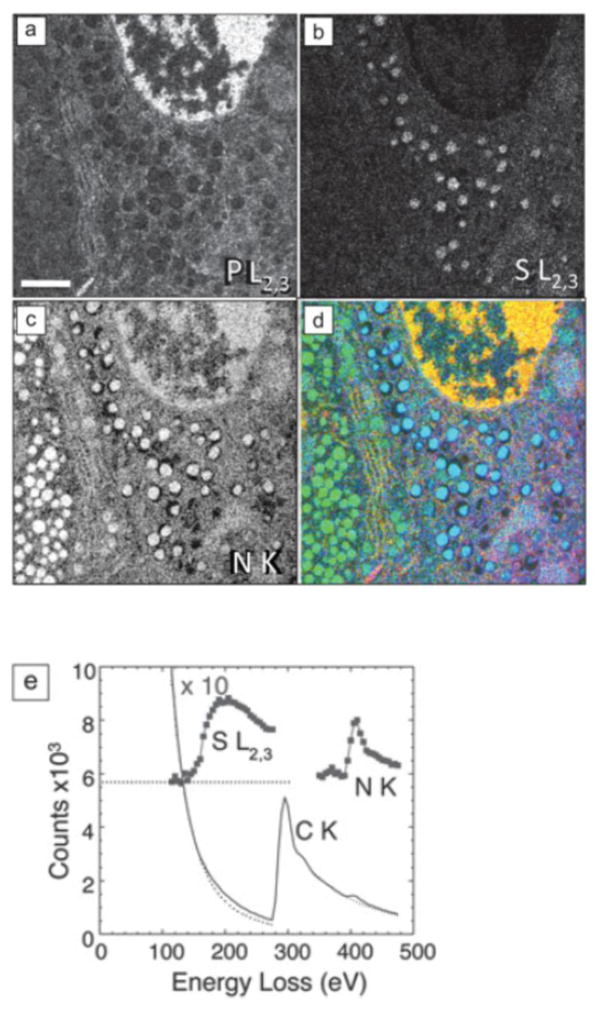
One method for obtaining quantitative elemental distributions from large regions of cells and tissues is to acquire a series of energy-selected images successively over a range of energy losses containing core-edges of interest, i.e., EFTEM spectroscopic imaging. Although this is inefficient in terms of the incident electron dose required to obtain a signal of a specific size, the acquisition is fast since the signal from ~106 pixels is read out in parallel. In this case, the signal is determined by the width of the energy-selecting slit, which is typically set to the required spectral energy channel width. For example, this approach has been applied to map the composition of hormone-containing secretory granules in mouse pancreatic islets of Langerhans, which are aggregates of endocrine cells that secrete hormones to control blood glucose levels [25, 26]. Figure 3 shows the phosphorus L2,3 edge, sulfur L2,3 edge, and nitrogen K edge maps from a sectioned region of an islet containing portions of an insulin-secreting β-cell (right), and a glucagon-secreting α-cell (left). Nitrogen serves as a general marker for protein, nucleic acid and other biological molecules, and is observed at high concentrations in both α-cells and β-cells, as well as in the nucleus. Phosphorus is concentrated in the DNA-containing chromatin of the β-cell nucleus as well as lesser amounts in membranes and ribosomes, but absent in the endocrine hormones. Only the β-cell secretory granules, and not the α-cell secretory granules, contain high levels of sulfur. The ratio of sulfur-to-nitrogen, which can be determined by quantitating the EELS spectra extracted from arbitrary regions of the specimen, can provide information about the fraction of amino acids that contain sulfur. Analysis of the EELS data extracted from a β-cell secretory granule in Figure 3(e) gives a sulfur-to-nitrogen ratio of 0.11, a value that is consistent with the nominal value of 0.097 for insulin, which contains 6 cysteine residues.
Figure 3.
EFTEM spectrum-imaging of unstained section of mouse pancreatic islet of Langerhans, obtained with GIF100 Imaging Filter. (a) Phosphorus L2,3 edge image showing high P in chromatin of cell nucleus. (b) Sulfur L2,3 edge image showing high S in secretory granules of insulin-producing beta cell, but low S in secretory granules of neighboring glucagon-producing alpha cell. (c) Nitrogen K edge image shows strong N signal in granules of both cell types, as well as in β-cell nucleus. (d) Overlay of the elemental maps with P (red), S (blue), and N (green); scale bar = 1 μm. (e) Extracted EELS from beta cell secretory granule can be analyzed quantitatively to give an atomic ratio of sulfur-to-nitrogen of 0.11±0.01:1. From G. Goping, et al. [25], and M.A. Aronova, and R.D. Leapman [26].
