Abstract
The mu-opioid receptor (MOR) is the primary target of methadone and buprenorphine. The primary neuronal transcript of the OPRM1 gene, MOR-1, contains a ~13kb 3′ untranslated region with five common haplotypes in European-Americans. We analyzed the effects of these haplotypes on the percentage of opioid positive urine tests in European-Americans (n = 582) during a 24-week, randomized, open-label trial of methadone or buprenorphine/naloxone (Suboxone) for the treatment of opioid dependence. A single haplotype, tagged by rs10485058, was significantly associated with patient urinalysis data in the methadone treatment group. Methadone patients with the A/A genotype at rs10485058 were less likely to have opioid-positive urine drug screens than those in the combined A/G and G/G genotypes group (Relative Risk = 0.76, 95% confidence intervals = 0.73–0.80, p = 0.0064). Genotype at rs10485058 also predicted self-reported relapse rates in an independent population of Australian patients of European descent (n = 1215) who were receiving opioid substitution therapy (p = 0.003). In silico analysis predicted that miR-95-3p would interact with the G, but not the A allele of rs10485058. Luciferase assays indicated miR-95-3p decreased reporter activity of constructs containing the G, but not the A allele of rs10485058, suggesting a potential mechanism for the observed pharmacogenetic effect. These findings suggest that selection of a medication for opioid dependence based on rs10485058 genotype might improve outcomes in this ethnic group.
Introduction
Opioid dependence is a significant public health issue in the United States and globally 1. More than 4.5 million Americans were current non-medical users of heroin or opioid analgesics in 2013 (National Survey on Drug Use and Health). The vast majority of these individuals were users of prescription painkillers, who outnumbered current heroin users more than 6:1, and dependence on opioid analgesics accounts for an estimated $55.7 billion in annual societal costs in the US 2. Susceptibility to opioid dependence includes both an environmental and a genetic component. Twin and families studies have estimated that opioid dependence is 40–60% heritable 3; however, few causal genetic variants have been identified 4.
The biological mechanisms underlying opioid dependence are intrinsically linked to the opioid receptor family of proteins, which activate downstream signaling in the presence of both endogenous peptides and exogenous opioids. The μ-opioid receptor (MOR), encoded by the OPRM1 gene, is responsible for mediating the rewarding effects of opioids and the role of the murine homolog, Oprm1, has been extensively studied in mouse models of opioid dependence 5–11.
FDA-approved treatment for opioid dependence frequently consists of opioid substitution therapy (OST) with medications such as methadone and buprenorphine 12. Methadone is a full MOR agonist; in contrast, buprenorphine is both a partial MOR agonist and a kappa-opioid receptor antagonist 13. Buprenorphine may also be compounded with the MOR antagonist naloxone to reduce the likelihood of injection of the medication. A significant number of patients treated with OST cease treatment and/or relapse to drug use 12, 14, 15. A meta-analysis of outcome data from OST studies indicates that a significant percentage of opioid dependent patients treated with OST do not sustain abstinence from illicit opioid use 12. Understanding the factors that predict reductions in illicit opioid use during treatment would allow clinicians to make more informed therapeutic decisions, selecting medications that are most likely to benefit individual patients.
Several studies have found evidence of genetic variants that are associated with the efficacy of treatments for drug dependence, where efficacy is defined as either reduction in drug use or prevention of relapse 16–22. These associations make these variants potentially useful as prospective biomarkers. Unfortunately there have only been a small number of studies examining the pharmacogenetics of opioid dependence treatment. Oneda, et al (2011) identified a haplotype block in arrestin beta 2 (ARRB2) that was associated with response to methadone 23. Our group has observed associations between several SNPs in the delta-opioid receptor gene (OPRD1) and outcomes during treatment for opioid dependence 20, 21. All of these findings require confirmation in additional populations before they can be used in a clinical setting. Thus, there is currently no FDA-approved pharmacogenetic basis for selecting a specific OST medication.
The primary OPRM1 transcript is MOR-1, which is abundant in neurons and composed of exons 1, 2, 3 and 4. The MOR-1 3′ untranslated region (UTR) can measure ~13.6 kb long, which is unusually large compared to the typical 3′ UTRs (~500 bp) of human protein coding genes 24. The functional importance of this large UTR is unclear. 3′ UTRs frequently contain regulatory elements that influence translation of the mRNA by recruiting RNA-binding proteins or microRNAs (miRNAs) 25. MiRNAs are small non-coding RNAs that can bind complementary sequences in mRNAs, resulting in increased degradation or decreased translation of the mRNA 26, 27. We hypothesized that the extended 3′ UTR of the MOR-1 transcript may be a significant target for post-transcriptional regulation by miRNAs. Since MOR is intimately involved in the mechanisms of opioid dependence, we analyzed the effects of SNPs in the MOR-1 3′ UTR on the percentage of opioid positive urine drug screens in a randomized, open-label trial of methadone and buprenorphine/naloxone for the treatment of opioid dependence.
