Abstract
Genetic susceptibility and environmental factors (such as stress) can interact to affect the likelihood of developing a mood disorder. Stress-induced changes in the hippocampus have been implicated in mood disorders and mutations in several genes have now been associated with increased risk, such as brain-derived neurotrophic factor (BDNF). The hippocampus has important anatomical subdivisions, and pyramidal neurons of the vulnerable CA3 region show significant remodeling after chronic stress, but the mechanisms underlying their unique plasticity remain unknown. This study characterizes stress-induced changes in the in vivo translating mRNA of this cell population using a CA3-specific EGFP reporter fused to the L10a large ribosomal subunit (EGFPL10a). RNA-sequencing after isolation of polysome-bound mRNAs allows for cell-type specific, genome-wide characterization of translational changes after stress. The data demonstrated that acute and chronic stress produced unique translational profiles and that the stress history of the animal can alter future reactivity of CA3 neurons. CA3-specific EGFPL10a mice were then crossed to the stress-susceptible BDNF Val66Met mouse line to characterize how a known genetic susceptibility alters both baseline translational profiles and the reactivity of CA3 neurons to stress. Not only did Met allele carriers exhibit distinct levels of baseline translation in genes implicated in ion channel function and cytoskeletal regulation, but also activated a stress response profile that was highly dissimilar from wild type mice. Closer examination of genes implicated in the mechanisms of neuroplasticity, such as the NMDA and AMPA subunits and the BDNF pathway, revealed how wild type mice were able to upregulate many of these genes in response to stress, but Met allele carriers failed to do so. These profiles provide a roadmap of stress-induced changes in a genetically homogenous population of hippocampal neurons and illustrate the profound effects of gene-environment interactions on the translational profile of these cells.
Keywords: stress, hippocampus, gene expression, RNA-Seq, BDNF-Val66Met
Introduction
The hippocampus is a brain region essential for learning and memory, and has been implicated in mood disorder etiology1. Additionally, the hippocampus is highly responsive to stress, which results in impaired learning and memory and increased anxiety and depressive-like behaviors in rodents2, 3. These behavioral outcomes have been linked to stress-induced changes in neuronal structure and function in the hippocampus4. Given that environmental stress is known to contribute to the onset and progression of mental illnesses such as bipolar disorder, depression and post-traumatic stress disorder (PTSD) in humans, studying the molecular changes underlying stress-induced plasticity in the hippocampus has important translational relevance for understanding these disorders.
Pyramidal neurons of the CA3 region of hippocampus have a well-established role in spatial memory5 and are of particular interest for mood disorders and stress susceptibility because they show pronounced dendritic retraction in response to chronic stress6, 7, as well as decreased spine density8. These structural changes correlate with impairments in memory tasks and defects in long term potentiation (LTP)9. Stress-induced remodeling of CA3 neurons and effective LTP depends on several signaling pathways including glucocorticoids, glutamate and brain-derived neurotrophic factor (BDNF)10.
BDNF has been implicated in neuroplasticity and mood disorders11, where increased levels have been observed in rodent hippocampus after antidepressant treatments12 and heterozygous knockout mice show attenuation of chronic stress-induced remodeling of CA3 pyramidal neurons13. A common human single nucleotide polymorphism (SNP) in the BDNF gene, which results in an amino acid change from a valine to a methionine at position 66 (Val66Met) has been associated with both memory impairments and increased risk of depression and anxiety-related disorders14–16. Importantly, human BDNF Met allele carriers exposed to stress were found to have decreased hippocampal volume and an increased likelihood of developing depression17, 18. Genetically modified knock-in mice with the Val66Met allele not only recapitulate the anxiety and depression-like phenotypes19, but also show increased stress reactivity and impaired learning and memory after chronic stress20, making them an important genetic model of increased stress susceptibility.
This study seeks to evaluate gene-environment interactions between the BDNF Val66Met allele and chronic stress on gene translation in CA3 pyramidal neurons of adult male mice. Several studies have characterized whole hippocampal gene expression in response to stress21–23 and in BDNF Val66Met mice24, but no studies have examined the stress by genotype interaction in a cell type-specific manner using high-throughput techniques. This study uses a transgenic mouse line to isolate in vivo translating mRNA from a genetically homogenous population of CA3 pyramidal neurons. These expression profiles provide a refined map of stress-induced changes at the molecular level that are unique to CA3 pyramidal neurons, which are distinctly vulnerable to chronic stressors4, 25, and will help unravel the mechanisms underlying the plasticity and damage-vulnerability of these cells in response to stress.
Materials and Methods
Animals
Gprin3-BacTRAP transgenic mice were generated in the Heintz Lab as previously described and backcrossed onto the C57/Bl6 background strain26. BDNF Val66Met knock-in mice were generated as previously described19 and were backcrossed more than 12 generations to the C57Bl/6 background prior to starting experiments. To generate mice carrying both the bacTRAP transgene and the BDNF Val66Met knock-in allele, offspring from several mating pairs from each line were crossed together and subsequently maintained as their own independent line within the colony to maintain genetic homogeneity. To control for litter specific effects (such as the quality of maternal care) mice in each group were selected from across multiple litters. Animals were group housed (n=4–5) in standard cages (28.5×17×13cm) and were kept on a 12-h light-dark cycle (lights off 1800h) in a temperature-controlled room maintained at 21±2°C. Food and water were available ad libitum. The Rockefeller University Institutional Animal Care and Use Committee approved all experimental procedures involving animals.
Stress Paradigm & Tissue Collection
Male mice, 7–12 weeks old, were used for all conditions. Acute Forced Swim Test (FST) stress was performed for 6 mins in 2L of RT water in a 4L beaker. Mice were removed and dried and allowed to recover for 1hr in the home cage before sacrificing by cervical dislocation. CRS began at 7–8 weeks of age and was carried out in 50mL conical tubes for 2hr/d starting at 10:00h for 21 consecutive days. Control mice were left undisturbed. On day 22 mice were sacrificed (CRS) or subjected to forced swim (CRS+FST). All groups had unstressed, age-matched controls that were sacrificed concurrently. Mice were immediately decapitated, brains were removed and hippocampus was fresh dissected and prepared for TRAP protocol.
TRAP and RNA-Seq Analyses
Translating Ribosome Affinity Purification (TRAP) was performed as described in Heiman et al., 201427. Briefly, hippocampal tissue from Gprin3–EFGPL10 mice (n=6 mice for each TRAP-seq, n=2 for qRT-PCR) was dissected out and placed in ice-cold dissecting buffer (HBSS, 2.5mM HEPES-KOH, 35mM Glucose, 4mM NaHCO3, 100μg/mL cycloheximide) Tissue was manually homogenized in homogenization buffer (10mM HEPES-KOH, 150mM KCl, 5mM MgCl2, 0.5mM DTT, protease inhibitors, RNasin and Superasin RNAse inhibitors, 100ug/mL cycloheximide) and centrifuged at 2,000×g for 10 min at 4°C, and supernatants were incubated with Nonidet P-40 for 5 min on ice before a 15-min spin at 20,000×g at 4°C. Supernatant was applied to Streptavidin MyOne T1 Dynabeads (Invitrogen no. 65601) that had been incubated with biotinylated protein-L (Pierce no. 29997) and α-EGFP antibodies (19C8 and 19F7 from the MSKCC antibody core facility) overnight at 4°C in 0.15M KCl buffer containing (10mM HEPES, 0.15M KCl, 5mM MgCl, 1% NP40, 0.05mM DTT, RNasin RNAase inhibitor, 100μg/mL cycloheximide). After immunoprecipitation, unbound fractions were saved, and beads were washed five times with 0.35 M KCl wash buffer (as above with 0.35M KCl) before beads were resuspended in lysis buffer (Stratagene Absolutely RNA Nanoprep Kit no. 400753). Unbound fraction was isolated using Qiagen Lipid RNA isolation kit (cat#74804) using a Qiacube (cat#9001292) per the manufacturer’s instructions. Quantification and RNA integrity were determined by using a Bioanalyzer (Agilent), and only samples with RNA Integrity Numbers greater than eight (out of 10) were used for sequencing.