Applications of STEM-EELS
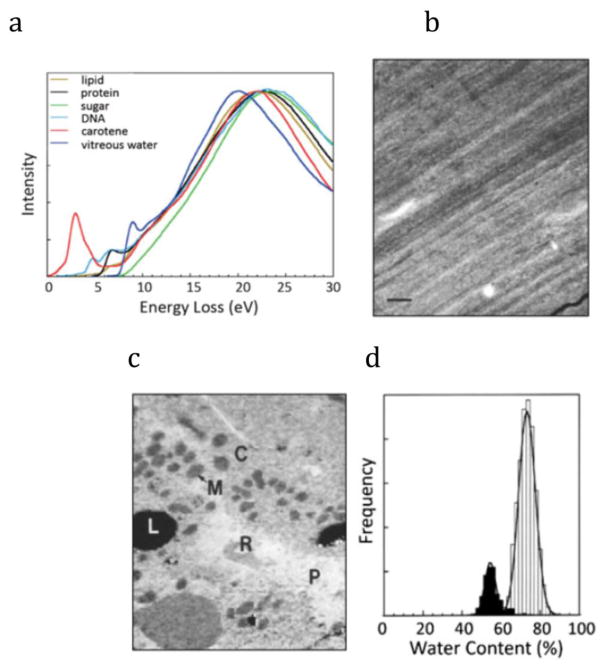
Extraction of chemical information from fine structure in the valence or core-edge spectra requires consideration of the radiation sensitivity of biological compounds and the subtle differences in EELS fine structure associated with compositional variations (Figure 4a) [27]. To map the distributions of biological compounds in cells therefore entails highly efficient and precise collection of spectral data at each pixel in an image, i.e., parallel detection of the spectrum in the STEM-EELS mode. Figure 4a shows that there are significant differences between the valence loss spectra between frozen water and protein, which can be exploited to derive water maps. For example, a study on cryosectioned rat hepatocytes showed that it is feasible to quantify water content in different subcellular organelles [27]. The dark-field STEM image of the frozen hydrated section in Figure 4b shows only compression artifacts caused by the cutting process. After acquiring a 128 × 128 pixel STEM-EELS spectrum-image, deriving the single scattering distributions, and fitting reference spectra at each pixel using a least squares approach, it was possible to obtain the distribution of water and protein, as shown in Figure 4c, in which regions containing mitochondria, cytoplasm, red blood cells, and plasma are all evident. Water content can be quantified at each pixel and expressed as a histogram (Figure 4d), which shows values in the cytoplasm of ~75% and in the mitochondria of ~55%. Other laboratories have also determined water distributions using this approach [28, 29].
Figure 4.
STEM-EELS imaging of frozen hydrated specimens at a beam energy of 100 keV using a Gatan PEELS interfaced to a VG Microscopes HB501 STEM. (a) Low-loss spectra up to an energy loss of 30 eV from major chemical constituents of cells; these can be used to fit spectra from cryosectioned cells to give quantitative compositional information; (b) Low-dose dark-field STEM of frozen hydrated liver cryosection showing no contrast apart from deformation lines; scale bar = 1 μm; (c) Water map of hepatocytes generated by multiple least squares fitting of water and protein reference spectra at each pixel revealing: mitochondria (M), cytoplasm (C), red blood cells (R), plasma (P) and lipid droplets (L); (d) Water content histogram for 2700 pixels of cytoplasm (light bars) and 500 pixels of mitochondria (dark bars) in hepatocyte, showing approximately Gaussian peaks with half width ~5%. From S. Sun et al. [27].
Other studies have been performed to map the uptake of the anti-cancer drug, doxorubicin in human cancer cells by means of a modified Zeiss EM902 EFTEM to obtain information from intense spectral features at an energy loss ~3-eV [30]. Recent work by Rez et al. [31] has shown that it is possible to extract extremely low energy loss spectra from biological molecules by using a monochromated Nion STEM100 and a high-resolution Gatan Enfinium EELS to record vibrational spectra at an energy resolution of approximately 15 meV. In those experiments, spectra were acquired from guanine crystals by placing the electron probe in an aloof position several nanometers outside the specimen. It this way is was possible to avoid specimen damage entirely [31, 32].