Materials and Methods
Participants and Procedures
The primary analysis was performed using data from the Starting Treatment with Agonist Replacement Therapy (START) clinical trial. The methodology, including a diagram of participant flow, and primary outcomes for the START trial have been previously described 28. Briefly, individuals were recruited for treatment at federally licensed opioid treatment programs in the United States between May 2006 and October 2009 and randomly assigned to 24 weeks of open-label Suboxone (buprenorphine/naloxone; hereafter “buprenorphine”) or methadone treatment. Institutional review boards at participating sites approved the study and oversight was provided by the NIDA Clinical Trials Network Data Safety and Monitoring Board. All patients provided informed consent for the study. Patients met DSM-IV-TR criteria for opioid dependence and were at least 18 years of age. Ethnicity was determined by self-report.
A flexible dosing approach was used, with a wide range allowed in both induction dosing and subsequent maintenance dosing. Buprenorphine ranged from a 2 to 8 mg initial dose to a maximum dose of 32 mg. The mean maximum daily dose for buprenorphine patients analyzed in this study was 24.8 ± 8.3 mg. The maximum initial dose of methadone was limited to 30 mg and the dose could be increased by 10mg increments with no specific maximum. The mean maximum daily dose for methadone patients analyzed in this study was 99.6 ± 46.0 mg. Participants came to the clinic daily for observed dosing except on Sundays and holidays or when take-home medications were permitted by local regulations. Urine drug samples were tested weekly for opioids and other illicit drugs using standard methods. Samples testing positive for methadone were counted as positive for individuals in the buprenorphine group, but not for individuals in the methadone group.
SNP Selection and Genotyping
The HaploView expectation-maximization algorithm was used to estimate haplotype frequency in the MOR-1 3′ UTR (chromosome 6: 154,439,890–154,453,502) of European-Americans (CEU population) from the 1000 Genomes Project data set 29. Five common haplotype blocks (frequency > 5%) were identified in the MOR-1 3′ UTR of the European-American population. These haplotype blocks can be identified by genotypes at four SNPs: rs671531, rs558948, rs645027, and rs10485058. A diagram of the OPRM1 gene with SNP locations is shown in Figure 1. African-Americans in the START trial were not included in the haplotype analysis because of insufficient statistical power due to small sample size (n = 77).
Figure 1.
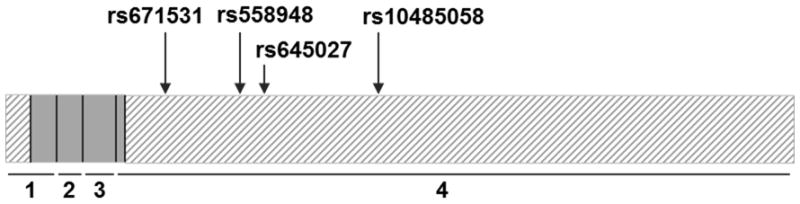
Diagram of the MOR-1 transcript with the location of the 4 SNPs genotyped for this study. Coding portions of exons are indicated by gray boxes. Untranslated regions are indicated by boxes with diagonal lines. Exons are numbered below the transcript. SNP and exon data were taken from the February 2009 build of the human genome in UCSC Genome Browser (www.genome.ucsc.edu).
SNP genotyping was performed using an ABI 7900 Thermocycler and Taqman® SNP Genotyping Assays (ThermoFisher Scientific, Waltham, MA, USA) as previously described 30.
Statistical Analysis
For each SNP, deviation from Hardy-Weinberg Equilibrium was assessed by chi-square analysis. All SNPs were in Hardy-Weinberg Equilibrium (p≥0.05). The percentage of opioid positive urines in the methadone treatment group compared to that of the buprenorphine treatment group was analyzed by Chi-square test. Haplotype analysis was used to analyze the associations between the genotype in the five common EA haplotypes and treatment outcome, defined as the percentage of opioid positive urine drug screens over 24 weeks, in the methadone and buprenorphine groups separately. A power calculation for the analysis is presented in Supplemental Table 2. Since the treatment outcome data was not normally distributed (Kolmogorov-Smirnov p < 0.01), haplotype analyses were performed with the software package PLINK v1.07 using a linear regression with permutation testing 31. Empirical p-values were calculated by 10,000 sample permutations. P-values were corrected for multiple testing using the false discovery rate (FDR) procedure with the cut-off for statistical significance after correction set to p≤0.05 32.
A Generalized Estimating Equation (GEE) is a quasi-likelihood based method that produces population averaged estimates for repeated binary outcomes 33. A GEE was used to investigate the associations of rs10485058 genotype and repeated urinalysis outcomes from week 1 to week 24, adjusting for the effects of time, age, sex, maximum dose, and whether or not subjects had injected opioids in the last 30 days. Injection status was included in the model because it has been previously associated with increased opioid use during treatment in the START trial 34. Estimates are reported as relative risks with 95% confidence intervals. Due to the small number of patients with the rs10485058 G/G genotype (n=3), individuals with the G/G genotype were combined with those with the rs10485058 A/G genotype in the GEE analysis. Urinalysis outcomes for both treatment groups were analyzed separately.
More urine drug screens were missing in the buprenorphine patients than the methadone patients (25.0% vs 12.7%, chi-square p < 0.0001). A higher proportion of patients in the buprenorphine group had at least one missing test compared to the proportion in the methadone group (74.2% vs 59.4%, chi-square p = 0.0001). All patients included in the pharmacogenetic analysis had available data for at least one urine drug screen during the treatment phase of the trial. None of the polymorphisms analyzed in this study were associated with the percentage of missing urine drug screens in either medication group (data not shown). Urine drug screens missed by patients were counted as unknowns and excluded from all analyses.
