Significance
Early life stress (ELS) is an important risk factor for schizophrenia. Our study shows that ELS in mice increases the levels of histone-deacetylase (HDAC) 1 in brain and blood. Although altered Hdac1 expression in response to ELS is widespread, increased Hdac1 levels in the prefrontal cortex are responsible for the development of schizophrenia-like phenotypes. In turn, administration of an HDAC inhibitor ameliorates ELS-induced schizophrenia-like phenotypes. We also show that Hdac1 levels are increased in the brains of patients with schizophrenia and in blood from patients who suffered from ELS, suggesting that the analysis of Hdac1 expression in blood could be used for patient stratification and individualized therapy.
Keywords: schizophrenia, histone-deacetylases, early life stress, personalized medicine, HDAC inhibitor
Abstract
Schizophrenia is a devastating disease that arises on the background of genetic predisposition and environmental risk factors, such as early life stress (ELS). In this study, we show that ELS-induced schizophrenia-like phenotypes in mice correlate with a widespread increase of histone-deacetylase 1 (Hdac1) expression that is linked to altered DNA methylation. Hdac1 overexpression in neurons of the medial prefrontal cortex, but not in the dorsal or ventral hippocampus, mimics schizophrenia-like phenotypes induced by ELS. Systemic administration of an HDAC inhibitor rescues the detrimental effects of ELS when applied after the manifestation of disease phenotypes. In addition to the hippocampus and prefrontal cortex, mice subjected to ELS exhibit increased Hdac1 expression in blood. Moreover, Hdac1 levels are increased in blood samples from patients with schizophrenia who had encountered ELS, compared with patients without ELS experience. Our data suggest that HDAC1 inhibition should be considered as a therapeutic approach to treat schizophrenia.
Schizophrenia is a complex neuropsychological disorder that affects ∼1% of the world’s population (1). It is characterized by positive symptoms, such as delusions and hallucinations, and by negative phenotypes, including impaired cognitive function and social abilities (2, 3). A number of genes have been associated with the risk to develop schizophrenia (4–6). In addition to genetic predisposition, environmental factors, such as urbanicity (7), obstetric complications (8), or exposure to early life stress (ELS) (9, 10), are known to increase the risk of developing schizophrenia. Such genome–environment interactions are mediated by epigenetic processes, including DNA methylation (DNAme) or histone modifications (11). Especially the role of histone acetylation has gained substantial interest in translational neuroscience, which is due to the fact that inhibitors of histone deacetylases (HDACs) enhance cognitive function and ameliorate pathogenesis in a number of neurodegenerative and neuropsychiatric diseases (12, 13). The human genome encodes 11 zinc-dependent HDACs that are grouped into three classes. The emerging picture suggests that mainly class I HDACs might be suitable targets to treat brain diseases (12, 14). HDAC inhibitors are also discussed as novel targets to treat schizophrenia (15–17). In fact, valproate given in combination with atypical antipsychotics shows beneficial effects in preclinical (18) and clinical (19) studies. These data have to be interpreted with care, however, because besides its action on HDACs, valproate affects many other cellular processes (20). Postmortem analysis of human brain tissue suggested that Hdac1 levels are elevated in the prefrontal cortex and hippocampus of patients with schizophrenia (21, 22). It was therefore surprising that mutant mice either lacking or overexpressing neuronal Hdac1 from early developmental stages exhibit no cognitive phenotype (23). The lack of a phenotype might be due to compensatory mechanisms during development, because manipulating HDAC1 in the adult brain has been shown to affect specific forms of cognitive function (14, 24).
In this study, we investigated the role of HDAC1 in the pathogenesis of schizophrenia. We confirm that Hdac1 levels are increased in the prefrontal cortex and hippocampus from patients with schizophrenia and show that increased Hdac1 expression in mice and humans is caused by ELS. Moreover, ELS induces schizophrenia-like phenotypes in mice. These phenotypes were rescued by systemic administration of the HDAC inhibitor MS-275 (Entinostat). In turn, overexpression of Hdac1 in the medial prefrontal cortex (mPFC) led to impaired synaptic plasticity, short-term memory, and prepulse inhibition of the startle response (PPI). Although Hdac1 levels were also increased in the hippocampus of patients with schizophrenia, manipulating hippocampal HDAC1 levels had no effect on schizophrenia-like phenotypes, suggesting that adverse early life events cause a general increase in Hdac1 expression. Indeed, we observed increased Hdac1 levels in blood samples from ELS mice and in patients with schizophrenia who had experienced ELS. Our data show that HDAC inhibition could represent a suitable therapeutic approach to treat schizophrenia and, moreover, suggest that measuring Hdac1 levels in blood samples may allow patient stratification and individualized therapy.
Results
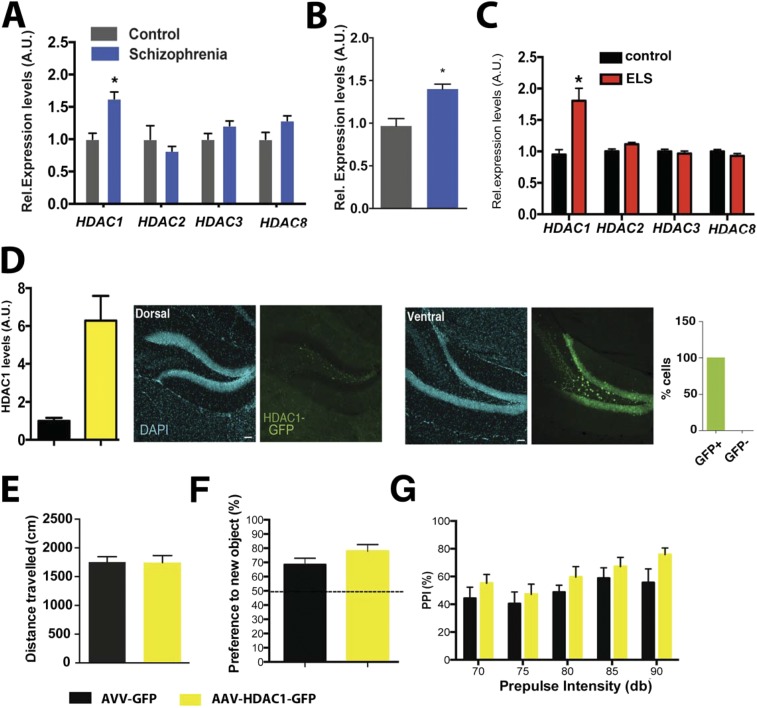