Samples were prepared for sequencing by the Rockefeller University Genomics Core Facility using the TrueSeq RNA Sample Preparation Kit v2 (Illumina) and barcoded to allow samples to be multiplexed within a flow cell lane. Barcoded cDNA libraries were sequenced on an Illumina HiSeq 2500 in a single lane to obtain 100-bp single-end reads at an approximate sequencing depth of 25–35 million reads per sample. Raw reads were trimmed to remove sequencing artifacts (10bp from 5′ end) and filtered to remove low quality reads (read with a QC < 20 in more than 10% of bases were discarded) before alignment to mouse genome (mm10) using TopHat228. Differential expression analysis was conducted with Strand NGS software (Agilent), in which DESeq29 was used to quantify transcript reads and obtain Z scores and fold change values for individual genes. Genes with p<0.001, Benjamini-Hochberg corrected and fold change greater than 1.5 were selected for further analysis. Differences in integrated read density were visualized against the mouse genome by using Strand NGS or IGV (Broad Institute). GO categories were manually curated from results of DAVID functional annotation cluster tool30, 31.
Immunohistochemistry
For immunohistochemistry, animals were killed by deep anesthesia with pentobarbital (100 mg/kg), followed by transcardial perfusion with heparinized saline and then 4% (wt/vol) paraformaldehyde (PFA) using a peristaltic perfusion pump (Fisher Scientific). Brains were postfixed for 24 h in 4% PFA before sinking in 30% (wt/vol) sucrose for 3 d. Finally, brains were flash-frozen on dry ice and stored at −80°C until they were cut using a cryostat into 40μm-thick sections, which were then stored −80°C before being processed for immunohistochemistry. Immunohistochemical staining was carried out on 3–4 serial 40μm coronal sections from the rostrocaudal extent of the hippocampus as follows: sections were washed 3 × 10 min in 0.1 M PBS solution (pH 7.4), blocked in 0.5% BSA in 0.1 M PBS with 0.25% Triton X-100 for 1h before incubation in primary antibody. Antibodies NeuN (1:1,000; MAB377, Chemicon) and GFAP (1:1,000; AB7260 Abcam) were diluted in the blocking solution overnight at 4 °C. On the following day, sections were rinsed in 0.1 M PBS (3 × 10 min) before being incubated with species appropriate secondary antibody (Alexa568 gt@Rb A11011, Alexa568 gt@ms A11004; Life Technologies) diluted 1:1000 in PBS for 1hr at RT. After three washes in 0.1 M PBS (3 × 10 min), slides were coverslipped using Vectashield with DAPI (H-1200, Vector Laboratories). The immunoreactivity for each cover-slipped section was assessed by a Nikon ECLIPSE 90i microscope, equipped with the digital camera DS-Fi1.
Preamplification and qRT-PCR
A separate cohort of animals was used for qRT-PCR validation of RNA-seq data. TRAP mRNA from two Gprin3-BAC-TRAP mice were pooled per replicate, and a total of four biological replicates were prepared per group (Control and FST). RNA was isolated as described above and reverse transcribed using AB Superscript reverse transcriptase, 100ng of RNA was added to each reaction. Primers were designed and verified for specificity using UCSC genome browser and NCBI Primer blast tools. Primers were purchased from IDT and resuspended to a stock concentration of 100uM in TE. Final primer pool contained all primers at concentration of 50nM in nuclease free H2O. Preamplification was carried out using Taqman preamp master mix as described by the manufacturer instructions (cat#4369016) and final product was diluted 1:5 in H2O for use in qRT-PCR reactions. SYBR green master mix was used with all primers and run on a Quantstudio 12k flex thermocycler. Fold change was calculated using the 2ΔΔCT method and normalized to GAPDH levels32.
Results
Isolation of in vivo translating mRNA from CA3 pyramidal neurons
Pyramidal neurons of the CA3 region are uniquely vulnerable to the effects of stress4, 25. In this study, the response of CA3 neurons to stressors is characterized using the Translating Ribosome Affinity Purification (TRAP) technique, a powerful approach to interrogate translating mRNAs from genetically defined cell populations in vivo26, 33. TRAP has been used successfully to identify cell type specific translational changes in a variety of experimental conditions including behavioral and pharmacological manipulations26, 34, 35. To study expression changes in CA3 neurons of the hippocampus in response to stress using the TRAP methodology, transgenic mice were generated that expressed an EGFPL10a ribosomal fusion protein under control of the Gprin3 promoter (Gprin3-BacTRAP). Reference in situ hybridization data (Allen brain atlas, www.brain-map.org) and BAC-EGFP reporter mice (www.gensat.org) demonstrate that Gprin3 expression is restricted to the CA3 region in the hippocampus. Expression of the EGFPL10a transgene was localized primarily in the cell bodies of CA3 pyramidal neurons, confirming these previous findings (Fig. 1A). Double immunohistochemical labeling for the EGFP and either the neuronal marker NeuN or the glial marker GFAP confirmed that cells expressing the transgene were neuronal (Fig. 1B).
Figure 1. Gprin3-BacTRAP mice allow for the isolation of translating mRNA from CA3 neurons.