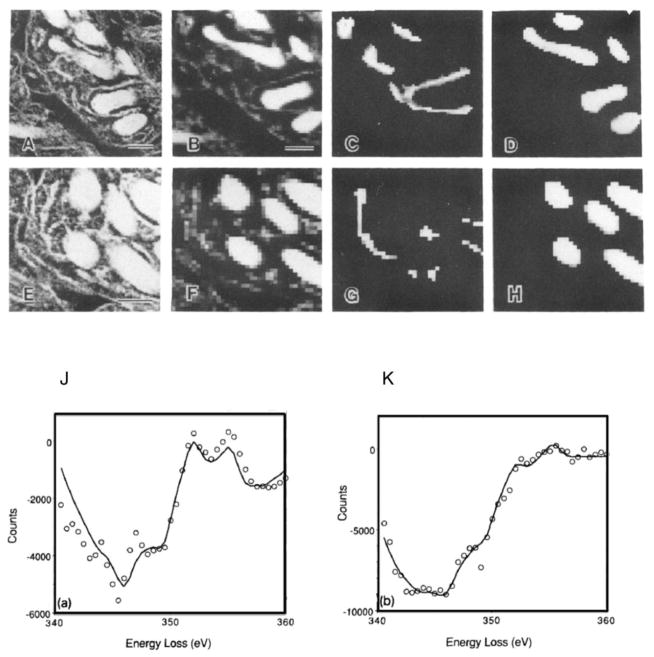
The core-edge signals in the electron energy loss spectrum provide quantitative information about endogenous elements in cells. Although major elements, such as nitrogen, phosphorus and sulfur, are often tightly bound to large macromolecules, enabling specimens to be prepared by conventionally fixation and embedding, as illustrated in the EFTEM measurements in Figure 3, other elements are diffusible and can only be retained in specimens prepared by rapid freezing, cryosectioning or focused ion beam milling, and freeze-drying. Of particular importance is calcium, which acts as a ubiquitous second messenger transmitting extracellular signals to intracellular targets, either by direct entry of Ca++ through the plasma membrane or by release of Ca++ from intracellular stores such as endoplasmic reticulum and mitochondria. The function of many cellular proteins depends on the binding of calcium, which is often present at very low overall concentration, of order 1 millimole per liter, or a few tens of atomic parts per million. To detect such low calcium concentrations often requires delivery of between 109 and 1010 incident electrons into each position of the probe on the specimen. In such measurements, the small signal can be swamped by fluctuations in the background due to channel-to-channel gain variations of the detector. As originally shown by Shuman and Somlyo [33, 34], the calcium signal becomes visible when the spectrum is deflected by a few electron volts across the detector and subtracted from the unshifted spectrum. This “difference spectrum” acquisition is illustrated by the measurement of calcium in dendrites of cryosectioned mouse cerebellar cortex in regions of endoplasmic reticulum and mitochondria (Figure 5) [35]. Here, the nitrogen maps (Figure 5b and 5f) are computed and used to segment the two compartments so that the spectra can be summed in each separately. The resulting spectra are fitted with reference spectra, and quantified to give an endoplasmic reticulum calcium concentration of 4.9±0.4 mmol/kg dry wt., and a mitochondrial calcium concentration of 1.4±0.4 mmol/kg dry wt, in good agreement with earlier measurements.
Figure 5.
Spectrum-imaging of two neuronal dendrites in the vicinity of the Ca L2,3 edge, together with dark-field images (A, E) obtained using the Gatan 666 Parallel EELS attached to a VG Microscopes HB501 STEM. Background-subtracted nitrogen K-edge maps (B, F) reveal location of mitochondria and membranes of endoplasmic reticulum. Very weak signals are detected when these nitrogen maps are segmented according to compartment: endoplasmic reticulum (C, G) and mitochondria (D, H); Bars = 200 nm. Spectra at each pixel were acquired in the difference mode with a 6 eV shift to reduce noise due to channel gain variations in the photodiode array. Multiple-least-squares fit (solid curves) of filtered reference spectra for segmented spectrum-image data (circles) in endoplasmic reticulum (J) and mitochondria (K). Analysis of the Ca L2,3 edge signal showed that the ER calcium concentration was 4.9±0.4 mmol/kg dry wt., and the mitochondrial calcium concentration was 1.4±0.4 mmol/kg dry wt. From R.D. Leapman et al. [35].
