Significance
Cellular senescence is important for the maintenance of tissue homeostasis. Dysregulation of senescence is linked to many human diseases, such as cancer, premature aging, and age-related diseases. Although DNA damage response has been linked to senescence, the underlying mechanism is unknown. Here we show that cGAS is essential for the senescence phenotypes, including expression of inflammatory genes. This finding reveals a molecular mechanism of cellular senescence and suggests that modulation of cGAS activity may be a new strategy to treat senescence-associated human diseases that potentially include cancer, neurodegenerative diseases, cardiovascular diseases, and aging.
Keywords: cGAS, senescence, DNA damage, DNA sensing, cancer
Abstract
Cellular senescence is a natural barrier to tumorigenesis and it contributes to the antitumor effects of several therapies, including radiation and chemotherapeutic drugs. Senescence also plays an important role in aging, fibrosis, and tissue repair. The DNA damage response is a key event leading to senescence, which is characterized by the senescence-associated secretory phenotype (SASP) that includes expression of inflammatory cytokines. Here we show that cGMP-AMP (cGAMP) synthase (cGAS), a cytosolic DNA sensor that activates innate immunity, is essential for senescence. Deletion of cGAS accelerated the spontaneous immortalization of mouse embryonic fibroblasts. cGAS deletion also abrogated SASP induced by spontaneous immortalization or DNA damaging agents, including radiation and etoposide. cGAS is localized in the cytoplasm of nondividing cells but enters the nucleus and associates with chromatin DNA during mitosis in proliferating cells. DNA damage leads to accumulation of damaged DNA in cytoplasmic foci that contain cGAS. In human lung adenocarcinoma patients, low expression of cGAS is correlated with poor survival. These results indicate that cGAS mediates cellular senescence and retards immortalization. This is distinct from, and complementary to, the role of cGAS in activating antitumor immunity.
Cellular senescence is a state of irreversible cell cycle arrest triggered by various types of cellular and environmental stress, such as telomere shortening, oncogene activation, and DNA damage (1, 2). Senescence appears to be an antiproliferation process that limits the growth of damaged cells and acts as a potent barrier to tumorigenesis (3, 4). Senescence is characterized by several unique features, including enlarged and flattened cell morphology (5), increased senescence-associated β-galactosidase (SA-β-Gal) activity (6, 7), and in some cell types, a widespread change in chromatin modification, known as senescence-associated heterochromatin foci (SAHF) (7). At the molecular level, the p53-p21WAF1 and pRb-p16INK4a tumor suppressor pathways have been reported to be key mechanisms that control the execution and maintenance of senescence (8, 9). In addition to these cellular features, senescent cells also undergo massive changes in the expression of genes that are thought to affect the tissue microenvironment (5). Senescent cells secrete a variety of soluble factors including inflammatory cytokines, growth factors, and proteases; such senescence-associated secretory phenotype (SASP) is a hallmark of senescence (10–12).
Components of SASP not only serve as a marker of senescence, but also participate in the senescence process (13). Interleukin 6 (IL6) and IL8, two key components of SASP, reinforce the senescence growth arrest in neighboring cells (14, 15). Additionally, these cytokines and other secreted factors attract immune cells, leading to the elimination of senescent cells (16). Given these important functions, SASP is regulated at both transcriptional and epigenetic levels, such as by nuclear factor κB (NF-κB), CCAAT/enhancer-binding protein β (C/EBPβ), bromodomain-containing protein 4 (BRD4), lysine methyltransferase MLL1 and G9A (14, 15, 17–21). Importantly, activation of DNA damage response (DDR) is required for the induction and maintenance of senescence (10). However, the precise regulatory mechanism directly linking the DDR to SASP remains unknown.
cGMP-AMP (cGAMP) synthase (cGAS) is a cytosolic DNA sensor that is activated by binding to double-stranded DNA, including microbial and self-DNA (22, 23). Cytosolic DNA triggers cGAS to produce the second messenger cGAMP (23), which binds and activates the adaptor protein STING (also known as MITA, MPYS, and TMEM173). STING then recruits TANK-binding kinase 1 (TBK1) and IκB kinase to activate IFN regulatory factor 3 (IRF3) and NF-κB, respectively, leading to the production of type I interferons and inflammatory cytokines (22, 23).
Interestingly, DNA damage by irradiation or chemicals such as etoposide or cytosine arabinoside (Ara-C), is linked to type I IFN signaling (24–26). These DNA damaging agents lead to accumulation of cytosolic DNA and activate the IRF3 pathway. Moreover, production of type I interferons by DNA damage leads to amplification of DNA damage response and induces cellular senescence (26). How DNA damage induces type I IFN responses is still not well understood.
In this study, we show that cGAS is essential for cellular senescence. We found that mouse embryonic fibroblasts (MEFs) from cGas−/− mice displayed reduced signs of senescence and underwent faster spontaneous immortalization compared with MEFs from WT mice. Deletion of cGAS in different mouse or human cells abrogated the expression of senescence-associated inflammatory genes in response to DNA damaging agents, including etoposide and ionizing irradiation. Interestingly, we found that cGAS is associated with chromatin during mitosis, suggesting a potential role of cGAS in regulating cell cycle. In response to DNA damaging agents, cGAS and DNA form cytoplasmic foci. Bioinformatics analyses of cGAS expression in human cancer patients revealed that lower expression of cGAS is strongly correlated with decreased survival of lung adenocarcinoma patients.
Results
cGAS Impedes Spontaneous Immortalization of MEFs.
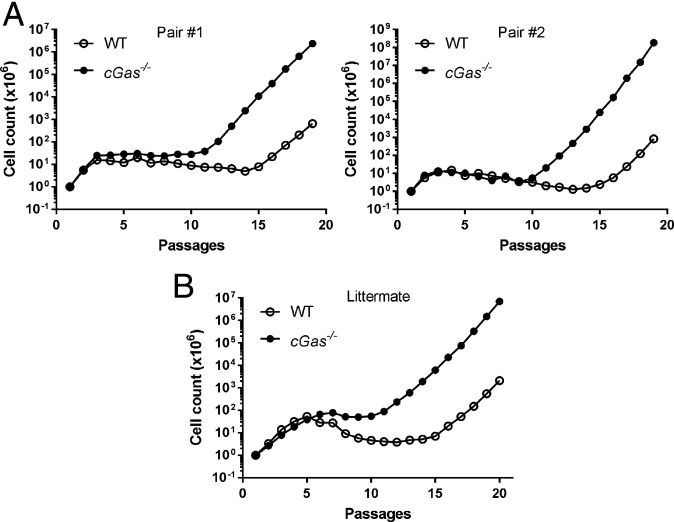
Normal cells divide until they reach a senescence phase as a result of telomere shortening or when cells are under stress (27, 28). Some cells, such as MEFs, can be immortalized (i.e., divide indefinitely) through serial passage of the cells in vitro (29). During this spontaneous immortalization process, all cells enter a senescence phase after several passages and then a few cells grow up and become immortalized (30, 31). In the course of studying cytosolic DNA sensing, we and others previously found that some transformed cells such as HEK293T cells and some immortalized MEF cells lost the ability to induce type I interferons in response to transfected DNA (22, 32, 33). These observations led us to speculate that DNA sensing by cGAS may be a barrier to cellular immortalization or transformation. To test this possibility, we isolated embryos from WT and cGas−/− mice and cultured the embryonic fibroblasts in vitro in atmospheric O2 (20%) through serial passages according to a modified 3T3 protocol (29). The result showed that cGas−/− MEFs became immortalized at passages 11–12, whereas WT MEFs were not immortalized until passages 15–16 (Fig. 1A, Left). Similar results were obtained from the embryos of another pair of WT and cGas−/− mice (Fig. 1A, Right). We also compared the embryos of different genotypes in a cGas+/− mother that was crossed to a cGas+/− male. As shown in Fig. 1B, cGAS KO MEFs became immortalized as early as passage 11, whereas MEFs from the WT littermate embryo did not get immortalized until passage 16. These results indicate that cGAS inhibits spontaneous immortalization of primary MEFs.