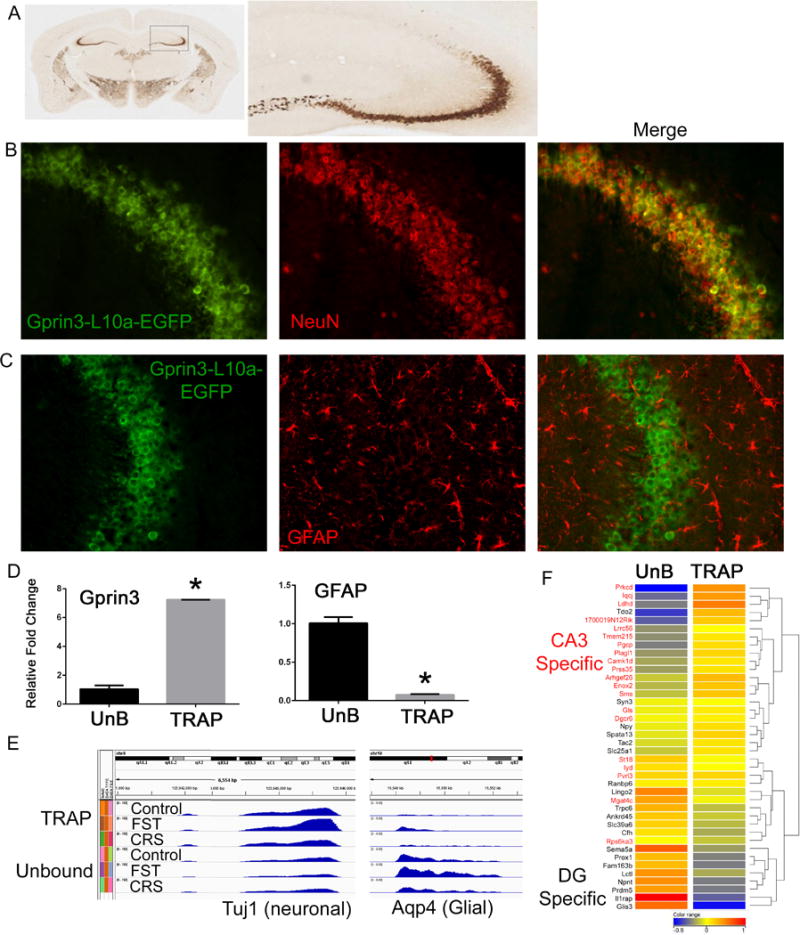
A) Immunohistochemical labeling for the EGFP fusion proteins shows expression in several brain regions, but in hippocampus the labeling is primarily in the CA3 region. B–C) Double labeling for either NeuN or GFAP confirm that Gprin3-EFGP-L10a positive cells are neuronal. D) qRT-PCR from TRAP-IP and Unbound (UnB) mRNA isolations demonstrates significant enrichment for Gprin3 in the TRAP fraction and the glial specific marker Gfap in the unbound fraction. E) Read density plots from RNA-Seq data of the canonically neuronal gene Tuj1 and the glial gene Aqp4 demonstrate enrichment of neuronal genes in the TRAP fraction and glial genes in the unbound fraction across stress conditions (FST= forced swim test; CRS= Chronic Restraint Stress). F) Heatmap of genes expressed in either dentate gyrus (black) or CA3 (red) shows that the TRAP-IP is highly enriched in CA3 specific genes, whereas dentate specific genes are enriched in the unbound fraction. (*p<0.05)
Anti-EGFP immunoprecipitations (IPs) were performed on dissected hippocampi from Gprin3-bacTRAP mice to purify polysomes and isolate polysome-bound mRNA from CA3 neurons. qRT-PCR was performed to quantify Gprin3 and Gfap mRNA levels in either the immunoprecipitated (TRAP) or supernatant (unbound) fractions to validate that the RNA isolated by the TRAP protocol reflected enrichment from the targeted cell population. There was a 7-fold enrichment of Gprin3 and conversely a nearly 80% depletion of Gfap, a glial marker, in the TRAP fraction compared to the unbound sample (Fig. 1D). TRAP and unbound mRNA was subjected to RNA-sequencing (RNA-Seq), allowing for the characterization of differential expression of all mRNAs undergoing translation. Examination of canonically neuronal (Tuj1) or glial (Aqp4) genes in the sequencing data similarly confirmed enrichment of neuronal genes in the TRAP fraction (Fig. 1E). Unbiased clustering across all genes and samples showed that the TRAP samples clustered independently from the unbound samples, demonstrating consistency of the method across multiple experiments (Supp. Fig 1). To further confirm that mRNA isolated by TRAP was primarily from CA3 neurons, both a fine structure and differential expression search were conducted using the Allen Brain Atlas to identify genes enriched in either CA3 pyramidal neurons or the dentate gyrus (DG). The top hits from each region were manually curated to confirm the gene expression pattern, and a heatmap illustrating the clustering of RNA-seq results for 20 genes from either CA3 or DG revealed significant enrichment for CA3-specific genes, and depletion of DG-specific genes, in the Gprin3 TRAP IPs (Fig. 1F).
Acute and chronic stress differentially alter the translational profiles of CA3 neurons
Gprin3-BacTRAP mice were subjected to stress conditions identical to those described in Gray et al., (2014), in which the animals received either a naïve forced swim stress (FST), chronic restraint stress for 2h/day for 21 days (CRS), or a combination of CRS followed by a FST on day 22. TRAP mRNA was collected for RNA-seq to characterize patterns of stress-induced differential gene expression. After a naïve FST, 2,440 genes were identified as both significantly differentially expressed (p<0.001, FDR corrected) and exhibiting a greater than 1.5 fold change by RNA-Seq. Several immediate early genes identified as significantly increased or decreased after FST in the TRAP-Seq data were validated by qRT-PCR, with fold-change levels showing a significant correlation (R^2=0.79; p<0.01) between the two methods (Fig. 2A). The identification of numerous immediate early genes, such as Fos and Arc is consistent with whole hippocampal studies of acute stress-induced neuronal activation. Further, 617 genes identified as changed by FST in either the TRAP or unbound fractions by RNA-Seq were identical to those changed on whole hippocampal microarrays21. Studies comparing concordance rates of differential expression between microarray and RNA-Seq technologies have identified rates between 70–75% when using identical mRNA samples36, 37. The RNA-Seq data from acute FST has a 60% concordance rate with the previous publication, despite coming from an entirely different cohort of mice, which demonstrates that this is a highly reliable representation of gene expression changes. Importantly, the TRAP-Seq identified genes that were not significant on the microarray platform, demonstrating that the cellular specificity of this technique can improve detection sensitivity.
Figure 2. Translational changes in CA3 neurons reveal unique activation profiles in response to stress.
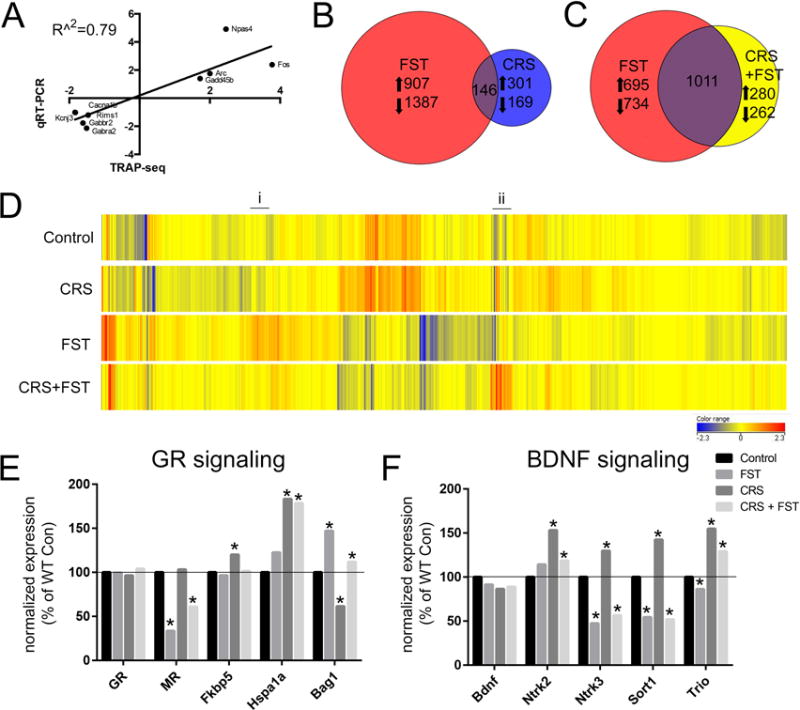
A) Scatter plot illustrating the correlation between the fold changes from RNA-seq and qRT-PCR for 9 genes differentially regulated by FST. B) Venn diagram illustrating that the number of genes differentially expressed after an acute stress (FST= forced swim test, red circle) were distinct from the genes that were changed after exposure to chronic stress (CRS= chronic restraint stress, blue circle). C) Venn diagram showing that exposure to a novel acute stressor after three weeks of chronic restraint (CRS+FST, yellow circle) activated significantly different genes than after a naïve exposure to the same FST. D) Heatmap illustrating expression level changes of known GR-regulated transcripts from Polman et al., (2013). Transcripts upregulated only in response to naïve FST are highlight with (i) and transcripts upregulated only when exposed to FST after CRS are highlighted with (ii). E) Levels of the glucocorticoid receptor and selected chaperone genes. F) Levels of Bdnf and selected downstream signaling molecules. (*p<0.001; Benjamin-Hochberg corrected for FDR)
After CRS, 616 genes were significantly different from unstressed controls using the same criteria (p<0.001 FDR; FC>1.5). Only 146 of the same genes were identified as changed after either CRS or FST in the TRAP fractions (Fig. 2B), which indicates that acute and chronic stress result in distinct translational changes in CA3 neurons.