We started our analysis by measuring Hdac1 levels in postmortem tissue from control individuals and patients with schizophrenia. Our data reveal that Hdac1 mRNA (Fig. S1A) and protein level (Fig. S1B) were elevated in the prefrontal cortex (Brodmann area 9) of patients with schizophrenia. The expression of other class I HDACs was not affected (Fig. S1A). These data confirm previous results showing that Hdac1 is up-regulated in postmortem brain samples from patients with schizophrenia (21, 22) and provide further evidence that HDAC1 might play a role in the pathogenesis of neuropsychiatric diseases. Because deletion or overexpression of Hdac1 in all neurons of the mouse brain from prenatal stages did not cause any behavioral changes (23), we speculated that elevated HDAC1 levels in patients with schizophrenia may be due to environmental risk factors that drive Hdac1 expression in the postnatal brain, thereby limiting the effect of compensatory processes. One environmental risk factor that has repeatedly, although not exclusively, been linked to the pathogenesis of schizophrenia is ELS (25, 26), which can be modeled in rodents (27). Thus, we hypothesized that ELS might increase Hdac1 expression. To induce ELS, we subjected mice to an adapted maternal separation paradigm (hereafter simply referred to as ELS) (28) and performed behavior testing when animals were 4 mo old [postnatal day 120 (PND120)]. Mice that did not undergo the ELS protocol were used as a control group. Explorative behavior assayed in the open field test did not differ significantly (Fig. 1A). Basal anxiety, as measured in the elevated plus maze test, was also similar across groups (Fig. S2). Next, we measured short-term memory via the novel object recognition paradigm and observed that mice subjected to ELS performed significantly worse compared with the control group (Fig. 1B). Long-term memory was not affected (Fig. S2). We also tested PPI, which is impaired in patients with schizophrenia and is considered to be a reliable measure of schizophrenia-like phenotypes in rodents (29). PPI was significantly impaired in the ELS group, whereas the basic startle response was similar across groups (Fig. 1C). In conclusion, these data show that the ELS protocol we used leads to schizophrenia-like phenotypes in mice. Next, we measured Hdac1 mRNA and protein levels in the mPFC of 4-mo-old mice that were exposed to ELS. Compared with the control group, we found that Hdac1 levels were significantly increased, whereas other class I HDACs were not affected (Fig. 1 D and E). We hypothesized that ELS-induced changes of HDAC1 levels may result from a failure to control Hdac1 expression during postnatal brain development. In line with this idea, we observed that Hdac1 levels decrease in the postnatal brain and that Hdac1 expression is similar between control and ELS mice at PND34 (Fig. S3 A and B), a time point when explorative behavior, short-term memory, and PPI are still normal in mice exposed to ELS (Fig. S3 D–F). To explore the mechanisms contributing to ELS-induced increased Hdac1 expression, we focused on DNAme. ELS has been associated with DNAme changes that alter the ability of transcription factors, such as glucocorticoid receptors (GRs), to bind DNA (30). The Hdac1 gene contains a GR binding site within the first exon that could potentially be subjected to DNAme. We first performed methylated DNA immunoprecipitation (MeDIP), followed by next-generation sequencing (NGS), from the mPFC of wild-type mice (n = 5) to define methylated DNA regions (data available via the Gene Expression Omnibus database, accession no. GSE90150) and observed that the GR-binding site within the first exon of Hdac1 is methylated (Fig. S4). Next, we used MeDIP, followed by quantitative PCR (qPCR), to quantify DNAme at the GR-binding site of the Hdac1 gene in the mPFC from control mice and mice that were subjected to ELS (tissue was collected at PND120). We found a highly significant decrease of DNAme in the ELS group (Fig. 1F). Although no changes in DNAme were detected at a non–GR-binding region of the Hdac1 gene (Fig. 1F and Fig. S4), these data do not exclude the possibility that ELS causes DNAme changes at other genomic loci, including additional GR-binding sites.
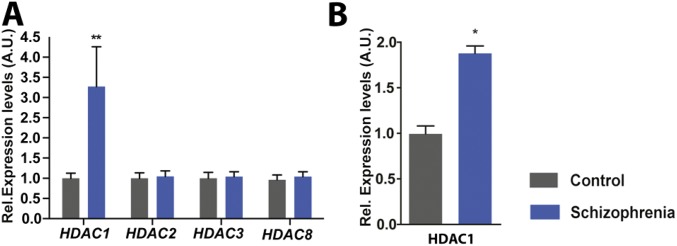
Fig. S1.
Expression of HDAC1 is elevated in patients with schizophrenia. (A) qPCR analysis from the human prefrontal cortex revealed a highly significant up-regulation of Hdac1 mRNA in patients with schizophrenia (n = 10) compared with control individuals (n = 11; **P = 0.009, unpaired t test). Levels of other class I HDACs were not affected. (B) Immunoblot analysis confirmed elevated HDAC1 protein levels in the prefrontal cortex from patients with schizophrenia and control individuals (n = 9 per group; *P = 0.0128, unpaired t test). Error bars indicate SEM. A.U., arbitrary units; Rel., relative.
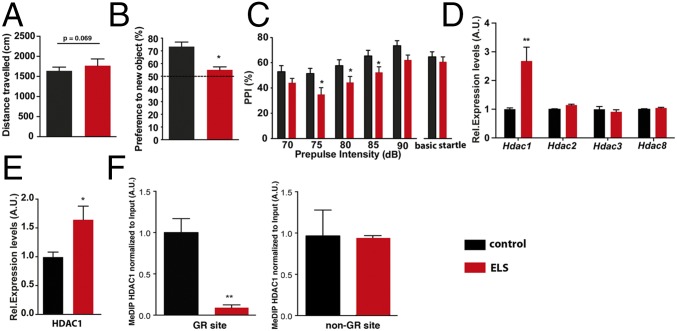
Fig. 1.
Expression of HDAC1 is elevated in mice exposed to ELS. (A) Mice subjected to ELS show normal explorative behavior (n = 15 per group), although there was a trend for locomotor hyperactivity (P = 0.069, unpaired Student’s t test). (B) Short-term memory analyzed via the novel object recognition paradigm revealed impairment in ELS-exposed mice (*P < 0.05, unpaired Student’s t test). (C) PPI was impaired in ELS mice. Two-way ANOVA of repeated measurements showed a significant effect of ELS [F(4,41) = 5.341, *P = 0.0259] and a significant effect of prepulse intensity [F(4,164) = 2.123]. A Bonferroni post hoc test revealed significant impaired PPI at 75 dB (P = 0.0089), 80 dB (P = 0.0356), and 85 dB (P = 0.0389). (D) Mice were subjected to ELS, and the prefrontal cortex (PFC) was isolated at PND120. qPCR analysis showed that Hdac1 mRNA is up-regulated in ELS-exposed mice compared with the control group (n = 6 per group; **P < 0.01, unpaired Student’s t test). Other class I HDACs were not affected. A.U., arbitrary units; Rel., relative. (E) Immunoblot analysis (n = 5 per group) confirmed elevated HDAC1 protein levels in the PFC from ELS-exposed mice. Tissue was isolated at PND120 (*P < 0.05, unpaired Student’s t test). (F, Left) MeDIP followed by qPCR shows that ELS leads to reduced DNAme of the Hdac1 gene at the GR-binding site (**P < 0.01, unpaired Student’s t test; n = 5 per group). Tissue was isolated at PND120. (F, Right) MeDIP followed by qPCR revealed no difference when a non-GR DNAme site within the Hdac1 gene was tested. Error bars indicate SEM.
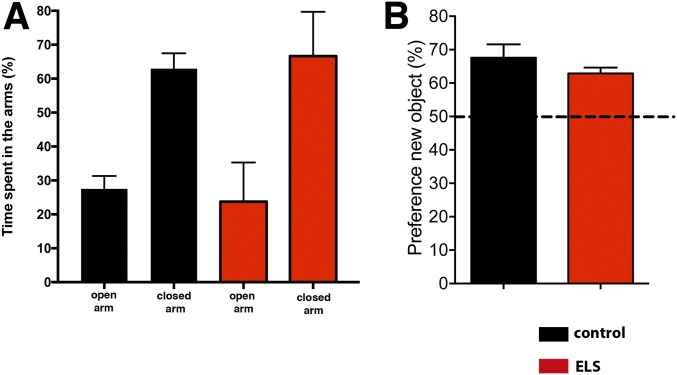
Fig. S2.