Fig. 1.
cGAS deletion accelerates spontaneous immortalization of MEFs. (A) Growth curve of MEFs derived from the embryos of two pairs of WT and cGas−/− mice during the course of 20 serial passages according to a modified 3T3 protocol. (B) Similar to A except that MEFs were from littermate WT and cGas-/− embryos in the same cGas+/− mother that was crossed to a cGas+/− male.
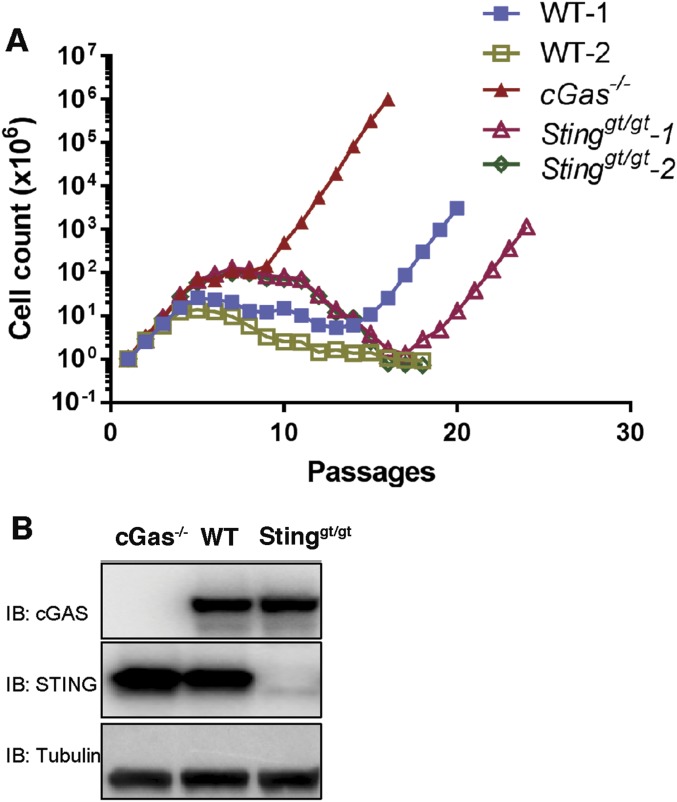
To determine whether STING plays a role in spontaneous immortalization, we obtained embryos from two Sting goldenticket (Stinggt/gt) mice, which harbor a point mutation that confers a null phenotype (34). Surprisingly, unlike MEFs from a cGas−/− mouse that resumed growth at passage 10, MEFs from a Stinggt/gt mouse resumed growth at passage 18, whereas MEFs from another Stinggt/gt mouse never became immortalized (Fig. S1A; the genotypes of the MEF cells were confirmed by immunoblotting shown in Fig. S1B). In parallel experiments, MEFs from a WT mouse resumed growth at around passage 15, but MEFs from another WT mouse never resumed growth. These results suggest that deletion of STING does not accelerate spontaneous immortalization, implying that cGAS may regulate cellular immortalization through a STING-independent mechanism. We therefore focus this study on the role of cGAS in immortalization and senescence.
Fig. S1.
cGas but not Sting deletion accelerates spontaneous immortalization of MEFs. (A) Growth curve of MEFs from the embryos of WT, cGas−/−, and Stinggt/gt mice. (B) Western blotting of cell lysates from indicated MEF cells with cGAS- or STING-specific antibodies.
cGAS Is Required for the Senescence Phenotypes of MEFs During Spontaneous Immortalization.
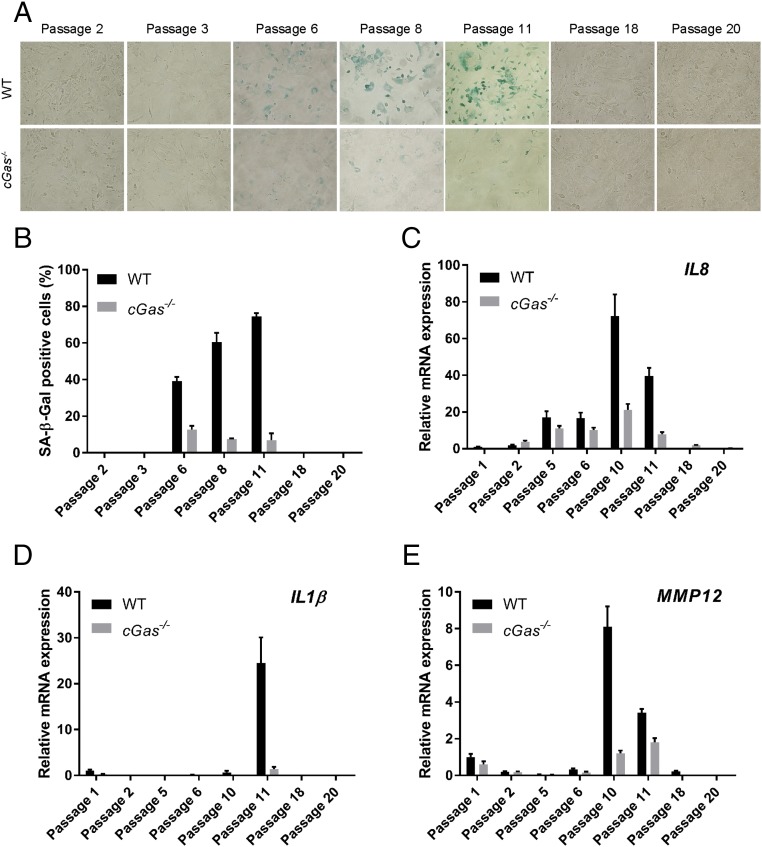
The faster immortalization of cGas−/− MEFs compared with WT MEFs led us to investigate whether cGAS plays a role in the senescence of cells during serial passages. A commonly used method to detect senescence cells is SA-β-Gal staining (28). As shown in Fig. 2A, β-Gal positive cells appeared in passages 6–11 in the culture of WT MEF cells, followed by a decline in later passages, correlating with the growth kinetics of these cells (Fig. 1). In contrast, very few senescent cells were detected in the time course of spontaneous immortalization of cGas−/− MEFs (Fig. 2 A and B).
Fig. 2.
cGAS deletion abrogates the senescence phenotypes of MEFs during spontaneous immortalization. (A) WT and cGas-/− MEFs at indicated passages were analyzed by SA-β-Gal staining. (B) Quantification of the β-Gal positive cells shown in A. (C–E) qRT-PCR analyses of the indicated SASP genes. Error bars indicate SEs of triplicate measurements. All results in this and other figures are representative of at least two independent experiments.