Replication Analysis
The Comorbidity and Trauma Study (CATS) has been previously described 35, 36. Briefly, opioid dependent patients were recruited from OST clinics in the greater Sydney area of Australia between 2003 and 2008, a period during which the vast majority of the sample were likely to be either currently receiving, or to have previously received, methadone treatment. Ethics approvals were obtained from the institutional review boards of University of New South Wales, Washington University in Saint Louis, Queensland Institute of Medical Research and the relevant NSW area heath service. The current analyses were restricted to individuals who met DSM-IV criteria for opioid dependence. During the interview, opioid dependent individuals were asked if they ever had a period of abstinence from opioids. Those endorsing such a period were asked if they ever had a subsequent relapse. Genotyping was performed using the Illumina Human660W-Quad BeadChip at the Johns Hopkins Center for Inherited Disease Research (CIDR) and principal components were identified using the smartpca program in the Eigensoft 3.0 package 37. An additive model was used in logistic regression analysis to examine the effect of rs10485058 genotype on relapse in 1215 patients of European descent, while controlling for age, sex, and two principal components.
In Silico microRNA Prediction
The RegRNA website (http://regrna.mbc.nctu.edu.tw/html/prediction.html) was used to identify prospective microRNA (miRNA) binding sites. The analysis was performed twice with the 200bp sequence flanking the rs10485058 locus: once with A allele of the SNP and once with the G allele.
Luciferase Assays
The 50bp genomic regions containing either the A or G allele of rs10485058 were cloned into the 3′ UTR of firefly luciferase in the pmirGLO dual-luciferase miRNA target expression vector (Promega, Madison, Wisconsin, USA). BE(2)C neuroblastoma cells (ATCC CRL-2268) were obtained from ATCC and the lot used for these experiments tested negative for mycoplasma contamination and was authenticated by STR analysis. Cells were cultured in 24-well plates in a 1:1 ratio of F-12 and Eagle’s Minimum Essential Medium with 10% fetal bovine serum. Cells were transfected at 60–80% confluency using Lipofectamine 3000 (ThermoFisher Scientific, Waltham, MA, USA). Transfections consisted of 167 ng of the A allele construct, the G allele construct, or the empty vector control, combined with 16.7 pmol of one of three mirVana miRNA mimics: miR-95-3p, miR-2053, or negative control #1 (ThermoFisher Scientific, Waltham, MA, USA). Mock transfections of miRNA mimics without plasmid were performed as controls to ensure the absence of background luminescence. All transfections were performed in triplicate in each of three independent experiments, for a total of nine replicates.
At 24 hrs post-transfection, cells were lysed for 15 minutes in 200uL of passive lysis buffer (Promega, Madison, Wisconsin, USA). The activities of firefly and Renilla luciferases were measured for each lysate on a TD-20/20 Luminometer using the dual-luciferase reporter assay system (Promega, Madison, Wisconsin, USA). The ratio of firefly:Renilla activity was used as the outcome data for each sample. To account for any inherent differences in luciferase activity unrelated to the miRNA mimics, the ratios for the rs10485058-A allele, rs10485058-G allele and the empty vector constructs were normalized to the negative control #1 mimic samples for each respective construct. To account for any off-target effect of the mimics on the pmiR-Glo vector itself, rather than on the rs10485058 locus, the resultant ratios were further normalized to the average of the empty vector control for each miRNA mimic. Data were analyzed by one-way ANOVA and post-hoc Tukey HSD tests using JMP v12.0.
Results
Demographics and Genotyping
DNA samples and urinalysis data were available for 582 European-American participants who enrolled in the genetics study of the START clinical trial. The mean age, sex percentages, and average outcome for the methadone and buprenorphine treatment groups are provided in Table 1. The percentage of opioid positive urine samples for patients treated with methadone (36.0%; 2134 positive tests out of 5931 total) was larger than that of patients treated with buprenorphine (31.7%; 1704 positive test out of 5381 total; p < 0.0001). In addition, buprenorphine patients were more likely to never submit an opioid positive urine during the trial than methadone patients (27.8% vs 17.3%, p = 0.0025). These data suggest that buprenorphine may be more effective than methadone in reducing illicit opioid use in this population.
Table 1.
Demographic information and treatment outcomes for European-American patients with opioid dependence treated with either methadone or buprenorphine. SD = standard deviation.
| Methadone | Buprenorphine | |
|---|---|---|
| Number (% male) | 283 (63.3%) | 299 (72.9%) |
| Mean age ± SD | 35.6 ± 10.6 | 36.0 ± 11.2 |
| Mean maximum dose ± SD (mg) | 99.6 ± 46.0 | 24.8 ± 8.3 |
| % opioid positive urine drug screens (positive urines/total urines) | 36.0% (2134/5931) | 31.7% (1704/5381) |
The minor allele frequencies in the two treatment groups were compared for each SNP to determine if the randomization process had altered the frequency of any of the variants in either the methadone or buprenorphine patient populations (Supplemental Table 1). The minor allele frequency of one SNP, rs645027, was significantly different between the two treatment groups (p = 0.007). No significant differences were observed between the methadone and buprenorphine groups for the other analyzed variants.
