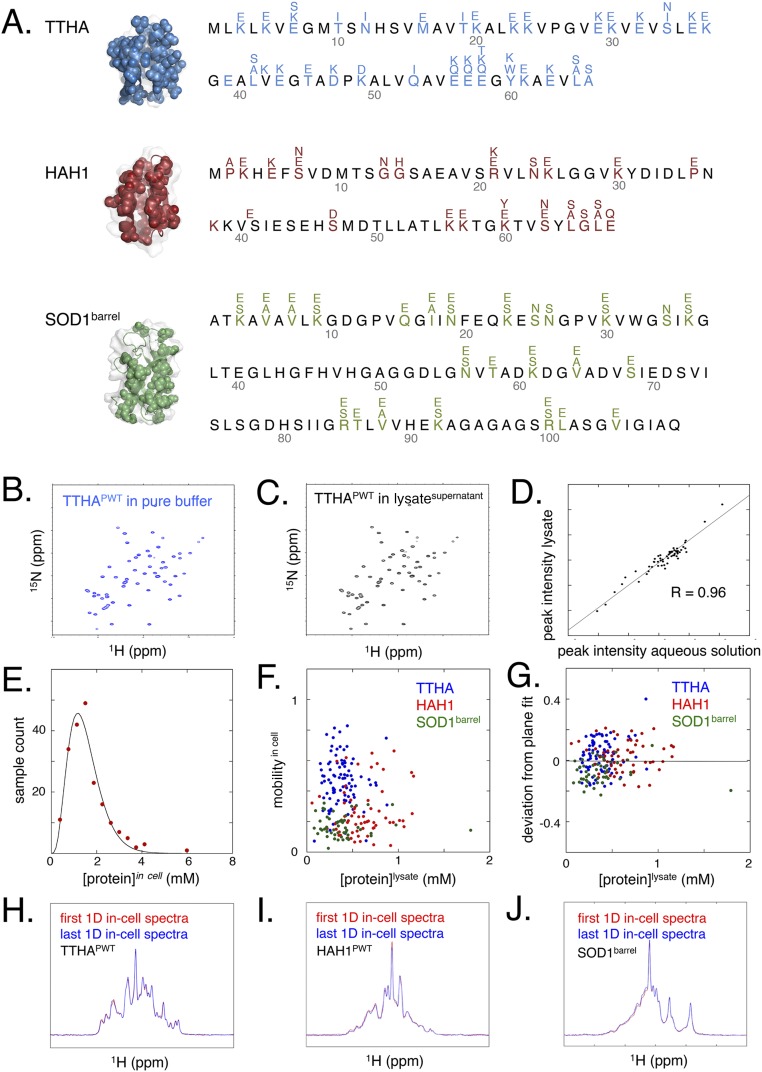
Significance
This study shows that the diffusive motions of proteins in live cells are by no means without control but follow simplistic physical−chemical rules that can be quantified and optimized through surface composition. Most strikingly, human proteins are observed to stick to the “foreign” environment of bacterial cells, whereas the bacterial analogue moves around freely. Even so, the human proteins can predictably be transformed to bacterial behavior with a few structurally benign surface mutations, and, conversely, the bacterial protein can be made to stick. The findings have not only fundamental implications for how protein function is controlled at the physical−chemical level but can also be used to adjust protein motion in Escherichia coli at will.
Keywords: in-cell NMR, protein surface properties, intracellular diffusion
Abstract
How proteins sense and navigate the cellular interior to find their functional partners remains poorly understood. An intriguing aspect of this search is that it relies on diffusive encounters with the crowded cellular background, made up of protein surfaces that are largely nonconserved. The question is then if/how this protein search is amenable to selection and biological control. To shed light on this issue, we examined the motions of three evolutionary divergent proteins in the Escherichia coli cytoplasm by in-cell NMR. The results show that the diffusive in-cell motions, after all, follow simplistic physical−chemical rules: The proteins reveal a common dependence on (i) net charge density, (ii) surface hydrophobicity, and (iii) the electric dipole moment. The bacterial protein is here biased to move relatively freely in the bacterial interior, whereas the human counterparts more easily stick. Even so, the in-cell motions respond predictably to surface mutation, allowing us to tune and intermix the protein’s behavior at will. The findings show how evolution can swiftly optimize the diffuse background of protein encounter complexes by just single-point mutations, and provide a rational framework for adjusting the cytoplasmic motions of individual proteins, e.g., for rescuing poor in-cell NMR signals and for optimizing protein therapeutics.
Despite considerable progress in mapping out how proteins interact functionally through structure and evolved interfaces (1–3), there is yet little known about how proteins interact nonspecifically upon random diffusive encounters (4–10). Although these nonspecific “quinary” (11) interactions are typically weak and short-lived, they are still expected to affect function because of their sheer numbers: Under crowded cellular conditions, they compete with specific binding (6–8), control diffusion (12), and skew structural stability (5, 13–19). The question is then to what extent this dynamic background of nonspecific interactions is biologically controlled and optimized. Part of the answer is hinted by the tendency of soluble proteins, nucleic acids, and membranes to carry a repulsive net-negative charge (20, 21). Such basal level of repulsion between the cellular components is further indicated by a matching surplus of positive counter ions like K+, Mg2+, Ca2+, and Zn2+, which greatly exceeds the concentration of negative dittos HCO3−, Cl−, SO42−, and PO42− (22). However, proteins expose also positive, polar, and hydrophobic moieties that operate against the net-negative charge repulsion by engaging in attractive interactions upon diffusive encounters. The strength and duration of these attractive interactions depend on the proteins’ detailed surface composition, relative orientations, and ability to adapt complementary shapes. Following Elcock’s estimate for the Escherichia coli cytoplasm, each protein experiences at all times approximately five putative interaction partners in its immediate cellular environment (8). Sometimes, mutual fits enable strong functional binding (1, 2), but, most often, the proteins just separate after a brief tête-à-tête (3), in search of higher-affinity partners. A key detail is that the effect of this quinary interplay on protein stability varies with protein identity and type of host cell (5, 9, 13–19, 23–26). The “crowding” effect is thus not limited to steric exclusion (27) but has also a decisive dependence on sequence composition and the details of the quinary interactions. In this study, we examine to what extent these ubiquitous background interactions are biologically tunable and can be accounted for (Fig. 1). The results show that the quinary interactions are critically sensitive to surface mutation and that proteins from divergent organisms respond very differently to the E. coli cytoplasm: Although the bacterial protein moves relatively freely in the bacterial interior, the human homolog tends to stick. Even so, the proteins can readily be tuned to any desired in-cell mobility by just a few structurally benign surface mutations, following a universal dependence on their macroscopic surface properties. As such, our findings present the physical−chemical code for quinary interactions in the E. coli cytoplasm with numerous implications for functional protein design and deciphering the functional evolution of proteomes.
Fig. 1.
Measurements of the cytoplasmic motions of three evolutionary divergent proteins by in-cell NMR: bacterial TTHA, human HAH1, and human SOD1barrel. Isotope-labeled proteins were generated directly in the E. coli cytoplasm by overexpression. The intact cells were then carefully washed and transferred to NMR tubes for detection of internal protein motions. Our observable is the protein’s rotation correlation time, i.e., how freely it tumbles around, which is a sensitive measure of the interactions experienced in the crowded E. coli interior. In essence, NMR spectra with narrow cross-peaks show that the proteins tumble unrestricted, and broadened-out spectra show that the proteins get stuck. By point mutation, we gradually tune the three proteins between these extremes to map out the physical−chemical code for the cytoplasmic crosstalk (Table S1 and Fig. S1).
Experimental Protocol
Proteins.
To assure that the proteins mainly report on the nonspecific in-cell encounters, possible specific interactions were mitigated by removal of the active sites and binding sites (SI Materials and Methods). For bacterial TTHA1718 [Protein Data Bank (PDB) 2ROE] (28) and human HAH1 (PDB 1TL4) (29), this process involved mutational substitution of the metal-binding ligands, and, for SOD1, truncation of the active-site loops that also ruptures the dimer interface (PDB 4BCZ) (30–32). The resulting pseudo-wild-type proteins are denoted TTHAPWT, HAH1PWT, and SOD1barrel, and are listed in Table S1 and Fig. S1 together with 127 analyzed surface mutations.
Table S1.
Parameters of all mutational variants of TTHA, HAH1, and SOD1barrel
| Protein | mobilityin cell* | Leakage† | ‡ | chargedensity§ | hydrophobicitySASA¶ | hydrophilicityGuy# | ||p|||| |
| TTHAPWT | 0.54 ± 0.02 | 0.02 | 1 ± 0 | 0.87 | 0.58 | 0.32 | 18.83 |
| TTHAPWT | 0.5 ± 0.02 | 0.03 | 1 ± 0 | 0.87 | 0.58 | 0.32 | 18.83 |
| TTHAL41A/L65A | 0.43 ± 0.01 | 0.01 | 1 ± 0 | 0.87 | 0.57 | 0.38 | 18.83 |
| TTHAL41A/L65A | 0.51 ± 0.01 | 0.02 | 1 ± 0 | 0.87 | 0.57 | 0.38 | 18.83 |
| TTHAE57Q | 0.41 ± 0.01 | 0.01 | 0.67 ± 0.01 | 0.54 | 0.58 | 0.33 | 18.81 |
| TTHAE57Q | 0.5 ± 0 | 0.03 | 0.67 ± 0.02 | 0.54 | 0.58 | 0.33 | 18.81 |
| TTHAL41S | 0.5 ± 0.01 | 0.05 | 1 ± 0 | 0.87 | 0.57 | 0.36 | 18.83 |
| TTHAY60W | 0.49 ± 0.02 | 0.02 | 0.96 ± 0.03 | 0.83 | 0.59 | 0.32 | 18.83 |
| TTHAY60W | 0.58 ± 0.01 | 0.04 | 0.96 ± 0.03 | 0.83 | 0.59 | 0.32 | 18.83 |
| TTHAY60W | 0.58 ± 0 | 0.01 | 0.96 ± 0.03 | 0.83 | 0.59 | 0.32 | 18.83 |
| TTHAY60K | 0.39 ± 0.03 | 0.04 | 0.69 ± 0.01 | 0.56 | 0.58 | 0.36 | 11.88 |
| TTHAY60K | 0.47 ± 0.01 | 0.02 | 0.69 ± 0.01 | 0.56 | 0.58 | 0.36 | 11.88 |
| TTHAL65S/A66S | 0.43 ± 0.01 | 0 | 1.05 ± 0.05 | 0.92 | 0.58 | 0.37 | 18.83 |
| TTHAL65S/A66S | 0.56 ± 0.01 | 0.04 | 1.05 ± 0.05 | 0.92 | 0.58 | 0.37 | 18.83 |
| TTHAL41A | 0.54 ± 0.01 | 0.03 | 1.05 ± 0.05 | 0.92 | 0.58 | 0.35 | 18.83 |