Basal anxiety and long-term memory in mice exposed to ELS. (A) Mice subjected to ELS were tested at PND120 for basal anxiety in the elevated plus maze test. Although both groups spent more time in the closed arms of the maze, there was no significant difference between groups when comparing the time spent in open and closed arms (n = 10 per group). (B) Mice subjected to ELS were tested at PND120 in the novel object recognition paradigm for long-term memory. To this end, animals were trained with two identical objects and tested for memory performance 24 h later in the presence of a novel object. There was no significant difference between groups. Error bars indicate SEM.
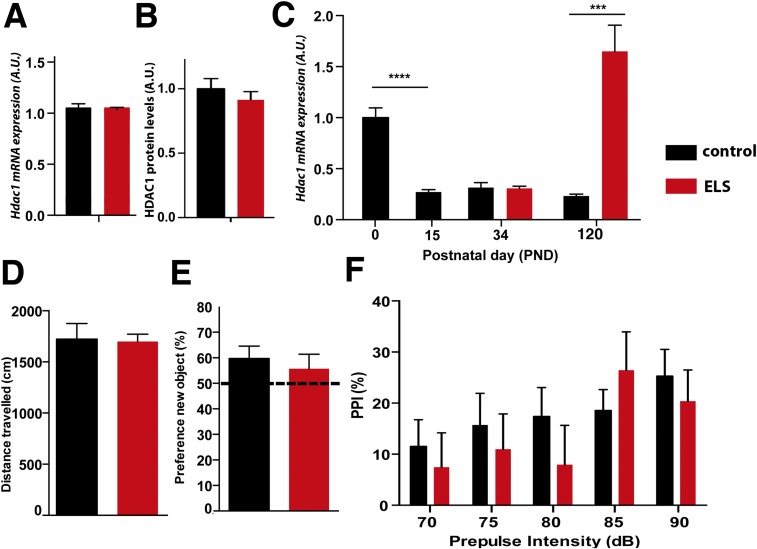
Fig. S3.
Postnatal increase of HDAC1 levels after exposure to ELS. (A) Hdac1 levels in the prefrontal cortex (PFC) were assayed in mice subjected to ELS (n = 7) or in control mice (n = 6) at PND34. HDAC1 mRNA levels were not affected. (B) Similarly, HDAC1 protein levels were not affected. (C) Hdac1 mRNA was analyzed via qPCR in mice at PND0 and PND15 and at PND34 and PND120 in mice subjected to ELS (n = 6 per group) and control conditions (n = 6 per group). Hdac1 levels decreased significantly from PND0 to PND15 (***P < 0.001, unpaired t test), whereas levels increased at PND120 upon ELS (***P < 0.001, ****P < 0.0001; unpaired t test). (D) Mice were subjected to ELS or a control condition (n = 9 per group), and explorative behavior was analyzed at PND34. No difference was observed between groups. Similar data were obtained when mice were tested for short-term memory (E) or PPI of the startle response (F). Error bars indicate SEM.
Fig. S4.
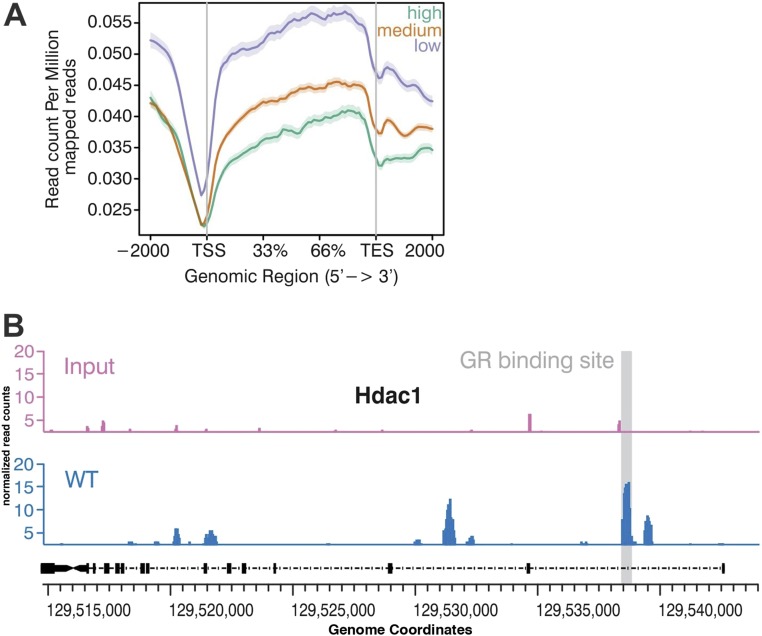
MeDIP followed by NGS in wild-type mice. (A) mPFC was isolated from 3-mo-old wild-type mice (n = 5) and subjected to MeDIP followed by NGS. We plotted DNAme across high-, medium-, and low-expressed genes, as defined by corresponding RNA-sequencing data (details are provided in Materials/Subjects and Methods). As expected DNAme was high in low-expressed genes and low in high-expressed genes. The dataset is available via the Gene Expression Omnnibus database (accession no. GSE90150). (B) DNAme and corresponding input signal at the Hdac1 gene. The GR-binding side within the first exon is shown as a gray-shadowed qPCR primer to study DNAme for the Hdac1 genes after ELSs were designed for this region (Fig. 1). TES, transcription end site; TSS, transcription start site.
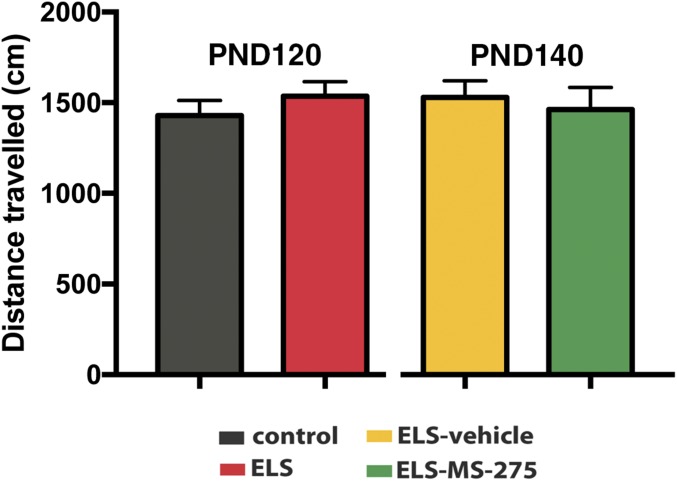
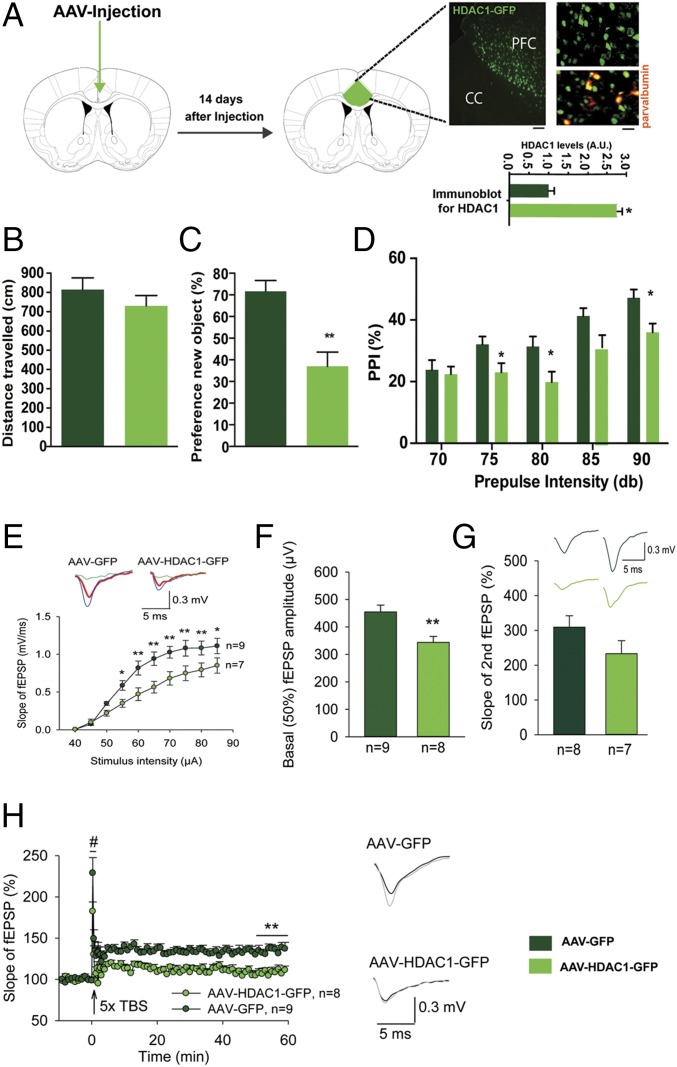
To test the role of HDAC1 more directly, we decided to use a pharmacological approach. Therefore, mice were subjected to our ELS paradigm (Fig. 2A). In agreement with our previous data, mice subjected to ELS displayed impaired PPI when tested at PND120 (Fig. 2B). Subsequently, mice of the ELS group were randomly assigned into one group that received daily i.p. injections of the class I HDAC inhibitor MS-275 for 10 d (ELS–MS-275 group). The other group of mice received vehicle injections (ELS-vehicle group). At PND140, mice of both groups were again subjected to PPI testing. PPI was significantly increased in MS-275–treated mice (Fig. 2C), although the basic startle response (Fig. 2D) and explorative behavior (Fig. S5) were not affected. An additional group of animals was subjected to the same experimental approach and tested for short-term memory in the novel object recognition paradigm. We first confirmed impaired short-term memory in the ELS group tested at PND120 (Fig. 2E). Treatment with MS-275 ameliorated this phenotype when animals were trained again in the novel object recognition test at PND140 (Fig. 2E). Because MS-275 is not a specific inhibitor of HDAC1, we sought to provide genetic evidence that HDAC1 can cause schizophrenia-like phenotypes in mice. Therefore, we used a viral-mediated approach to overexpress HDAC1 [HDAC1-adeno-associated virus (AAV)] under the control of the synapsin promoter, thereby restricting expression to neuronal cells (14). Although endogenous HDAC1 is moderately expressed in neurons of the prefrontal cortex (23) (Fig. S6), injection of HDAC1-AAV particles into the mPFC of mice caused a two- to threefold overexpression of HDAC1 protein (Fig. 3A and Fig. S6), which closely mimicked the situation seen in human patients with schizophrenia and in mice subjected to ELS (Fig. 1 and Fig. S1). Thus, we injected