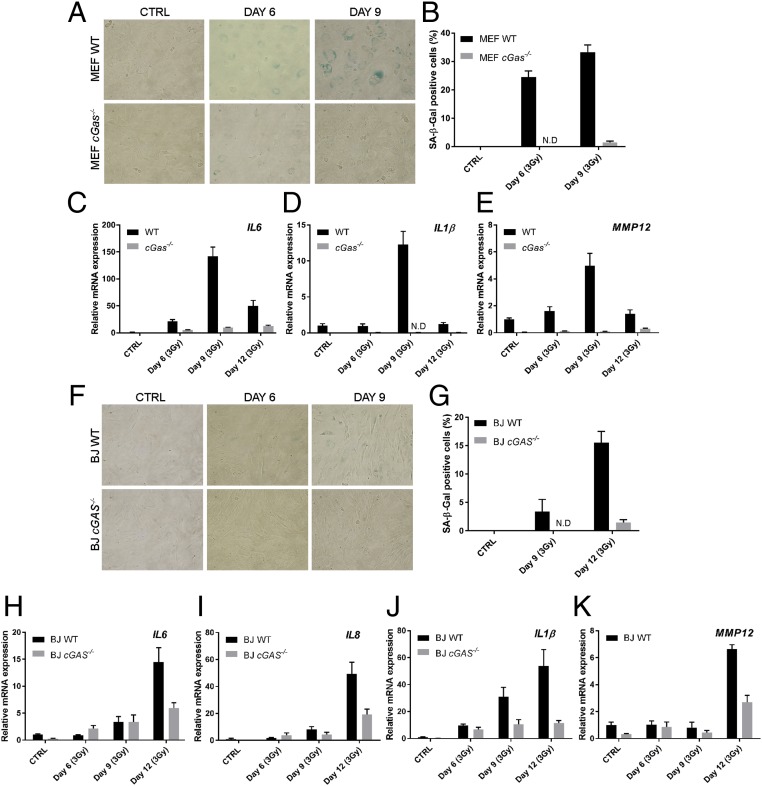
Another hallmark of senescent cells is SASP, which can be assessed by measuring the expression levels of SASP genes, including those involved in inflammation and cell cycle arrest. Quantitative reverse transcription-PCR (qRT-PCR) analysis of three SASP genes, including interleukin 8 (IL8), IL1β, and metalloprotease 12 (MMP12) revealed that these genes were expressed during the middle passages (passages 6–11) of WT MEFs (Fig. 2 C–E), correlating with their senescent states as measured by the β-Gal assay (Fig. 2A). In contrast, the expression of these SASP genes was significantly reduced in cGas−/− MEFs. These results indicate that cGAS is required for MEF cells to acquire the senescence phenotypes during spontaneous immortalization.
cGAS Is Essential for Cellular Senescence Induced by DNA Damage.
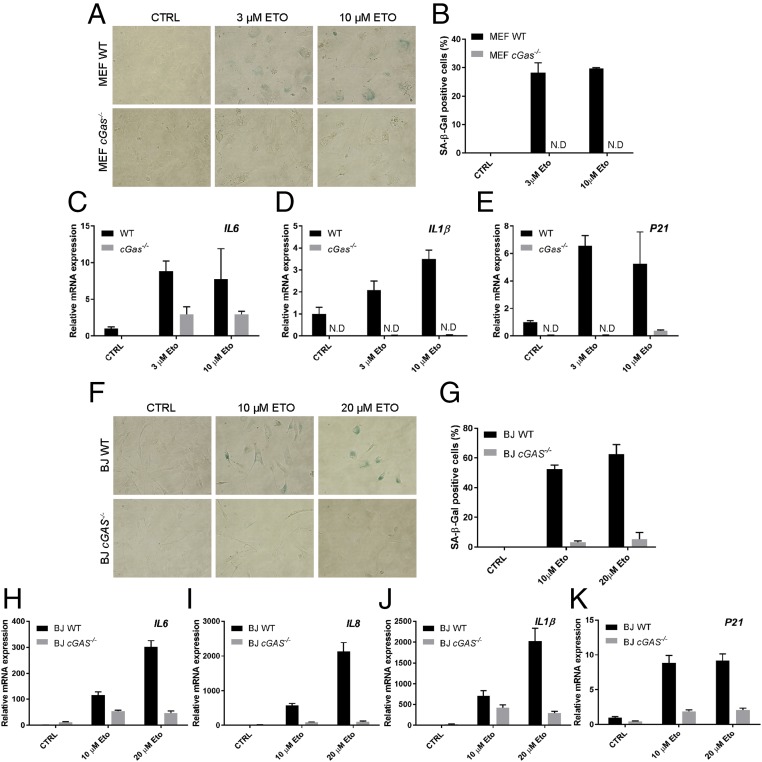
Senescence can be induced by cellular and environmental stress, such as DNA damage. The topoisomerase inhibitor etoposide is a cytotoxic anticancer agent that induces DNA damage and cellular senescence (35, 36). To determine whether cGAS plays a role in etoposide-induced cellular senescence, we treated immortalized MEFs with different concentrations of etoposide and then performed β-Gal staining and SASP gene analyses. About 30% of the WT MEF cells became β-Gal positive on day 6 after treatment with etoposide (Fig. 3 A and B). In contrast, virtually no β-Gal positive cells were detected in cGas−/− MEFs. Consistently, the SASP genes including IL6, IL1β, and the cyclin-dependent kinase (CDK) inhibitor p21 were induced by etoposide in WT, but not cGas−/−, MEF cells (Fig. 3 C–E).
Fig. 3.
cGAS deletion abrogates the senescence phenotypes induced by etoposide. (A and B) WT and cGas−/− MEFs were treated with the indicated concentrations of etoposide (Eto) for 24 h followed by 5 d of culturing in normal media before cells were stained by SA-β-Gal (A) and quantified (B). (C–E) Aliquots of the MEF cells treated with Eto were analyzed for the expression of SASP genes by qRT-PCR. p21 is a cell cycle-dependent kinase inhibitor and a senescence marker. (F–K) Similar to A–E, except that WT and cGas−/− BJ cells were used. Error bars indicate SEs of triplicate measurements. Analyses of additional SASP genes are shown in Fig. S2 H–M. ND, not detectable.
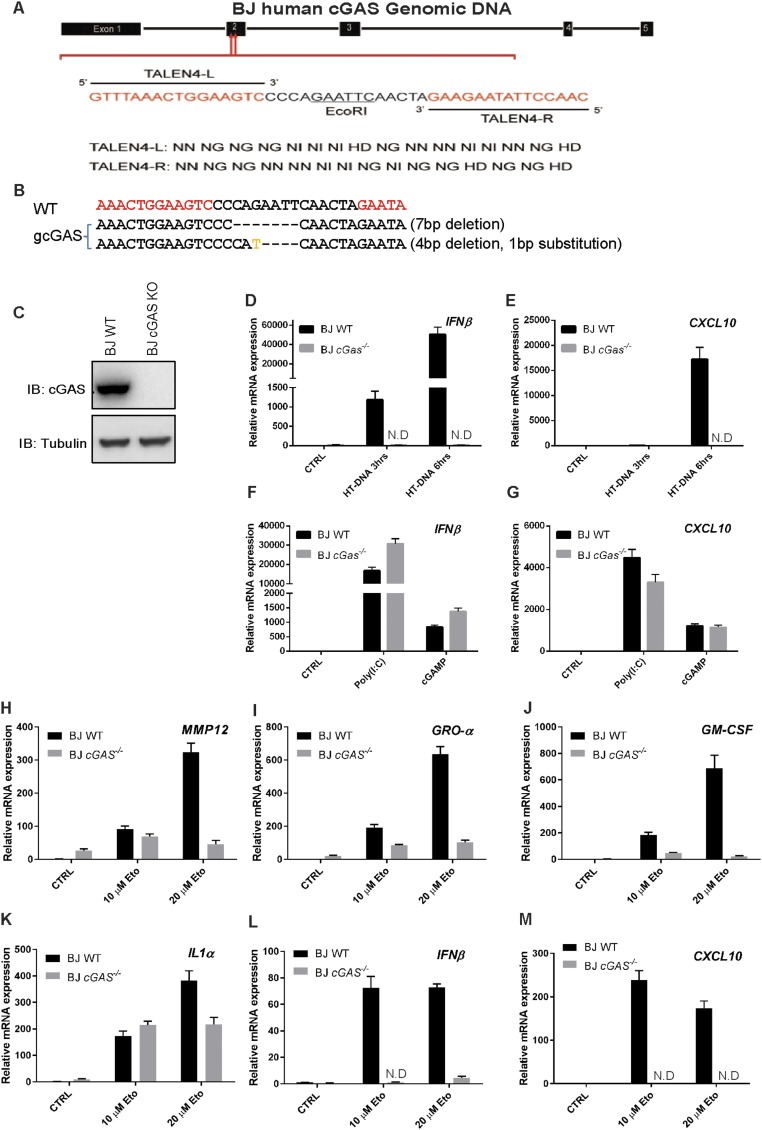
To study senescence in human cells, we used the human fibroblast cell line BJ that has been immortalized with telomerase; this cell line has been used extensively for senescence studies (11, 37). For our studies, we generated a cGas-deficient BJ cell line using the transcription activator-like effector nuclease (TALEN) technology (Fig. S2 A–C). As expected, this cell line failed to induce IFNβ or CXCL10 in response to stimulation by herring testis DNA (HT-DNA) but the induction of these cytokines by poly[I:C] or cGAMP was normal (Fig. S2 D–G). SA-β-Gal staining showed that more than 50% of the WT BJ cells were β-Gal positive after treatment with etoposide, whereas fewer than 5% of the cGas−/− cells became senescent in the same experiment (Fig. 3 F and G). Consistently, expression of SASP genes, including IL6, IL8, IL1β, and p21, was induced by etoposide in WT but not cGas−/− BJ cells (Fig. 3 H–K). We further examined a larger panel of SASP genes, including MMP12, GROα (CXCL1), IL1α, GM-CSF, IFNβ, and CXCL10 (Fig. S2 H–M). With the exception of IL1α, all SASP genes that we have examined depended on cGAS for their expression in response to etoposide.