| TTHAL41A | 0.7 ± 0.01 | 0.02 | 1.05 ± 0.05 | 0.92 | 0.58 | 0.35 | 18.83 |
| TTHAL65A | 0.69 ± 0.01 | 0.02 | 1.05 ± 0.05 | 0.92 | 0.57 | 0.35 | 18.83 |
| TTHAL65A | 0.45 ± 0.01 | 0.01 | 1.05 ± 0.05 | 0.92 | 0.57 | 0.35 | 18.83 |
| TTHAK37E | 0.64 ± 0.01 | 0.02 | 1.76 ± 0.19 | 1.63 | 0.56 | 0.31 | 38.55 |
| TTHAK37E | 0.59 ± 0.01 | 0.08 | 1.76 ± 0.19 | 1.63 | 0.56 | 0.31 | 38.55 |
| TTHAK24E | 0.69 ± 0.01 | 0.03 | 1.77 ± 0.13 | 1.64 | 0.57 | 0.31 | 14.39 |
| TTHAK24E | 0.60 ± 0.01 | 0.02 | 1.77 ± 0.13 | 1.64 | 0.57 | 0.31 | 14.39 |
| TTHAK24E | 0.68 ± 0.01 | 0.12 | 1.77 ± 0.13 | 1.64 | 0.57 | 0.31 | 14.39 |
| TTHAK3E | 0.56 ± 0.01 | 0.06 | 1.59 ± 0.03 | 1.46 | 0.57 | 0.31 | 25.34 |
| TTHAK3E | 0.49 ± 0.01 | 0.01 | 1.59 ± 0.03 | 1.46 | 0.57 | 0.31 | 25.34 |
| TTHAK3E | 0.58 ± 0.01 | 0.04 | 1.59 ± 0.03 | 1.46 | 0.57 | 0.31 | 25.34 |
| TTHAK20E | 0.79 ± 0.01 | 0 | 1.62 ± 0.02 | 1.49 | 0.57 | 0.31 | 17.23 |
| TTHAK20E | 0.77 ± 0.01 | 0.12 | 1.62 ± 0.02 | 1.49 | 0.57 | 0.31 | 17.23 |
| TTHAK23E | 0.75 ± 0.01 | 0.01 | 1.68 ± 0.06 | 1.55 | 0.57 | 0.31 | 8.44 |
| TTHAK23E | 0.67 ± 0.01 | 0.08 | 1.68 ± 0.06 | 1.55 | 0.57 | 0.31 | 8.44 |
| TTHAK23E | 0.67 ± 0.01 | 0.05 | 1.68 ± 0.06 | 1.55 | 0.57 | 0.31 | 8.4 |
| TTHAM16E | 0.83 ± 0.01 | 0.01 | 1.4 ± 0.15 | 1.27 | 0.56 | 0.37 | 20.98 |
| TTHAM16E | 0.81 ± 0.01 | 0.07 | 1.4 ± 0.15 | 1.27 | 0.56 | 0.37 | 20.98 |
| TTHAT45E | 0.61 ± 0.01 | 0.01 | 1.37 ± 0.07 | 1.27 | 0.57 | 0.34 | 10.83 |
| TTHAT45E | 0.53 ± 0.01 | 0.04 | 1.37 ± 0.07 | 1.24 | 0.57 | 0.34 | 10.83 |
| TTHAE58K | 0.18 ± 0.01 | 0.05 | 0.45 ± 0.01 | 0.32 | 0.58 | 0.34 | 21.62 |
| TTHAE58K | 0.14 ± 0.03 | 0 | 0.45 ± 0.01 | 0.32 | 0.58 | 0.34 | 21.62 |
| TTHAE58K | 0.12 ± 0.04 | 0.04 | 0.45 ± 0.01 | 0.32 | 0.58 | 0.34 | 21.62 |
| TTHAE7S | 0.48 ± 0.02 | 0.02 | 0.68 ± 0.06 | 0.55 | 0.58 | 0.32 | 8.66 |
| TTHAE7S | 0.36 ± 0.01 | 0.04 | 0.68 ± 0.06 | 0.55 | 0.58 | 0.32 | 8.66 |
| TTHAY60W/E57K | 0.28 ± 0.02 | 0.01 | 0.42 ± 0.03 | 0.29 | 0.59 | 0.33 | 24.27 |
| TTHAY60W/E57K | 0.27 ± 0.02 | 0.03 | 0.42 ± 0.03 | 0.29 | 0.59 | 0.33 | 24.27 |
| TTHAE32K | 0.19 ± 0.01 | 0.02 | 0.54 ± 0.09 | 0.41 | 0.58 | 0.34 | 29.7 |
| TTHAE32K | 0.25 ± 0.01 | 0.02 | 0.54 ± 0.09 | 0.41 | 0.58 | 0.34 | 29.7 |
| TTHAD47K | 0.34 ± 0.01 | 0.01 | 0.4 ± 0.03 | 0.27 | 0.58 | 0.34 | 36.72 |
| TTHAD47K | 0.35 ± 0.01 | 0.03 | 0.4 ± 0.03 | 0.27 | 0.58 | 0.34 | 36.72 |
| TTHAD47K | 0.34 ± 0.01 | 0.04 | 0.4 ± 0.03 | 0.27 | 0.58 | 0.34 | 36.72 |
| TTHAE29K | 0.25 ± 0.01 | 0.02 | 0.51 ± 0.03 | 0.38 | 0.58 | 0.34 | 43.72 |
| TTHAE29K | 0.36 ± 0.01 | 0.03 | 0.51 ± 0.03 | 0.38 | 0.58 | 0.34 | 43.72 |
| TTHAK61E | 0.45 ± 0.01 | 0.01 | 1.62 ± 0.12 | 1.49 | 0.57 | 0.31 | 37.9 |
| TTHAK61E | 0.47 ± 0.01 | 0.03 | 1.62 ± 0.12 | 1.49 | 0.57 | 0.31 | 37.9 |
| TTHAK49D | 0.5 ± 0.01 | 0.03 | 1.73 ± 0.16 | 1.60 | 0.58 | 0.31 | 27.13 |
| TTHAK49D | 0.58 ± 0.02 | 0.05 | 1.73 ± 0.16 | 1.60 | 0.58 | 0.31 | 27.13 |
| TTHAE43K | 0.27 ± 0.01 | 0.06 | 0.5 ± 0.08 | 0.37 | 0.58 | 0.34 | 39.12 |
| TTHAE43K | 0.36 ± 0.03 | 0.02 | 0.5 ± 0.08 | 0.37 | 0.58 | 0.34 | 39.12 |
| TTHAE56K | 0.46 ± 0.01 | 0.05 | 0.44 ± 0.02 | 0.31 | 0.58 | 0.34 | 17.8 |
| TTHAE56K | 0.43 ± 0.01 | 0.03 | 0.44 ± 0.02 | 0.31 | 0.58 | 0.34 | 17.8 |
| TTHAE36k. | 0.32 ± 0.03 | 0 | 0.43 ± 0.03 | 0.30 | 0.57 | 0.34 | 22.13 |
| TTHAE36K | 0.27 ± 0.01 | 0.03 | 0.43 ± 0.03 | 0.30 | 0.57 | 0.34 | 22.13 |
| TTHAE7K | 0.28 ± 0.02 | 0.03 | 0.39 ± 0.03 | 0.26 | 0.58 | 0.34 | 2.37 |
| TTHAE7K | 0.39 ± 0.01 | 0.01 | 0.39 ± 0.03 | 0.26 | 0.58 | 0.34 | 2.37 |
| TTHAE39K | 0.39 ± 0.01 | 0.02 | 0.44 ± 0.03 | 0.31 | 0.58 | 0.34 | 17.01 |
| TTHAE39K | 0.42 ± 0.01 | 0.03 | 0.44 ± 0.03 | 0.31 | 0.58 | 0.34 | 17.01 |
| TTHAE63K | 0.41 ± 0.01 | 0.02 | 0.5 ± 0.05 | 0.37 | 0.59 | 0.34 | 15.95 |
| TTHAE63K | 0.44 ± 0.01 | 0.02 | 0.5 ± 0.05 | 0.37 | 0.59 | 0.34 | 15.95 |
| TTHAK30E/K61E | 0.58 ± 0.01 | 0.01 | 1.75 ± 0.05 | 1.62 | 0.57 | 0.3 | 17.27 |
| TTHAK30E/K61E | 0.58 ± 0.02 | 0.02 | 1.75 ± 0.05 | 1.62 | 0.57 | 0.3 | 17.27 |
| TTHAK5E | 0.48 ± 0.01 | 0.01 | 1.45 ± 0.03 | 1.32 | 0.57 | 0.31 | 32.28 |
| TTHAK5E | 0.41 ± 0.01 | 0.01 | 1.45 ± 0.03 | 1.32 | 0.57 | 0.31 | 32.28 |
| TTHAK30E | 0.6 ± 0.01 | 0.04 | 1.41 ± 0.12 | 1.28 | 0.57 | 0.31 | 8.98 |
| TTHAK30E | 0.59 ± 0.01 | 0.13 | 1.41 ± 0.12 | 1.28 | 0.57 | 0.31 | 8.98 |
| TTHAY60W/E57Q | 0.48 ± 0.01 | 0.06 | 0.65 ± 0.02 | 0.52 | 0.58 | 0.32 | 18.81 |
| TTHAY60W/E57Q | 0.41 ± 0.01 | 0.02 | 0.65 ± 0.02 | 0.52 | 0.58 | 0.32 | 18.81 |
| TTHAY60W/E57Q | 0.50 ± 0.01 | 0.06 | 0.65 ± 0.02 | 0.52 | 0.58 | 0.32 | 18.81 |
| TTHAY60W/E58K | 0.28 ± 0.04 | 0.16 | 0.65 ± 0.02 | 0.52 | 0.58 | 0.32 | 17.49 |
| TTHAY60W/E56Q | 0.34 ± 0.01 | 0.03 | 0.65 ± 0.02 | 0.52 | 0.58 | 0.32 | 15.84 |
| TTHAE56Q/E58Q | 0.22 ± 0.01 | 0.04 | 0.44 ± 0.02 | 0.31 | 0.58 | 0.33 | 19.06 |
| TTHAS34I | 0.75 ± 0 | 0.12 | 0.98 ± 0.01 | 0.85 | 0.57 | 0.29 | 18.83 |
| TTHAN12I | 0.31 ± 0.01 | 0.1 | 1.03 ± 0.03 | 0.90 | 0.58 | 0.29 | 18.83 |
| TTHAT19I | 0.37 ± 0.01 | 0.03 | 0.98 ± 0 | 0.85 | 0.57 | 0.3 | 18.83 |
| TTHAQ53I | 0.38 ± 0.01 | 0.02 | 0.98 ± 0.01 | 0.85 | 0.59 | 0.28 | 18.83 |
| TTHAT10I | 0.33 ± 0.01 | 0.05 | 1.02 ± 0.02 | 0.89 | 0.58 | 0.3 | 18.83 |
| TTHAT45I | 0.44 ± 0.01 | 0.02 | 1.12 ± 0.09 | 0.99 | 0.59 | 0.3 | 18.83 |
| TTHAE57K | 0.44 ± 0.01 | 0.01 | 0.47 ± 0.02 | 0.34 | 0.58 | 0.34 | 24.27 |
| TTHAE58Q | 0.45 ± 0.01 | 0.03 | 0.68 ± 0.01 | 0.55 | 0.58 | 0.33 | 17.49 |
| TTHAS34N | 0.51 ± 0.01 | 0.06 | 1.12 ± 0.09 | 0.99 | 0.56 | 0.32 | 18.83 |
| TTHAE56Q | 0.51 ± 0.01 | 0.07 | 0.68 ± 0.01 | 0.55 | 0.58 | 0.33 | 15.85 |
| TTHAL41S/L65S/A66S | 0.42 ± 0.01 | 0.02 | 1 ± 0 | 0.87 | NA | 0.4 | 18.83 |
| TTHAL41S/L65S/A66S | 0.49 ± 0.01 | 0.01 | 1 ± 0 | 0.87 | NA | 0.4 | 18.83 |
| TTHAE56K/E57K/E58T | 0.08 ± 0.07 | 0.13 | 0 | −0.13 | NA | 0.33 | 44.62 |
| TTHAE56K/E57K/E58T | 0.07 ± 0.02 | 0.16 | 0 | −0.13 | NA | 0.33 | 44.62 |
| HAH1PWT | 0.11 ± 0.03 | 0.08 | 0.18 ± 0.01 | −0.07 | 0.58 | 0.31 | 54.26 |
| HAH1PWT | 0.16 ± 0.05 | 0.02 | 0.18 ± 0.01 | −0.07 | 0.58 | 0.31 | 54.26 |
| HAH1P36E/K38E | 0.2 ± 0.01 | 0.09 | 1.1 ± 0.1 | 0.82 | 0.58 | 0.3 | 69.09 |
| HAH1P36E/K38E | 0.2 ± 0.01 | 0.16 | 1.1 ± 0.1 | 0.82 | 0.58 | 0.3 | 69.09 |
| HAH1K60E/S63E | 0.29 ± 0.01 | 0.03 | 1.2 ± 0 | 0.92 | 0.58 | 0.3 | 42.09 |
| HAH1R21E/K25E | 0.66 ± 0.01 | 0 | 1.6 ± 0.1 | 1.31 | 0.57 | 0.27 | 43.45 |
| HAH1R21E/K25E | 0.65 ± 0 | 0.18 | 1.6 ± 0.1 | 1.31 | 0.57 | 0.27 | 43.45 |
| HAH1K60E/K56E | 0.27 ± 0.02 | 0.05 | 1.47 ± 0.04 | 1.18 | 0.56 | 0.28 | 31.42 |
| HAH1K60E/K56E | 0.34 ± 0.01 | 0.15 | 1.47 ± 0.04 | 1.18 | 0.56 | 0.28 | 31.42 |
| HAH1K60E/K57E | 0.58 ± 0 | 0.08 | 1.5 ± 0.1 | 1.21 | 0.56 | 0.28 | 24.07 |
| HAH1K60E/K57E | 0.54 ± 0 | 0.19 | 1.5 ± 0.1 | 1.21 | 0.56 | 0.28 | 24.07 |
| HAH1N24S | 0.16 ± 0.01 | 0.14 | 0.22 ± 0 | −0.04 | 0.58 | 0.31 | 54.26 |
| HAH1N24S | 0.11 ± 0.01 | 0.12 | 0.22 ± 0 | −0.04 | 0.58 | 0.31 | 54.26 |
| HAH1R21K | 0.18 ± 0.02 | 0.04 | 0.22 ± 0.02 | −0.04 | 0.58 | 0.3 | 54.26 |
| HAH1R21K | 0.15 ± 0.01 | 0.07 | 0.22 ± 0.02 | −0.04 | 0.58 | 0.3 | 54.26 |
| HAH1G13N | 0.09 ± 0.03 | 0.05 | 0.2 ± 0.02 | −0.06 | 0.58 | 0.31 | 54.26 |
| HAH1G13N | 0.09 ± 0.03 | 0 | 0.2 ± 0.02 | −0.06 | 0.58 | 0.31 | 54.26 |
| HAH1G14H | 0.05 ± 0.04 | 0.22 | 0.29 ± 0.01 | 0.03 | 0.58 | 0.29 | 54.26 |
| HAH1G14H | 0.07 ± 0.02 | 0.22 | 0.29 ± 0.01 | 0.03 | 0.58 | 0.29 | 54.26 |
| HAH1P2A | 0.26 ± 0.01 | 0.05 | 0.23 ± 0.01 | −0.03 | 0.56 | 0.29 | 54.26 |
| HAH1P2A | 0.20 ± 0.01 | 0.04 | 0.23 ± 0.01 | −0.03 | 0.56 | 0.29 | 54.26 |
| HAH1L65A/L67A | 0.28 ± 0 | 0.08 | 0.22 ± 0.02 | −0.04 | 0.58 | 0.35 | 54.26 |
| HAH1L65A/L67A | 0.25 ± 0.01 | 0.04 | 0.22 ± 0.02 | −0.04 | 0.58 | 0.35 | 54.26 |
| HAH1L65S/L67S | 0.21 ± 0.02 | 0.02 | 0.22 ± 0.02 | −0.04 | 0.57 | 0.37 | 54.26 |
| HAH1L65S/L67S | 0.2 ± 0.01 | 0.07 | 0.22 ± 0.02 | −0.04 | 0.57 | 0.37 | 54.26 |
| HAH1R21E/K30E | 0.58 ± 0.01 | 0.12 | 1.45 ± 0.05 | 1.16 | 0.57 | 0.27 | 59.3 |
| HAH1R21E/K30E | 0.62 ± 0 | 0.07 | 1.45 ± 0.05 | 1.16 | 0.57 | 0.27 | 59.3 |
| HAH1R21E/K30E | 0.5 ± 0.01 | 0.01 | 1.45 ± 0.05 | 1.16 | 0.57 | 0.27 | 59.3 |
| HAH1R21E/K38E | 0.56 ± 0 | 0.07 | 1.5 ± 0 | 1.21 | 0.59 | 0.27 | 48.48 |
| HAH1R21E/K38E | 0.52 ± 0.01 | 0.02 | 1.5 ± 0 | 1.21 | 0.59 | 0.27 | 48.48 |