HDAC1-AAV particles into the prefrontal cortex of mice at 2 mo of age. Mice injected with GFP-AAV particles served as controls. Immunohistochemical analysis suggested that 82.6% of the neurons within the transfected region of the mPFC exhibited increased HDAC1 expression (Fig. S6). At 3 mo of age, animals were subjected to behavioral testing. Although explorative behavior assayed in the open field test was similar across groups (Fig. 3B), we found that short-term memory in the novel object recognition task was significantly impaired in the HDAC1-AAV group compared with the GFP-AAV control group (Fig. 3C). Most importantly, sensory-motor gating function measured via PPI was also impaired in the HDAC1-AAV group (Fig. 3D). In conclusion, these data provide strong evidence that elevated levels of HDAC1 in neurons of the mPFC contribute to schizophrenia-like phenotypes. To gain further mechanistic insight, we decided to assess whether overexpression of HDAC1 would affect synaptic transmission and plasticity. As described before, 2-mo-old mice were injected with either HDAC1-AAV or GFP-AAV into the mPFC and were subjected to analysis at 3 mo of age to allow direct comparison with the results obtained by behavioral analysis. We first recorded evoked-field excitatory postsynaptic potentials (fEPSPs) in layer II through layer V pyramidal cell synapses in acute mPFC slices from AAV-GFP– or AAV-HDAC1-GFP–injected mice. To investigate the effects of HDAC1 expression on basal excitatory synaptic transmission, stimulus–response curves were calculated for slope of fEPSPs in layer V of the mPFC. Two-way ANOVA with repeated measures detected statistically significant differences in the stimulus–response curves between AAV-GFP and AAV-HDAC1-GFP mice (Fig. 3E). Accordingly, the basal (50%) fEPSP amplitude in AAV-HDAC1-GFP slices (343 ± 22 μV) was significantly smaller compared with AAV-GFP controls (455 ± 25 μV; Fig. 3F). The paired-pulse ratio at a 50-ms interpulse interval, a measure of short-term plasticity, was not significantly different between the two groups (Fig. 3G), suggesting that the deficit in synaptic transmission is due to postsynaptic defects rather than presynaptic defects.
Fig. 2.
HDAC inhibitor MS-274 rescues ELS-induced impairment in PPI. (A) Experimental design. (B) Mice (n = 33) were subjected to ELS (ELS–PPI-1 group). PPI was significantly impaired at PND120 compared with a control group (n = 34). Two-way ANOVA of repeated measurements (RM) showed a significant effect of ELS [F(1,66) = 5.213, P = 0.025]. A Bonferroni post hoc test revealed significantly impaired PPI (*P < 0.05) at 75–85 dB. (C) Ten days after behavioral characterization, ELS–PPI-1 mice were assigned to two groups. One group of ELS mice (n = 8) was injected i.p. daily for 10 d with MS-275 (12.5 mg/kg, ELS–MS275–PPI-2 group), whereas the other group of ELS mice received vehicle injections (ELS-vehicle–PPI-2 group, n = 14). Twenty-four hours after the last injection, mice were subjected to a second PPI test (PPI-2). MS-275 treatment improved PPI performance compared with the vehicle group. Two-way ANOVA of RM showed a significant effect of MS-275 treatment [F(1,21) = 4.917, P = 0.0378], and the Bonferroni post hoc test revealed significantly improved PPI responses at 70–80 dB (*P < 0.05) in MS-275–treated mice. (D) Startle response was not affected. (E, Left) At PND120, novel object recognition learning is impaired in mice subjected to ELS (n = 18) compared with a control group (n = 11; ***P < 0.001, unpaired Student’s t test). (E, Right) Mice subjected to ELS but treated with MS-275 from PND130–140 (n = 9) show improved novel object recognition memory compared with a vehicle control group (n = 9; ***P < 0.001; unpaired Student’s t test). Error bars indicate SEM.
Fig. S5.
Explorative behavior in mice exposed to ELS and treated with MS-275. Mice were subjected to ELS as described in Fig. 2 and tested for explorative behavior in the open field test. There was no difference between groups. As described in Fig. 2 mice of the ELS group were then separated into two groups, treated with MS-275 or vehicle for 10 d, and then tested again for explorative behavior. Again, no difference between groups was observed. Please note that the data were obtained from the same animals that subsequently were tested in the novel object recognition test (details are provided in Fig. 2). Error bars indicate SEM.
Fig. S6.