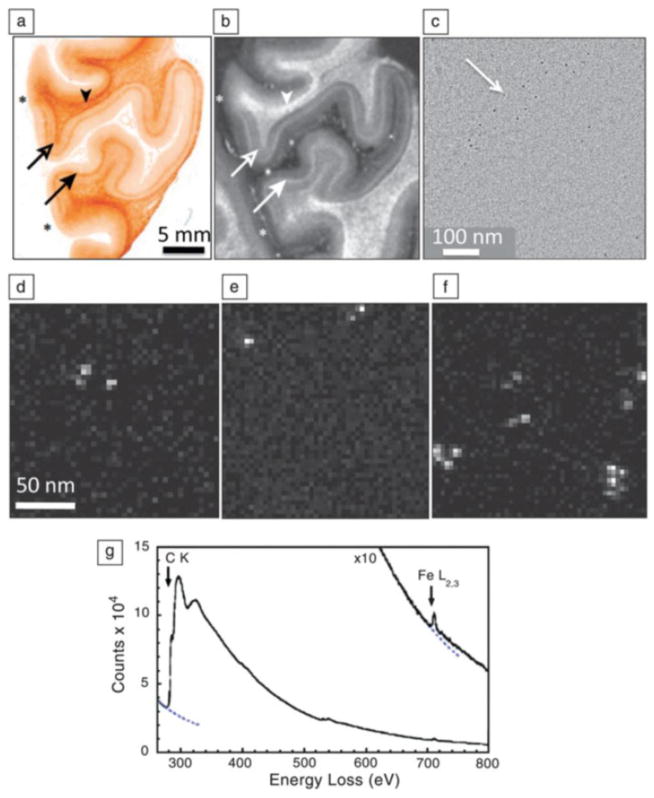
STEM-EELS has also proven useful for analyzing the distribution of intrinsic superparamagnetic iron in the human brain to determine whether this element contributes toward contrast in magnetic resonance medical imaging [36]. Figures 6a and 6b show, respectively, an optical image (stained for iron) and a magnetic resonance image of a paraffin section of post-mortem human visual cortex, with a specific region called the Line of Gennari indicated by the double arrows. Phase contrast TEM (Figure 6c) reveals small punctate structures that are attributed to the iron cores of ferritin molecules. This is confirmed by STEM-EELS spectrum images in small sub-regions (Figures 6d–f), which show that the punctate structures are composed of iron, and a quantitative analysis of extracted spectra (Figure 6g) reveals that they contain 1740±580 atoms, consistent with the known iron complement of ferritin cores [36].
Figure 6.
Application of scanning transmission electron microscope-electron energy-loss spectroscopy (STEM-EELS) to explain contrast observed in brain magnetic resonance images (MRI) in terms of iron concentrations. (a) Optical micrograph of post-mortem human visual cortex treated with Perl stain for iron, showing elevated iron in the region of the line of Gennari (arrows). (b) Corresponding MRI, also showing contrast in the line of Gennari (arrows). Image widths in (a) and (b) are the same, and asterisks indicate boundaries of region of visual cortex. (c) Phase-contrast transmission electron microscopy of unstained section in the region of the line of Gennari showing electron-dense particles. (d–f) STEM-EELS iron maps obtained from randomly selected areas of an unstained specimen in the vicinity of the line of Gennari, showing particles with high Fe content. (g) Typical EELS extracted from one of the Fe-containing particles reveals a strong Fe L2,3 edge; quantitative analysis showed that the particles contained on average 1740 ± 580 Fe atoms, consistent with the iron cores of ferritin molecules; dotted lines indicate extrapolated background intensity. From M. Fukunaga et al. [36].
Applications to Bionanotechology
EFTEM and STEM-EELS are becoming standard tools for characterizing synthetic bionanoparticles in the size range from about 1 nm to 100 nm, which are designed for potential use in medical diagnostic or therapeutic applications, i.e., the field of nanomedicine. Such particles can incorporate multiple functional moieties to provide both diagnostic and therapeutic capabilities, i.e., “theranostic” nanoparticles.
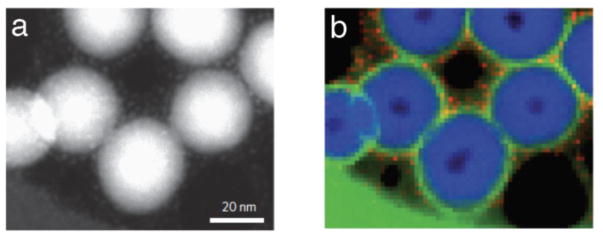
For example, hybrid nanocomplexes have been developed as magnetic resonance (MR) labels to image neural stem cells, bone marrow stromal cells and natural killer T-cells that have potential for cancer therapy [37]. Here, we used EFTEM spectroscopic imaging to analyze these nanocomplexes, which contain a mixture of three federally approved constituents, heparin (H), protamine (P), and iron-containing ferumoxytol (F) (Figure 7) [38]. It was found that the ferumoxytol nanoparticles surround a soft core about 200 nm in diameter, composed of sulfur-containing heparin and nitrogen-containing protamine. By quantifying the GIF spectrum images to determine the ratio of sulfur-to-nitrogen in the HPF nanocomplex cores, it was found that number of negative charges on the heparin balanced the number of positive charges on the protamine, indicating that the charge balance is an important factor in producing stable nanoparticles.