| HAH1S63N | 0.13 ± 0.03 | 0.08 | 0.22 ± 0.02 | −0.04 | 0.58 | 0.3 | 54.26 |
| HAH1K3E | 0.32 ± 0.05 | 0.11 | 0.66 ± 0.01 | 0.39 | 0.58 | 0.29 | 76.39 |
| HAH1K60Y | 0.21 ± 0.01 | 0.05 | 0.61 ± 0.01 | 0.34 | 0.57 | 0.27 | 46.24 |
| HAH1P36E | 0.19 ± 0.01 | 0.04 | 0.58 ± 0 | 0.32 | 0.56 | 0.31 | 58.48 |
| HAH1P36E | 0.14 ± 0.01 | 0.04 | 0.58 ± 0 | 0.32 | 0.56 | 0.31 | 58.48 |
| HAH1S63E | 0.23 ± 0.01 | 0.04 | 0.5 ± 0.03 | 0.24 | 0.58 | 0.31 | 53.99 |
| HAH1S63E | 0.2 ± 0.01 | 0.04 | 0.5 ± 0.03 | 0.24 | 0.58 | 0.31 | 54 |
| HAH1S7E | 0.21 ± 0.01 | 0.01 | 0.53 ± 0.04 | 0.26 | 0.58 | 0.31 | 56.51 |
| HAH1S7E | 0.19 ± 0.02 | 0.05 | 0.53 ± 0.04 | 0.26 | 0.58 | 0.31 | 56.51 |
| HAH1S47D | 0.13 ± 0.02 | 0.06 | 0.57 ± 0.03 | 0.30 | 0.57 | 0.31 | 59.84 |
| HAH1S47D | 0.11 ± 0.03 | 0.06 | 0.57 ± 0.03 | 0.30 | 0.57 | 0.31 | 59.84 |
| HAH1K57E | 0.18 ± 0.02 | 0.12 | 0.75 ± 0.01 | 0.48 | 0.56 | 0.29 | 35.5 |
| HAH1K57E | 0.37 ± 0.01 | 0.06 | 0.75 ± 0.01 | 0.48 | 0.56 | 0.29 | 35.5 |
| HAH1K57E | 0.39 ± 0.01 | 0.06 | 0.75 ± 0.01 | 0.48 | 0.56 | 0.29 | 35.5 |
| HAH1K56E | 0.23 ± 0.01 | 0.06 | 0.73 ± 0.02 | 0.46 | 0.56 | 0.29 | 42.5 |
| HAH1K56E | 0.23 ± 0.02 | 0.04 | 0.73 ± 0.02 | 0.46 | 0.56 | 0.29 | 42.5 |
| HAH1K56E | 0.29 ± 0.01 | 0.03 | 0.73 ± 0.02 | 0.46 | 0.56 | 0.29 | 42.5 |
| HAH1K57E/K56E | 0.6 ± 0.01 | 0.12 | 1.35 ± 0.05 | 1.07 | 0.56 | 0.28 | 30.33 |
| HAH1K57E/K56E | 0.43 ± 0.01 | 0.04 | 1.35 ± 0.05 | 1.07 | 0.56 | 0.28 | 30.33 |
| HAH1K60E | 0.24 ± 0.02 | 0.08 | 0.85 ± 0.05 | 0.58 | 0.57 | 0.29 | 39.51 |
| HAH1K60E | 0.19 ± 0.02 | 0.07 | 0.85 ± 0.05 | 0.58 | 0.57 | 0.29 | 39.51 |
| HAH1K38E | 0.22 ± 0.02 | 0.03 | 0.74 ± 0.02 | 0.47 | 0.59 | 0.29 | 60.59 |
| HAH1R21E | 0.4 ± 0.01 | 0.05 | 0.77 ± 0.05 | 0.50 | 0.58 | 0.28 | 43.66 |
| HAH1K30E | 0.16 ± 0.01 | 0.19 | 0.73 ± 0.07 | 0.46 | 0.56 | 0.29 | 66.16 |
| HAH1K30E | 0.16 ± 0.02 | 0.04 | 0.73 ± 0.07 | 0.46 | 0.56 | 0.29 | 66.16 |
| HAH1K25E | 0.32 ± 0.01 | 0.04 | 0.85 ± 0.01 | 0.58 | 0.57 | 0.29 | 46.17 |
| HAH1K25E | 0.34 ± 0.02 | 0.05 | 0.85 ± 0.01 | 0.58 | 0.57 | 0.29 | 46.17 |
| HAH1S7N | 0.17 ± 0.01 | 0.08 | 0.2 ± 0.03 | −0.06 | 0.58 | 0.30 | 54.26 |
| HAH1E68Q | 0.11 ± 0.02 | 0.08 | 0** | −0.25 | 0.57 | 0.31 | 40.73 |
| HAH1E5K | 0.04 ± 0.02 | 0.12 | 0** | −0.25 | 0.59 | 0.31 | 41.72 |
| HAH1L65S/G66S/L67S | 0.2 ± 0.01 | 0.09 | 0.21 ± 0.04 | −0.05 | NA | 0.37 | 54.26 |
| SOD1barrel | 0.14 ± 0.01 | 0.01 | 0.79 ± 0.01 | 0.30 | 0.60 | 0.29 | 45.66 |
| SOD1barrel | 0.11 ± 0.02 | 0.05 | 0.79 ± 0.01 | 0.30 | 0.60 | 0.29 | 45.66 |
| SOD1R100E | 0.31 ± 0.01 | 0.05 | 1.22 ± 0.01 | 0.63 | 0.60 | 0.28 | 41.03 |
| SOD1K3E | 0.15 ± 0.01 | 0.09 | 1.22 ± 0.02 | 0.63 | 0.61 | 0.29 | 50.82 |
| SOD1K92E/R85E | 0.3 ± 0.01 | 0.16 | 1.53 ± 0.03 | 0.88 | 0.6 | 0.27 | 40.03 |
| SOD1K92E/R85E | 0.31 ± 0.01 | 0.17 | 1.53 ± 0.03 | 0.88 | 0.6 | 0.27 | 40.03 |
| SOD1R85E | 0.21 ± 0.01 | 0.13 | 1.21 ± 0 | 0.63 | 0.61 | 0.28 | 36.47 |
| SOD1K92E | 0.14 ± 0.02 | 0.12 | 1.21 ± 0.03 | 0.63 | 0.60 | 0.29 | 53.13 |
| SOD1K92E | 0.15 ± 0.02 | 0.2 | 1.21 ± 0.03 | 0.63 | 0.60 | 0.29 | 53.13 |
| SOD1K92S | 0.15 ± 0.02 | 0.05 | 0.98 ± 0.01 | 0.45 | 0.6 | 0.28 | 47.08 |
| SOD1K23S | 0.12 ± 0.01 | 0.03 | 1 ± 0.01 | 0.46 | 0.61 | 0.28 | 54.34 |
| SOD1K23S | 0.08 ± 0.01 | 0.19 | 1 ± 0.01 | 0.46 | 0.61 | 0.28 | 54.34 |
| SOD1K30S | 0.08 ± 0.02 | 0.07 | 0.99 ± 0.01 | 0.45 | 0.61 | 0.28 | 53.02 |
| SOD1K30S | 0.09 ± 0.02 | 0.17 | 0.99 ± 0.01 | 0.45 | 0.61 | 0.28 | 53.02 |
| SOD1K36E | 0.13 ± 0.02 | 0.12 | 1.24 ± 0.06 | 0.65 | 0.6 | 0.29 | 48.31 |
| SOD1R100S | 0.2 ± 0.01 | 0.1 | 0.91 ± 0.06 | 0.39 | 0.6 | 0.28 | 40.7 |
| SOD1K9E/K36E | 0.24 ± 0.01 | 0.1 | 1.62 ± 0.07 | 0.98 | 0.6 | 0.28 | 43.12 |
| SOD1R85S | 0.16 ± 0.01 | 0.1 | 0.98 ± 0 | 0.45 | 0.6 | 0.28 | 39.93 |
| SOD1R85S | 0.15 ± 0.02 | 0.07 | 0.98 ± 0 | 0.45 | 0.61 | 0.28 | 39.93 |
| SOD1K92E/R100E | 0.28 ± 0.02 | 0.06 | 1.56 ± 0.06 | 0.90 | 0.6 | 0.27 | 63.91 |
| SOD1K3S | 0.17 ± 0.02 | 0.08 | 0.99 ± 0.01 | 0.45 | 0.61 | 0.28 | 46.31 |
| SOD1K61E | 0.09 ± 0.02 | 0.14 | 1.23 ± 0.05 | 0.64 | 0.61 | 0.29 | 68.31 |
| SOD1K92S/R100S | 0.27 ± 0.01 | 0.08 | 1.26 ± 0.03 | 0.67 | 0.6 | 0.26 | 46.97 |
| SOD1K9S | 0.21 ± 0.01 | 0.1 | 1 ± 0.09 | 0.46 | 0.6 | 0.28 | 36.52 |
| SOD1K9S | 0.11 ± 0.01 | 0.24 | 1 ± 0.09 | 0.46 | 0.6 | 0.28 | 36.52 |
| SOD1K36S | 0.13 ± 0.02 | 0.07 | 1.04 ± 0.05 | 0.49 | 0.60 | 0.28 | 45.32 |
| SOD1V5E/V7E | 0.32 ± 0.02 | 0.02 | 1.26 ± 0.03 | 0.67 | 0.59 | 0.35 | 35.42 |
| SOD1Q15E/N19E | 0.22 ± 0.02 | 0.02 | 1.28 ± 0.03 | 0.68 | 0.6 | 0.3 | 40.59 |
| SOD1N56E/T58E | 0.23 ± 0.02 | 0.03 | 1.26 ± 0.02 | 0.67 | 0.60 | 0.31 | 61.71 |
| SOD1L101E/V105E | 0.1 ± 0.01 | 0.06 | 0.81 ± 0.01 | 0.31 | 0.59 | 0.35 | 31.13 |
| SOD1S34N | 0.16 ± 0.02 | 0.08 | 1.02 ± 0.02 | 0.47 | 0.60 | 0.29 | 45.66 |
| SOD1K36E/K61E | 0.15 ± 0.02 | 0.12 | 1.65 ± 0.02 | 0.97 | 0.6 | 0.28 | 81.93 |
| SOD1K92E/R100S | 0.28 ± 0.02 | 0.03 | 1.46 ± 0.01 | 0.82 | 0.60 | 0.27 | 56.76 |
| SOD1K92E/R85S | 0.27 ± 0.04 | 0.06 | 1.44 ± 0.02 | 0.81 | 0.6 | 0.27 | 45.87 |
| SOD1K9S/K36E | 0.14 ± 0.04 | 0.06 | 1.48 ± 0.04 | 0.84 | 0.6 | 0.27 | 44.33 |
| SOD1N19S | 0.03 ± 0.04 | 0.2 | 0.74 ± 0.05 | 0.26 | 0.61 | 0.29 | 45.66 |
| SOD1K23E | 0.13 ± 0.01 | 0.07 | 1.1 ± 0 | 0.54 | 0.61 | 0.29 | 67.21 |
| SOD1K30E | 0.08 ± 0.03 | 0.1 | 1.06 ± 0 | 0.51 | 0.60 | 0.29 | 61.6 |
| SOD1V64A | 0.11 ± 0.02 | 0.2 | 0.75 ± 0.07 | 0.27 | 0.6 | 0.31 | 45.66 |
| SOD1K61S | 0.12 ± 0.02 | 0.2 | 0.93 ± 0.04 | 0.41 | 0.6 | 0.28 | 54.5 |
| SOD1K9E | 0.19 ± 0.02 | 0.1 | 1.16 ± 0.08 | 0.59 | 0.6 | 0.29 | 29.15 |
| SOD1I17A | 0.16 ± 0.02 | 0.19 | 0.74 ± 0.03 | 0.26 | 0.6 | 0.31 | 45.66 |
| SOD1V5AV7A | 0.10 ± 0.03 | 0.09 | 0.78 ± 0.05 | 0.29 | 0.62 | 0.33 | 45.66 |
| SOD1N26S | 0.2 ± 0.01 | 0.06 | 0.75 ± 0.05 | 0.27 | 0.61 | 0.29 | 45.66 |
| SOD1K92S/R85E | 0.31 ± 0 | 0.15 | 1.2 ± 0 | 0.62 | 0.6 | 0.27 | 35.09 |
| SOD1V64E/S68E | 0.14 ± 0 | 0.22 | 1.1 ± 0 | 0.54 | 0.6 | 0.33 | 58.69 |
| SOD1T86E/V88E | 0.20 ± 0.03 | 0.15 | 1.06 ± 0.03 | 0.51 | 0.6 | 0.33 | 36.18 |
| SOD1N56S | 0.13 ± 0.02 | 0.1 | 0.78 ± 0.05 | 0.29 | NA | 0.29 | 45.66 |
| SOD1S25N | 0.2 ± 0.01 | 0.06 | 0.73 ± 0.04 | 0.25 | NA | 0.29 | 45.66 |
| SOD1V88A | 0.16 ± 0.02 | 0.2 | 0.69 ± 0.04 | 0.22 | NA | 0.31 | 45.66 |
| SOD1Q15E/I17E/N19E | 0.18 ± 0.01 | 0.08 | 1.42 ± 0.01 | 0.79 | NA | 0.32 | 40.26 |
Calculated as the ratio of in-cell peak heights and lysate peak heights. The error is estimated from the noise-to-signal ratio of the NMR spectra.
Defined by the ratio of supernatant peak intensity to in-cell peak intensity. Data points with protein leakage of >10% are highlighted in boldface.
Reference molecule is for all values TTHAPWT.
From relative mobility scaled by hydrodynamic radii, Eq. 5.
From in silico mutagenesis and hydrophobicity calculations (SI Materials and Methods). NA, data not available.
Calculated from Eq. 7.
Calculated from the PDB files and Eq. 8.
HAH1E68Q and HAH1E5K do not migrate at all on the native gel.
Fig. S1.
(A) Structures, sequences, and mutations of the analyzed proteins TTHA [PDB 2ROE (28)], HAH1 [PDB 1TL4 (29)], and SOD1barrel [PDB 4BCZ (30)]. (B and C) The HMQC spectrum of TTHAPWT in (B) pure buffer is very similar to that in (C) lysatesupernatant. (D) The peak heights in dilute buffer and the lysate supernatant at the same protein concentration are very similar and correlate with R = 0.96, underlining that the lysatesupernatant conditions correspond to the “free” rotational tumbling observed in pure buffer. (E) Intracellular concentrations of overexpressed protein show a skewed normal distribution, with average 1.6 mM and SD ± 0.8 mM. (F) Although the intracellular concentration of overexpressed protein is relatively high at 1.6 ± 0.8 mM, there is no apparent correlation between expression levels and mobilityin cell. (G) Likewise, there is no correlation between expression levels and deviation from the fitted plane in Fig. 6. (H–J) Temporal stability of the 1D 15N-filtered NMR spectra during the acquisition of mobilityin cell. The overlap of spectra obtained at the start and end of acquisition shows that the levels and properties of overexpressed protein remain the same during the course of the experiment.
Experimental Strategy.