HDAC1 expression in the prefrontal cortex. (A–C) Representative images showing overexpression of HDAC1-GFP in the mPFC 4 wk after animals were injected with AAV-HDAC1-GPF particles. (A) Immunofluorescence showing DAPI staining in the mPFC. (B) Same brain section showing HDAC1-GFP staining (green). (C) Merged image showing the overlap of DAPI and HDAC1-GFP signal. (Scale bar, 50 μm.) (D) Number of HDAC1-GFP–expressing cells per analyzed brain section was calculated using ImageJ software (NIH). A total of 82.6% of all analyzed cells showed HDAc1-GFP expression. (E) HDAC1 expression in the mPFC of a 3-mo-old wild-type mouse. (Scale bar, 50 μm.) (F) Representative images showing HDAC1 immunofluorescence in the mPFC of 3-mo-old wild-type mice and costaining with the neuronal marker NeuN. (Scale bar, 80 μm.) (G) Representative images showing costaining of HDAC1 and Pvalb in the mPFC. (Scale bar, 5 μm.) The data indicate that HDAC1 is expressed in the majority of the cortical neurons, including Pvalb-positive cells. Arrowheads indicate HDAC1- and Pvalb-positive cells.
Fig. 3.
Increasing HDAC1 levels in the mPFC causes schizophrenia-like phenotypes. (A) AVV-HDAC1-GFP particles were injected into the PFC. Robust nuclear HDAC1-GFP expression in neurons was detected 14 d after injection. The image shows low-magnification (Left) and high-magnification (Right) images illustrating HDAC1-GFP immunoreactivity. CC, corpus callosum. (Scale bars: Left, 50 μm; Right, 10 μm.) Costaining with parvalbumin (red) shows expression of HDAC1-GFP in inhibitory neurons. Because HDAC1-GFP is expressed via the synapsin promoter, its expression is restricted to neurons. *P < 0.05. (B) Explorative behavior was similar when comparing mice injected with AAV-HDAC1-GFP (n = 6) versus AAV-GFP (n = 6). (C) Short-term memory measured in the novel object recognition test was impaired in AAV-HDAC1-GFP-mice (n = 8) compared with the control group (n = 8; **P = 0.0031, unpaired Student’s t test). (D) PPI was impaired in AAV-HDAC1-GFP mice. Two-way ANOVA of repeated measures showed a significant effect of virus expression [F(4,27) = 5.640, P = 0.0249] and a significant effect of prepulse intensity [F(4,108) = 2.123, P = 0.0829]. A Bonferroni post hoc test revealed significantly impaired PPI in HDAC1-GFP mice at 75 dB (*P = 0.0432507), 80 dB (*P = 0.010581), 85 dB (*P = 0.0242066), and 90 dB (*P = 0.0242066). (E) Relationships between stimulus intensity and slope of evoked fEPSPs. A two-way ANOVA with repeated measures revealed significant effects of HDAC1 [F(1,126) = 7.261, P = 0.017], stimulus intensity [F(9,126) = 126.225, P < 0.001] and interaction between HDAC1 and stimulus intensity [F(9,126) = 5.203, P < 0.001]. A Holm–Sidak post hoc test revealed significantly reduced fEPSP slope in AAV-HDAC1-GFP–injected mice at 55 μA (*P = 0.026), 60 μA (**P = 0.003), 65 μA (**P = 0.001), 70 μA (**P = 0.002), 75 μA (**P = 0.003), 80 μA (**P = 0.008), and 85 μA (*P = 0.018). (Left Inset) Representative examples, which are averages of three to five fEPSPs evoked at 45 μA (green), 50 μA (red), and 55 μA (blue) in AAV-GFP– and AAV-HDAC1-GFP–injected mice. (Right Inset) Note that AAV-HDAC1-GFP–expressing slices displayed a detectable population spike already at 50 μA (red thick trace). (F) AAV-HDAC1-GFP–expressing brain slices showed reduced basal excitatory transmission (**P = 0.004, unpaired Student’s t test). (G) Unaltered PPF (P = 0.149, unpaired Student’s t test), measured as the ratio between the slopes of second and first fEPSPs (%) at a 50-ms interstimulus interval in AAV-HDAC1-GFP–injected mice. (Inset) Representative examples of PPF of fEPSPs in AAV-GFP–expressing (dark green) and AAV-HDAC1-GFP–expressing (light green) slices. (H, Left) HDAC1-GFP–expressing mice showed impaired LTP (50–60 min after TBS; **P = 0.001, unpaired Student’s t test) and a tendency for a decrease in STP (#P = 0.05, unpaired Student’s t test). The mean slope of fEPSPs recorded during a baseline period of 10 min before TBS was taken was 100%, and an arrow indicates delivery of TBS. (H, Right) Representative examples of fEPSPs, which are averages of 30 fEPSPs recorded during 10 min before (black, baseline) or 50–60 min after (gray) induction of LTP, respectively. Data represent measures from eight to nine slices recorded from five AAV-GFP–expressing mice and seven to eight slices from four AAV-HDAC1-GFP–expressing mice. The experiments described in B–H were performed 4 wk after injection. Error bars indicate SEM.
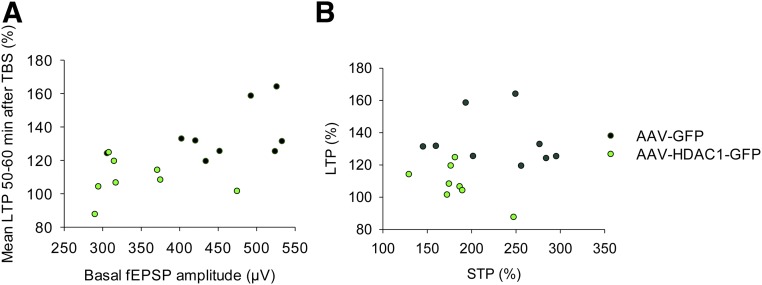
Analysis of synaptic plasticity elicited by repetitive theta-burst stimulation (TBS) revealed that the levels of long-term potentiation (LTP) recorded in the mPFC of AAV-HDAC1-GFP mice (108.23 ± 4.05%) were significantly lower compared with AAV-GFP mice (134.65 ± 5.23%; Fig. 3H). Also, for short-term potentiation (STP), there was a strong tendency for reduction in AAV-HDAC1-GFP–injected mice (182.43 ± 11.42%) versus AAV-GFP–injected mice (229.31 ± 18.50%; Fig. 3H). Notably, neither AAV-HDAC1-GFP nor AAV-GFP slices displayed a significant correlation between LTP and basal fEPSP slope, or between LTP and STP, as determined by analysis of Pearson’s and Spearman’s coefficients of correlation (Fig. S7). Thus, our data indicate that overexpression of neuronal HDAC1 leads to multiple independent deficits in synaptic transmission and plasticity.
Fig. S7.
Synaptic plasticity in the mPFC of mice overexpressing neuronal HDAC1. (A) No significant correlation was observed between LTP and basal fEPSP slope in slices obtained from mice that were injected with AAV-HDAC1-GFP or AAV-GFP. Analysis was performed 4 wk after viral injection. (B) Similarly, no significant correlation was observed between LTP and STP. Analysis was performed testing for Pearson’s and Spearman’s coefficients of correlation.