Fig. S2.
cGAS deletion abrogates etoposide-induced senescence phenotypes of BJ cells. (A) Targeted mutation of exon 2 of human cGAS gene in BJ cells using TALEN technology. (B) Sequence verification of disruption of both alleles of the cGas gene in a BJ clone. (C) Western blotting of extracts of the cGAS−/− BJ cell clone with a human cGAS antibody. (D–G) cGas−/− BJ cells were stimulated with HT-DNA, poly[I:C] or cGAMP as indicated, then total RNA was isolated for qRT-PCR. (H–M) WT and cGAS−/− BJ cells were treated with the indicated concentrations of etoposide (Eto) for 24 h, followed by culturing in normal media for 5 d. Total cellular RNA was prepared from the cells to measure SASP gene expression by qRT-PCR. ND, not detectable.
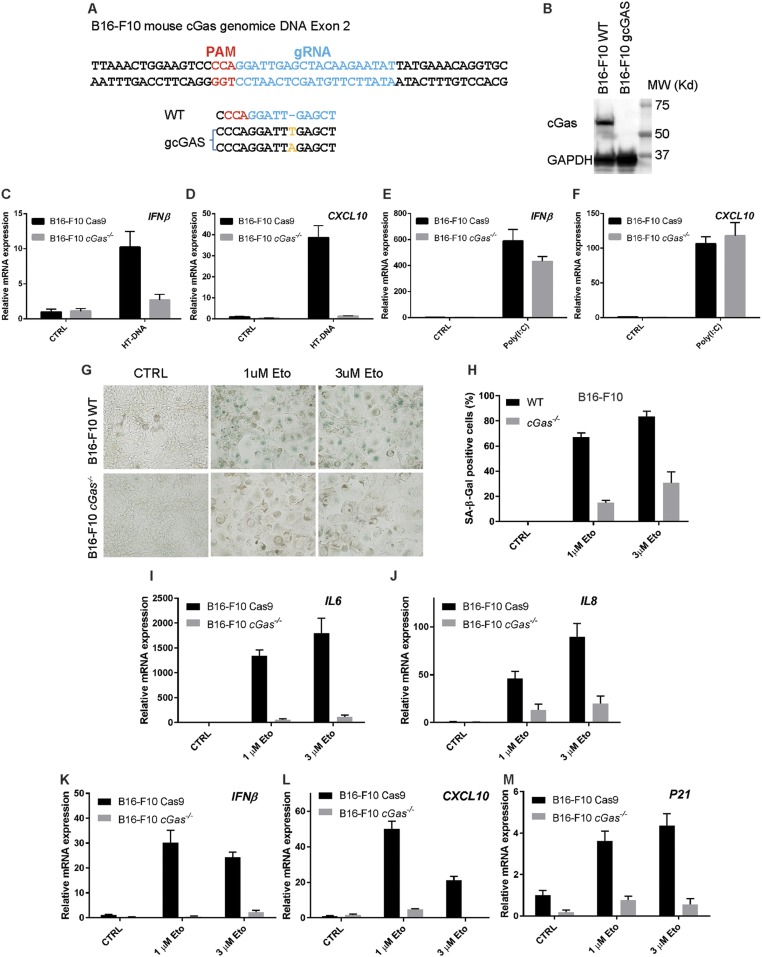
To extend our findings to a tumor cell line, we used the mouse B16F10 melanoma cells, which express cGAS and STING and have a functional DNA sensing pathway. We generated a cGas−/− B16F10 cell line using the CRISPR technology (Fig. S3 A and B); this cell line is defective in the induction of IFNβ and CXCL10 in response to DNA transfection but the induction of these genes by poly[I:C] was normal (Fig. S3 C–F). In response to etoposide treatment, more than 60% of WT B16F10 cells stained positive for β-Gal, whereas the number of senescent cells significantly decreased when cGas was deleted (Fig. S3 G and H). Consistent with these results, cGAS was essential for the expression of SASP genes, including IL6, IL8, IFNβ, CXCL10, and p21, in response to etoposide treatment (Fig. S3 I–M).
Fig. S3.
cGAS is essential for etoposide-induced senescence of B16F10 cells. (A) Sequence verification of disruption of the cGAS gene in a B16F10 clone generated by CRISPR. (B) Western blotting of l from lysates of WT and cGas−/− B16F10 cells. (C and D) WT and cGas−/− B16F10 cells were stimulated by HT-DNA transfection, then the induction of IFNβ and CXCL10 RNA was measured by qRT-PCR. (E and F) WT and cGas−/− B16F10 cells were stimulated by transfection with poly[I:C] and the induction of IFNβ and CXCL10 RNA was measured by qRT-PCR. (G and H) WT and cGas−/− B16F10 cells were treated with the indicated concentrations of etoposide for 24 h before growing in normal culture media for 5 d. Cells were stained with β-Gal (G) and the results quantified (H). (I–M) Total cellular RNA was isolated for qRT-PCR analyses of SASP genes.
cGAS Is Essential for Cellular Senescence Induced by Ionizing Radiation.
Ionizing radiation (IR) is well known to induce cellular senescence by causing double-stranded DNA breaks (11, 38, 39). To investigate the role of cGAS in IR-induced senescence, we irradiated WT and cGas−/− MEF cells with X-ray at 3 Gy followed by several days of culture before cells were stained with β-Gal. Approximately 25% and 33% of WT MEFs were stained positive with β-Gal on day 6 and 9, respectively, after IR (Fig. 4 A and B). In contrast, very few cGas−/− MEFs were stained positive for β-Gal. Moreover, expression of the SASP genes IL6, IL1β, and MMP12, which was significantly induced by IR in WT MEFs, was barely detectable in cGas−/− MEFs (Fig. 4 C–E).
Fig. 4.
cGAS deletion abrogates the senescence phenotypes induced by ionizing radiation. (A and B) WT and cGas−/− MEFs were irradiated with 3 Gy X-ray and then cultured in normal media for the indicated days before cells were stained with SA-β-Gal (A) and quantified (B). (C–E) Aliquots of the MEF cells were analyzed for expression of the indicated SASP genes. (F–K) Similar to A–E, except that BJ cells were used. Error bars indicate SEs of triplicate measurements. Analyses of additional SASP genes are shown in Fig. S4 C–F. ND, not detectable.