The proteins (Table S1 and Fig. S1) were produced and analyzed in E. coli BL21(DE3)pLysS cells as follows (SI Materials and Methods). Overexpression was induced by 1-thio-β-d-galactopyranoside (IPTG) in isotope-enriched medium and sustained for 4 h, yielding intracellular concentrations of 1.6 ± 0.8 mM N15-labeled protein (SI Materials and Methods and Fig. S1). The culture was then nondisruptively centrifuged at 800 × g for 8 min to obtain a pellet of intact, viable cells. One gram of this cell pellet was carefully dissolved in 1 mL of M9 buffer, transferred to an NMR tube, and subjected to in-cell spectral analysis, i.e., samplein cell. For control of protein leakage, the samplein cell was extracted from the NMR tube and centrifuged at 2,400 × g for 2 min to obtain a cell-free supernatant of the intervening M9 buffer, i.e., samplesupernatant. The pelleted cells were then resuspended to the original sample volume with M9 buffer, lysed to set free the intracellular target protein, and centrifuged at 17,000 × g for 10 min to remove cell debris and larger macromolecular species. This decrowded lysate, which represents a much more dilute background than the cytoplasm (SI Controls, Fig. S1, and Table S2), was finally used as a control for internalized protein, i.e., lysatesupernatant. An advantage of this protocol is that the amount of labeled protein in the NMR detection volume is kept the same in samplein cell and lysatesupernatant, allowing estimates of dynamic changes upon cell rupture from changes in peak heights alone (Results and SI Controls).
Table S2.
E. coli expression levels
| [protein]lysate | Millimolars | [protein]lysate | Millimolars | [protein]lysate | Millimolars | [protein]lysate | Millimolars |
| TTHAPWT | 0.25 | TTHAK49D | 0.17 | HAH1N24S | 0.72 | SOD1barrel | 0.45 |
| TTHAPWT | 0.19 | TTHAK49D | 0.39 | HAH1N24S | 0.66 | SOD1R100E | 0.54 |
| TTHAL41A/L65A | 0.49 | TTHAE43K | 0.26 | HAH1R21K | 0.75 | SOD1K3E | 0.58 |
| TTHAL41A/L65A | 0.41 | TTHAE43K | 0.45 | HAH1R21K | 0.32 | SOD1K92E/R85E | 0.49 |
| TTHAE57Q | 0.29 | TTHAE56K | 0.75 | HAH1G13N | 0.45 | SOD1K92E/R85E | 0.67 |
| TTHAE57Q | 0.30 | TTHAE56K | 0.53 | HAH1G13N | 0.48 | SOD1R85E | 0.38 |
| TTHAL41S | 0.44 | TTHAE36k. | 0.32 | HAH1G14H | 0.73 | SOD1K92E | 0.22 |
| TTHAY60W | 0.41 | TTHAE36K | 0.18 | HAH1G14H | 0.52 | SOD1K92E | 0.16 |
| TTHAY60W | 0.39 | TTHAE7K | 0.50 | HAH1P2A | 0.52 | SOD1K92S | 0.15 |
| TTHAY60W | 0.33 | TTHAE7K | 0.30 | HAH1P2A | 0.67 | SOD1K23S | 0.70 |
| TTHAY60K | 0.22 | TTHAE39K | 0.29 | HAH1L65A/L67A | 0.52 | SOD1K23S | 0.46 |
| TTHAY60K | 0.19 | TTHAE39K | 0.29 | HAH1L65A/L67A | 1.15 | SOD1K30S | 0.34 |
| TTHAL65S/A66S | 0.40 | TTHAE63K | 0.37 | HAH1L65S/L67S | 1.01 | SOD1K30S | 0.44 |
| TTHAL65S/A66S | 0.36 | TTHAE63K | 0.43 | HAH1L65S/L67S | 0.44 | SOD1K36E | 0.34 |
| TTHAL41A | 0.16 | TTHAK30E/K61E | 0.15 | HAH1R21E/K30E | 0.56 | SOD1R100S | 0.34 |
| TTHAL41A | 0.27 | TTHAK30E/K61E | 0.23 | HAH1R21E/K30E | 0.43 | SOD1K9E/K36E | 0.43 |
| TTHAL65A | 0.44 | TTHAK5E | 0.49 | HAH1R21E/K30E | 1.17 | SOD1R85S | 0.56 |
| TTHAL65A | 0.16 | TTHAK5E | 0.30 | HAH1R21E/K38E | 0.70 | SOD1R85S | 0.29 |
| TTHAK37E | 0.32 | TTHAK30E | 0.41 | HAH1R21E/K38E | 1.14 | SOD1K92E/R100E | 0.19 |
| TTHAK37E | 0.42 | TTHAK30E | 0.25 | HAH1S63N | 0.50 | SOD1K3S | 0.34 |
| TTHAK24E | 0.36 | TTHAY60W/E57Q | 0.68 | HAH1K3E | 0.06 | SOD1K61E | 0.30 |
| TTHAK24E | 0.38 | TTHAY60W/E57Q | 0.48 | HAH1K60Y | 0.45 | SOD1K92S/R100S | 0.29 |
| TTHAK24E | 0.33 | TTHAY60W/E57Q | 0.48 | HAH1P36E | 0.59 | SOD1K9S | 0.41 |
| TTHAK3E | 0.41 | TTHAY60W/E58K | 0.19 | HAH1P36E | 0.55 | SOD1K9S | 0.70 |
| TTHAK3E | 0.59 | TTHAY60W/E56Q | 0.41 | HAH1S63E | 0.57 | SOD1K36S | 0.42 |
| TTHAK3E | 0.49 | TTHAE56Q/E58Q | 0.62 | HAH1S63E | 0.94 | SOD1V5E/V7E | 0.19 |
| TTHAK20E | 0.36 | TTHAS34I | 0.87 | HAH1S7E | 1.05 | SOD1Q15E/N19E | 0.24 |
| TTHAK20E | 0.22 | TTHAN12I | 0.87 | HAH1S7E | 0.31 | SOD1N56E/T58E | 0.22 |
| TTHAK23E | 0.29 | TTHAT19I | 0.41 | HAH1S47D | 0.35 | SOD1L101E/V105E | 0.38 |
| TTHAK23E | 0.41 | TTHAQ53I | 0.51 | HAH1S47D | 0.35 | SOD1S34N | 0.22 |
| TTHAK23E | 0.26 | TTHAT10I | 0.47 | HAH1K57E | 0.41 | SOD1K36E/K61E | 0.19 |
| TTHAM16E | 0.44 | TTHAT45I | 0.76 | HAH1K57E | 0.53 | SOD1K92E/R100S | 0.17 |
| TTHAM16E | 0.30 | TTHAE57K | 0.70 | HAH1K57E | 0.14 | SOD1K92E/R85S | 0.09 |
| TTHAT45E | 0.51 | TTHAE58Q | 0.46 | HAH1K56E | 0.41 | SOD1K9S/K36E | 0.08 |
| TTHAT45E | 0.46 | TTHAS34N | 0.37 | HAH1K56E | 0.62 | SOD1N19S | 0.23 |
| TTHAE58K | 0.29 | TTHAE56Q | 0.29 | HAH1K56E | 0.70 | SOD1K23E | 0.40 |
| TTHAE58K | 0.25 | TTHAL41S/L65S/A66S | 0.43 | HAH1K57E/K56E | 0.33 | SOD1K30E | 0.39 |
| TTHAE58K | 0.42 | TTHAL41S/L65S/A66S | 0.35 | HAH1K57E/K56E | 0.42 | SOD1V64A | 0.30 |
| TTHAE7S | 0.39 | TTHAE56K/E57K/E58T | 0.65 | HAH1K60E | 0.49 | SOD1K61S | 0.12 |
| TTHAE7S | 0.18 | TTHAE56K/E57K/E58T | 0.08 | HAH1K60E | 0.73 | SOD1K9E | 0.33 |
| TTHAY60W/E57K | 0.26 | HAH1PWT | 0.37 | HAH1K38E | 0.86 | SOD1I17A | 0.42 |
| TTHAY60W/E57K | 0.24 | HAH1PWT | 0.22 | HAH1R21E | 0.82 | SOD1V5AV7A | 0.22 |
| TTHAE32K | 0.32 | HAH1P36E/K38E | 0.63 | HAH1K30E | 0.61 | SOD1N26S | 0.39 |
| TTHAE32K | 0.45 | HAH1P36E/K38E | 0.91 | HAH1K30E | 0.73 | SOD1K92S/R85E | 0.93 |
| TTHAD47K | 0.43 | HAH1K60E/S63E | 0.52 | HAH1K25E | 0.41 | SOD1V64E/S68E | 1.79 |
| TTHAD47K | 0.25 | HAH1R21E/K25E | 0.95 | HAH1K25E | 0.67 | SOD1T86E/V88E | 0.14 |
| TTHAD47K | 0.34 | HAH1R21E/K25E | 0.71 | HAH1S7N | 0.66 | SOD1N56S | 0.27 |
| TTHAE29K | 0.24 | HAH1K60E/K56E | 0.79 | HAH1E68Q | 1.06 | SOD1S25N | 0.36 |
| TTHAE29K | 0.52 | HAH1K60E/K56E | 0.48 | HAH1E5K | 0.56 | SOD1V88A | 0.22 |
| TTHAK61E | 0.26 | HAH1K60E/K57E | 0.77 | HAH1L65S/G66S/L67S | 0.91 | SOD1Q15E/I17E/N19E | 0.97 |
| TTHAK61E | 0.24 | HAH1K60E/K57E | 0.83 | SOD1barrel | 0.43 |
Protein concentrations ([protein]lysate) were determined from the NMR signal intensity of the decrowded sample lysates (lysatesupernatant), using standard NMR signal intensities at [protein] = 100 μM as references. The intracellular protein concentrations were subsequently estimated from [protein]in cell = [protein]lysate/φ, where φ ≈ 0.29 is the E. coli volume fraction of the NMR sample (samplein cell).
Results
Bacterial and Human Homologs Respond Differently to the E. coli Cytoplasm.
For reference, we acquired first the in-cell NMR spectra of the pseudo-wild-type variants of bacterial TTHA (TTHAPWT), human HAH1 (HAH1PWT), and the human SOD1 barrel (SOD1barrel) (Fig. 1). Analysis shows that E. coli cells expressing bacterial TTHAPWT (samplein cell) display a highly dispersed Heteronuclear Multiple Quantum Coherence (HMQC) spectrum with well-resolved cross-peaks, characteristic for a rapidly tumbling and fully folded structure (Fig. 2). Subsequent tests of leakage (samplesupernatant) reveal typically very low amounts of labeled material in the intervening medium (SI Controls, Table S1, and Fig. S2), indicating that the signal indeed stems from protein in the cytoplasm (Fig. 2). Finally, the supernatant of the lysed cells (lysatesupernatant) yields a spectrum analogous to that in samplein cell, but with a slight sharpening of the HMQC cross-peaks (Fig. 2). The lysatesupernatant spectrum is further indistinguishable from that obtained in pure buffer, showing that the low levels of endogenous E. coli proteins remaining in the lysate supernatant (∼10 mg/mL; SI Controls and Fig. S1) have negligible impact on the analysis: lysatesupernatant mimics a pure-buffer control. Taken together, these results show that the E. coli cytoplasm has limited, but yet measurable, impact on the dynamic behavior of bacterial TTHAPWT and that these restrictions ease upon cell lysis. With the human analog HAH1PWT, however, the situation is distinctly different. The in-cell spectrum of this protein is broadened out to the extent that it completely disappears, using the same data contour level settings as for TTHAPWT. What remain visible in samplein cell are only the distinct cross-peaks of the background of labeled metabolites that always accumulate during overexpression in labeled growth medium (Fig. 2). The cause of such global line broadening for a globular protein is normally decreased rotational mobility (33–35). Conspicuously, the folded HAH1PWT spectrum subsequently restores to high resolution upon cell lysis in lysatesupernatant, indicating that the protein is just reversibly restricted by the cytoplasm and readily sets free upon cell lysis (Fig. 2). We observe the very same behavior for the human SOD1barrel (Fig. 2): Although the protein is previously found to move freely in human cells (5, 31), it seems to stick in the E. coli cytoplasm.
Fig. 2.
In-cell HMQC spectra showing that the bacterial TTHAPWT moves relatively unrestricted in the E. coli cytoplasm, whereas the human HAH1PWT and SOD1barrel get restricted by intracellular interactions, i.e., the protein motions become significantly retarded. The behavior, however, is readily reversed by point mutation of surface charges. (A) The well-resolved in-cell spectrum of TTHAPWT indicates rapid motion in the cytoplasm, whereas the broadened-out spectra of HAH1PWT and SOD1barrel show that the in-cell motions are restricted (samplein cell, intact cells). The cross-peaks of the latter spectra are largely from the background of small metabolites. (B) Corresponding spectra after cell lysis, showing that all three proteins move freely upon removal of the cytoplasmic restrictions (lysatesupernatant, decrowded lysates). (C) The in-cell motions respond readily to mutation of surface charge, allowing reversal of the pseudo-wild-type behavior. Reduction of negative charge causes the TTHAE58K spectrum to broaden out. Conversely, addition of negative charge brings the spectra of HAH1K58E and SOD1R100E to high resolution.
Fig. S2.
(A−C) Representative NMR spectra of (A) TTHAPWT, (B) HAH1PWT, and (C) SOD1barrel used for determining mobilityin cell and leakage. The reduction in tumbling freedom was quantified by comparing the signal intensity from the samplein cell and the lysatesupernatant sample, and the leakage was quantified by the intensity of the samplesupernatant. (D) Overlaid lysate spectra obtained by sonication alone (red) and by chemical treatment with Triton X in combination with sonication (blue). The almost identical spectra show that sonication alone results in near-complete release of cytosolic soluble proteins. (E−G) Representative HMQC spectra for (E) TTHAPWT, (F) HAH1R21E, and (G) SOD1R100E. The spectra show single folded states in both the samplein cell (colored spectra) and the lysatesupernatant (black). (H and I) Protein stability was further confirmed by chevron analysis for the pseudo-wild-type variants and selected surface mutants. (J) The line broadening, quantified by mobilityin cell, shows no systematic variation along the sequence or projected on the structure (not shown). The homogenous structural distribution suggests that the line broadening mainly stems from reduction in rotational correlation time. The dotted line corresponds to the determined mobilityin cell value (Table S1).
In-Cell NMR Spectra Can Be Made to Appear and Vanish by Single-Point Mutations.