Figure 7.
EFTEM spectrum-image from plastic sections of hybrid nanocomplexes containing heparin, protamine, and ferumoxytol, which are being used for magnetic labeling of stem cells. (a) Fe L2,3 edge image; (b) overlay of Fe L2,3 edge image (red), S L2,3 edge image (blue) and N K edge image (green) shows that the iron-containing ferumoxytol nanoparticles surround a soft core composed of sulfur-containing heparin and nitrogen-containing protamine. The two components in the core co-localize due to association between the negatively charged heparin and the positively charged protamine. From L.H. Bryant et al. [38].
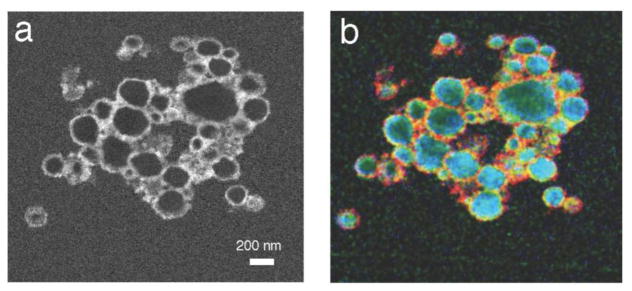
Hybrid nanoparticles containing both inorganic and organic components are sensitive to beam damage, in which case it is often preferable to use the more efficient STEM-EELS approach. For example, van Schooneveld et al. [39] have used a VG HB501 STEM equipped with a Gatan 666 PEELS to characterize nanoparticles consisting of CdSe/ZnS quantum dots (optical probe) inside silica shells, which are covered with a hydrophobic layer of octadecane and, finally, an outer layer that includes, among other components, lipids and paramagnetic gadolinium atoms (MRI probe). Van Schooneveld et al. [39] were able to identify all the constituents of the nanoparticle (Figure 8). In addition, by analysis of subtle variations in the carbon K-edge spectral fine structure, they were able to identify different types of chemical bonding in the organic layers that are added during various stages of the nanoparticle’s preparation.
Figure 8.
Spatially resolved element STEM-EELS analysis of hybrid silica nanoparticles containing quantum dots and coated with lipid that bind gadolinium; HAADF image of the lipid-coated nanoparticles (a); composite color map showing the location of different elements: red, blue and green indicate gadolinium (N4,5 edge), silicon (L2,3 edge) and carbon atoms (C K edge), respectively. From M.M. van Schooneveld et al. [39].
Future Directions
There are a number of exciting possibilities for applying EELS and EFTEM to unravel the complex ultrastructure in cells and tissues. For example, recently, a group led by Roger Tsien and Mark Ellisman at the University of California, San Diego, have developed multicolor EFTEM probes containing rare earth elements that can be used to label multiple specific cellular proteins in cells. In 2008, Roger Tsien won the Chemistry Nobel prize (together with Osamu Shimomura and Martin Chalfie) for developing genetically encoded green fluorescent protein (GPF), which has revolutionized studies of the biology of living cells in the optical microscope, and the rare earth EFTEM probes were conceived as a way to extend the GFP labeling approach to a finer scale in the electron microscope. In this recent work, the University of California San Diego team developed a genetically encoded photoactivated, singlet oxygen generator that, on activation, polymerizes the compound diaminobenzidine (DAB) that is chelated with a rare earth element [40]. After sample preparation for electron microscopy, the rare earth atoms are retained in close proximity to the location of the original genetic tag. In their recent paper published in Cell Chemical Biology, Adams et al. genetically tagged the complementary DNA for an important brain protein PKM-ζ involved in long-term potentiation, i.e., memory, with DNA for fluorescent protein and a singlet oxygen generator [40]. In these experiments, transfected cultured rat neurons were stimulated with a compound to induce long-term potentiation, followed by addition of cerium-chelated DAB, then photoactivation to generate singlet oxygen species and hence DAB polymerization, and finally the specimens were prepared for electron microscopy. In their experiments, Adams et al. used an in-column Omega filter on a JEOL JEM-3200EF TEM and an Ultrascan 4000 CCD detector or a Direct Electron DE-12 detector. They found cerium deposited at the post-synaptic membrane in stimulated neurons but not in the controls, demonstrating that PKM-ζ had indeed been produced following stimulation [40]. This result suggests that EFTEM could play an important role in mapping specific proteins in cells and tissues on the scale of a few tens of nanometers rather than the approximately ten times large scale of optical fluorescence microscopy.