To reveal how a history of chronic stress can alter translational reactivity to a novel stressor, mice were subjected to a novel swim stress 24h after the final CRS exposure (CRS+FST). While over 1,000 of the significantly altered genes from CRS+FST mice were also changed by naïve FST, there were over 500 genes unique to the CRS+FST condition, demonstrating that the stress history of the animals alters the molecular reactivity of CA3 neurons (Fig. 2C). Acute swim stress results in significant elevation of circulating corticosterone and activation of the glucocorticoid receptor (GR) in the hippocampus (reviewed in38). Previous studies using genome-wide methods have identified GR-responsive genes in the rodent hippocampus39. Using these data, expression levels of known GR-responsive genes were plotted across stress conditions. Consistent with an acute elevation of corticosterone, an unbiased clustering of GR-responsive genes based on expression levels revealed that FST and CRS+FST were the most dissimilar from control and CRS (Fig. 2D). However, numerous differences are still apparent between FST and CRS+FST (Fig. 2D(i) & (ii)), which demonstrates that there are certain known GR-responsive genes that are no longer activated or inactivated after CRS in response to an elevation of corticosterone.
Interestingly, these changes were observed despite the fact that no difference was detected in the levels of GR (Nr3c1) itself across stress conditions (Fig. 2E). However, significant differences in MR (Nr3c2), as well as three known chaperone proteins Fkbp5, Hsp70 (Hspa1a), and Bag1 were observed across the stress conditions (Fig. 2E). Previous studies have shown that changes in the GR-MR balance or differences in the levels of these chaperones are sufficient to alter the levels of GR-dependent transcripts40–42. Acute and chronic stress has been associated with changes in BDNF mRNA levels in hippocampus43, 44, and a non-significant decrease is observed here in the translational fraction of CA3 neurons (Fig. 2F). However, significant changes were observed in the neurotrophin receptors TrkB and TrkC (Ntrk2/Ntrk3), which were both increased with CRS, as were neurotrophin associated molecules, sortilin1 (Sort1) and Trio, which will be discussed below.
BDNF Val66Met mice have a unique translational profile and response to stress in CA3 neurons
To identify changes in the translational profiles of CA3 neurons associated with a genetic susceptibility to mood disorders, Gprin3-BacTRAP mice were crossed with BDNF Val66Met mice, a translationally relevant model of a human polymorphism that has been correlated with anxiety and depression16, 19. TRAP was performed on the hippocampus of Gprin3-bacTRAP mice and that were either heterozygous (BDNFMet/+) or homozygous (BDNFMet/Met) for the Met allele, and purified mRNA was used for RNA-Seq. Examination of reads mapped to the Bdnf locus confirmed the presence of the point mutation in both genotypes (Supp. Fig. 2) and revealed decreases in levels of Bdnf exons 4, 6, and 9 in CA3 neurons of BDNFMet/+ and BDNFMet/Met mice (Supp. Fig. 3), which is consistent with previous in situ hybridization results45. Differential expression analysis identified 1,420 genes that were different in heterozygous mice and 3,736 differentially expressed genes in homozygous mice for the Met allele compared to WT controls (Fig. 3A; p<0.001 FDR corrected; FC > 1.5). Over 60% of the genes identified as differentially expressed in heterozygotes were also present in homozygotes (Fig. 3A). Gene ontology analysis of differentially expressed genes from the BDNFMet/+ mice identified enrichment of gene clusters involved in transmembrane structure, cellular homeostasis, and ion transport (Table 1).
Figure 3. BDNF-Val66Met mice exhibit a unique translational profile in response to stress.
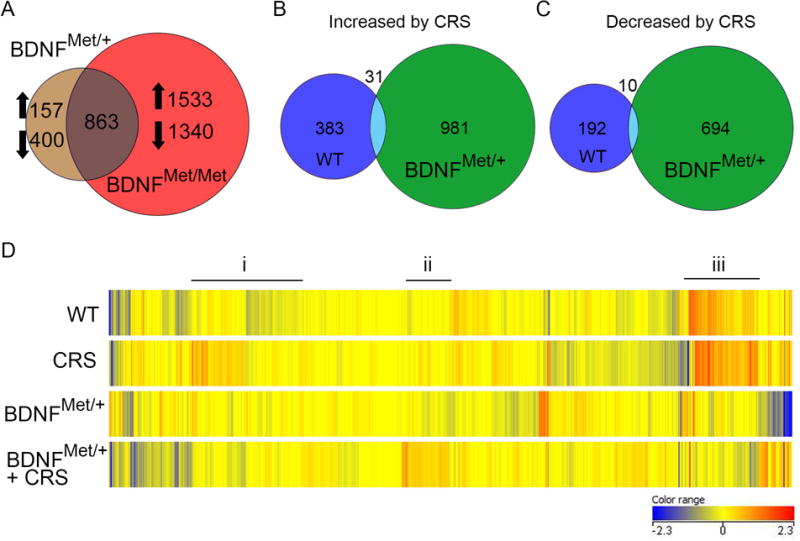
A) Venn diagram showing that similar genes were differentially expressed in both the BDNFMet/+ and BDNFMet/Met mice compared to WT. B,C) Venn diagram illustrating that BDNFMet/+ mice exposed to CRS exhibited numerous unique expression changes that were not present in WT mice after CRS. D) Heatmap of expression level changes in GR-regulated transcripts revealed i) genes increased by CRS in WT mice that did not show the same change with CRS in BDNFMet/+, ii) genes increased by CRS in BDNFMet/+ that were primarily unchanged with stress in WT mice and iii) genes that were different by genotype at baseline and changed by stress in BDNFMet/+.
Table 1.
Enrichment of GO Pathways in genes changes in BDNFMet/+ CA3 Neurons
| Rank | Common GO Term for Cluster | Enrichment Score | Selected Example Genes |
|---|---|---|---|
| 1 | Transmembrane & Membrane | 9.66 | APP, CREB3, CREB3L1, GRIN2A, GPR56, GRM5, L1CAM, NCAM2, NLGN3, SLC1A2, SLC1A3 |
| 2 | Glycoproteins & N-linked Glycosylation | 5.71 | CLCN3, FGFRL1, GABRA1, GRIK2, IL1RAP, NCAM2, NLGN3, NPY2R, ROR1, TGF1B |
| 3 | Lysosomes | 5.13 | CTSB, CLCN7, LAMP1, LAMP2, MT1, NPC1, NOS1, TMEM9, TRIM23 |
| 4 | Cellular Homeostasis | 4.87 | ANK1, BCL2, GRIK2, GRIN2a, IGF1, NR3C2, S100B, SGK1, TGFB1 |
| 5 | Cell-Cell Adhesion | 3.56 | ARHGAP6, ASTN1, CELSR2, CNTN1, CSTN2, L1CAM, MCAM, NCAM2, NELL1, NELL2, NLGN3 |
| 6 | Ion Transport | 3.51 | CACNA1C, CLCN3, GABRA1, GABRA2, KCNQ3, KCNV1, SLC1A2, SLC1A3, TRPC5 |
| 7 | Neuronal projection | 3.12 | APC, BACE1, GABBR2, IQGAP1, KIF5A, NCAM2, NOS1, NRCAM, SHANK2, TGFB1 |
| 8 | Cytoskeletal Components | 3.06 | ACTN1, ACTN2, ACTN4, ANK1, IQGAP2 MARCKS, MYH10, PARD6B |
| 9 | Exocytosis | 3.02 | CPLX2, EXOC4, EXOC5, EXOC7 NR3C2, RAPGEF4, RIMS1, RIMS3, RIMS4, SYN3 |
| 10 | Ca+ Ion Transport | 2.96 | CACNA1B, CACNA1C, CACNA1E, CACNA2D1, GRIN2A, GRIN3A, KCNQ3, SLC9A6, SLC9A7, |
To further explore how Met allele carriers respond to chronic stress, double heterozygous Gprin3-bacTRAP/BDNF Val66Met mice were exposed to CRS for translational profiling to identify expression changes in CA3 associated with increased stress susceptibility. BDNFMet/+ exhibited greater transcriptional reactivity to CRS than WT mice, with over 1,700 genes identified as changing with stress in BDNFMet/+ compared with over 600 in WT mice (Fig. 3B,C). Interestingly, there was very little overlap among genes changed by stress in WT mice and BDNFMet/+, suggesting that even one copy of the Met allele was sufficient to substantially alter the translational profile of CA3 neurons after chronic stress. BDNF and GR interact to produce synergistic changes in gene expression46. Therefore, levels of the GR-responsive genes from Polman et al. (2013) were investigated in BDNFMet/+ mice (Fig. 3D). Examination of the heatmap revealed several important patterns. First, there were GR-responsive genes that were increased by CRS in WT mice, but not in BDNFMet/+ (Fig. 3Di). Second, several genes were increased by stress in BDNFMet/+ and not in WT mice (Fig. 3Dii). Lastly, numerous GR-responsive genes had a difference in baseline levels by genotype, which were not changed by stress (Fig. 3Diii).