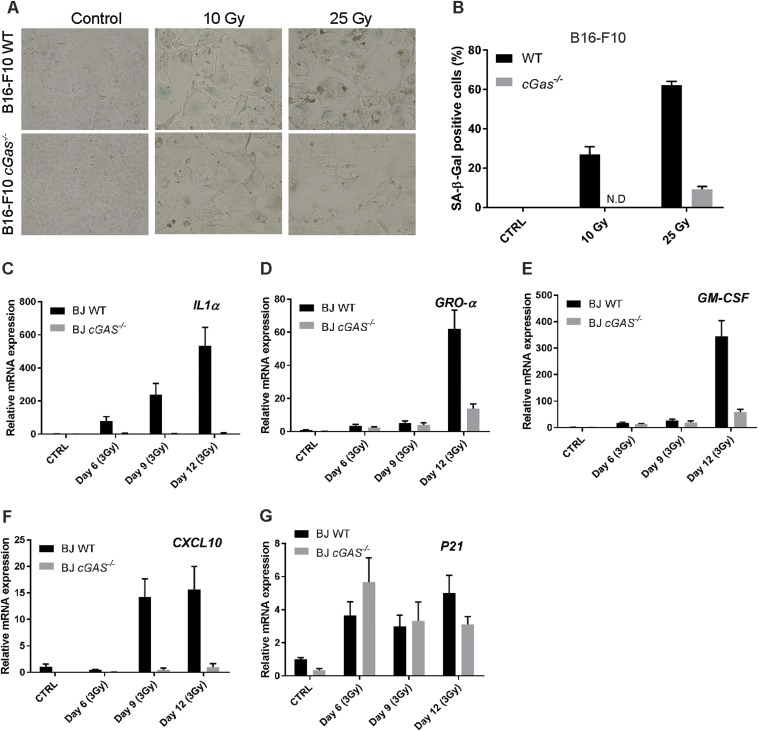
IR also induced senescence in WT BJ and B16F10 cells as evidenced by significant number of β-Gal positive cells after IR (Fig. 4 F and G and Fig. S4 A and B). In both cell types, deletion of cGAS abrogated the appearance of β-Gal positive cells. cGAS−/− BJ cells also expressed significantly lower levels of SASP genes after IR compared with WT cells (Fig. 4 H–K and Fig. S4 C–F). However, p21 was only weakly induced by IR and this induction appeared normal in cGas−/− BJ cells (Fig. S4G). Overall, our results show that cGAS is required for cellular senescence and the induction of most SASP genes by DNA damaging agents in human and mouse cells.
Fig. S4.
cGAS deletion abrogates the senescence phenotypes induced by IR. (A and B) WT and cGas−/− B16F10 cells were irradiated with the indicated IR dosages. At 7 d after radiation, cells were stained with β-Gal (A) and the blue cells quantified (B). (C–G) WT and cGas−/− BJ cells were irradiated as described in Fig. 4, and the expression of the indicated SASP genes was measured by qRT-PCR. ND, not detectable.
cGAS Is Cytoplasmic in Nondividing Cells but Accumulates on Chromatin During Mitosis in Proliferating Cells.
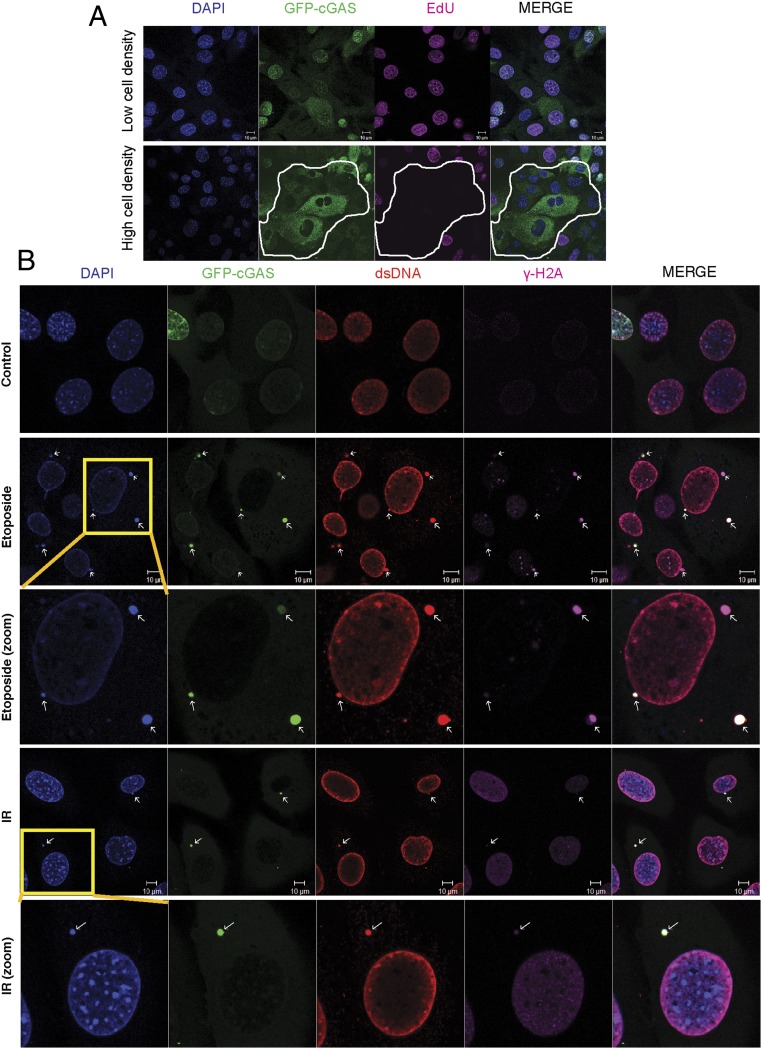
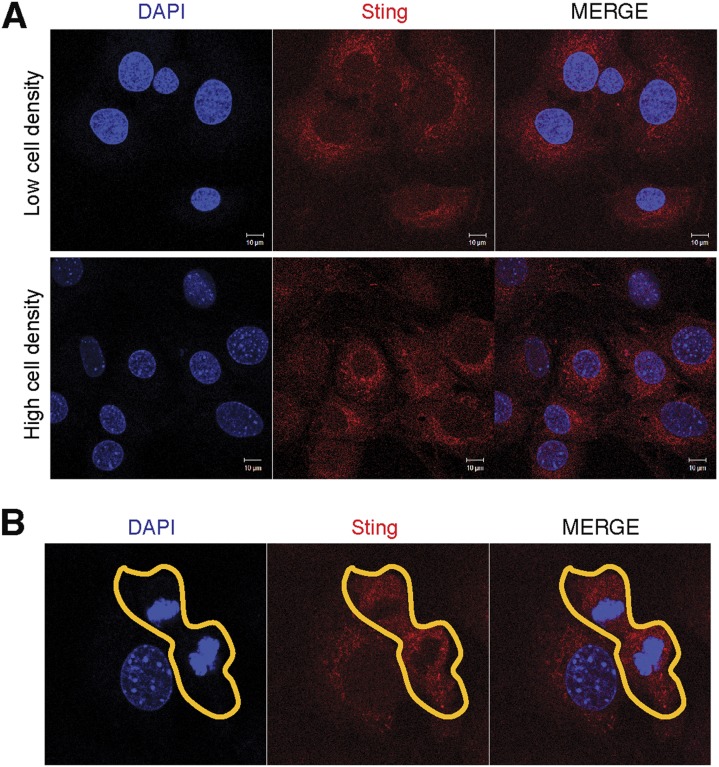
To investigate the mechanism by which cGAS regulates cellular senescence, we stably expressed GFP-cGAS in a cGas−/− MEF cell line and performed cell imaging. Interestingly, we found that in cells growing at low density, GFP-cGAS was mainly detected in the nucleus, whereas in cells growing at high density, GFP-cGAS was predominantly localized in the cytoplasm (Fig. 5A). Because cells growing at low density divide faster than those at higher density, we tested whether the nuclear localization of cGAS is associated with rapidly dividing cells by detecting incorporation of the nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU), which can be conjugated to a fluorescent dye through the click chemistry. Indeed, EdU positive cells contained GFP-cGAS in the nucleus, whereas EdU negative cells contained GFP-cGAS in the cytoplasm (Fig. 5A). In contrast to cGAS, STING was localized in the cytoplasm in cells at both high and low densities (Fig. S5A). Strikingly, live cell imaging showed that cGAS entered the nucleus and associated with the chromatin during mitosis (Movie S1). In contrast, STING remained in the cytoplasm of dividing cells (Fig. S5B).