Pharmacogenetics
To determine if there were any significant associations between haplotypes in the MOR-1 3′ UTR and treatment outcome in either the methadone or buprenorphine patient populations, a linear regression with permutation testing was used to analyze the effect of haplotype status on the percentage of opioid positive urine tests over 24 weeks. One haplotype, tagged exclusively by the G allele of rs10485058, was significantly associated with treatment outcome in methadone patients (p = 0.0025), while none of the haplotypes had significant associations with outcome in buprenorphine patients (Table 2). Since the G allele of rs10485058 is the only marker that tags the significant haplotype, all subsequent analyses simply use rs10485058 genotype as a proxy for the presence or absence of the haplotype. In the methadone treatment group, patients with the AA genotype at rs10485058 had significantly fewer opioid positive urine drug screens over 24 weeks (33.4 ± 32.3%) than patients in the combined AG and GG genotypes group (48.6 ± 33.0%, p = 0.0025). This association remained significant after correction for multiple testing to reflect the analysis of five independent haplotypes.
Table 2.
Analysis of associations between common MOR-1 3′UTR haplotypes and treatment outcome in opioid dependent European-Americans. Only haplotypes with a frequency greater than 5% in the European-American population were analyzed. The genotype for each SNP is provided for each of the five common haplotypes. P-values were generated by linear regression for the individual treatment groups.
| Haplotype Alleles | ||||||
|---|---|---|---|---|---|---|
| Haplotype # | rs671531 | rs558948 | rs645027 | rs10485058 | Methadone P-value | Buprenorphine P-value |
| 1 | G | C | A | A | 0.30 | 0.34 |
| 2 | A | T | A | A | 0.20 | 0.51 |
| 3 | G | C | A | G | 0.0025 | 0.27 |
| 4 | G | C | G | A | 0.34 | 0.33 |
| 5 | A | C | A | A | 0.35 | 0.81 |
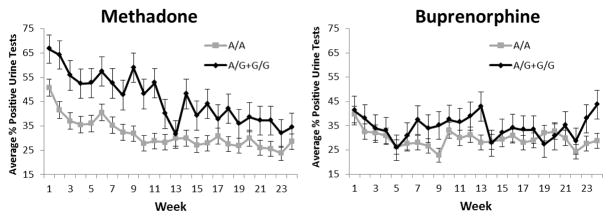
Generalized estimating equations were used to further investigate the associations of the significant haplotype and repeated urinalysis outcomes from week 1 to week 24, adjusting for the effects of age, sex, maximum dose and whether the patient had injected opioids in the last 30 days (“injection status”). The buprenorphine and methadone treatment groups were analyzed separately. Again no effect was observed in the buprenorphine group (p > 0.05) (Figure 2b). Methadone patients with the A/A genotype, however, were less likely to have opioid-positive urine drug screens than those in the combined A/G and G/G genotypes group (Relative Risk = 0.76, 95% confidence intervals = 0.73–0.80, p = 0.0064) (Figure 2a). As previously described34, injection status was significantly associated with the percentage of opioid-positive urine drug screens (p = 8.49 × 10−14). No effects of age, sex, or maximum dose were observed (p > 0.05).
Figure 2.
Weekly urinalysis data for European-Americans based on rs10485058 genotype. Patients were treated for opioid dependence with either methadone or buprenorphine for 24 weeks. Weekly urine drug screens were administered for the presence of opioids other than the one prescribed. The average percentage of opioid positive urine tests during each week is provided for individuals with the A/A genotype or the A/G and G/G genotypes. The A/G and G/G genotypes were combined due to the low number of G/G individuals (n=3). Time, age, sex, maximal dose, and injection status were used as covariates. A) Methadone patients with the A/A genotype (n=204) were less likely to have opioid-positive urine drug screens than patients with the A/G or G/G genotypes (n=67) (Relative Risk = 0.76, 95% confidence intervals = 0.73–0.80, p = 0.0064). No effect of genotype was observed in the buprenorphine group.
Effect of Genotype on Self-Reported Relapse in an Independent Cohort
The Comorbidity and Trauma Study (CATS) collected self-reported data on ever having had a relapse after a period of abstinence and genotypes from 1215 Australian opioid dependent individuals of European descent. To determine the effect of rs10485058 on relapse in this independent cohort, an additive model was used in logistic regression analyses of the CATS data. The analysis controlled for the effects of age and sex, as well as two principal components previously identified in the data set. The genotype at rs10485058 predicted never having had a relapse after achieving abstinence; in an additive model, the A allele was found to be significantly associated with never having relapsed (p = 0.003). These results are consistent with the analysis in the START study, in which the A/A genotype was associated with reduced opioid use.
In vitro microRNA Analysis
Analysis of putative miRNA binding sites using the RegRNA website indicated that miR-95-3p is predicted to bind to the G allele of rs10485058 but not the A allele (Figure 3). This difference in predicted binding is due to lack of perfect complementarity between the miRNA seed sequence and the rs10485058 locus when the A allele is present. Another miRNA (miR-2053) was predicted to bind both alleles (Figure 3). Luciferase activity from rs10485058 constructs transfected into BE(2)C neuroblastoma cells showed a significant 30% reduction in the G allele samples compared to empty vector control in the presence of miR-95-3p (p < 0.0001), while no reduction was observed in the A allele samples (Figure 3). These data suggest that miR-95-3p negatively regulates the G allele of rs10485058 but not the A allele. A smaller, but still statistically significant, reduction in luciferase activity was observed when the miR-2053 mimic was co-transfected alongside either the G allele (p = 0.0022) or the A allele (p = 0.040) (Figure 3).