Together these data suggest that the genetic susceptibility to mood disorders conferred by the Met allele results in both an altered baseline state of gene expression, but more importantly the mRNA profile of these cells in response to stress is almost entirely distinct from WT mice. Interestingly, many of the same genes changed by acute stress in wild type mice overlap with genes that show baseline difference in BDNFMet/+ mice (Supp. Fig. 4). This further demonstrates how genetic susceptibility and environmental stress can interact to produce a unique cellular response and may share common mechanisms.
Genes implicated in LTP and neuroplasticity are altered in stress and BDNF-Val66Met mice
Pyramidal neurons of the CA3 regions are primarily glutamatergic47 and stress is known to regulate the expression of glutamate receptor genes in the hippocampus48. Glutamatergic signaling is essential for LTP, which is mediated by NMDA and AMPA receptors and necessary for hippocampal spatial memory49. Therefore, changes in the levels of genes encoding NMDA and AMPA receptors were investigated.
Both NMDA and AMPA receptors showed significantly increased translation with chronic stress, particularly the AMPA subunits Gria1, Gria2, and Gria3, and the NMDA subunits Grin1 (Fig. 4A,B). Previous studies have characterized electrophysiological defects in LTP/LTD in BDNF Val66Met mice50. These defects suggest that there may be baseline differences in the expression of NMDA and AMPA receptors subunits. In CA3 neurons, both BDNFMet/Met and BDNF+/Met mice showed significant decreases in the NMDA receptor subunit genes Grin1 and Grin2a (NR2a), but increases in Grin2c (Fig. 4A,B). Most interestingly, BDNFMet/+ mice exposed to CRS failed to show the same increases in AMPA and NMDA subunits observed in WT mice.
Figure 4. Changes in translation of LTP and BDNF-related genes in CA3 pyramidal neurons.
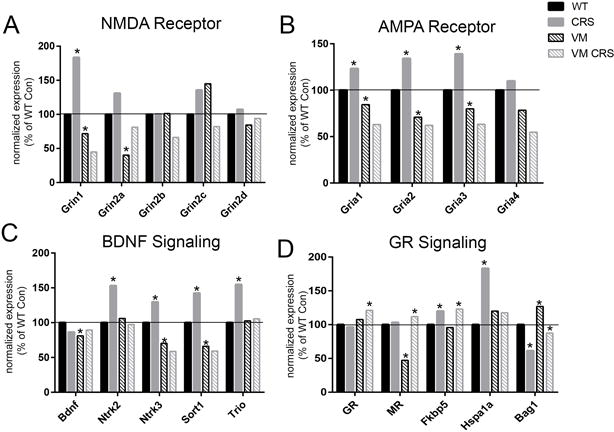
A) Changes in expression of the NMDA receptor subunits from the TRAP fraction in response to acute stress (FST), chronic stress (CRS), or heterotypic stress (CRS+FST) plotted as a percentage change from control levels normalized to 100. B) Levels of AMPA receptor subunit expression as a percent of control in response to stress. C) Changes in the relative expression levels of BDNF receptors and downstream signaling molecules after stress. D) BDNF-related gene expression levels in BDNF+/Met and BDNFMet/Met mice normalized to wildtype levels. (*p<0.001; Benjamin-Hochberg corrected for FDR)
BDNF signaling is also known to facilitate LTP51, 52. A modest decrease in total BDNF mRNA levels was observed in the TRAP fraction in response to both acute and chronic stress in WT mice (Fig. 2F), and significant decreases in total BDNF mRNA, as well as the coding exon 9 of the Bdnf gene, were observed in the TRAP fraction from both the BDNFMet/Met and BDNFMet/+ mice (Fig. 4C; Supp. Fig. 3A). The neurotrophin receptors for BDNF, TrkB (Ntrk2) and TrkC (Ntrk3), were significantly increased by chronic stress, however, TrkC showed a rapid downregulation in response to acute stress (Fig. 4C). Sortilin1 (Sort1), a protein involved in trafficking of BDNF, was similarly decreased in response to acute stress, but increased by chronic stress. Because the expression levels of another neurotrophin receptor, p75 (Ngfr), were too low for reliable detection in the CA3 TRAP fractions, it’s downstream signaling protein, Trio, was examined. Trio has been linked to the neuroplastic effects of BDNF53, 54 and its mRNA was significantly increased in response to chronic stress in the TRAP-seq results (Fig. 4C). Carriers of the Met allele showed the most significant decreases in the expression of Sort1, and BDNFMet/Met mice also had significantly decreased levels of Trio (Fig. 4C).
Hormone receptor levels were also altered by the interaction of the BDNF Val66Met mutation with stress. GR was only increased in the BDNFMet/+ mice subjected to CRS (Fig. 4D) compared with unstressed WT levels. Whereas MR was significantly decreased by genotype in BDNFMet/+ mice and showed an increase only in BDNFMet/+ mice in response to CRS, but did not increase in WT mice exposed to CRS. Unlike observations from the NMDA & AMPA receptors, the GR chaperones, Fkbp5 and Bag1 showed similar increases in levels after CRS, irrespective of the Met allele. Hsp70 family gene, Hspa1a (Hsp72), was significantly increased by CRS in WT mice, but was unchanged by CRS in Met allele carriers.
These gene expression changes reveal a potential mechanism underlying the impaired hippocampal LTP observed in BDNF Met allele mice50. Further, the differences in the translational response between WT and BDNF Met allele carriers to chronic stress provides substantial insight into the increased stress sensitivity of the BDNF Met allele mice.
Discussion
The present study used cell-type specific translational profiling to identify stress-induced changes in genes undergoing active translation in CA3 pyramidal neurons of the hippocampus. This cell population has a specialized role in learning and memory, and is particularly vulnerable to chronic stress and over-stimulation, as in seizures4, 25, 55. These results advance our understanding of the molecular changes that underlie the unique structural and electrophysiological plasticity occurring in this especially vulnerable subpopulation of hippocampal neurons in response to stress as well as with genetic manipulation of the BDNF gene.