Fig. 5.
cGAS enters the nucleus in proliferating cells and associates with damaged DNA in the cytosol in response to DNA damage. (A) MEF cells stably expressing GFP-cGAS were grown at low (8 × 104 per well of a 12-well plate) or high (3 × 105 per well of a 12-well plate) densities as indicated. The cells were cultured in the presence of EdU for 12 h and then EdU on the DNA was labeled and detected by fluorescence microscopy. Highlighted area indicates Edu-negative cells that contained GFP-cGAS predominantly in the cytoplasm. (B) GFP-cGAS MEF cells were treated with etoposide or IR and then stained with DAPI or antibodies specific for dsDNA or γ-H2AX. Zoomed cells highlight colocalization of cGAS with DNA and γ-H2AX in the cytoplasmic DNA foci.
Fig. S5.
cGAS, but not STING, associates with the chromatin during mitosis. (A) WT MEF cells were seeded at low (2.5 × 104 per well of a 12-well plate) or high (1 × 105 per well of a 12-well plate) densities. The cells were cultured for 2 d before staining with a STING antibody followed by immunofluorescence microscopy. (B) Immunofluorescent staining of STING in a dividing cell.
DNA Damage Leads to Accumulation of Cytosolic DNA and Its Association with cGAS.
Our findings that cGAS is important for the induction of SASP genes suggest that DNA may associate with cGAS in the cytosol to activate the enzyme in response to DNA damage. Etoposide treatment led to significant DNA damage as evidenced by staining with an antibody against phosphorylated H2AX (γ-H2AX), a marker of DNA damage (Fig. 5B). Staining of the same cells with DAPI and a dsDNA-specific antibody revealed that the damaged DNA accumulated in the cytoplasm. Remarkably, virtually all cytoplasmic DNA foci contained GFP-cGAS (Fig. 5B). Similarly, ionizing radiation led to formation of cytoplasmic DNA foci that colocalized with GFP-cGAS (Fig. 5B). These results indicate that DNA damage leads to accumulation of damaged DNA in the cytoplasm, leading to cGAS activation and expression of SASP genes. Furthermore, our findings that cGAS is associated with chromatin DNA in the nucleus during the cell cycle and that cGAS is associated with damaged DNA in the cytoplasmic foci raise an interesting possibility that cGAS may regulate cell cycle and senescence through mechanisms that are both STING dependent and STING independent.
Low Expression of cGAS Is Correlated with Poor Survival Outcome in Human Lung Adenocarcinoma.
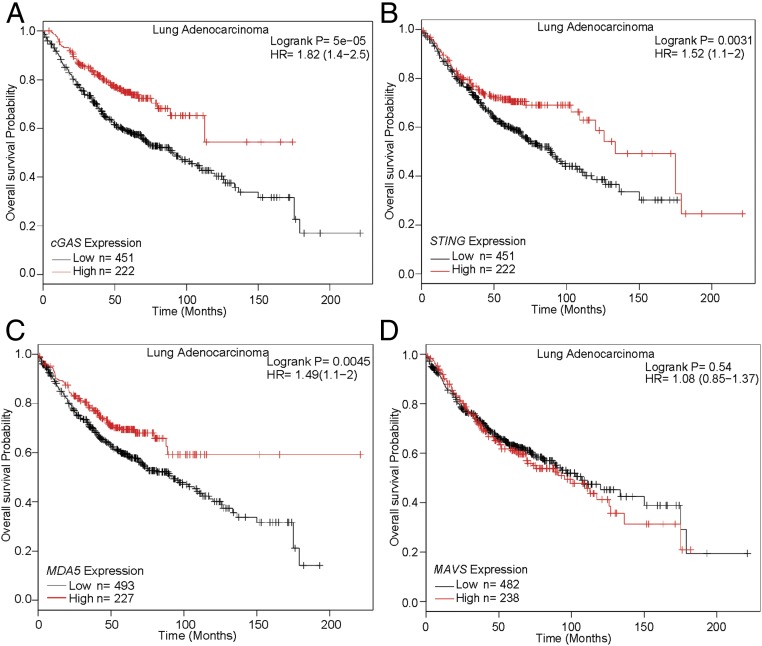
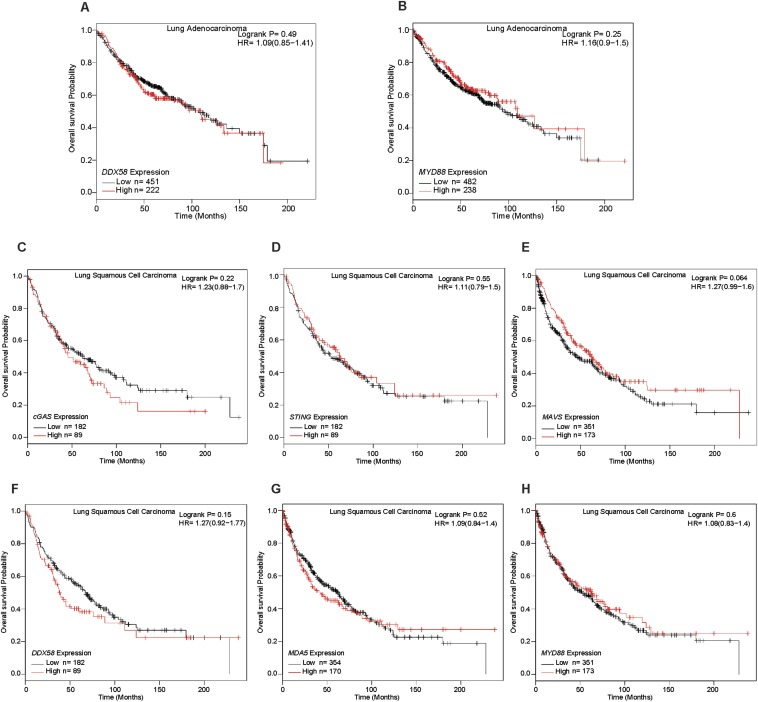
It has been shown that senescence is associated with the premalignant stages of tumorigenesis, but is largely absent in malignant tumors (40, 41). This observation suggests that senescence may act as a barrier to tumor progression. Our findings that cGAS is required for senescence and our previous findings that cGAS is important for antitumor immunity predict that cGAS may be a tumor suppressor under some circumstances (42). To test this possibility, we carried out bioinformatics analyses of publicly available gene expression datasets, including The (human) Cancer Genome Atlas (TCGA), the Gene Expression Omnibus (GEO), and the Cancer Biomedical Informatics Grid (caBIG). As shown in Fig. 6A, Kaplan–Meier analysis showed that lower expression of cGAS was strongly correlated with worse survival of lung adenocarcinoma patients (Fig. 6A). We also found that low expression of STING and MDA5 was correlated with decreased survival of lung adenocarcinoma patients (Fig. 6 B and C), whereas no statistically significant correlation was found between the expression levels of MAVS, RIG-I (also known as DDX58), and MyD88 and the survival of these cancer patients (Fig. 6D and Fig. S6 A and B). For the lung squamous cell carcinoma patients, none of the innate immunity genes that we have examined showed a statistically significant correlation with patient survival (Fig. S6 C–H). These analyses support the tumor-suppressing functions of cGAS in lung adenocarcinoma.