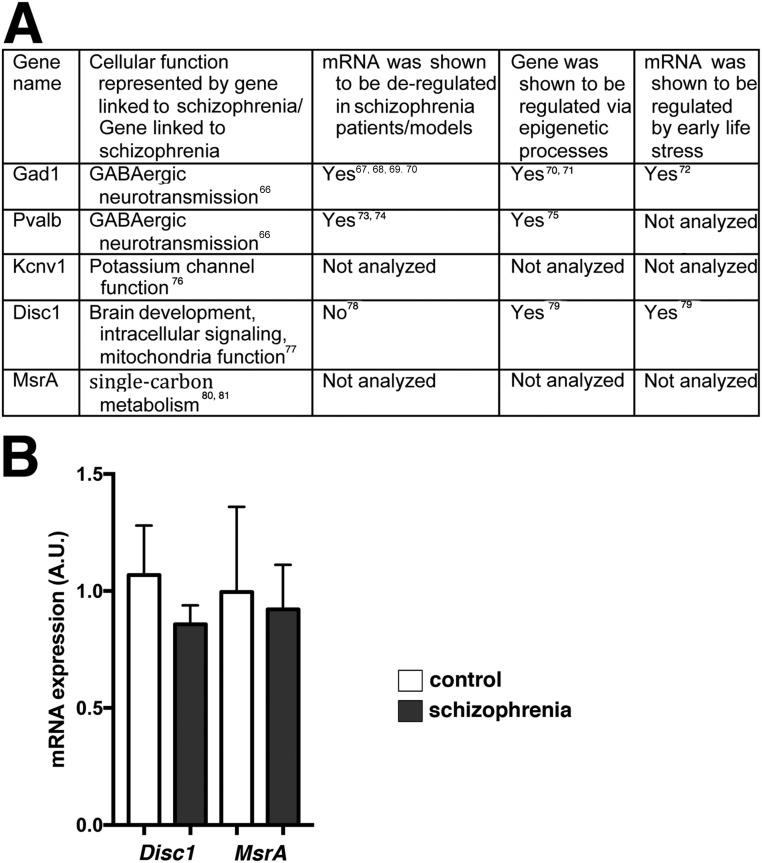
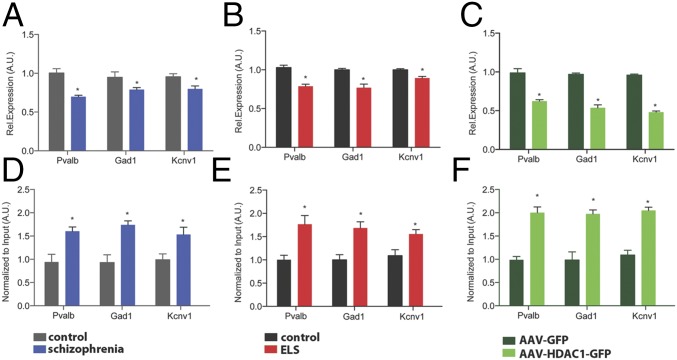
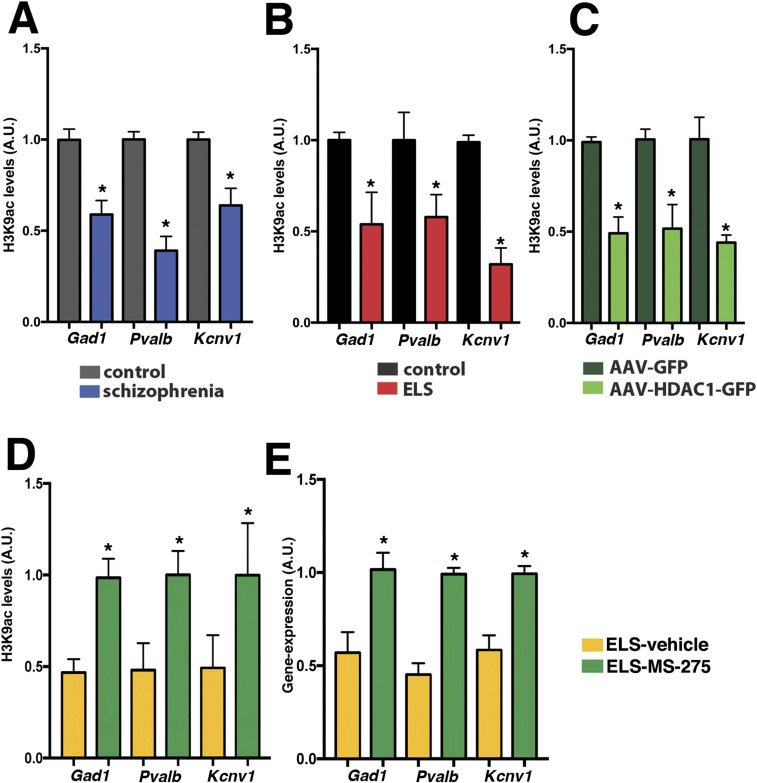
Next, we aimed to explore the impact of HDAC1 on gene expression using ChIP analysis. To this end, we first selected genes that had been linked to schizophrenia (Fig. S8). We found that the expression of Gad1, a gene encoding glutamate decarboxylase 1, Pvalb, which encodes parvalbumin, and the Kcnv1 gene, which encodes potassium channel subfamily V member 1, were decreased in the prefrontal cortex from patients with schizophrenia (Fig. 4A). A similar reduction of expression was observed in the mPFC of mice subjected to ELS and in animals that overexpressed HDAC1 (Fig. 4 B and C). Therefore, we were able to perform ChIP analysis to measure the levels of HDAC1 bound to the Gad1, Pvalb, and Kcnv1 promoters in human patients with schizophrenia, in mice subjected to ELS, and in mice overexpressing HDAC1. Consistent with the down-regulation of the Gad1, Pvalb, and Kcnv1 genes, we observed significantly increased HDAC1 binding at the promoter regions of these genes in human patients with schizophrenia (Fig. 4D) and in the corresponding animal models (Fig. 4 E and F). In agreement with this observation, H3K9 acetylation was reduced within the Gad1, Pvalb, and Kcnv1 gene promoters (Fig. S9). Moreover, reduced H3K9ac and gene expression after ELS exposure was ameliorated in mice treated with MS-275 (Fig. S9). These data do not exclude the possibility that other mechanisms, such as DNAme, contribute to the down-regulation of these genes after ELS exposure. Indeed, we observed that DNAme at the Pvalb promoter was increased after ELS (Fig. S10). The same trend was also observed in mice overexpressing HDAC1, however, which is in line with previous findings showing that HDAC1 can form complexes with DNA methyltransferases (31, 32). At least in the case of Pvalb, these data suggest that increased HDAC1 function acts in concert with DNAme changes to regulate Pvalb expression. DNAme most likely also contributes to ELS-induced changes in gene expression that are independent of HDAC1 action, an issue that needs to be addressed in future research.
Fig. S8.
Selection of genes for HDAC1 ChIP analysis. (A) To test the impact of HDAC1 on gene expression directly, we decided to use an “educated guess” approach. Thus, we performed a literature search for genes that have been linked to schizophrenia or represent pathways linked to schizophrenia, were shown to be deregulated in patients with schizophrenia patients/models, and/or were affected by ELS. Our rational was to find genes down-regulated in the postmortem tissue samples from patients with schizophrenia who included in our study and that were also down-regulated in our experimental settings. We reasoned that such genes would be suitable examples to test the impact of HDAC1 on gene expression via ChIP analysis. On this basis, we initially selected five genes for qPCR analysis of postmortem tissue. In addition to Gad1, Pvalb, and Kcnv1, which are described in the main text (Fig. 4), we selected Disrupted in schizophrenia (Disc1) and methionine sulfoxide reductase A (MsrA), which have been linked to schizophrenia via genetic studies. (B) RT-qPCR analysis was performed using RNA from postmortem tissue (prefrontal cortex) of patients with schizophrenia and control individuals (same material as shown in Fig. 4). No significant difference in gene expression was observed for Disc1 and MsrA (Student’s t test). Both genes have been linked to schizophrenia via genetic studies. In addition, one study analyzed the expression of Disc1 in human brain tissue from patients with schizophrenia and found no difference (14). Our data confirm this observation. To the best of our knowledge, MsrA mRNA expression has not been tested before in brain tissue from patients with schizophrenia. Error bars indicate SEM.