The question is then, what allows TTHAPWT to tumble relatively freely in the E. coli interior whereas HAH1PWT and SOD1barrel seem to get stuck? Because the effect unlikely arises from specific binding to biological partners but rather from diffuse quinary interactions with the molecular background (5, 9), we start by investigating the role of protein charge (21). As illustrated in Fig. 3, the electrostatic potentials of the TTHAPWT, HAH1PWT, and SOD1barrel surfaces are quite different. First out was the mutation HAH1K57E, which replaces the positive surface charge K57 of the human protein with the negative counterpart E57 of the bacterial TTHAPWT. The mutation has negligible impact on protein folding and stability (SI Controls and Fig. S2), but it increases the formal net charge of HAH1 by −2. As a result, the in-cell cross-peaks of HAH1 sharpen up radically and reveal a high-resolution HMQC spectrum of the folded state, analogous to that of TTHAPWT (Fig. 2). Cytoplasmic localization was verified by the samplesupernatant, which revealed no trace of leakage (Table S1). Whatever dynamic restrictions are experienced by the pseudo-wild-type protein in the E. coli cytoplasm appear to be largely counteracted by the point mutation K57E. Similarly, the mutation R100E induces distinct cross-peak sharpening of SOD1barrel, albeit that the spectral restoration is not as prominent as observed for HAH1K57E (Fig. 2). Boosting protein’s net-negative charge seems thus to be an efficient means to increase cytoplasmic mobility and rescue poor in-cell spectra. As proof of principle, we finally designed a protein to stick to the E. coli interior by decreasing the formal net charge of TTHAPWT with the construct TTHAE58K. The result is a nearly complete loss of the in-cell NMR signal (Table S1 and Fig. 2).
Fig. 3.

The electrostatic potentials of the TTHA, HAH1, and SOD1barrel surfaces as calculated by the software PyMol. Despite the bacterial TTHA and human HAH1 being homologous, their electrostatic surface patterns are very different. The projections of the structures to the left follow those in Fig. 1.
Strategy for Quantification of In-Cell Mobility.
To establish whether the ordered response to net charge alterations indeed relates to unspecific electrostatic repulsion, or stems from serendipitous obstruction of functional binding sites, we set out to quantify the effect. Under the present conditions, the NMR line width () scales with the protein correlation time (τc), which, in turn, is mainly determined by three components,
| [1] |
where τr is the rotational correlation time, τl is the local dynamics correlation time, and τexch is the correlation time of any putative chemical exchange. Because the protein variants in this study are fully folded and maintain, in all cases, fixed 3D structures in the E. coli cytoplasm (SI Controls, Table S3, and Fig. S2), the contribution from local motions (τl) is negligible (36). Likewise, the uniform distribution of the line-broadening effect seen for all amino acids of the proteins (SI Controls and Fig. S2) indicates that the change in chemical exchange (τexch) upon transfer into the cells is small, as this would preferentially affect moieties on the protein surfaces. Hence, the line-broadening effect is global, i.e., the relative change in line width is similar for all cross-peaks. Consequently, the main modulator of NMR line width () can here be ascribed to the rotation correlation time (τr), in accordance with previous in-cell NMR studies (37, 38),
| [2] |
where VH is the hydrodynamic volume and η is the apparent viscosity. Experimentally, we determined the change in (Eqs. 1 and 2) upon transfer into the cytoplasm from the ratio of the cross-peak heights (h) in the lysatesupernatant and the samplein cell spectra according to
| [3] |
using the property that hin cell/hlysate = when protein concentration is kept constant. For simplicity, we refer tentatively to as change in intracellular ‘mobility’ (37). A value of = 1 means that the in-cell mobility is similar to that in pure water, i.e., = 4 ns for TTHA (28) and HAH1 (29) and 7 ns for SOD1barrel (32), whereas = 0 means that the in-cell cross-peaks have broadened beyond detection (i.e., 60 ns).
Table S3.
Kinetic parameters and protein stabilities from in vitro controls
| Protein | * | mf,* M−1 | * | mu,* M−1 | † | ‡ kcal/mol |
| TTHAPWT | 1.11 ± 0.02 | −0.97 ± 0.02 | −3.33 ± 0.04 | 0.61 ± 0.01 | 4.44 ± 0.06 | 6.04 ± 0.08 |
| TTHAE57K | 1.06 ± 0.03 | −0.87 ± 0.01 | −1.96 ± 0.02 | 0.59 ± 0.01 | 3.02 ± 0.05 | 4.11 ± 0.07 |
| HAH1PWT | 2.68 ± 0.03 | −0.49 ± 0.02 | 0.27 ± 0.05 | 0.22 ± 0.01 | 2.41 ± 0.08 | 3.28 ± 0.11 |
| HAH1K57E | 2.56 ± 0.02 | −0.54 ± 0.01 | 0.51 ± 0.03 | 0.16 ± 0 | 2.05 ± 0.05 | 2.79 ± 0.07 |
| SOD1barrel | −0.28 ± 0.03 | −0.78 ± 0.01 | −4.03 ± 0.04 | 0.27 ± 0.01 | 3.75 ± 0.07 | 5.1 ± 0.1 |
Quantification of Protein Net Charge.
Because estimates of residue charge from model compound values fail to account for shifts in protonation state of titratable groups (i.e., shifted pKa values), induced by folding or mutation (39), we opted for analysis by electrophoretic mobility (40). Following standard protocols, each protein construct i (Table S1) was run on a native gel along with the TTHAPWT reference (SI Materials and Methods and Fig. S4). The measured mobility ratio () relates here to the protein net charge ratio () according to (40)
| [4] |
where Z is the protein net charge, E is the applied electrical field, is the friction coefficient, and is the hydrodynamic radius = , where is the gel density. To obtain proportionality with in-cell interaction, was converted to solvent-accessible surface area (SASA = 2), yielding a dimensionless measure of the surface charge density ratio () between construct i and the TTHAPWT reference,
| [5] |
where is the observed gel mobility ratio (Eq. 5) and is the radius-to-area conversion factor. The latter was determined by diffusion NMR, yielding = 1, 0.98, and 0.79 for TTHAPWT, HAH1PWT, and SOD1barrel, respectively (Fig. S4 and Table S4). Because some proteins in Table S1 tend to lose net negative charge at pH values below 7, thus halting or reversing their electrophoretic mobility, the native gels had to be run according to standard protocols at pH 8.3 (SI Materials and Methods), where all constructs remain net negative. As a control, the eletrophoretically determined values of Zi/Zref agree well with those calculated from model compound pKA values (Fig. S4). Finally, to account also for the partial histidine protonation observed under intracellular conditions at pH 6.5 to 6.7 (SI Controls), we added the chargedensity offsets 0.13, 0.25, and 0.32 for TTHA, HAH1, and SOD1barrel, respectively (SI Materials and Methods). Although this minor correction falls largely within the data scatter, it is added for completeness of analysis.
Fig. S4.
(A) Native gel migration profiles, showing the variation in migration length due to single surface mutations. Surface charge density (Eq. 5) is determined by dividing the migration length of the mutant (Rfi) to the reference molecule TTHAPWT (Rfref, lane 2), according to Eq. 4. (B) Native gel migration was measured for each protein variant in lysatesupernatant, and comparison of migration in lysate and pure buffer shows no significant difference. The results show that, even in a more complex solution, the native gel mainly reports on surface charge density and hydrodynamic radius of the target proteins. (C−E) Signal attenuation profiles from PFG NMR diffusion determination of translational diffusion coefficient for (C) TTHAPWT, (D) HAH1PWT, and (E) SOD1barrel. Data for SOD1barrel are from ref. 32. All experiments were performed at 298 K in 100% D2O. The fitted lines are least square fits for the Stejskal−Tanner equation with correction for nonlinear gradient field (66). (F) Plot of size-corrected electrophoretic mobility ( × rHi/rHref = ) vs. net charge calculated from model compound pKA values (Zi). (G and H) Docking of isobutene to the surface of SOD1barrel. The simulation shows a substantial reorganization of the protein surface layer upon binding the isobutene moiety.
Table S4.
Hydrodynamic dimensions of TTHAPWT, HAH1PWT, and SOD1barrel determined by PFG NMR diffusion measurements
In-Cell Mobility Shows Common Dependence on Net Charge Density.
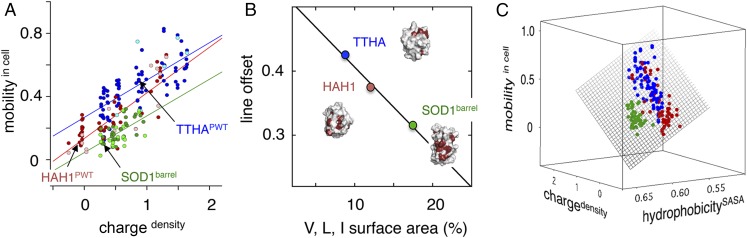
Analysis of a comprehensive set of mutations, including alterations of both surface charge and hydrophobicity (Table S1), shows that the bacterial protein TTHA undergoes a progressive increase of in-cell mobility upon increasing the net-negative charge (Fig. 4). Despite considerable scatter in the vs. plot (Eqs. 3 and 5), the result is statistically significant with R = 0.73. A similarly scattered dependence on is found for the surface mutations of the human homolog HAH1 (R = 0.84), but, in this case, the plot is overall offset to lower values (Table S1 and Fig. 4). The trend is completed by the surface mutations of SOD1barrel, adding a third—again seemingly parallel—data set below that of HAH1 (R = 0.61) (Table S1 and Fig. 4). On the whole, the results are consistent with net-negative repulsion being a fundamental factor in modulating in-cell solubility and protein−protein interactions (20–22). However, the discrete offsets in the vs. plots in Fig. 4 indicate that there are more factors than surface net charge at play in controlling the in-cell mobility. Contributions from such additional factors are also emphasized by the conspicuous data scatter, which is clearly outside the errors of the individual measurements (Fig. 4). The data spread for the whole data set as measured by linear-fit deviation per point, i.e., the residual sum squared (RSS), is RSS = 0.015 (SI Controls, Fig. S3, and Table S5).
Fig. 4.
First step toward quantification of mutational effects on in-cell motion. (A) The observed in-cell motions (; Eq. 3) display an overall scattered increase upon mutational increase of the negative charge density (; Eq. 5). Even so, the three proteins show differences by being offset in the plot. Given the relatively small measurement errors, this finding indicates that other factors than surface charge density are at play in controlling the in-cell motion. Notably, the experiments with indicated leakage (lightly colored markers) do not stand out in the correlations, suggesting that this leakage occurs during cell handling after in-cell detection (SI Controls). (B) Plot of in-cell motion vs. exposure of the hydrophobic side chains V, L, and I. The simplistic estimate of surface hydrophobicity accounts for the fitted line offsets in A at = 1. (C) Plot with refined estimate of hydrophobic surface area-based energy minimization of the mutant structures (hydrophobicitySASA; Eq. 6). The plot includes the entire data set in Table S1 and is extended to three dimensions, i.e., vs. [; hydrophobicitySASA]. Upon accounting for the surface hydrophobicity variation, the data in A convert to a well-defined plane. Although the fit is overall improved, the parameter hydrophobicitySASA captures poorly the details of individual point mutations because of considerable local surface rearrangements in the energy minimizations.
Fig. S3.
(A−D) Results and residuals from fits of data to linear or planar models. (A and B) Linear fit to the complete set of mobilityin cell vs. chargedensity data shows a strong systematic deviation in the residuals. In particular, the systematic deviation is between the three proteins, and only HAH1 data fall onto the line. (C) Increasing the dimensionality to three dimensions by adding hydrophilicityGuy as a parameter improves the fit significantly, whereas (D) adding ||p|| as an independent parameter in 4D only marginally improves the fit. (E) The corresponding distributions of residuals shown together with the best Gaussian fit, to highlight the narrowing in distribution widths by extending the fit to higher dimensions. (F−I) Morphological effects of E. coli induced by of overexpression. Transmission electron micrographs of E. coli cells overexpressing (F) TTHAPWT, (G) HAH1PWT, and (H) SOD1barrel, compared with (I) nonoverexpressing cells. The general morphology is very similar for all overexpressing bacteria, and only very small differences in morphology are seen between the overexpressing and nonoverexpressing cells.
Table S5.
Residual squared sum from data fits to line and expanded plane models
| Independent variables | RSSall* | RSSall/point | RSSsub† | RSSsub/point |
| chargedensity | 2.98 | 0.015 | 2.14 | 0.014 |
| [chargedensity; hydrophobicitySASA] | 2.24 | 0.012 | 1.75 | 0.011 |
| [chargedensity; hydrophilicityGuy] | 2.10 | 0.011 | 1.45 | 0.009 |
| [chargedensity; hydrophilicityGuy; ||p||] | 1.79 | 0.009 | 1.25 | 0.008 |
All data points are included, n = 193.
Corresponds to the subset with very low leakage, n = 155.
Notes on Variability, Protein Leakage, and Self-Interaction.
As a measure of experimental variability, the data set in Fig. 4 contains—without any exclusions—all measurements performed, including the subset where the supernatant controls revealed some extent of leakage. The latter constitute 38 of a total of 203 experiments, and, on average, the indicated leakage was 16% (Table S1). To single out these experiments, the statistics of the analysis are given both for the total data set (RSSall) and for the data set where the leakage set is excluded (RSSsub), where the unit is in “per point,” i.e., RSS total/number of data points. As seen below, however, the difference between RSSall and RSSsub is very small (Table S5). The explanation seems to be that the detected leakage is generally not present during the determination of in samplein cell, but occurs during the handling of fatigued cells in the supernatant preparation. In support of this possibility, (i) the supernatant sample in some cases show distinct narrow cross-peaks that are missing in samplein cell, (ii) repeated measurements with varying degrees of leakage yield still small variations in , and (iii) the samplein cell signal is, in all cases, stable over the acquisition time and not gradually increasing as would be expected from on-going leakage (Fig. S1). Second, to examine whether the results show any dependence on expression level, we plotted mobilityin cell and deviation from fit vs. intracellular concentration of overexpressed protein (Fig. S1). The controls reveal no self-interaction between the overexpressed proteins, i.e., the mobilityin cell appears to stem from interaction with endogenous cellular components.
Surface Hydrophobicity: A Matter of Depth.