In the application above, the rare earth elements are imaged using relatively weak M4,5 edges situated at energy losses from around 800 eV to 1400 eV, even though the peak-to-background is high due to the presence of strong resonances (white lines) at the edge threshold. It is therefore likely that direct electron detectors with single electron sensitivity will prove useful for this type of application, and the new generation of post-column Quantum GIF filters is now available with such detectors.
For certain applications, especially ones involving detection of near-trace levels of the important signaling ion calcium, STEM-EELS imaging provides higher sensitivity than EFTEM. Here, recent advances in EELS performance offer strong advantages. For example, the Gatan Quantum Dual EELS system, a natural progression from Ondrej’s original design, enables as many as 1,000 parallel spectra to be read out per second. This allows larger areas of a sample to be imaged in a reasonable time, an important factor in the context of biological specimens, where regions of interest can be sparsely distributed, such as synapses in brain. This new spectrum-imaging capability is also expected to offer higher sensitivity for imaging trace levels of exogenous elements such as manganese that are used as contrast agents in magnetic resonance imaging.
In addition, the improved capabilities of STEM-EELS also makes it possible to acquire low-loss spectrum-images containing large numbers of pixels in the low dose regime, which will enable specific chemical information to be extracted much more rapidly from frozen hydrated specimens or suitably prepared embedded preparations. This is likely to increase the range of applications of this technique, including the mapping of water in frozen hydrated cryosections, imaging the distributions of compounds localized in specific organelles, as well as imaging functionalized bionanoparticles.
Conclusion
The purpose of this brief account has been to present a historical account of Ondrej Krivanek’s contributions to the development of EELS and EFTEM with specific application to the life sciences with an emphasis on elemental and chemical mapping of cells and tissues. There was not sufficient space here to discuss the use of EFTEM to filter out inelastic scattering and hence to improve image quality in cryo-electron microscopy of macromolecular assemblies, or in electron tomography of frozen hydrated samples through elimination of chromatic aberration effects resulting from energy spread due to multiple inelastic scattering [41]. Our most important conclusion is that today’s electron energy loss spectrometers, which were originally developed by Ondrej, enable a wide range of biological studies. STEM-EELS spectrum imaging provides detection limits of a few atoms and a sensitivity of few parts-per-million. In this regard, operation at lower accelerating voltage (e.g., 60 kV) might be advantageous in minimizing knock-on beam damage to light atoms in biological specimens. The application of the post-column or in-column EFTEMs, when coupled with direct electron detectors, enables weak core-edge signals to be imaged over large regions of biological specimens, and thus opens up a number of exciting applications, such as the localization of genetically tagged proteins by the deposition of specific elemental labels.
Highlights.
Ondrej Krivanek developed the first commercial EELS and post-column imaging filter.
EELS and imaging filters have extended their reach in biological as well as materials research.
Applications of EELS have included studies in cell biology, neurobiology and bio-nanotechnology.