Figure 3.
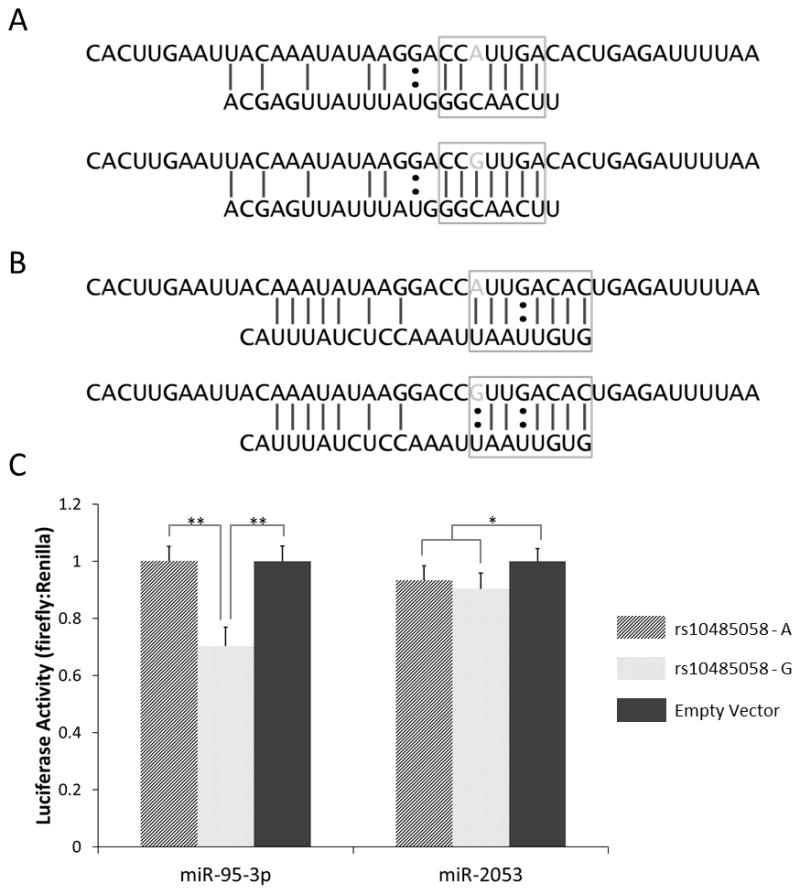
Predicted binding of miR-95-3p (A) and miR-2053 (B) to the genomic region containing the rs10485058 variant. RegRna (http://regrna.mbc.nctu.edu.tw/html/prediction.html) was used for in silico miRNA binding prediction. The top sequences represent mRNA, while the bottom sequences represent the respective miRNAs. The gray bases in the mRNA sequences indicate the rs10485058 allele. The seed sequence is boxed in each diagram. Perfect binding within the seed sequence area of miR-95-3p is predicted when the G allele is present, but not when the A allele is present. (C) Luciferase activity in BE(2)C cells co-transfected with mimics of miR-95-3p or miR-2053 and pmiR-Glo dual-luciferase constructs containing the rs10485058 locus in the 3′ UTR of firefly luciferase is shown. Luciferase activities were measured and the ratios of firefly:Renilla luciferase, normalized to the empty vector control, were graphed. Data are expressed as mean ± standard deviation (n = 9). * p < 0.05; ** p < 0.001
Discussion
We have identified an association between a haplotype tagged by rs10485058 in the MOR-1 3′ UTR and opioid positive urine drug screens in opioid dependent European-Americans treated with methadone. Furthermore, genotype at the rs10485058 variant also predicted self-reported relapse rates in an independent population of Australian patients of European descent who were receiving opioid substitution therapy. Together these data suggest that rs10485058 genotype is associated with ability of methadone to reduce opioid use in opioid dependent patients (i.e “methadone efficacy”), whether measured by urinalysis or by self-reported relapse. The limited number of previously observed pharmacogenetic effects in the dependence field have not yet been paired with mechanistic data. We hypothesized that rs10485058 might disrupt miRNA binding and provide in vitro evidence that a luciferase construct carrying the G allele of the variant is negatively regulated by miR-95-3p, whereas a construct with the A allele is not. Although the specific cell type(s) remain unknown, miR-95 is expressed in human brain 38. Nonetheless, these data suggest that the G allele of rs10485058 might be bound by miR-95-3p in vivo, resulting in reduced expression of MOR protein in individuals carrying that allele. Conversely, people with the AA genotype would have naturally higher levels of MOR than carriers of the G allele.
A difference in MOR expression might therefore explain the observed effect of rs10485058 genotype on methadone efficacy in treating opioid dependence. It is also possible that opioid use or withdrawal alters the level of miR-95-3p expression in the brain. Although opioid treatment alters expression of certain miRNAs in mouse models and in cells39, 40, there is currently little supporting evidence for such regulation in human brain samples and no studies have specifically addressed miR-95-3p. If opioids do affect expression of miR-95-3p, the effect of rs10485058 genotype on MOR expression may only be present in opioid dependent patients and not healthy controls. Analyses of MOR-1 transcript and protein expression in postmortem brain samples from control and opioid dependent individuals may help link rs10485058 genotype to the suspected phenotype. Positron emission tomography (PET) studies could also be used to image MOR availability in living brain and identify associations with rs10485058 genotype. Additional studies are certainly necessary to further evaluate this hypothesis.