Fig. 4.
Elevated HDAC1 levels affect the expression of genes linked to schizophrenia. (A) qPCR analysis reveals elevated expression of the genes encoding Pvalb, Gad1, and Kcnv1 in the prefrontal cortex of patients with schizophrenia (n = 9 per group; *P < 0.05, unpaired Student’s t test) and in the mPFC of mice subjected to ELS compared with the control group (n = 5 per group; *P < 0.05, unpaired Student’s t test). Similar data were obtained in mice subjected to ELS (B; n = 5 per group; *P < 0.05, unpaired Student’s t test) and in mice that received AAV-HDAC1-GFP injections compared with the HDAC-GFP control group (C; n = 5 per group; *P < 0.05, unpaired Student’s t test). (D) ChIP analysis reveals increased HDAC1 levels at the gene promoters of Pvalb, GAD1, and Kcnv1 in the prefrontal cortex of patients with schizophrenia and compared with controls (n = 8 per group; *P < 0.05, unpaired Student’s t test). Similar data were obtained in the mPFC of mice subjected to ELS (n = 5) compared with the corresponding control group (E; n = 5 per group; *P < 0.05, unpaired t test) and in mice that received AAV-HDAC1-GFP injections compared with the HDAC1-GFP group (F; n = 5 per group; *P < 0.05, unpaired Student’s t test). Tissue from the ELS group and corresponding control group was isolated at PND120. Tissue from AAV-injected mice was harvested 4 wk after injection. Error bars indicate SEM.
Fig. S9.
H3K9 acetylation is decreased in patients with schizophrenia in response to ELS and HDAC1 overexpression and is ameliorated by MS-275 treatment. (A) ChIP analysis reveals decreased H3K9ac levels at the gene promoters of Pvalb, Gad1, and Kcnv1 in the prefrontal cortex (PFC) of patients with schizophrenia compared with controls (n = 8 per group; *P < 0.05, unpaired Student’s t test). (B) Similar data were obtained in the mPFC of mice subjected to ELS (n = 5) compared with the corresponding control group (n = 5 per group; *P < 0.05, unpaired Student’s t test). (C) In mice that received AAV-HDAC1-GFP injections, decreased H3K9ac levels (PFC) were observed at the gene promoters of Pvalb, Gad1, and Kcnv1 compared with the HDAC1-GFP group (n = 5 per group; *P < 0.05, unpaired Student’s t test). Tissue from the ELS and corresponding control group was isolated at PND120. Tissue from AAV-injected mice was harvested 4 wk after injection. (D) ChIP analysis reveals increased H3K9ac at the gene promoters of Pvalb, Gad1, and Kcnv1 when measured in the mPFC of mice subjected to ELS and treated with MS-275 from PND120–140. (n = 5 per group; *P < 0.05 versus vehicle, unpaired Student’s t test). (E) qPCR analysis reveals that Pvalb, Gad1, and Kcnv1 mRNA levels in the mPFC are increased in mice subjected to ELS and treated with MS-275 from PND120–140 compared with the vehicle group (n = 5 per group; *P < 0.05, unpaired Student’s t test). Error bars indicate SEM.
Fig. S10.
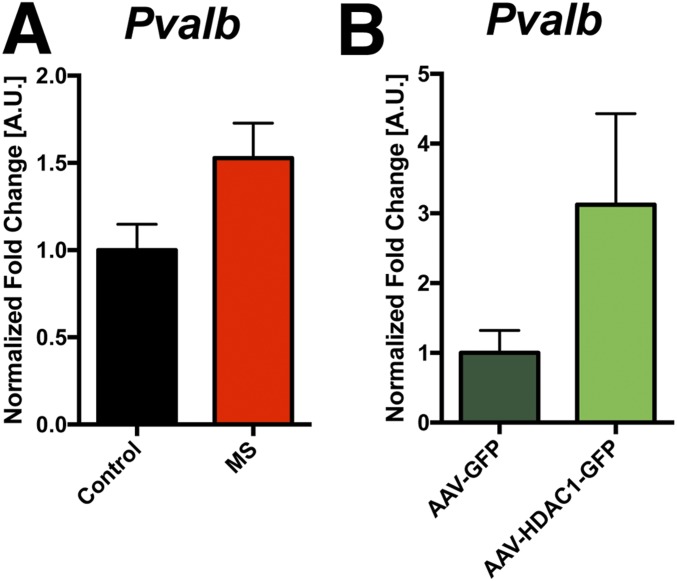
DNAme of the Pvalb gene in response to ELS and HDAC1 overexpression. (A) Mice were subjected to ELS, and mPFC was isolated at PND120. MS, maternal separation. Mice not subjected to ELS served as a control group (n = 5 per group). MeDIP-qPCR was performed to measure DNAme at the promoter region of the Pvalb gene. Increased DNAme was detected in the ELS group (P = 0.06, Student’s t test). This difference is considered significant using an alpha level of 0.1 but just failed to reach significance if the more common alpha level of 0.05 is chosen. (B) Mice were injected with AAV-GFP or AAV-HDAC1-GFP (n = 5 per group), and mPFC tissue was isolated 4 wk later. MeDIP-qPCR for the promoter region of the Pvalb gene was performed as described in A. Similar to ELS, we detected increased DNAme in the AAV-HDAC1-GFP group (P = 0.06, Student’s t test). This difference is considered significant using an alpha level of 0.1 but just failed to reach significance if the more common alpha level of 0.05 is chosen. These data suggest that DNAme likely contributes to the decreased expression of Pvalb in response to ELS and suggest that HDAC1 acts in concert with the DNAme machinery. We could not detect DNAme in the selected promoter regions of Gad1 and Kcnv4. Error bars indicate SEM.
To test if the observed phenotypes are specific to the mPFC, we also analyzed HDAC1 levels in the hippocampus of patients with schizophrenia. Similar to the data observed in the cortex, Hdac1 mRNA and protein levels were elevated in patients with schizophrenia (Fig. S11 A and B). Similarly, exposing mice to ELS caused elevated hippocampal Hdac1 levels (Fig. S11C). We had previously shown that overexpression of HDAC1 in the dorsal hippocampus of mice did not affect PPI or short-term memory (14). The possibility remained, however, that increased expression of HDAC1 in the ventral hippocampus, which is linked to regulation of mood and anxiety, and moreover directly projects to the mPFC (33, 34), may contribute to schizophrenia-like phenotypes in mice. To test this possibility, we injected AAV-HDAC1-GFP or AAV-GFP particles into the ventral hippocampus of mice and performed behavioral testing 4 wk later. Immunohistochemical analysis suggested that 95% of the hippocampal neurons within the infected area express HDAC1-GFP (Fig. S11D). Of note, increased levels of HDAC1 in the ventral hippocampus did not affect explorative behavior, short-term memory, or PPI (Fig. S11 E–G). These data suggest that increased expression of HDAC1 in patients with schizophrenia might be a common phenomenon that is not restricted to one brain region. Nevertheless, the detrimental phenotypes seem to be specifically linked to the action of HDAC1 in the prefrontal cortex and do not involve hippocampal function.