Fig. 6.
cGAS expression levels positively correlate with better survival of human lung adenocarcinoma patients. Kaplan–Meier curves were generated from the public microarray databases of human patients using online software (kmplot.com/analysis/). Groups of patients with high (top one-third) expression levels of the indicated genes cGAS (A), STING (B), MDA5 (C), and MAVS (D), were compared with those with low expression levels (bottom one-third). The Affymetrix microarray ID for each gene is shown in Materials and Methods. Statistical significance was determined using the log-rank test. HR, hazard ratio.
Fig. S6.
Kaplan–Meier analysis of gene expression microarray and survival data of human lung cancer patients. Data analysis of lung adenocarcinoma (A and B) and squamous cell carcinoma (C–H) was carried out online at kmplot.com/analysis/. For comparisons between groups of high and low expression, the high expression group is defined as those having the top one-third expression values and the low expression group represents those with the bottom one-third expression values. The Affymetrix microarray ID for each gene is shown in Materials and Methods. Statistical significance was determined using the log-rank test. HR, hazard ratio.
Discussion
Since its formal description more than 50 years ago (27), cellular senescence has been extensively studied and found to play a critical role in cancer, aging, and age-related diseases (2). Cellular senescence can be induced by DNA damage, telomere shortening, oxidative stress, and oncogenes. Interestingly, all of these senescence-inducing conditions impinge on DNA directly or indirectly. Here we showed that the DNA sensor cGAS is essential for cellular senescence during spontaneous immortalization or in response to DNA damaging agents. We found that damaged DNA is associated with cGAS in the cytoplasm and that deletion of cGAS abrogated SASP gene expression and other markers of cellular senescence. These results reveal cGAS as an important molecular link between DNA damage, SASP gene expression, and senescence. This conclusion is consistent with the previous reports that cGAS is activated by dsDNA independently of the DNA sequence and that cGAS activation by endogenous DNA causes autoimmune diseases in mice lacking the DNase Trex1 or DNaseII (22, 43, 44).
Surprisingly, we found that Stinggt/gt MEFs, which have been shown to lack the STING protein and are completely defective in the cytosolic DNA signaling pathway, were more resistant to spontaneous immortalization than cGas−/− MEFs (Fig. S1B). The simplest interpretation of these data is that cGAS has additional functions that are independent of STING. Indeed, we found that cGAS is present in the cytoplasm of nondividing cells but becomes associated with chromatin DNA during mitosis of rapidly dividing cells. These findings raise the interesting possibility that cGAS may regulate cell cycle and cellular senescence through mechanisms yet to be unveiled. Nevertheless, cGAS-dependent expression of SASP genes likely contributes to the establishment and maintenance of cellular senescence.
In response to DNA damage, cytoplasmic DNA foci appear and they contain cGAS and γ-H2AX, suggesting that damaged DNA fragments are associated with cGAS. It has been shown that oxidatively damaged DNA (e.g., 8-hydroxyguanosine generated by UV) could still bind and activate cGAS (45). This finding is consistent with the crystal structure of the cGAS:DNA complex, which shows that cGAS binds to the sugar phosphate backbone but not bases of DNA (46). Oxidized DNA is more resistant to degradation by nucleases such as Trex1 (45). Thus, damaged DNA may accumulate in the cytoplasmic foci and recruit cGAS. It remains to be determined whether cGAS is activated by the DNA in the foci, although the induction of inflammatory genes suggests that this is the case. The composition and fate of the cytoplasmic DNA foci also require further investigation. It is possible that such foci represent micronuclei, which are commonly detected in cells exposed to genotoxic compounds. It has been shown that cytoplasmic DNA generated during senescence can be cleared by the lysosome, likely through an autophagy pathway (47).
Whereas cGAS clearly plays an important role in cellular senescence, it should be noted that cGas-deficient mice appear to be healthy in a barrier facility (48). We have not observed a significant increase in spontaneous tumors in our cGas−/− mice even though some of these mice are more than 20 months old. Because there are multiple barriers for a cell to become a malignant cancer cell, removal of cGAS alone may not be sufficient to cause spontaneous tumors. However, it will be very interesting to test whether cGAS deletion promotes tumor development driven by oncogenes such as Ras, which is known to induce senescence. In this context, we recently reported that cGas-deficient mice are refractory to the antitumor effect of immune checkpoint blockade in a syngeneic tumor transplant model (42), indicating that cGAS is required for generating intrinsic antitumor immunity. It remains to be determined whether cGAS has a cell-autonomous function in impeding the transformation of a normal cell into a cancer cell. Consistent with the tumor-suppressive functions of the cGAS-STING pathway, many tumor cell lines have lost the expression of cGAS and/or STING (22, 49). Our analyses of the public database show that low expression levels of cGAS and STING are correlated with the poor survival of human lung adenocarcinoma patients. The low expression of cGAS in advanced metastatic tumors may also explain why these tumor cells lose the senescence phenotypes (40, 41). However, the correlation between cGAS expression and patient survival does not apply to all tumor types. In addition to lung squamous cell carcinoma, no statistically significant correlation between cGAS expression and patient survival was observed in ovarian and gastric cancers (kmplot.com/analysis; data not shown). Moreover, in breast cancer, higher levels of cGAS expression appear to correlate with poor patient survival, although such correlation is less significant in estrogen receptor positive or negative patients (data not shown). It is not clear why cGAS expression levels correlate particularly well with the survival of lung adenocarcinoma patients, but it is interesting to note that smoking, which induces DNA damage, is a major etiological agent of this type of cancer.
Recent studies have provided strong evidence that senescence has a causal role in aging and age-related diseases and that genetic deletion of senescent cells increases the lifespan and ameliorates age-related pathologies in mice (50, 51). It would be very interesting to determine whether cGAS plays a role in normal aging as well as age-related diseases in animal models. If so, cGAS inhibitors may be used for treating not only autoimmune diseases but also a variety of age-related diseases, including atherosclerosis and neurodegenerative diseases.
Materials and Methods
Cell Lines.
Human foreskin fibroblast BJ/TERT cells, spontaneously immortalized MEFs, and B16F10 cells were cultured in DMEM supplemented with 10% FBS and penicillin and streptomycin (GIBCO).
cGas−/− BJ cells were generated using the TALEN method. TALEN constructs were designed using the free online tool Mojo Hand (www.talendesign.org/mojohand_main.php), and assembled using the Golden Gate TALEN and TAL Effector Kit (Addgene, 1000000016), according to published protocols (52). The TALEN constructs were designed to target exon 2 of the human cGAS genomic locus, which encodes most of the catalytic domain of cGAS. Assembled TALENs were transfected into BJ cells using electroporation (Amaxa) following the manufacturer’s protocol and the cells were then maintained at 30 °C, 5% CO2 for 3 d before splitting to isolate single colonies. The single cell clone screening was performed as previously described (52). Briefly, genomic DNA was extracted from each cell clone, and DNA fragments flanking the TALEN target sites were amplified using specific primers (5′ primer: GTCCCACTCCCAGAGGGTAT, 3′ primer: TGAGAATGGGAGTGTACATTGG). The PCR product was then digested with EcoRI and resolved on 2% agarose gel. Resistance to digestion indicates that deletion had occurred at the EcoRI sites, thus single cell clones containing the uncleavable DNA were expanded for further analyses. DNA sequencing was performed after TA cloning of DNA from each of seven single cell clones. More than 12 TA cloned plasmids for each single cell clone were sequenced. Deletion of cGAS was further confirmed by Western blotting using a human cGAS specific antibody.
cGas−/− B16F10 cells was generated with the CRISPR technology. The sgRNA sequence of cGas was designed based on the Human CRISPR Knockout Pooled Library (GeCKO v2) (53). This sequence, ATATTCTTGTAGCTCAATCC, was cloned into lentiCRISPR v2 (AddGene, 52961), which was cotransfected with packaging plasmids pMD2.G and psPAX2 (Addgene, plasmids 12259 and 12260) into HEK293 cells to generate lentivirus. Briefly, HEK293T cells growing at 80% confluency on a 6-well plate were transfected with Lipofectamine 2000 (Life Technologies) together with 1 μg lentiCRISPR, 0.25 μg pMD2.G and 0.75 μg psPAX2. After 6 h, cells were cultured in fresh DMEM plus 10% FBS. The virus was harvested at 24 h and filtered with 0.45 μm low protein binding membrane (Millipore). B16F10 cells were infected with the virus at a 1:1 (virus:medium) ratio and selected with 1 μg/mL puromycin for another 3 d. Single cells were seeded on 96-well dishes with fresh medium. Clones were expanded and evaluated for knockout status by Western blotting with an antibody against human cGAS. DNA in the clones lacking the cGAS protein was sequenced after TA cloning.