The differential effect of rs10485058 alleles in the methadone and buprenorphine arms of the START trial is notable and may be related to the different mechanisms of the two medications. Whereas both methadone and buprenorphine have agonist activity at MOR, there are several key differences. Methadone is a full agonist of MOR, but buprenorphine is only a partial MOR agonist with additional antagonism at the kappa opioid receptor that may be relevant to opioid dependence treatment (reviewed in 13). Since buprenorphine has MOR-independent mechanisms, differences in MOR expression or function associated with rs10485058 genotype may have larger effects in methadone patients than those prescribed buprenorphine. Methadone may be less effective in situations where MOR is downregulated, due to reduced activation of MOR and subsequent increased withdrawal. Buprenorphine also has a significantly higher affinity for MOR and will displace other opioids, including methadone, from the receptor 41. Buprenorphine may, therefore, be less affected by downregulated MOR since it will out-compete other opioids for the smaller number of receptors.
Our data collectively suggest that rs10485058 is predictive of methadone efficacy in treating opioid dependence and that the two alleles of the polymorphism are differentially affected by miR-95-3p. However, certain caveats to these analyses should be addressed. The START trial was not specifically designed for this retrospective pharmacogenetic analysis, which analyzes a subset of total patients, and was randomized only for the primary study. This means that factors including age and sex may not be completely randomized across the genetics cohort and could potentially affect the results. Confirmation of the findings described in this study will require a prospective pharmacogenetic study with appropriate randomization. The START trial also did not collect socioeconomic information (e.g. household income, level of education, etc) or data on lifetime history of opioid dependence treatment. These factors may be associated with continued opioid use during treatment and any future studies would do well to include them if possible.
Another caveat is that the CATS did not collect data on participants’ current and past opioid agonist treatment. As a result, the sample includes unknown percentages of both methadone and buprenorphine patients. However, it is important to recognize that the CATS assessment asked about ever having relapsed after a period of abstinence and, based on the time period of data collection, it is likely that an overwhelming majority of the sample had had a trial of methadone treatment 42. Since the initial effect was observed in methadone and not buprenorphine patients, it would be preferable to replicate the effect in a solely methadone-treated population. However, since we did not find any effect in the START trial buprenorphine patients initially, it may actually be more notable that a significant effect of rs10485058 was still observed in the mixed CATS population. Retrospective identification of methadone patients in the CATS cohort or future studies in other methadone populations will be helpful in further supporting the observed phenotype and confirming the specificity of the effect to methadone treatment.
An additional limitation of this replication sample is the use of self-reported abstinence in the CATS study instead of urinalysis data. Although self-reports of illicit drug use are frequently used in dependence research, there is the potential for patients to misrepresent their drug use. However, self-report is generally reliable when there are no disincentives for being honest and in this study all participants were assured that their responses would be de-identified and confidential 43. It is possible but unlikely that the observed association between rs10485058 genotype and relapse rate is the result of inaccurate self-report data that correlates with genotype by chance. Importantly for our analysis, the lack of urinalysis data also prevents replication of the association between rs10485058 and positive urine drug screens in methadone patients. Longitudinal urinalysis data may be more useful as an outcome measurement than a binary abstinence metric given the increased emphasis on “harm reduction” in addiction treatment. Due to these caveats, the pharmacogenetic effect of rs10485058 on methadone treatment outcome is a promising finding but still requires further replication in additional independent populations.
Supplementary Material
Acknowledgments
Main START study funding came from the National Institute on Drug Abuse through the Clinical Trials Network (CTN) through a series of grants provided to each participating node: the Pacific Northwest Node (U10 DA01714), the Oregon Hawaii Node (U10 DA013036), the California/Arizona Node (U10 DA015815), the New England Node (U10 DA13038), the Delaware Valley Node (U10 DA13043), the Pacific Region Node (U10 DA13045), and the New York Node (U10 DA013046). Suboxone for the START trial was provided by Reckitt-Benckiser Pharmaceuticals (now Indivior Inc.). Dr. Berrettini was supported by the Delaware Valley Node (U10 DA13043) and by R21 DA036808. Dr. Crist was supported by NIDA grant K01 DA036751 and pilot project funding through the Veterans Integrated Service Network (VISN) 4 Mental Illness Research, Education, and Clinical Center (MIRECC). Funding support for the Comorbidity and Trauma Study (CATS) was provided by the National Institute on Drug Abuse (R01 DA17305); GWAS genotyping services at the Center for Inherited Disease Research (CIDR) at The Johns Hopkins University were supported by the National Institutes of Health [contract N01-HG-65403].
We thank Elisia Clark, Emre Karatas, Alison Lai, and Wint Thu Saung for contributions to the genotyping of the START population.
Footnotes
Conflict of Interest:
Dr. Degenhardt has received educational grants from Mundipharma International and Indivior Inc. Dr. Saxon receives funding from MedicaSafe and has received compensation from UpToDate Inc., Indivior Inc., and Alkermes Inc. Dr. Ling is a consultant for Indivior Inc., US WorldMed Inc., and Cerecor Inc and also has research support from the Patient-Centered Outcomes Research Institute, Indivior Inc., Alkermes Inc., and Brauburn Pharmaceuticals Inc. The other authors declare no conflicts of interest.