As second modulator of in-cell mobility, we examined hydrophobicity. The trend is evident already at the level of the TTHAPWT, HAH1PWT, and SOD1barrel structures, where the hydrophobic surface area as measured simply by exposure of V, L, and I methyl groups sums up to 8%, 12%, and 17%, respectively (Fig. 4). Moreover, the plot of vs. this fractional exposure seems to account for the protein-specific offsets in the vs. data (Fig. 4). To quantify the effect more strictly, we estimated, for each mutant, the change in hydrophobic exposure from energy minimizations of the TTHAPWT, HAH1PWT, and SOD1barrel structures (SI Materials and Methods) according to
| [6] |
where is the hydrophobic SASA and is the total SASA (SI Materials and Methods). The resulting plot of vs. [; hydrophobicitySASA] reveals a plane that captures the in-cell motion for all three proteins (Fig. 4). In essence, the offsets in the vs. plots (Fig. 4) are orderly displaced in the added hydrophobicitySASA dimension, with an accompanying reduction of the scalar-fit deviations from RSSall = 0.015 and RSSsub = 0.014 to RSSall = 0.012 and RSSsub = 0.011 (Table S5). When it comes to the detailed effects of point mutations, however, the hydrophobicitySASA measure appears relatively insensitive. Although we at mutant level generally discern decreased in-cell mobility upon increasing side-chain hydrophobicity, the resulting change in hydrophobicitySASA can go either way because of local repacking (Table S1). Analogous surface adjustment is often seen upon binding of designed ligands (41, 42) and is here illustrated computationally by docking isobutane (valine mimic) to the SOD1barrel: The ligand becomes almost entirely engrossed in the surface, driven by contacts that are buried in the crystal structure (Fig. S4). In an attempt to quantify this dynamically accessible surface area (DASA), we assumed crudely that all residues that are not buried in the protein’s hydrophobic core are free to interact with the cellular environment upon dynamic rearrangement (Fig. 5). Second, we assigned a hydrophilicity to each of the residues in the DASA layer according to their Guy solvation energies (43), (Table S1). In essence, is a statistical term describing the depth distribution of the different residues in protein crystal structures, which also correlates with the residue water-to-octanol partition free energies (43). Normalization to surface area was finally obtained by dividing the sum of the Guy solvation energies with the number of residues in the DASA layer (nDASA) construct according to
| [7] |
Interestingly, the plot vs. [; hydrophilicityGuy] yields an alternative plane with RSSall = 0.011 and RSSsub = 0.009 (Fig. 6 and Table S5), even though the values of hydrophilicityGuy and hydrophobicitySASA are themselves poorly correlated (R = −0.32; Table S1). Consistently, the RSS deviations decrease yet another step to RSSall = RSSsub = 0.009 upon including both parameters in the fit. Although hydrophobicitySASA and hydrophilicityGuy have similar weight in improving the overall correlation, the latter captures better the effects of point mutation. In the hydrophilicityGuy plot, the individual mutants of TTHA and HAH1 spread out and mix across the plane (Fig. 6 and Movie S1). Whether this indicates that the protein surfaces are indeed plastic by exposing buried material upon in-cell encounters, or simply reflects that the Guy potential—by including also the solvation energies of polar and charged residues—captures properties complementary to hydrophobicitySASA remains to be found out. Even so, the very convergence of experimental data from three unrelated proteins into a single plane shows that the quinary in-cell interactions—despite their detailed complexity—follow common and simplistic rules, governed by the proteins’ macroscopic surface properties (Fig. 6). Moreover, the data in Fig. 6 indicate that the cellular components responsible for the in-cell retardation carry repulsive negative charge and attractive surface hydrophobicity. Species matching these characteristics are, e.g., other proteins, membranes, and nucleic acid material.
Fig. 5.
Alternative method for determining the exposure of hydrophobic surface area. To account for the notion that protein surfaces are not perfectly “fixed” but, to some extent, adjust upon contact with external ligands (41, 42), we assumed that all residues outside the hydrophobic cores (transparent structural regions) are free to interact with the environment via dynamic rearrangement. The atoms in this dynamic layer define the DASA, which is considerably more extensive than the crystallographic SASA. For quantification and comparison with in-cell mobility data, we finally derived the Guy solvation energies (43) of each residue in the DASA layers to obtain the macroscopic parameter hydrophilicityGuy (Eq. 7). The final relation between in-cell motion and hydrophilicityGuy is shown in Fig. 6 and Movie S1.
Fig. 6.
The high-dimensional relation between in-cell motion and the proteins’ macroscopic surface properties. Wild-type proteins are denoted by ■ (black solid squares), pseudo-wild-type variants are denoted by • (black solid circles), and mutants are denoted by ❍ (colored solid circles), color-coded as in previous figures. TTHA variants are in blue, HAH1 variants are in red, and SOD1barrel variants are in green. Data are from Table S1. Rotations of plots are shown in Movies S1 and S2. (A) A 3D plot of the experimentally measured (Eq. 3) vs. the surface net charge density (; Eq. 5) and the surface hydrophilicity (hydrophilicityGuy; Eq. 7). (B) Corresponding plot of (Eq. 3) vs. the surface net charge density (; Eq. 5) and the electric dipole moment (||p||; Eq. 8). The correlations in A and B call attention to the strikingly systematic response of the quinary in-cell interactions to point mutation. Also, the correlations appear generic because they capture all protein constructs, including those of different evolutionary origin.
Enhancing the Precision One Step Further: The Protein Dipole Moment.
In some analogy with frustrated spin-glass systems (44), charged proteins possess electric dipole moments that are expected to influence their motions under crowded in-cell conditions. Upon mutation of surface charges, these dipole moments undergo distinct changes with possible impact on (Eq. 3). The magnitude of the dipole moment (||p||) for each protein construct in Table S1 was calculated from the charge positions in the PDB structures of TTHAPWT, HAH1PWT, and SOD1barrel according to (45)
| [8] |
where qi is the elemental charge at position ri, and r is the protein center of mass, treating the R/K and E/D side chains as positive and negative point charges, respectively. The histidines were omitted from the calculation because their pKA values are not yet known. Conveniently, because ||p|| is a general vector in space, it does not need normalization to protein surface area to scale with in-cell interaction, as required for the protein net charge Z (Eq. 5) and the Guy solvation energies (Eq. 7). The impact of ||p|| on in-cell motions emerges clearly in a plot of vs. [; ||p||], which defines a plane (RSSall = 0.010 and RSSsub = 0.009) similar to that of vs. [; hydrophilicityGuy] (Fig. 6 and Movie S2). In other words, the dipolar interactions that influence in-cell protein motion are on par with hydrophilicityGuy. Upon final extension of the correlation to four dimensions, i.e., vs. [; hydrophilicityGuy; ||p||], the scalar fit deviations decrease just slightly to RSSall = 0.009 and RSSsub = 0.008, indicating that we have reached the limit of the data scatter (Fig. S3 and Table S5).
Discussion
The Nature of In-Cell Encounters.
Although the question of how a given protein senses and moves in the intracellular compartment of E. coli may at first seem intricate, our results indicate that this process conforms to simplistic physical−chemical rules. Measured by the rotational freedom (Eq. 3), the intracellular motions () of 127 mutant constructs fall coarsely on a common plane in the protein property space (Fig. 6). The parameters of this property space are, moreover, easy to deduce for any protein with known structure, i.e., the surface net charge density (; Eq. 5), the surface hydrophilicity (hydrophilicityGuy; Eq. 7), and the charge dipole moment (||p||; Eq. 8). Out of these properties, the net charge density is the most decisive (SI Controls), consistent with findings in earlier in-cell NMR studies (38, 46) and the notion that net charge repulsion generically assures dispersion of the cellular components (21, 47, 48). The lack of correlation between the intracellular concentration of overexpressed protein (1.6 ± 0.8 mM; SI Materials and Methods) and (Fig. S1) indicates further that the dynamic changes are not from self-interactions but stem from quinary interactions with other cytoplasmic components. Because the in-cell measurements only report on the specific isotope-labeled construct being overexpressed, however, it is not yet possible to determine which components of the E. coli cytoplasm cause the in-cell retardation; it could be neighboring soluble proteins, cytoskeleton structures, proteomembrane surfaces, nucleotide material (49), or any species presenting repulsive negative charge and attractive surface hydrophobicity. In any case, the overall morphology of the bacteria is similar when overexpressing the different proteins (SI Controls and Fig. S3), suggesting that the intracellular environment remains unperturbed. However, it is apparent, from the NMR spectral features, that the underlying quinary encounters are short-lived and diffusive, in agreement with previous observations (5, 37, 38, 50). As the correlations in Fig. 6 concur with the average properties of the mutated proteins, we further conclude that the motions are mainly controlled nonspecifically by diffusive encounters with the “bulk” cellular background, i.e., the quinary crosstalk (15). By no means does this exclude that the data in Fig. 6 also involve in-cell interactions that rely on the site-specific details of the protein surfaces, but, as yet, these seem to be within the data scatter.
Implications for Functional Optimization and Protein Design.
In view of the rapid divergence of surface composition in protein evolution (51), it is interesting that the human HAH1PWT and SOD1barrel, which are seen to tumble freely in mammalian cells (31, 52), tend to get stuck in the E. coli cytoplasm (Fig. 2). The bacterial protein TTHAPWT, on the other hand, yields high-resolution HMQC spectra in E. coli from the very start (Fig. 2). Whether these differences in cytoplasmic mobility indeed stem from evolutionary divergence, i.e., mammalian proteins are not fit for E. coli, or simply reflect the natural span of intracellular mobilities is not yet clear. Even so, it is evident from the ease with which our model proteins can be moved across the mobility span (Fig. 6) that the quinary crosstalk is readily open to optimization. One conceivable gain of such optimization is in molecular search (12). At one extreme, very long-lived encounter complexes will increase the chance of two functional partners finding their right orientations by Brownian surface diffusion (3), but also slow down the shift to alternative partners when the match is wrong. Encounter complexes that are very swift will conversely allow probing of more putative partners, but with the risk that correct fits are missed by premature dissociation. Regardless of what the answer may be, the quinary interactions stand out as a significant part of the intracellular crosstalk and provide also the ever-present background for in vivo function (10). It is further evident from the results in Figs. 2–5 and previous studies (9, 46) that the quinary interactions, despite their complexity, at some level follow simplistic macroscopic rules. As such, the correlations in Fig. 6 not only add physical−chemical detail to the effects of intracellular crowding (4, 12, 27, 53) but also provide a tool for rational protein surface design. Applications can include optimization of target proteins for in-cell NMR detection (54), surface optimization of protein therapeutics (55), and mutational examination of the yet poorly understood relation between protein motion, spatial localization, and function (7, 56, 57). The message stands clear and simple to test: Can any protein be tuned to desired rotational motion in E. coli. by mutational tweaking of surface net charge density (Eq. 5), surface hydrophilicity (Eqs. 6 and 7), and the electric dipole moment (Eq. 8)?
Materials and Methods
Protein Engineering.
Mutagenesis, expression, and purification of HAH1, TTHA, SOD1barrel, and their variants were as in refs. 5 and 32, where the encoding genes were subcloned into the vector pET3a (GenScript) (SI Materials and Methods).
Overexpression and In-Cell NMR Sample Preparation.
The proteins (Table S1 and Fig. S1) were produced and measured in E. coli cells. Before induction, BL21(DE3)pLysS cells (Thermo Fisher Scientific) were grown overnight at 37 °C in 200 mL of LB. The cells were then harvested and resuspended in M9 medium with labeled 15NH4Cl as sole nitrogen source, and overexpression was induced for 4 h (SI Materials and Methods and Fig. S1). The culture was then nondisruptively centrifuged at 800 × g for 8 min to obtain a pellet of intact, viable cells. One gram of this cell pellet was dissolved in 1 mL of M9 buffer, transferred to an NMR tube, and subjected to in-cell spectral analysis (SI Materials and Methods). All experiments were performed at 37 °C unless otherwise stated.
NMR Spectroscopy.
NMR experiments were performed on a Bruker Avance 500- or 700-MHz spectrometer equipped with a triple-resonance cryogenically cooled probe head. In-cell and in vitro spectra were obtained by 1D and 2D 1H-[15N]-band-selective optimized flip-angle short-transient heteronuclear multiple quantum coherence (1H-[15N]-SOFAST HMQC) pulse schemes (58, 59) with 32 scans, 64 increments for the 2D experiment and 1,024 scans for 1D experiment. The relaxation delay was set to 0.2 s, and the acquisition time was to 40 ms. Hydrodynamic radii were determined by pulsed field gradient (PFG) NMR diffusion experiments, where the z-gradient strength was calibrated by the known diffusion coefficient of α-cyclodextrin and HDO in 99.6% D2O (60) (SI Materials and Methods).
Mobility Determination on Native Gel.
To quantify net charge density, 2 μL to 5 μL of lysate samples of the various protein variants (Table S1) were loaded on precasted gels (Bio-Rad), with TTHAPWT as references () and HAH1PWT as reference control. For the SOD1barrel variants, we used, additionally, SOD1barrel for benchmarking/verification of normalization. Running time was 90 min to 210 min, at 125 V and 4 °C, and the running buffer was 25 mM Tris plus 192 mM Glycine at pH 8.3 (SI Materials and Methods).
SI Materials and Methods
Mutagenesis, Expression, and Purification.
Mutagenesis, expression, and purification of HAH1, TTHA, SOD1barrel, and their variants were as in refs. 5 and 32, where the encoding genes were subcloned into the vector pET3a (GenScript). To remove functional binding of metals and prevent disulphide linking, the involved cysteine ligands were replaced by serine. For TTHA, the replacements were C11S/C14S, and, for HAH1, they were C12S/C15S/C41S. The resulting pseudo-wild-type constructs are denoted HAH1PWT and TTHAPWT.
NMR Sample Preparations and Experimental Conditions.
Before induction, the E. coli BL21 (DE3) pLysS cells (Thermo Fisher Scientific) were grown overnight at 37 °C in 200 mL of LB medium in the presence of 100 μg/mL of carbenicillin and 34 μg/mL of chloramphenicol. The cells were then harvested by centrifugation (8 min at 800 × g) and resuspended in M9 medium to OD600 ≈ 1.5 with labeled 15NH4Cl as sole nitrogen source, and protein expression was induced by 0.5 mM IPTG. All experiments were performed in M9 buffer at 37 °C unless otherwise stated.
Intracellular Concentration of Overexpressed Proteins.
The cytoplasmic concentration of 15N-labeled protein ([protein]in cell) following 4 h of overexpression was estimated as follows. First, the protein concentration in the NMR detection volume ([protein]lysate) was determined from the peak intensities of decrowded cell lysate (lysatesupernatant), using reference spectra obtained at fixed concentrations of [protein] = 100 μM. Subsequently, the intracellular protein concentration was calculated from [protein]in cell = [protein]lysate/φ, where φ ≈ 0.29 is the E. coli volume fraction of samplein cell. The volume fraction φ ≈ 0.29 was calculated from the concentration of cells in the samplein cell, derived from the number of cells in 1 g of cell pellet using optical density at 600 nm (OD600) ≈ 109 cells per milliliter per OD (58), and the volume of the individual E. coli cells, i.e., 1 μm3 per cell (59). Based on the [protein]lysate values in Table S2, the [protein]in cell were estimated to 1.6 ± 0.8 mM (Fig. S1E) for the various overexpressed constructs in this study. There is no correlation between [protein]in cell and mobilityin cell (Fig. S1 F and G).