Analysis of the CA3-specific translational profiles supports several conclusions. First, similar to findings in whole hippocampus, chronic and acute stress result in highly distinct molecular changes within this cell population (Fig. 2B). Second, exposure to a novel stressor after a history of chronic stress results in a different expression profile than observed upon naïve exposure to the same stressor (Fig. 2C). Third, many of the changes occur in known GR responsive genes (Fig. 2E), although we have shown previously that glucocorticoid treatment does not mimic the effect of acute stress or vice versa21. Comparisons with previous mRNA studies from whole hippocampus will help identify novel regulatory mechanisms as well as the cellular or regional specificity of the stress response of specific genes. Lastly, mice with a genetic predisposition to increased stress susceptibility have a unique baseline translational profile in CA3 neurons (Fig. 3A; Table 1) and neurons from BDNF Met allele mice respond to chronic stress by activating a highly distinct set of genes from WT mice (Fig. 3B,C). Many of these differentially responsive genes in pyramidal neurons are known to be implicated in LTP and memory formation (Fig. 4). The patterns of stress-induced gene translation in Met allele mice identified potential mechanisms underlying their increased susceptibility to anxiety-like behavior19 and stress-induced impairments in learning and memory and depressive-like behavior20.
TRAP-Seq improves stress-induced expression profiling
Several previous studies have examined gene expression changes in response to stress in either whole hippocampal tissue21–23 or using laser capture to isolate specific subregions56. These techniques both yield a heterogeneous mix of cell types that display highly varied gene expression profiles, a common problem when studying brain tissue. Using the Gprin3 bacTRAP reporter line allows for the interrogation of gene expression specifically in highly vulnerable CA3 pyramidal neurons. The present data also provide a more refined characterization of stress-induced changes than previous microarray studies by selecting for poly-adenylated mRNAs undergoing active translation. TRAP mRNA results have been shown to provide a vastly more accurate molecular profile than whole tissue assays33. Further, expression profiling by RNA-seq provides an increased dynamic range over microarrays and allows for the identification of novel transcripts and isoforms57. Together, TRAP mRNA purification and RNA-sequencing represent a significant advance over previous studies of stress-induced changes in hippocampal gene expression.
Given both the increased cellular specificity of the TRAP-IP and quantitative resolution of RNA-seq, differences that were previously undetectable in whole hippocampal lysates can be identified. For example, previous reports in BDNFMet/+ mice did not show a baseline difference in levels of BDNF exon 9 mRNA in whole hippocampal qRT-PCR experiments20. In the TRAP-seq data, a decrease in both total BDNF transcript density (Fig. 4C) and specifically the levels of BDNF exon 9 were detectable in both BDNFMet/+ and BDNFMet/Met mice (Supp. Fig3. A). Conversely, the ability to isolate the actively translating fraction of mRNA from a specific cell population has also resulted in subtle differences from previous data that hint at additional cellular mechanisms in the regulation of certain genes. Several studies have reported changes in hippocampal BDNF mRNA with stress43, 44, but there have also been studies reporting no difference58, 59. In the present data, no difference was found after acute or chronic stress in the WT TRAP-Seq fraction (Fig. 2F). One possibility is that previous studies observed changes in transcription of Bdnf that is not immediately undergoing translation. Multiple studies of BDNF have shown it to be highly dynamic, with varying results across stress paradigms, timing and species in hippocampus60. Future work will be necessary to resolve these questions about BDNF regulation and other stress-induced genes as the field moves forward.
Despite some differences between this data and previous studies, these results largely confirm genes changed by stress in hippocampus, with 617 genes identified as significantly different after FST by both RNA-Seq fractions and microarray as one example. In addition to the changes observed in CA3, the cell specificity of this data can be further validated against whole hippocampal microarray data by examining changes in the unbound fraction of genes whose expression is restricted to other hippocampal subregions. For example, Btg1 expression is highly specific to the dentate gyrus (Allen Brain Atlas, www.brain-map.org). This gene was significantly decreased by FST in both the unbound RNA-Seq results and whole hippocampal microarray studies, but showed no difference in the TRAP IPs since it is not expressed in CA3 pyramidal neurons. Importantly, CA3 pyramidal neurons represent a subset of cells in the hippocampus, and changes occurring in this population after stress may be unobservable in whole tissue lysate. Therefore, the TRAP provides a more reliable list of candidate genes for further study. These data will provide the foundation for numerous future experiments interrogating the mechanisms underlying stress-induced neuroplasticity of CA3 pyramidal neurons by allowing us to link molecular changes with those changes already observed in LTP and morphology.
Translational profiling of Gene-Environment interactions
After demonstrating that chronic stress leads to a remodeling of the translational stress response in CA3 neurons, we sought to evaluate the interaction of the BDNF Val66Met SNP with this chronic stress paradigm. This SNP leads to altered trafficking of the BDNF mRNA to dendrites61 and diminishes the activity dependent release of BDNF protein16, 19. BDNF release at the synapse is required for forms of early and late LTP, the former involving post translational modification of downstream signaling targets and the latter involving protein translation62. BDNF release can lead to phosphorylation of NMDA receptor subunits as well as insertion of AMPA receptors into the postsynaptic membrane, thus increasing synaptic strength. BDNF signaling through TrkB also leads to phosphorylation of synaptic proteins such as Rims163, which are important in neurotransmitter release. BDNF action in hippocampal neurons also leads to elevated protein levels of the NMDA receptor subunits NR1, NR2A and NR2B, and increased local translation of AMPA receptor subunits64, 65. Thus, the diminished activity dependent secretion of BDNF, combined with the impaired dendritic localization of BDNF mRNA in the Val66Met animals provides a plausible link between the altered translational profile observed in CA3 neurons in these mice and LTP deficits shown previously50.
BDNF Val66Met mice also show an exaggerated response to a seven-day chronic stress, with larger CORT elevation, disruptions in HPA axis and greater memory impairment20. This suggests that impairments in activity dependent secretion impair the ability of the neurons to undergo stress-induced plasticity that is normally observed in CA3 neurons and is dependent on GR signaling4. Recent studies have shown that BDNF activation of the TrkB receptor leads to phosphorylation of GR. When these phosphorylation residues are mutated, mice display a lack of the chronic stress-induced structural plasticity in cortical neurons that is observed in WT66. The authors also show that GR signaling without BDNF signaling in vitro activates a different set of genes than when both systems are activated together46, suggesting that BDNF-GR crosstalk is important in the structural remodeling observed during chronic stress, and could be a mechanism underlying the lack of plasticity in BDNF Val66Met mice. These results point to a model where CA3 neurons in BDNF Met allele carriers lack the necessary plasticity to provide an adaptive hippocampal circuitry, which leads to impaired negative HPA feedback and memory deficits. A pathway analysis of genes changed in BDNFMet/+ compared to wildtype mice demonstrated a preponderance of ion channel and neurotransmission-related genes (Table 1), such as glutamate (Grin2a, Grin3a, Grm5) and GABA (Gabra1) receptor subunits, as well as the Ca+ channels (Cacna1b, 1c, 1e), which are critically involved in LTP and other forms of neuroplasticity. Additionally, in response to chronic stress, WT mice show up regulated translation of Grin1, and Gria1, 2, and 3, consistent with previous reports that have shown an upregulation of Grin1 and Gria1 after chronic stress67–69, whereas BDNFMet/+ mice not only have lower basal levels of these genes, but the level do not change in response to stress.
Interestingly, the baseline changes in BDNF Val66Met mice showed considerable overlap with genes changed by acute stress in wild type mice (1,066 gene), and far less overlap with genes changed by CRS in wild type mice (121 genes; Supp. Fig. 4). This suggests that even in the absence of stress the Met allele confers many of the same changes that are occurring after acute stress, and therefore the highly divergent response of BDNFMet/+ mice to CRS may be accounted for by the fact that they are starting from a baseline similar to a stressed wild type mouse (“pre-stressed”). However, there were over 300 genes changed that were unique to the unstressed BDNFMet/+ mice that are equally as likely to account for their altered stress reactivity. Therefore, future experiments will seek to further characterize both of these overlapping and non-overlapping sets to better identify the genes that confer increased risk for mood disorders in this translational model.