MEF cells stably expressing GFP-cGAS were generated by infecting cGas−/− MEF cells with lentiviruses expressing human cGAS fused to eGFP at the N terminus and a hygromycin B-resistant gene. The drug-resistant cells were used for this study.
MEF Immortalization.
cGas−/− mice on a C57BL/6 background were generated and maintained as described previously (48). Stinggt/gt mice were purchased from The Jackson Laboratory (34). MEFs were generated from embryonic day 13.5 (E13.5) embryos of WT and mutant mice under the normal culture condition that includes 20% oxygen and 5% CO2 (29). For spontaneous immortalization, we followed a modified 3T3 protocol by seeding 1 × 106 cells in a 10-cm dish every 3.5 d. Aliquots of the cells at indicated passages were expanded for continued passages, frozen in liquid nitrogen, or analyzed by β-Gal staining and qRT-PCR. Mice were maintained in the animal research center of the University of Texas Southwestern Medical Center according to protocols approved by the Institutional Animal Care and Use Committee.
Senescence Assays.
Senescence was induced by spontaneous immortalization, etoposide treatment, or irradiation. Senescent cells were identified by a senescence-associated β-galactosidase kit (Cell Signaling, 9860) according to the manufacturer’s protocol. For senescence induced by spontaneous immortalization, MEFs at passages 2, 3, 6, 8, 11, 18, and 20 were harvested and fixed for β-Gal staining. For etoposide-induced senescence, immortalized MEFs were treated with 3 μM or 10 μM etoposide for 24 h and cultured in normal medium for another 5 d. The same treatment with etoposide was performed on BJ (10 μM or 20 μM) and B16F10 cells (1 μM or 3 μM). For ionizing radiation experiments, BJ cells and MEFs were irradiated with 3 Gy by the X-ray irradiator. The cells were harvested on days 6, 9, and 12 for SA-β-Gal assay. B16F10 cells were irradiated with 10 Gy or 25 Gy to induce senescence as described previously (39).
RNA Isolation and qRT-PCR.
Total cellular RNA was isolated with TRIzol reagent according to the manufacturer’s instructions (Invitrogen). cDNA was synthesized from the purified RNA using iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed using a SYBR Green Supermix PCR kit (Bio-Rad). PCR primers used are listed in Dataset S1.
Fluorescence Microscopy.
MEF cells stably expressing GFP-cGAS were treated with etoposide or irradiation followed by immunostaining with an antibody against dsDNA or γ-H2AX according to published protocols (24). Briefly, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 for 15 min at room temperature. After blocking with 6% BSA in PBS, cells were stained with a primary antibody against dsDNA (Santa Cruz Biotechnology, HYB331-01) or phosphohistone H2AX (γ-H2AX, Ser-139; Cell Signaling Technology, 9718), followed by a goat anti-mouse IgG conjugated with Alexa Fluor 568 or 633. Nuclei were stained with DAPI in the mounting medium (Vectashield). Images were captured using a Zeiss LSM700 confocal microscope and processed with a Zeiss LSM image browser.
For live cell imaging, GFP-cGAS MEFs were grown on a four-chambered cover glass (Lab-Tek II, 155382) at a density of 40,000 cells per chamber (∼50% confluency) in 5% CO2 and 20% O2 at 37 °C, and the green fluorescence was captured every 5 min with a Nikon A1R confocal microscope for 20 h.
EdU Labeling of Proliferating Cells.
MEFs were labeled with EdU (10 μM) using the Click -iT EdU Alexa Fluor 647 Imaging Kit (Thermo Fisher Scientific, C10340). Briefly, after overnight incubation with EdU, cells were fixed for 10 min in 3.7% paraformaldehyde in PBS, permeabilized with 0.5% Triton X-100 in PBS for 20 min at room temperature. After blocking with 3% BSA in PBS, the cells were incubated for 30 min at room temperature with Click-iT reaction mixtures. After washing with PBS, nuclei were labeled by staining with DAPI and images were captured using a Zeiss LMS700 confocal microscope.
Kaplan–Meier Analysis of Lung Cancer Patients.
Kaplan–Meier curves were generated using publicly available microarray datasets of human lung adenocarcinoma and squamous cell carcinoma patients (54) (kmplot.com/analysis). Patients were divided according to the expression values of target genes, with expression values in the top one-third (∼33%) range grouped as high expressers and those in the bottom one-third range grouped as low expressers. The Affymetrix IDs for cGAS is 1559051, STING (also named TMEM173) is 224929, RIG-I (also known as DDX58) is 242961, MDA5 is 216020, MAVS is 220305, and MyD88 is 209124.
Supplementary Material
Acknowledgments
We thank Youtong Wu and Fenghe Du for generating the cGas−/− BJ cells and Xiang Chen for assistance in the maintenance of mouse colonies. This work was supported by grants from the Cancer Prevention and Research Institute of Texas (RP120718 and RP150498) and the Welch Foundation (I-1389). Z.J.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705499114/-/DCSupplemental.
References
- 1.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat Med. 2015;21:1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collado M, Serrano M. Senescence in tumours: Evidence from mice and humans. Nat Rev Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prieur A, Peeper DS. Cellular senescence in vivo: A barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–155. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J, d’Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 6.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coppé J-P, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 13.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acosta JC, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 15.Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasdemir N, et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016;6:612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien Y, et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing H, et al. Opposing roles of NF-κB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev. 2011;25:2137–2146. doi: 10.1101/gad.17620611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capell BC, et al. MLL1 is essential for the senescence-associated secretory phenotype. Genes Dev. 2016;30:321–336. doi: 10.1101/gad.271882.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi A, et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol Cell. 2012;45:123–131. doi: 10.1016/j.molcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 22.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan YY, Londoño D, Bouley R, Rooney MS, Hacohen N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Reports. 2014;9:180–192. doi: 10.1016/j.celrep.2014.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Härtlova A, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42:332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Yu Q, et al. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Reports. 2015;11:785–797. doi: 10.1016/j.celrep.2015.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 28.Parrinello S, et al. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherr CJ, DePinho RA. Cellular senescence: Mitotic clock or culture shock? Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 31.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 32.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Sauer JD, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62:1876–1883. [PubMed] [Google Scholar]
- 37.de Magalhães JP, Chainiaux F, Remacle J, Toussaint O. Stress-induced premature senescence in BJ and hTERT-BJ1 human foreskin fibroblasts. FEBS Lett. 2002;523:157–162. doi: 10.1016/s0014-5793(02)02973-3. [DOI] [PubMed] [Google Scholar]
- 38.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flor A, Doshi A, Kron S. Modulation of therapy-induced senescence by reactive lipid aldehydes. Cell Death Discov. 2016;2:16045–16054. doi: 10.1038/cddiscovery.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collado M, et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 41.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci USA. 2017;114:1637–1642. doi: 10.1073/pnas.1621363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao D, et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci USA. 2015;112:E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutières syndrome. J Immunol. 2015;195:1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gehrke N, et al. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity. 2013;39:482–495. doi: 10.1016/j.immuni.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Reports. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov A, et al. Lysosome-mediated processing of chromatin in senescence. J Cell Biol. 2013;202:129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia T, Konno H, Ahn J, Barber GN. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Reports. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Childs BG, et al. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354:472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.