Mobility Determination on Native Gel.
To quantify net charge density, 2 μL to 5 μL of lysate samples of the various protein variants (Table S1) were loaded on precasted gels (Bio-Rad), with TTHAPWT as references () and HAH1PWT as reference control. For the SOD1 variants, we used, additionally, SOD1barrel for benchmarking/verification of normalization. Running time was 90 min to 210 min, at 125 V and 4 °C, and the running buffer was 25 mM Tris plus 192 mM Glycine at pH 8.3. After staining and destaining, the mobility in millimeters of each protein variant (Rf) was determined and normalized according to Fig. S4 A and B and Eq. 4. The vertical electrophoresis cell was from Bio-Rad, and the power supply was from Thermo Fisher Scientific.
Correction for Partial Histidine Protonation.
Because the electrophoretic mobility at pH 8.3 (; Fig. S4A) does not report on the differences in histidine charge between TTHAPWT, HAH1PWT, and SOD1barrel under intracellular conditions at pH 6.5 to 6.7 (SI Controls), these were accounted for separately as follows. From the partial histidine protonation observed by E. coli in-cell NMR (SI Controls) and the structures in Fig. S1A, we estimate the total histidine charge (Histot) to be +0.5 for TTHA (H13), +1 for HAH1 (H4, H46), and +2 for SOD1barrel (H46, H48, H80, H90). These Histot values can, in turn, be translated to Δchargedensity for each protein according to
| [S1] |
where = −0.26 is the slope of the linear fit in Fig. S4F, and is the surface normalization by protein’s hydrodynamic radius (Fig. S4 C−E and Table S4). The values of Δchargedensity are −0.13, −0.25, and −0.32 for TTHA, HAH1, and SOD1, respectively, and have been used as offsets for the chargedensity (Eq. 5) throughout the manuscript.
Folding Kinetics.
Measurements of folding kinetics were as described in references (60, 61). In short, guanidinium chloride (AppliChem) and urea (MP Biomedicals) of ultrapure grade was used for protein denaturation. All kinetic measurements were performed in 20 mM pH 6.3 Mes buffer. The final concentration of SOD1barrel was 4 μM, whereas, for TTHAPWT, TTHAE57K, HAH1PWT, and HAH1K57E, it was increased to 20 μM to allow for tyrosine fluorescence detection. The observed relaxation rates, kobs, were fitted by standard procedures according to (62)
| [S2] |
where, kf and ku are the refolding and unfolding rate constants, respectively, and are their values at [denaturant] = 0 M, and mf and mu are the slopes used for extrapolation. Data analysis was with KaleidaGraph (Synergy Software).
NMR Spectroscopy.
NMR experiments were performed on a Bruker Avance 500- or 700-MHz spectrometer equipped with a triple-resonance cryogenically cooled probe head. The in-cell, as well as the in vitro, experiments were obtained using 1D and 2D HMQC pulse schemes (63, 64) with 32 scans, 64 increments for the 2D experiment and 1,024 scans for the 1D experiment. The relaxation delay was set to 0.2 s, and the acquisition time was set to 40 ms. Line broadening was measured from the sum of peak heights for resonances between 8.6 ppm and 9.4 ppm, and a normalized value of the in-cell retardation was obtained from the ratio of the peak heights in the lysate supernatant and the in-cell samples according to Eq. 3.
The hydrodynamic radii of TTHAPWT and HAH1PWT were determined by PFG NMR diffusion experiments, where the z-gradient strength was calibrated by the known diffusion coefficient of α-cyclodextrin and HDO in 99.6% D2O (65). The diffusion experiments were conducted by altering the effective gradient strength from 0.5 G/cm to 25.5 G/cm in 16 steps, and the attenuation signal was fitted to a modified Stejskal−Tanner equation (66) to account for nonlinearity of the gradient field.
Transmission Electron Microscopy.
The bacterial cells were fixed in 2.5% glutaraldehyde and 1% paraformaldehyde in 0.1 M phosphate buffer at pH 7.4, and incubated overnight at 4 °C. After centrifugation, the pellet was rinsed in 0.1 M phosphate buffer at pH 7.4, followed by postfixation in 2% osmium tetroxide in 0.1 M phosphate buffer at pH 7.4 for 2 h at 4 °C. The sample was then dehydrated in ethanol, followed by acetone, and finally embedded in LX-112 (Ladd). Ultrathin sections (∼50 nm to 60 nm) were cut by a Leica ultracut UCT (Leica). Sections were contrasted with uranyl acetate followed by lead citrate and examined in a Hitachi HT 7700 at 80 kV. Digital images were taken with a Veleta camera (Olympus Soft Imaging Solutions, GmbH). The transmission electron microscopy was performed at the Electron Microscopy Unit at Karolinska University Hospital, in Sweden.
Calculation of HydrophobicitySASA.
Hydrophobic SASA were calculated for each protein construct using the Parameter Optimised Surfaces (POPS) method (67). The values presented are the mean values calculated over 20 mutant structures. To obtain the mutant structures, we used the program Eris (68, 69). In each round of the Eris calculation, 20 structures of the native and mutant proteins were produced. Each structure was the result of 20 steps of Monte Carlo simulation repacking the side chains within 10 Å of the mutation site. From the 20 mutant and 20 native structures, the program also computed the mean change in free energy between the mutant and native structures using the Medusa force field (70). For each mutation, we performed 20 independent rounds of Eris calculations. We selected the round with the lowest mean change in free energy between mutant and native proteins. The 20 mutant structures within this round of the Eris calculation were used to calculate the solvent accessible surface areas, and in Eq. 6.
Docking of Isobutane to the SOD1 Surface.
The docking analysis was performed using MedusaDock, a docking program that simultaneously models the flexibility of the receptor and ligand (71). We performed 200 independent docking calculations and selected, finally, the structure with the lowest energy as evaluated by the Medusa force field (70) (Fig. S4 G and H).
SI Controls
Expression Levels, Cytoplasmic Protein Concentration, and Signal Stability over Time.
E. coli BL21 (DE3) pLysS cells were used as overexpression system. This strain has minimal background leakage expression (72), enabling us to keep the levels of nonlabeled protein low. After 4 h of expression, the cytoplasmic concentration of 15N-labeled protein was estimated to be 1.6 ± 0.8 mM (Fig. S1E) from the number of E. coli cells [OD600 ≈ 109 cells per milliliter per OD (58)], the E. coli volume [∼1 μm3 per cell (59)], and the peak intensity of the lysate NMR spectra, using reference spectra with known concentrations. We find no correlation (R = 0.22) between expression level and observed mobilityin cell (Fig. S1F), or between data deviation from the fitted plane and expression level (Fig. S2G and Table S2). The stability of the in-cell signal was verified by comparing the 1D HMQC spectrum recorded upon initiation of the experiments with that recorded at the end. In all cases, identical spectra suggested stable conditions (Fig. S1 H−J).
Cytoplasmic Environment Controls.
Some of the macroscopic properties seen to affect quinary interactions, e.g., surface charge density, the electric dipole moment, and hydrophilicityGuy, are intrinsically dependent on pH (46). Following the methodology in refs. 5 and 31, i.e., by comparing the in-cell 1H chemical shifts of TTHAPWT with in vitro NMR spectra at series of known pHs, we determined the pH of E. coli cytoplasm to be 6.7 ± 0.1. Moreover, in terms of chemical shifts, the in-cell spectra are superimposable with those of the lysate supernatants at pH = 6.5.
The observed retardation of molecular tumbling in the E. coli cytoplasm (mobilityin cell) is assumed to report on the surface properties of the proteins. However, it is conceivable that overexpression-induced differences in the cytoplasmic environment can also contribute to the observed effects. To pinpoint such putative morphological differences between E. coli overexpressing TTHAPWT, HAH1PWT, and SOD1barrel, we examined the bacterial interior by transmission electron microscopy. Bacterial cells overexpressing either TTHAPWT, HAH1PWT, or SOD1barrel were prepared using the same protocol as for the NMR experiments. As a control, we included cells without overexpressing vector. The bacteria were fixated and contrast enhanced as described in SI Materials and Methods. All preparations showed nondisrupted cells with little variation in morphologies (Fig. S3 G−I). The cells displayed, generally, signs of chromatin assemblies, but without significant differences between the samples. The control bacteria with no overexpression, however, displayed slightly less condensed interior, whereas the number of ribosomes seemed up-regulated in the overexpressing cells (Fig. S3 G−I). On this basis, we conclude that the overall morphology of the cytoplasmic compartment remains similar across the experiments and is unlikely to explain the observed differences in mobilityin cell.
Protein Leakage Controls.
To assure that the NMR signals stem from the overexpressed 15N-labeled proteins in the E. coli cytoplasm, rather than from proteins that have leaked out into the intervening medium during sample preparation, handling, or acquisition, leakage controls were conducted. Each in-cell NMR sample was collected and gently centrifuged after the NMR experiment to obtain a cell-free supernatant of the intervening medium (samplesupernatant). Representative 1D HMQC leakage spectra of TTHAPWT, HAH1PWT, and SOD1barrel are shown in Fig. S2 A−C. In all cases, the spectra of the samplesupernatant indicate small, but detectable, amounts of protein leakage (Fig. S2 A−C and Table S1). Data from samples with excessive leakage (>25%) were omitted from the analysis, and data from samples with protein leakage of >10% are annotated in boldface in Table S1. The latter data points are omitted in the subset denoted “RSSsub,” but are kept in the full analysis denoted “RSSall.” Interestingly, the data points from samples with indicated leakage do not systematically differ from those with a low degree of leakage (Figs. 4 and 6). Generally, protein leakage is expected to result in systematically too high values of mobilityin cell, because proteins that are freely moving outside the cells contribute to increase the NMR peak heights. Also, the level of leaked protein is expected to increase during the cause of the acquisition. As neither of these effects can be distinguished in the data sets, we conclude that the detected leakage occurs after the in-cell acquisition, i.e., during the preparation of the supernatant sample as a result of disturbing the analysis-stressed cells by pipetting, transfer, and centrifugation.
Protein Stability Controls.
The assumption that changes in line width reflect retardation of rotational motion is only valid for folded proteins where the transversal relaxation is dominated by global Brownian motions, rather than local side-chain dynamics (33–35). This assumption holds only for rigid, folded proteins (36). To assure that the proteins are indeed fully folded during analysis, the thermodynamic stabilities of all studied variants were validated in both the cytosol and the cell lysates (5, 13, 73). First, the in vitro stabilities of the pseudo-wild-type proteins were determined by standard chevron data analysis, verifying that all three proteins are fully folded and show archetypical two-state behavior (Table S3 and Fig. S2 H and I). Chevron analysis was also used to verify folded structures of the TTHA and HAH1 mutations with the largest changes in mobilityin cell (Fig. S2 H and I). For the rest of the 135 protein variants, the thermodynamic stability was verified directly from their respective in-cell NMR spectra (95% of which were 2D), where even small populations of unfolded material would be identified (Fig. S2 E−G). For eight variants of the SOD1barrel, however, the stability was affected to such an extent that the unfolded state was significantly populated, and these were omitted from the data set. The omitted variants were SOD1Q15E/I17E/N19E, SOD1K9E/R85E/K92E, SOD1R85E/K92E/K61E, SOD1K9E/R85E/K92E/K61E/R100E, SOD1K9E/K61E/R85E/K92E/K36E/R100E, SOD1K9E/K23E/K36E/K61E/R85E/K92E/R100E, SOD1K9E/K23E/K30E/K36E/K61E/R85E/K92E/R100E, and SOD1K3E/K9E/K23E/K30E/K36E/K61E/R85E/K92E/R100E.
Line-Broadening Controls.
Line broadening in NMR is not always due to retardation in global rotational dynamics; it can also result from chemical exchange on intermediate time scales (74). If such chemical exchange were to be the main cause of the in-cell line broadening, it would be biased to surface-exposed residues, as all of the variants in this study are fully folded in the E. coli cytoplasm (see above). The observed line broadening, however, is relatively uniformly distributed along the protein sequence, involving both the core and surface-exposed residues (Fig. S2J); this indicates that chemical exchange is not a major contributor to the in-cell line broadening.
Lysis Efficiency Control.
To further validate that the NMR signal stems from cytoplasmic protein localization, we measured the NMR signal recovery following cell lysis by sonication (10 × 19 s, 1-s pulses, 40% amplitude). After cell lysis, we removed large portions of the cellular debris by centrifugation, keeping the total soluble protein concentration constant. This sample is denoted lysatesupernatant and was analyzed with the same experimental setup as for the samplein cell. As the lysatesupernatant sample is used for quantification of the line broadening, it is crucial that the lysing protocol result in near-complete release of all cytosolic overexpressed protein. To test the lysate preparation protocol, we used Triton X-100 to further enhance cell disintegration. The samplein cell was divided into two equal parts, where one was treated with 1% (wt/wt) Triton X-100 to chemically disrupt the cell membrane (75), and the other was handled according to the original protocol. No difference in NMR signal intensity or spectral appearance of the lysatesupernatant from the two preparations was observed (Fig. S2D), suggesting that the sonication protocol results in near-full recovery of the cytosolic soluble protein material.
“Crowding Level” of the Lysate Supernatant.
As cell lysis releases not only the target proteins but the entire E. coli content, it needs to be established that the line width of the lysatesupernatant sample is not skewed by quinary interactions with coreleased proteins. Accordingly, the concentration of endogenous E. coli proteins in the lysatesupernatant sample was determined to be 10.3 ± 1 mg/mL (76), using the Bicinchoninic acid assay kit (Sigma-Aldrich). The lysate supernatant is thus much more diluted than the >300 mg/mL E. coli cytoplasm (77) and should have negligible effect on the NMR line widths. To more directly test possible lysate effects on the NMR line broadening, we also compared the lysatesupernatant spectrum with a spectrum recorded in dilute buffer (Fig. S1 B−D). No significant difference in line widths or spectral appearance was observed, indicating that neither interactions with endogenous protein nor self-interaction with the overexpressed protein affect the lysate supernatant sample line width.
Native Gel Mobility Control.