In conclusion, these data provide a blueprint of the cell-type specific changes occurring in vivo in CA3 pyramidal neurons in response to stress and in the presence of a genetic mutation that is known to alter stress susceptibility. These findings will support future studies seeking to alter the effects of stress through pharmacological or environmental manipulations, and may help others identify significant gene expressions changes in pathways beyond the glutamatergic and BDNF systems that are involved in remodeling of the neural circuitry underlying stress susceptibility.
Supplementary Material
Acknowledgments
This project was supported by NIH/F32 MH102065 to JDG, NIH/RO1 MH41256 and the Hope for Depression Research Foundation grant RGA#13-004 to BSM, the Pritzker Consortium and NIH/ R01 NS052819 to FSL, Brain & Behavior Research Foundation NARSAD Young Investigator Grant 21464, the van Ameringen Foundation and NIH/NINDS R01NS091722 to EFS.
Footnotes
CONFLICTS OF INTEREST: The authors have no conflicts of interest to declare.
References
- 1.Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct. 2008;213(1–2):93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conrad CD, Galea LA, Kuroda Y, McEwen BS. Chronic stress impairs rat spatial memory on the Y maze, and this effect is blocked by tianeptine pretreatment. Behav Neurosci. 1996;110(6):1321–1334. doi: 10.1037//0735-7044.110.6.1321. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan V, Nestler EJ. Animal models of depression: molecular perspectives. Curr Top Behav Neurosci. 2011;7:121–147. doi: 10.1007/7854_2010_108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEwen BS. Stress and hippocampal plasticity. Annu Rev Neurosci. 1999;22:105–122. doi: 10.1146/annurev.neuro.22.1.105. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert PE, Brushfield AM. The role of the CA3 hippocampal subregion in spatial memory: a process oriented behavioral assessment. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33(5):774–781. doi: 10.1016/j.pnpbp.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolley CS, Gould E, McEwen BS. Exposure to excess glucocorticoids alters dendritic morphology of adult hippocampal pyramidal neurons. Brain Res. 1990;531(1–2):225–231. doi: 10.1016/0006-8993(90)90778-a. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe Y, Gould E, McEwen BS. Stress induces atrophy of apical dendrites of hippocampal CA3 pyramidal neurons. Brain Res. 1992;588(2):341–345. doi: 10.1016/0006-8993(92)91597-8. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Dube CM, Rice CJ, Baram TZ. Rapid loss of dendritic spines after stress involves derangement of spine dynamics by corticotropin-releasing hormone. J Neurosci. 2008;28(11):2903–2911. doi: 10.1523/JNEUROSCI.0225-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JJ, Diamond DM. The stressed hippocampus, synaptic plasticity and lost memories. Nat Rev Neurosci. 2002;3(6):453–462. doi: 10.1038/nrn849. [DOI] [PubMed] [Google Scholar]
- 10.Popoli M, Yan Z, McEwen BS, Sanacora G. The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat Rev Neurosci. 2012;13(1):22–37. doi: 10.1038/nrn3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59(12):1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Nibuya M, Morinobu S, Duman RS. Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci. 1995;15(11):7539–7547. doi: 10.1523/JNEUROSCI.15-11-07539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magarinos AM, Li CJ, Gal Toth J, Bath KG, Jing D, Lee FS, et al. Effect of brain-derived neurotrophic factor haploinsufficiency on stress-induced remodeling of hippocampal neurons. Hippocampus. 2011;21(3):253–264. doi: 10.1002/hipo.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sen S, Nesse RM, Stoltenberg SF, Li S, Gleiberman L, Chakravarti A, et al. A BDNF coding variant is associated with the NEO personality inventory domain neuroticism, a risk factor for depression. Neuropsychopharmacology. 2003;28(2):397–401. doi: 10.1038/sj.npp.1300053. [DOI] [PubMed] [Google Scholar]
- 15.Verhagen M, van der Meij A, van Deurzen PA, Janzing JG, Arias-Vasquez A, Buitelaar JK, et al. Meta-analysis of the BDNF Val66Met polymorphism in major depressive disorder: effects of gender and ethnicity. Mol Psychiatry. 2010;15(3):260–271. doi: 10.1038/mp.2008.109. [DOI] [PubMed] [Google Scholar]
- 16.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 17.Elzinga BM, Molendijk ML, Oude Voshaar RC, Bus BA, Prickaerts J, Spinhoven P, et al. The impact of childhood abuse and recent stress on serum brain-derived neurotrophic factor and the moderating role of BDNF Val66Met. Psychopharmacology (Berl) 2011;214(1):319–328. doi: 10.1007/s00213-010-1961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatt JM, Nemeroff CB, Dobson-Stone C, Paul RH, Bryant RA, Schofield PR, et al. Interactions between BDNF Val66Met polymorphism and early life stress predict brain and arousal pathways to syndromal depression and anxiety. Mol Psychiatry. 2009;14(7):681–695. doi: 10.1038/mp.2008.143. [DOI] [PubMed] [Google Scholar]
- 19.Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314(5796):140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Wang DD, Wang Y, Liu T, Lee FS, Chen ZY. Variant brain-derived neurotrophic factor Val66Met polymorphism alters vulnerability to stress and response to antidepressants. J Neurosci. 2012;32(12):4092–4101. doi: 10.1523/JNEUROSCI.5048-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray JD, Rubin TG, Hunter RG, McEwen BS. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2014;19(11):1171–1178. doi: 10.1038/mp.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Yang N, Zuo P. cDNA microarray analysis of gene expression in the cerebral cortex and hippocampus of BALB/c mice subjected to chronic mild stress. Cell Mol Neurobiol. 2010;30(7):1035–1047. doi: 10.1007/s10571-010-9534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ejchel-Cohen TF, Wood GE, Wang JF, Barlow K, Nobrega JN, B SM, et al. Chronic restraint stress decreases the expression of glutathione S-transferase pi2 in the mouse hippocampus. Brain Res. 2006;1090(1):156–162. doi: 10.1016/j.brainres.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 24.Wang DD, Tian T, Dong Q, Xu XF, Yu H, Wang Y, et al. Transcriptome profiling analysis of the mechanisms underlying the BDNF Val66Met polymorphism induced dysfunctions of the central nervous system. Hippocampus. 2014;24(1):65–78. doi: 10.1002/hipo.22204. [DOI] [PubMed] [Google Scholar]
- 25.McEwen BS. Stress-induced remodeling of hippocampal CA3 pyramidal neurons. Brain Res. 2016;1645:50–54. doi: 10.1016/j.brainres.2015.12.043. [DOI] [PubMed] [Google Scholar]
- 26.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat Protoc. 2014;9(6):1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt EF, Warner-Schmidt JL, Otopalik BG, Pickett SB, Greengard P, Heintz N. Identification of the cortical neurons that mediate antidepressant responses. Cell. 2012;149(5):1152–1163. doi: 10.1016/j.cell.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ainsley JA, Drane L, Jacobs J, Kittelberger KA, Reijmers LG. Functionally diverse dendritic mRNAs rapidly associate with ribosomes following a novel experience. Nat Commun. 2014;5:4510. doi: 10.1038/ncomms5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9(1):e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, et al. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40(20):10084–10097. doi: 10.1093/nar/gks804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Kloet ER, Molendijk ML. Coping with the Forced Swim Stressor: Towards Understanding an Adaptive Mechanism. Neural Plast. 2016;2016:6503162. doi: 10.1155/2016/6503162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polman JA, de Kloet ER, Datson NA. Two populations of glucocorticoid receptor-binding sites in the male rat hippocampal genome. Endocrinology. 2013;154(5):1832–1844. doi: 10.1210/en.2012-2187. [DOI] [PubMed] [Google Scholar]