References
- 1.Degenhardt L, Charlson F, Mathers B, Hall WD, Flaxman AD, Johns N, et al. The global epidemiology and burden of opioid dependence: results from the global burden of disease 2010 study. Addiction. 2014;109(8):1320–1333. doi: 10.1111/add.12551. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaum HG, White AG, Schiller M, Waldman T, Cleveland JM, Roland CL. Societal costs of prescription opioid abuse, dependence, and misuse in the United States. Pain Med. 2011;12(4):657–667. doi: 10.1111/j.1526-4637.2011.01075.x. [DOI] [PubMed] [Google Scholar]
- 3.Yuferov V, Levran O, Proudnikov D, Nielsen DA, Kreek MJ. Search for genetic markers and functional variants involved in the development of opiate and cocaine addiction and treatment. Annals of the New York Academy of Sciences. 2010;1187:184–207. doi: 10.1111/j.1749-6632.2009.05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry. 2014;76(1):66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker A, Grecksch G, Kraus J, Loh HH, Schroeder H, Hollt V. Rewarding effects of ethanol and cocaine in mu opioid receptor-deficient mice. Naunyn-Schmiedeberg’s archives of pharmacology. 2002;365(4):296–302. doi: 10.1007/s00210-002-0533-2. [DOI] [PubMed] [Google Scholar]
- 6.Hall FS, Goeb M, Li XF, Sora I, Uhl GR. mu-Opioid receptor knockout mice display reduced cocaine conditioned place preference but enhanced sensitization of cocaine-induced locomotion. Brain research. 2004;121(1–2):123–130. doi: 10.1016/j.molbrainres.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Hummel M, Schroeder J, Liu-Chen LY, Cowan A, Unterwald EM. An antisense oligodeoxynucleotide to the mu opioid receptor attenuates cocaine-induced behavioral sensitization and reward in mice. Neuroscience. 2006;142(2):481–491. doi: 10.1016/j.neuroscience.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Matthes HW, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383(6603):819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 9.Sora I, Takahashi N, Funada M, Ujike H, Revay RS, Donovan DM, et al. Opiate receptor knockout mice define mu receptor roles in endogenous nociceptive responses and morphine-induced analgesia. Proc Natl Acad Sci U S A. 1997;94(4):1544–1549. doi: 10.1073/pnas.94.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berrettini WH, Ferraro TN, Alexander RC, Buchberg AM, Vogel WH. Quantitative trait loci mapping of three loci controlling morphine preference using inbred mouse strains. Nat Genet. 1994;7(1):54–58. doi: 10.1038/ng0594-54. [DOI] [PubMed] [Google Scholar]
- 11.Doyle GA, Schwebel CL, Ruiz SE, Chou AD, Lai AT, Wang MJ, et al. Analysis of candidate genes for morphine preference quantitative trait locus Mop2. Neuroscience. 2014;277:403–416. doi: 10.1016/j.neuroscience.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattick RP, Breen C, Kimber J, Davoli M. Buprenorphine maintenance versus placebo or methadone maintenance for opioid dependence. Cochrane Database Syst Rev. 2014;2:CD002207. doi: 10.1002/14651858.CD002207.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholls L, Bragaw L, Ruetsch C. Opioid dependence treatment and guidelines. J Manag Care Pharm. 2010;16(1 Suppl B):S14–21. doi: 10.18553/jmcp.2010.16.S1-B.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiellin DA, O’Connor PG, Chawarski M, Pakes JP, Pantalon MV, Schottenfeld RS. Methadone maintenance in primary care: a randomized controlled trial. JAMA. 2001;286(14):1724–1731. doi: 10.1001/jama.286.14.1724. [DOI] [PubMed] [Google Scholar]
- 15.Pani PP, Maremmani I, Pirastu R, Tagliamonte A, Gessa GL. Buprenorphine: a controlled clinical trial in the treatment of opioid dependence. Drug Alcohol Depend. 2000;60(1):39–50. doi: 10.1016/s0376-8716(99)00140-4. [DOI] [PubMed] [Google Scholar]
- 16.Anton RF, Oroszi G, O’Malley S, Couper D, Swift R, Pettinati H, et al. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry. 2008;65(2):135–144. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamorro AJ, Marcos M, Miron-Canelo JA, Pastor I, Gonzalez-Sarmiento R, Laso FJ. Association of micro-opioid receptor (OPRM1) gene polymorphism with response to naltrexone in alcohol dependence: a systematic review and meta-analysis. Addict Biol. 2012;17(3):505–512. doi: 10.1111/j.1369-1600.2012.00442.x. [DOI] [PubMed] [Google Scholar]
- 18.Lerman C, Wileyto EP, Patterson F, Rukstalis M, Audrain-McGovern J, Restine S, et al. The functional mu opioid receptor (OPRM1) Asn40Asp variant predicts short-term response to nicotine replacement therapy in a clinical trial. Pharmacogenomics J. 2004;4(3):184–192. doi: 10.1038/sj.tpj.6500238. [DOI] [PubMed] [Google Scholar]