Acknowledgments
The work was supported by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Colliex C, Jouffrey B. Inelastic scattering of electrons in a solid by excitation of deep atomic levels. 1. Energy loss spectra. Phil Mag. 1972;25:491–511. [Google Scholar]
- 2.Egerton RF. Inelastic scattering of 80 keV electrons in amorphous carbon. Phil Mag. 1975;31:199–215. [Google Scholar]
- 3.Isaacson M, Johnson D. The microanalysis of light elements using transmitted energy loss electrons. Ultramicroscopy. 1975;1:33–52. doi: 10.1016/s0304-3991(75)80006-4. [DOI] [PubMed] [Google Scholar]
- 4.Crewe AV, Isaacson M, Johnson D. A high resolution electron spectrometer for use in transmission scanning electron microscopy. Rev Sci Instrum. 1971;42:411–420. doi: 10.1063/1.1685116. [DOI] [PubMed] [Google Scholar]
- 5.Isaacson M. Interaction of 25 keV electrons with the nucleic acid bases, adenine, thymine, and uracil. I. Outer shell excitation. J Chem Phys. 1972;56:1803–1812. [Google Scholar]
- 6.Wittry DB. An electron spectrometer for use with the transmission electron microscope. J Phys D. 1969;2:1757–1767. [Google Scholar]
- 7.Krivanek OL, Gaskell PH, Howie A. Seeing order in ‘amorphous materials’. Nature. 1976;262:454–457. [Google Scholar]
- 8.Egerton RF, Philip JG, Turner PS, Whelan MJ. Modification of a transmission electron microscope to give energy-filtered images and diffraction patterns, and electron energy loss spectra. J Phys E: Sci Instrum. 1975;8:1033–1037. [Google Scholar]
- 9.Castaing R, Henry L. Filtrage magétique des vitesses en microscopie électronique. J Microscopie. 1964;3:133–152. [Google Scholar]
- 10.Curtis GH, Silcox J. A Wien filter for use as an energy analyzer with an electron microscope. Rev Sci Instrum. 1971;42:630–637. doi: 10.1063/1.1685189. [DOI] [PubMed] [Google Scholar]
- 11.Krivanek OL, Swann PR. Proc Quant Microanalysis with High Spatial Resolution. Metals Society; London: 1981. An advanced electron energy loss spectrometer; pp. 136–140. [Google Scholar]
- 12.Krivanek OL, Ahn CC, Keeney RB. Parallel detection electron spectrometer using quadrupole lenses. Ultramicroscopy. 1987;22:103–115. [Google Scholar]
- 13.Shuman H. Parallel recording of electron energy loss spectra. Ultramicroscopy. 1981;6:163–167. doi: 10.1016/0304-3991(81)90056-5. [DOI] [PubMed] [Google Scholar]
- 14.Jeanguillaume C, Colliex C. Spectrum-image: The next step in EELS digital acquisition and processing. Ultramicroscopy. 1989;28:252–257. [Google Scholar]
- 15.Hunt JA, Williams DB. Electron energy-loss spectrum-imaging. Ultramicroscopy. 1991;38:47–73. [Google Scholar]
- 16.Mory C, Colliex C. Elemental analysis near the single-atom detection level by processing sequences of energy-filtered images. Ultramicroscopy. 1989;28:339–346. [Google Scholar]
- 17.Leapman RD. Detecting single atoms of calcium and iron in biological structures by electron energy-loss spectrum-imaging. J Microsc. 2003;210:5–15. doi: 10.1046/j.1365-2818.2003.01173.x. [DOI] [PubMed] [Google Scholar]
- 18.Krivanek OL, Friedman SL, Gubbens AJ, Kraus B. An imaging filter for biological applications. Ultramicroscopy. 1995;59:267–282. doi: 10.1016/0304-3991(95)00034-x. [DOI] [PubMed] [Google Scholar]
- 19.Krivanek OL, Dellby N, Lupini AR. Towards sub-angstrom electron beams. Ultramicroscopy. 1999;78:1–11. [Google Scholar]
- 20.Batson PE, Dellby N, Krivanek OL. Sub-angstrom resolution using aberration corrected electron optics. Nature. 2002;418:617–620. doi: 10.1038/nature00972. [DOI] [PubMed] [Google Scholar]
- 21.Ottensmeyer FP, Andrew JW. High-resolution microanalysis of biological specimens by electron energy-loss spectroscopy and by electron spectroscopic imaging. J Ultrastruct Res. 1980;72:336–348. doi: 10.1016/s0022-5320(80)90069-6. [DOI] [PubMed] [Google Scholar]
- 22.Ottensmeyer FP. Electron spectroscopic imaging: Parallel energy filtering and microanalysis in the fixed-beam electron microscope. J Ultrastruct Res. 1984;88:121–134. doi: 10.1016/s0022-5320(84)80004-0. [DOI] [PubMed] [Google Scholar]
- 23.Bazett-Jones DP, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 24.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goping G, Pollard HB, Srivastava M, Leapman RD. Mapping protein expression in mouse pancreatic islets by immunolabeling and electron energy loss spectrum-imaging. Microsc Res Tech. 2003;61:448–456. doi: 10.1002/jemt.10294. [DOI] [PubMed] [Google Scholar]