To validate that the native gel mobility is not skewed by cellular contaminants, but mainly reflects the properties of the studied proteins, we compared native gel mobility of TTHAPWT, HAH1PWT, and SOD1barrel in E. coli lysate and in aqueous solution. The experiment reports on whether the intrinsic properties of the analyzed proteins, i.e., hydrodynamic radius and surface charge density, are the main determinant of gel mobility, or if interactions with other, comigrating lysate components interfere with the mobility. The results show that proteins have indistinguishable mobility in aqueous solution and in E. coli lysate (Table S1 and Fig. S4B), indicating that the lysate supernatant sample yields unbiased results.
Standardized Regression Coefficients.
Although the protein properties chargedensity, hydrophilicityGuy, hydrophobicitySASA, and ||p|| correlate orderly with the mobilityin cell, their relative importance/weight is difficult to quantify in a nonbiased way. To get an indication of the response on mobilityin cell by a given perturbation, we normalized the parameters as (78)
| [S3] |
where χi is any parameter i, <> is the average value, and σi is the SD of the parameter. Then, by fitting a hyperdimensional plane to the observed mobilityin cell using the normalized parameters as
| [S4] |
we obtain αi = δmobilityin cell /δχinorm, which gives an indication of the response on a small perturbation in the dimension i. Using this approach, we determined αi for all four parameters. The results show that chargedensity has the strongest impact, with αcharge = 0.11 ± 0.008, and the other independent parameters are αGuy = 0.03 ± 0.008, αSASA = −0.06 ± 0.008, and αdipole = −0.05 ± 0.007. We thus conclude that the chargedensity is the main effector on the cytoplasmic mobility.
Supplementary Material
Acknowledgments
We thank Håkan Wennerström for valuable discussions. We thank Kjell Hultenby (Karolinska University Hospital at Huddinge) for help with the transmission electron microscopy. Support was provided by the Swedish Research Council, Hjärnfonden, the Knut and Alice Wallenberg Foundation, the Magnus Bergwall Foundation, the Bertil Hållsten Foundation, and by National Institutes of Health Grant R01GM080742 (to N.V.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621227114/-/DCSupplemental.
References
- 1.Moreira IS, Fernandes PA, Ramos MJ. Hot spots—A review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber G, Keating AE. Protein binding specificity versus promiscuity. Curr Opin Struct Biol. 2011;21:50–61. doi: 10.1016/j.sbi.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 4.Ross JL. The dark matter of biology. Biophys J. 2016;111:909–916. doi: 10.1016/j.bpj.2016.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danielsson J, et al. Thermodynamics of protein destabilization in live cells. Proc Natl Acad Sci USA. 2015;112:12402–12407. doi: 10.1073/pnas.1511308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarrinpar A, Park SH, Lim WA. Optimization of specificity in a cellular protein interaction network by negative selection. Nature. 2003;426:676–680. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- 7.French JB, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science. 2016;351:733–737. doi: 10.1126/science.aac6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLOS Comput Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith AE, Zhou LZ, Gorensek AH, Senske M, Pielak GJ. In-cell thermodynamics and a new role for protein surfaces. Proc Natl Acad Sci USA. 2016;113:1725–1730. doi: 10.1073/pnas.1518620113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharyya S, et al. Transient protein-protein interactions perturb E. coli metabolome and cause gene dosage toxicity. eLife. 2016;5:e20309. doi: 10.7554/eLife.20309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConkey EH. Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc Natl Acad Sci USA. 1982;79:3236–3240. doi: 10.1073/pnas.79.10.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mika JT, Poolman B. Macromolecule diffusion and confinement in prokaryotic cells. Curr Opin Biotechnol. 2011;22:117–126. doi: 10.1016/j.copbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Dhar A, et al. Protein stability and folding kinetics in the nucleus and endoplasmic reticulum of eucaryotic cells. Biophys J. 2011;101:421–430. doi: 10.1016/j.bpj.2011.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo M, Xu Y, Gruebele M. Temperature dependence of protein folding kinetics in living cells. Proc Natl Acad Sci USA. 2012;109:17863–17867. doi: 10.1073/pnas.1201797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monteith WB, Cohen RD, Smith AE, Guzman-Cisneros E, Pielak GJ. Quinary structure modulates protein stability in cells. Proc Natl Acad Sci USA. 2015;112:1739–1742. doi: 10.1073/pnas.1417415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ignatova Z, et al. From the test tube to the cell: Exploring the folding and aggregation of a beta-clam protein. Biopolymers. 2007;88:157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inomata K, et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar M, Smith AE, Pielak GJ. Impact of reconstituted cytosol on protein stability. Proc Natl Acad Sci USA. 2013;110:19342–19347. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman I, Gelman H, Tai J, Gruebele M. The extracellular protein VlsE is destabilized inside cells. J Mol Biol. 2014;426:11–20. doi: 10.1016/j.jmb.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gitlin I, Carbeck JD, Whitesides GM. Why are proteins charged? Networks of charge-charge interactions in proteins measured by charge ladders and capillary electrophoresis. Angew Chem Int Ed Engl. 2006;45:3022–3060. doi: 10.1002/anie.200502530. [DOI] [PubMed] [Google Scholar]
- 21.Kurnik M, Hedberg L, Danielsson J, Oliveberg M. Folding without charges. Proc Natl Acad Sci USA. 2012;109:5705–5710. doi: 10.1073/pnas.1118640109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson O. Encyclopedia of Metalloproteins. Springer; New York: 2013. Cellular electrolyte metabolism; pp. 580–587. [Google Scholar]
- 23.Ghaemmaghami S, Oas TG. Quantitative protein stability measurement in vivo. Nat Struct Biol. 2001;8:879–882. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 24.Monteith WB, Pielak GJ. Residue level quantification of protein stability in living cells. Proc Natl Acad Sci USA. 2014;111:11335–11340. doi: 10.1073/pnas.1406845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhar A, Ebbinghaus S, Shen Z, Mishra T, Gruebele M. The diffusion coefficient for PGK folding in eukaryotic cells. Biophys J. 2010;99:L69–L71. doi: 10.1016/j.bpj.2010.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AE, Zhang Z, Pielak GJ, Li C. NMR studies of protein folding and binding in cells and cell-like environments. Curr Opin Struct Biol. 2015;30:7–16. doi: 10.1016/j.sbi.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Rivas G, Minton AP. Macromolecular crowding in vitro, in vivo, and in between. Trends Biochem Sci. 2016;41:970–981. doi: 10.1016/j.tibs.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakakibara D, et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature. 2009;458:102–105. doi: 10.1038/nature07814. [DOI] [PubMed] [Google Scholar]
- 29.Anastassopoulou I, et al. Solution structure of the apo and copper(I)-loaded human metallochaperone HAH1. Biochemistry. 2004;43:13046–13053. doi: 10.1021/bi0487591. [DOI] [PubMed] [Google Scholar]
- 30.Danielsson J, et al. Global structural motions from the strain of a single hydrogen bond. Proc Natl Acad Sci USA. 2013;110:3829–3834. doi: 10.1073/pnas.1217306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danielsson J, et al. Pruning the ALS-associated protein SOD1 for in-cell NMR. J Am Chem Soc. 2013;135:10266–10269. doi: 10.1021/ja404425r. [DOI] [PubMed] [Google Scholar]
- 32.Danielsson J, Kurnik M, Lang L, Oliveberg M. Cutting off functional loops from homodimeric enzyme superoxide dismutase 1 (SOD1) leaves monomeric β-barrels. J Biol Chem. 2011;286:33070–33083. doi: 10.1074/jbc.M111.251223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halle B, Wennerström H. Interpretation of magnetic resonance data from water nuclei in heterogeneous systems. J Chem Phys. 1981;75:1928–1943. [Google Scholar]
- 34.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 35.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 36.Lee AL, Wand AJ. Assessing potential bias in the determination of rotational correlation times of proteins by NMR relaxation. J Biomol NMR. 1999;13:101–112. doi: 10.1023/a:1008304220445. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Zhuravleva A, Gierasch LM. Exploring weak, transient protein−protein interactions in crowded in vivo environments by in-cell nuclear magnetic resonance spectroscopy. Biochemistry. 2011;50:9225–9236. doi: 10.1021/bi201287e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbieri L, Luchinat E, Banci L. Protein interaction patterns in different cellular environments are revealed by in-cell NMR. Sci Rep. 2015;5:14456. doi: 10.1038/srep14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveberg M, Arcus VL, Fersht AR. pKA values of carboxyl groups in the native and denatured states of barnase: The pKA values of the denatured state are on average 0.4 units lower than those of model compounds. Biochemistry. 1995;34:9424–9433. doi: 10.1021/bi00029a018. [DOI] [PubMed] [Google Scholar]
- 40.Cantor CR, Schimmel PR. Biophysical Chemistry, Part 2: Techniques for the Study of Biological Structure and Function. W. H. Freeman; New York: 1980. [Google Scholar]
- 41.Teague SJ. Implications of protein flexibility for drug discovery. Nat Rev Drug Discov. 2003;2:527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 42.Tsao D, Diatchenko L, Dokholyan NV. Structural mechanism of S-adenosyl methionine binding to catechol O-methyltransferase. PLoS One. 2011;6:e24287. doi: 10.1371/journal.pone.0024287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guy HR. Amino acid side-chain partition energies and distribution of residues in soluble proteins. Biophys J. 1985;47:61–70. doi: 10.1016/S0006-3495(85)83877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreiro DU, Komives EA, Wolynes PG. Frustration in biomolecules. Q Rev Biophys. 2014;47:285–363. doi: 10.1017/S0033583514000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antosiewicz J. Computation of the dipole moments of proteins. Biophys J. 1995;69:1344–1354. doi: 10.1016/S0006-3495(95)80001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen RD, Pielak GJ. Electrostatic contributions to protein quinary structure. J Am Chem Soc. 2016;138:13139–13142. doi: 10.1021/jacs.6b07323. [DOI] [PubMed] [Google Scholar]
- 47.Lang L, Kurnik M, Danielsson J, Oliveberg M. Fibrillation precursor of superoxide dismutase 1 revealed by gradual tuning of the protein-folding equilibrium. Proc Natl Acad Sci USA. 2012;109:17868–17873. doi: 10.1073/pnas.1201795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vendruscolo M, Dobson CM. Chemical biology: More charges against aggregation. Nature. 2007;449:555. doi: 10.1038/449555a. [DOI] [PubMed] [Google Scholar]
- 49.Frembgen-Kesner T, Elcock AH. Computer simulations of the bacterial cytoplasm. Biophys Rev. 2013;5:109–119. doi: 10.1007/s12551-013-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luh LM, et al. Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition. J Am Chem Soc. 2013;135:13796–13803. doi: 10.1021/ja405244v. [DOI] [PubMed] [Google Scholar]
- 51.Sasidharan R, Chothia C. The selection of acceptable protein mutations. Proc Natl Acad Sci USA. 2007;104:10080–10085. doi: 10.1073/pnas.0703737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banci L, et al. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat Chem Biol. 2013;9:297–299. doi: 10.1038/nchembio.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gnutt D, Ebbinghaus S. The macromolecular crowding effect−From in vitro into the cell. Biol Chem. 2016;397:37–44. doi: 10.1515/hsz-2015-0161. [DOI] [PubMed] [Google Scholar]
- 54.Freedberg DI, Selenko P. Live cell NMR. Annu Rev Biophys. 2014;43:171–192. doi: 10.1146/annurev-biophys-051013-023136. [DOI] [PubMed] [Google Scholar]
- 55.Leader B, Baca QJ, Golan DE. Protein therapeutics: A summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 56.Wirth AJ, Gruebele M. Quinary protein structure and the consequences of crowding in living cells: Leaving the test-tube behind. BioEssays. 2013;35:984–993. doi: 10.1002/bies.201300080. [DOI] [PubMed] [Google Scholar]
- 57.Srere PA. Macromolecular interactions: Tracing the roots. Trends Biochem Sci. 2000;25:150–153. doi: 10.1016/s0968-0004(00)01550-4. [DOI] [PubMed] [Google Scholar]
- 58.Schanda P, Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J Am Chem Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 59.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 60.Danielsson J, Jarvet J, Damberg P, Gräslund A. Two-site binding of beta-cyclodextrin to the Alzheimer Abeta(1-40) peptide measured with combined PFG-NMR diffusion and induced chemical shifts. Biochemistry. 2004;43:6261–6269. doi: 10.1021/bi036254p. [DOI] [PubMed] [Google Scholar]
- 61.Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kubitschek HE, Friske JA. Determination of bacterial cell volume with the Coulter Counter. J Bacteriol. 1986;168:1466–1467. doi: 10.1128/jb.168.3.1466-1467.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: Disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci USA. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: Parallels to precursors in amyloid disease. Proc Natl Acad Sci USA. 2006;103:10218–10223. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fersht AR. Optimization of rates of protein folding: The nucleation−condensation mechanism and its implications. Proc Natl Acad Sci USA. 1995;92:10869–10873. doi: 10.1073/pnas.92.24.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Damberg P, Jarvet J, Gräslund A. Accurate measurement of translational diffusion coefficients: A practical method to account for nonlinear gradients. J Magn Reson. 2001;148:343–348. doi: 10.1006/jmre.2000.2260. [DOI] [PubMed] [Google Scholar]
- 67.Cavallo L, Kleinjung J, Fraternali F. POPS: A fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Res. 2003;31:3364–3366. doi: 10.1093/nar/gkg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin S, Ding F, Dokholyan NV. Eris: An automated estimator of protein stability. Nat Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 69.Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 70.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLOS Comput Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ding F, Yin S, Dokholyan NV. Rapid flexible docking using a stochastic rotamer library of ligands. J Chem Inf Model. 2010;50:1623–1632. doi: 10.1021/ci100218t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abolliel AA, Zedan H. Synthesis, cloning and expression of a novel pre-miniproinsulin analogue gene in Escherichia coli. J Adv Res. 2015;6:663–671. doi: 10.1016/j.jare.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tai J, Dave K, Hahn V, Guzman I, Gruebele M. Subcellular modulation of protein VlsE stability and folding kinetics. FEBS Lett. 2016;590:1409–1416. doi: 10.1002/1873-3468.12193. [DOI] [PubMed] [Google Scholar]
- 74.Matsuo H, et al. Identification by NMR spectroscopy of residues at contact surfaces in large, slowly exchanging macromolecular complexes. J Am Chem Soc. 1999;121:9903–9904. [Google Scholar]
- 75.Cornett JB, Shockman GD. Cellular lysis of Streptococcus faecalis induced with triton X-100. J Bacteriol. 1978;135:153–160. doi: 10.1128/jb.135.1.153-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith PK, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 77.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 78.Bring J. How to standardize regression-coefficients. Am Stat. 1994;48:209–213. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.