- 19.Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, et al. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28(8):1546–1552. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- 20.Clarke TK, Crist RC, Ang A, Ambrose-Lanci LM, Lohoff FW, Saxon AJ, et al. Genetic variation in OPRD1 and the response to treatment for opioid dependence with buprenorphine in European-American females. Pharmacogenomics J. 2014;14(3):303–308. doi: 10.1038/tpj.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crist RC, Clarke TK, Ang A, Ambrose-Lanci LM, Lohoff FW, Saxon AJ, et al. An intronic variant in OPRD1 predicts treatment outcome for opioid dependence in African-Americans. Neuropsychopharmacology. 2013;38(10):2003–2010. doi: 10.1038/npp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kranzler HR, Covault J, Feinn R, Armeli S, Tennen H, Arias AJ, et al. Topiramate treatment for heavy drinkers: moderation by a GRIK1 polymorphism. Am J Psychiatry. 2014;171(4):445–452. doi: 10.1176/appi.ajp.2013.13081014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oneda B, Crettol S, Bochud M, Besson J, Croquette-Krokar M, Hammig R, et al. beta-Arrestin2 influences the response to methadone in opioid-dependent patients. Pharmacogenomics J. 2011;11(4):258–266. doi: 10.1038/tpj.2010.37. [DOI] [PubMed] [Google Scholar]
- 24.Ide S, Han W, Kasai S, Hata H, Sora I, Ikeda K. Characterization of the 3′ untranslated region of the human mu-opioid receptor (MOR-1) mRNA. Gene. 2005;364:139–145. doi: 10.1016/j.gene.2005.05.040. [DOI] [PubMed] [Google Scholar]
- 25.Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86(3):481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- 26.Vreugdenhil E, Berezikov E. Fine-tuning the brain: MicroRNAs. Front Neuroendocrinol. 2010;31(2):128–133. doi: 10.1016/j.yfrne.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R, Su B. Small but influential: the role of microRNAs on gene regulatory network and 3′UTR evolution. J Genet Genomics. 2009;36(1):1–6. doi: 10.1016/S1673-8527(09)60001-1. [DOI] [PubMed] [Google Scholar]
- 28.Saxon AJ, Ling W, Hillhouse M, Thomas C, Hasson A, Ang A, et al. Buprenorphine/Naloxone and methadone effects on laboratory indices of liver health: A randomized trial. Drug Alcohol Depend. 2013;128(1–2):71–76. doi: 10.1016/j.drugalcdep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Crist RC, Ambrose-Lanci LM, Vaswani M, Clarke TK, Zeng A, Yuan C, et al. Case-control association analysis of polymorphisms in the delta-opioid receptor, OPRD1, with cocaine and opioid addicted populations. Drug Alcohol Depend. 2013;127(1–3):122–128. doi: 10.1016/j.drugalcdep.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behavioural brain research. 2001;125(1–2):279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 33.Liang KYZ, SL Longitudinal data analysis using generalized linear models. Biometrika. 1986;(73):13–22. [Google Scholar]
- 34.Potter JS, Marino EN, Hillhouse MP, Nielsen S, Wiest K, Canamar CP, et al. Buprenorphine/naloxone and methadone maintenance treatment outcomes for opioid analgesic, heroin, and combined users: findings from starting treatment with agonist replacement therapies (START) J Stud Alcohol Drugs. 2013;74(4):605–613. doi: 10.15288/jsad.2013.74.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shand FL, Degenhardt L, Slade T, Nelson EC. Sex differences amongst dependent heroin users: histories, clinical characteristics and predictors of other substance dependence. Addict Behav. 2011;36(1–2):27–36. doi: 10.1016/j.addbeh.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson EC, Agrawal A, Heath AC, Bogdan R, Sherva R, Zhang B, et al. Evidence of CNIH3 involvement in opioid dependence. Mol Psychiatry. 2016;21(5):608–614. doi: 10.1038/mp.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, et al. Birth and expression evolution of mammalian microRNA genes. Genome Res. 2013;23(1):34–45. doi: 10.1101/gr.140269.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Z, Xu J, Xu M, Pasternak GW, Pan YX. Morphine regulates expression of mu-opioid receptor MOR-1A, an intron-retention carboxyl terminal splice variant of the mu-opioid receptor (OPRM1) gene via miR-103/miR-107. Mol Pharmacol. 2014;85(2):368–380. doi: 10.1124/mol.113.089292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng H, Chu J, Zeng Y, Loh HH, Law PY. Yin Yang 1 phosphorylation contributes to the differential effects of mu-opioid receptor agonists on microRNA-190 expression. J Biol Chem. 2010;285(29):21994–22002. doi: 10.1074/jbc.M110.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh SL, June HL, Schuh KJ, Preston KL, Bigelow GE, Stitzer ML. Effects of buprenorphine and methadone in methadone-maintained subjects. Psychopharmacology (Berl) 1995;119(3):268–276. doi: 10.1007/BF02246290. [DOI] [PubMed] [Google Scholar]
- 42.Burns L, Gisev N, Larney S, Dobbins T, Gibson A, Kimber J, et al. A longitudinal comparison of retention in buprenorphine and methadone treatment for opioid dependence in New South Wales, Australia. Addiction. 2015;110(4):646–655. doi: 10.1111/add.12834. [DOI] [PubMed] [Google Scholar]
- 43.Chan D. So why ask me? Are self-report data really that bad? In: Lance CE, Vandenberg RJ, editors. Statistical and Methodological Myths and Urban Legends: Doctrine, verity and Fable in the Organizational and Social Sciences. Taylor & Franci; New York: 2009. pp. 309–336. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.