- 26.Aronova MA, Leapman RD. Development of electron energy-loss spectroscopy in the biological sciences. MRS Bulletin. 2012;37:53–62. doi: 10.1557/mrs.2011.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun S, Shi S, Hunt JA, Leapman RD. Quantitative water mapping of cryosectioned cells by electron energy loss spectroscopy. J Microsc. 1995;177:18–30. doi: 10.1111/j.1365-2818.1995.tb03530.x. [DOI] [PubMed] [Google Scholar]
- 28.Sousa A, Aitouchen A, Libera M. Water mapping in hydrated soft materials. Ultramicroscopy. 2006;106:130–145. doi: 10.1016/j.ultramic.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 29.Yakovlev S, Misra M, Shi S, Firlar E, Libera M. Quantitative nanoscale water mapping in frozen-hydrated skin by low-loss electron energy-loss spectroscopy. Ultramicroscopy. 2010;110:866–876. doi: 10.1016/j.ultramic.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Barfels MMG, Jiang X, Heng YM, Arsenault AL, Ottensmeyer FP. Low energy loss electron microscopy of chromophores. Micron. 1998;29:97–104. doi: 10.1016/s0968-4328(98)00009-2. [DOI] [PubMed] [Google Scholar]
- 31.Rez P, Aoki T, March K, Gur D, Krivanek OL, Dellby N, Lovejoy TC, Wolf SG, Cohen H. Damage-free vibrational spectroscopy of biological materials in the electron microscope. Nat Commun. 2016;7:10945. doi: 10.1038/ncomms10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egerton RF. Vibrational-loss EELS and the avoidance of radiation damage. Ultramicroscopy. 2015;159:95–100. doi: 10.1016/j.ultramic.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Shuman H, Kruit P. Quantitative data processing of parallel recorded electron energy-loss spectra with low signal to background. Rev Sci Instrum. 1985;56:231–239. [Google Scholar]
- 34.Shuman H, Somlyo AP. Electron energy loss analysis of near-trace element concentrations of calcium. Ultramicroscopy. 1987;21:23–32. doi: 10.1016/0304-3991(87)90004-0. [DOI] [PubMed] [Google Scholar]
- 35.Leapman RD, Hunt JA, Buchanan RA, Andrews SB. Measurement of low calcium concentrations in cryosectioned cells by parallel-EELS mapping. Ultramicroscopy. 1993;49:225–234. doi: 10.1016/0304-3991(93)90229-q. [DOI] [PubMed] [Google Scholar]
- 36.Fukunaga M, Lia T-Q, van Gelderen P, de Zwarta JA, Shmuelia K, Yaoa B, Lee J, Maric D, Aronova MA, Zhang G, Leapman RD, Schenck JF, Merkle H, Duyn JH. Layer-specific variation of iron content in cerebral cortex as a source of MRI contrast. Proc Natl Acad Sci USA. 2010;107:3834–3839. doi: 10.1073/pnas.0911177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thu MS, Bryant LH, Coppola T, Jordan EK, Budde MD, Lewis BK, Chaudhry A, Ren J, Varma NRS, Arbab AS, Frank JA. Self-assembling nanocomplexes by combining ferumoxytol, heparin and protamine for cell tracking by magnetic resonance imaging. Nat Med. 2012;18:463–467. doi: 10.1038/nm.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant LH, Jr, Kim SJ, Hobson M, Milo B, Kovacs ZI, Jikiria N, Lewis BK, Aronova MA, Sousa AA, Zhang G, Leapman RD, Frank JA. Physicochemical characterization of ferumoxytol, heparin and protamine nanocomplexes for improved magnetic labeling of stem cells. Nanomedicine: Nanotechnology, Biology, Medicine. 2016 doi: 10.1016/j.nano.2016.07.011. http://dx.doi.org/10.1016/j.nano.2016.07.011. [DOI] [PMC free article] [PubMed]
- 39.van Schooneveld MM, Gloter A, Stephan O, Zagonel LF, Koole R, Meijerink A, Mulder WJM, de Groot FMF. Imaging and quantifying the morphology of an organic–inorganic nanoparticle at the sub-nanometre level. Nature Nanotech. 2010;5:538. doi: 10.1038/nnano.2010.105. [DOI] [PubMed] [Google Scholar]
- 40.Adams SR, Mackey MR, Ramachandra R, Palida Lemieux SF, Steinbach P, Bushong EA, Butko MT, Giepmans BNG, Ellisman MH, Tsien RY. Multicolor electron microscopy for simultaneous visualization of multiple molecular species. Cell Chemical Biology. 2016 doi: 10.1016/j.chembiol.2016.10.006. http://dx.doi.org/10.1016/j.chembiol.2016.10.006. [DOI] [PMC free article] [PubMed]
- 41.Koster AJ, Grimm R, Typke D, Hegerl R, Stoschek A, Walz J, Baumeister W. Perspectives of molecular and cellular electron tomography. J Struct Biol. 1997;120:276–308. doi: 10.1006/jsbi.1997.3933. [DOI] [PubMed] [Google Scholar]