- 40.Harris AP, Holmes MC, de Kloet ER, Chapman KE, Seckl JR. Mineralocorticoid and glucocorticoid receptor balance in control of HPA axis and behaviour. Psychoneuroendocrinology. 2013;38(5):648–658. doi: 10.1016/j.psyneuen.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S186–195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 42.Kullmann M, Schneikert J, Moll J, Heck S, Zeiner M, Gehring U, et al. RAP46 is a negative regulator of glucocorticoid receptor action and hormone-induced apoptosis. J Biol Chem. 1998;273(23):14620–14625. doi: 10.1074/jbc.273.23.14620. [DOI] [PubMed] [Google Scholar]
- 43.Marmigere F, Givalois L, Rage F, Arancibia S, Tapia-Arancibia L. Rapid induction of BDNF expression in the hippocampus during immobilization stress challenge in adult rats. Hippocampus. 2003;13(5):646–655. doi: 10.1002/hipo.10109. [DOI] [PubMed] [Google Scholar]
- 44.Smith MA, Makino S, Kvetnansky R, Post RM. Stress and glucocorticoids affect the expression of brain-derived neurotrophic factor and neurotrophin-3 mRNAs in the hippocampus. J Neurosci. 1995;15(3 Pt 1):1768–1777. doi: 10.1523/JNEUROSCI.15-03-01768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallei A, Baj G, Ieraci A, Corna S, Musazzi L, Lee FS, et al. Expression and Dendritic Trafficking of BDNF-6 Splice Variant are Impaired in Knock-In Mice Carrying Human BDNF Val66Met Polymorphism. Int J Neuropsychopharmacol. 2015;18(12) doi: 10.1093/ijnp/pyv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert WM, Xu CF, Neubert TA, Chao MV, Garabedian MJ, Jeanneteau FD. Brain-derived neurotrophic factor signaling rewrites the glucocorticoid transcriptome via glucocorticoid receptor phosphorylation. Mol Cell Biol. 2013;33(18):3700–3714. doi: 10.1128/MCB.00150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spruston N, Jonas P, Sakmann B. Dendritic glutamate receptor channels in rat hippocampal CA3 and CA1 pyramidal neurons. J Physiol. 1995;482(Pt 2):325–352. doi: 10.1113/jphysiol.1995.sp020521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartanusz V, Aubry JM, Pagliusi S, Jezova D, Baffi J, Kiss JZ. Stress-induced changes in messenger RNA levels of N-methyl-D-aspartate and AMPA receptor subunits in selected regions of the rat hippocampus and hypothalamus. Neuroscience. 1995;66(2):247–252. doi: 10.1016/0306-4522(95)00084-v. [DOI] [PubMed] [Google Scholar]
- 49.Lu W, Man H, Ju W, Trimble WS, MacDonald JF, Wang YT. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 2001;29(1):243–254. doi: 10.1016/s0896-6273(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 50.Ninan I, Bath KG, Dagar K, Perez-Castro R, Plummer MR, Lee FS, et al. The BDNF Val66Met polymorphism impairs NMDA receptor-dependent synaptic plasticity in the hippocampus. J Neurosci. 2010;30(26):8866–8870. doi: 10.1523/JNEUROSCI.1405-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92(19):8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, et al. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J Neurosci. 2002;22(5):1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deinhardt K, Kim T, Spellman DS, Mains RE, Eipper BA, Neubert TA, et al. Neuronal growth cone retraction relies on proneurotrophin receptor signaling through Rac. Sci Signal. 2011;4(202):ra82. doi: 10.1126/scisignal.2002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anastasia A, Deinhardt K, Chao MV, Will NE, Irmady K, Lee FS, et al. Val66Met polymorphism of BDNF alters prodomain structure to induce neuronal growth cone retraction. Nat Commun. 2013;4:2490. doi: 10.1038/ncomms3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Almeida L, Idiart M, Lisman JE. Memory retrieval time and memory capacity of the CA3 network: role of gamma frequency oscillations. Learn Mem. 2007;14(11):795–806. doi: 10.1101/lm.730207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datson NA, van den Oever JM, Korobko OB, Magarinos AM, de Kloet ER, McEwen BS. Previous history of chronic stress changes the transcriptional response to glucocorticoid challenge in the dentate gyrus region of the male rat hippocampus. Endocrinology. 2013;154(9):3261–3272. doi: 10.1210/en.2012-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Isgor C, Kabbaj M, Akil H, Watson SJ. Delayed effects of chronic variable stress during peripubertal-juvenile period on hippocampal morphology and on cognitive and stress axis functions in rats. Hippocampus. 2004;14(5):636–648. doi: 10.1002/hipo.10207. [DOI] [PubMed] [Google Scholar]
- 59.Kuroda Y, McEwen BS. Effect of chronic restraint stress and tianeptine on growth factors, growth-associated protein-43 and microtubule-associated protein 2 mRNA expression in the rat hippocampus. Brain Res Mol Brain Res. 1998;59(1):35–39. doi: 10.1016/s0169-328x(98)00130-2. [DOI] [PubMed] [Google Scholar]
- 60.Gray JD, Milner TA, McEwen BS. Dynamic plasticity: the role of glucocorticoids, brain-derived neurotrophic factor and other trophic factors. Neuroscience. 2013;239:214–227. doi: 10.1016/j.neuroscience.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, et al. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci U S A. 2009;106(38):16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leal G, Comprido D, Duarte CB. BDNF-induced local protein synthesis and synaptic plasticity. Neuropharmacology. 2014;76(Pt C):639–656. doi: 10.1016/j.neuropharm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 63.Simsek-Duran F, Lonart G. The role of RIM1alpha in BDNF-enhanced glutamate release. Neuropharmacology. 2008;55(1):27–34. doi: 10.1016/j.neuropharm.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 64.Caldeira MV, Melo CV, Pereira DB, Carvalho RF, Carvalho AL, Duarte CB. BDNF regulates the expression and traffic of NMDA receptors in cultured hippocampal neurons. Mol Cell Neurosci. 2007;35(2):208–219. doi: 10.1016/j.mcn.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 65.Caldeira MV, Melo CV, Pereira DB, Carvalho R, Correia SS, Backos DS, et al. Brain-derived neurotrophic factor regulates the expression and synaptic delivery of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits in hippocampal neurons. J Biol Chem. 2007;282(17):12619–12628. doi: 10.1074/jbc.M700607200. [DOI] [PubMed] [Google Scholar]
- 66.Arango-Lievano M, Lambert WM, Bath KG, Garabedian MJ, Chao MV, Jeanneteau F. Neurotrophic-priming of glucocorticoid receptor signaling is essential for neuronal plasticity to stress and antidepressant treatment. Proc Natl Acad Sci U S A. 2015;112(51):15737–15742. doi: 10.1073/pnas.1509045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiland NG, Orchinik M, Tanapat P. Chronic corticosterone treatment induces parallel changes in N-methyl-D-aspartate receptor subunit messenger RNA levels and antagonist binding sites in the hippocampus. Neuroscience. 1997;78(3):653–662. doi: 10.1016/s0306-4522(96)00619-7. [DOI] [PubMed] [Google Scholar]
- 68.Schwendt M, Jezova D. Gene expression of two glutamate receptor subunits in response to repeated stress exposure in rat hippocampus. Cell Mol Neurobiol. 2000;20(3):319–329. doi: 10.1023/A:1007062109386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calabrese F, Guidotti G, Molteni R, Racagni G, Mancini M, Riva MA. Stress-induced changes of hippocampal NMDA receptors: modulation by duloxetine treatment. PLoS One. 2012;7(5):e37916. doi: 10.1371/journal.pone.0037916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