Fig. S11.
HDAC1 levels in the ventral hippocampus are not linked to schizophrenia-like phenotypes. (A) Mild but significant increase of Hdac1 mRNA was observed in the hippocampus of patients with schizophrenia (n = 14) compared with control individuals (n = 14; *P = 0.015, unpaired Student’s t test). Other class I HDACs were not affected. (B) Similar results were obtained when HDAC1 protein levels were analyzed in patients with schizophrenia and control individuals (n = 5 per group; *P < 0.05, unpaired Student’s t test) by immunoblotting. (C) qPCR analysis showing increased Hdac1 levels in the hippocampus of mice subjected to ELS (PND120) compared with a control group (n = 5 per group; *P < 0.05, unpaired Student’s t test). (D, Left) Immunoblot analysis of the ventral hippocampus obtained from mice (n = 4 per group) injected with HDAC1-GFP. There was a significant increase of HDAC1 compared with the GFP control group (n = 5 per group; P < 0.05, unpaired Student’s t test). (D, Right) Representative images showing the dorsal (Left) and ventral (Right) hippocampal dentate gyrus region of mice injected with HDAC1-GFP. Note that HDAC1-GFP was exclusively expressed in the ventral hippocampus. The number of HDAC1-GFP–expressing cells per analyzed brain section was calculated using ImageJ software. Almost all of the cells of the analyzed region express HDAC1-GFP. (Scale bar, 100 μm.) (E) AAV-HDAC1-GFP (n = 8) or AAV-GFP (n = 8) particles were injected into the ventral hippocampus of mice. Overexpression of HDAC1 in the ventral hippocampus did not affect explorative behavior. (F) Similarly, there was no difference in working memory measured in the novel object recognition test. (G) PPI was similar in AAV-HDCA1-GFP (n = 8) or AAV-GFP (n = 8) injected mice. Error bars indicate SEM.
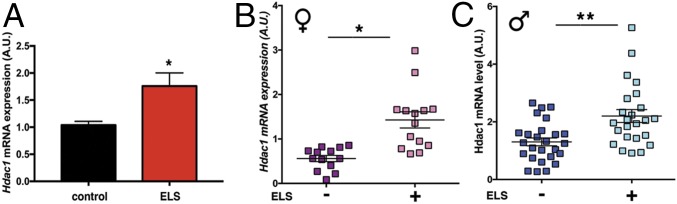
If this hypothesis is true, HDAC1 might be a suitable biomarker for patient stratification and individualized therapy. Thus, we speculated that increased Hdac1 levels might also be observed in the blood of mice subjected to ELS. Indeed, qPCR analysis revealed increased Hdac1 levels in blood samples (obtained at PND120) of mice subjected to ELS (Fig. 5A). Next, we set out experiments to test if our findings can be translated to human patients. To this end, we relied on an ongoing study on genotype–phenotype relationships and the neurobiology of the longitudinal course of major psychiatric disorders (www.kfo241.de and www.psycourse.de). From this cohort, we selected patients suffering from paranoid schizophrenia [Diagnostic and Statistical Manual, 4th Edition (DSM-IV) diagnosis code 295.30 (35)] who were diagnosed using a comprehensive phenotyping inventory (details are provided in Materials/Subjects and Methods) also assessing ELS (36, 37). Thus, we were able to classify patients with schizophrenia as either having experienced ELS (n = 38) or not (n = 39).
Fig. 5.
Hdac1 levels are increased in blood samples from patients with schizophrenia who were subjected to ELS. (A) qPCR analysis showing increased Hdac1 expression in blood isolated from ELS-exposed mice at PND120. Hdac1 levels were significantly increased in the ELS group (n = 15 per group; *P < 0.05, unpaired Student’s t test) (B) Increased Hdac1 expression in female patients with schizophrenia (n = 14) exposed to ELS compared with the no-ELS group (n = 13; *P = 0.0129, unpaired t test). (C) Increased Hdac1 expression in male patients with schizophrenia exposed to ELS (n = 24) compared with the no-ELS group (n = 24; **P = 0.001, unpaired t test). Error bars indicate SEM.
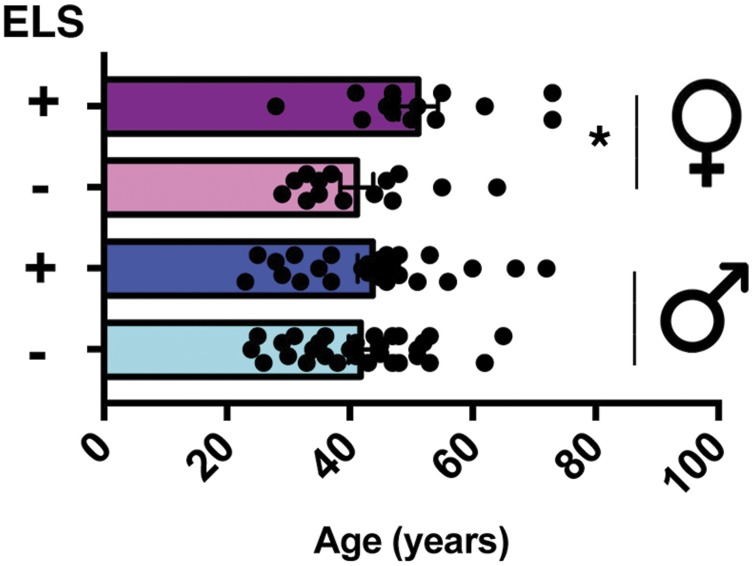
There was no significant age difference among male patients, although female patients with ELS were slightly older than female patients without ELS (Fig. S12). Next, we isolated RNA from all samples and performed qPCR analysis for Hdac1. Hdac1 mRNA was significantly increased in female (n = 14) and male (n = 24) patients with schizophrenia who encountered ELS compared with patients who were not exposed to ELS (nfemale = 13, nmale = 26) (Fig. 5 B and C).
Fig. S12.
HDAC1 levels in blood samples from patients with schizophrenia subjected to ELS. Age was similar in male patients with schizophrenia exposed (n = 24) or not exposed to ELS (n = 26). In female patients with schizophrenia, the ELS group (n = 14) was significantly older compared with the no-ELS group (n = 13; *P = 0.039, unpaired Student’s t test). Error bars indicate SEM.
Discussion
In this study, we show that Hdac1 mRNA and protein levels are increased in postmortem brain tissue, namely, in the hippocampus and prefrontal cortex of patients with schizophrenia. Our findings confirm previous studies that reported elevated Hdac1 expression in patients with schizophrenia (21, 22). Together with our observation that overexpression of HDAC1 in neurons of the mPFC of mice leads to schizophrenia-like phenotypes, these data provide solid evidence for a role of HDAC in the pathogenesis of schizophrenia. It has to be noted that no animal model recapitulates all of the complex phenotypes observed in human patients. For example, we did not observe locomotor hyperactivity, which is know to occur in human patients and some animal models (38). Our data suggest that the up-regulation of HDAC1 in patients with schizophrenia is due to environmental risk factors, especially ELS. It is noteworthy that ELS-induced HDAC1 expression may result from the deregulation of processes that control Hdac1 expression during normal brain development. This assumption is based on the finding that cortical HDAC1 expression normally decreases from PND0 to PND120, whereas ELS exposure appears to disturb this process. This observation is in agreement with previous studies indicating that exposure to environmental risk factors can cause long-lasting changes in DNAme and gene expression (39, 40). It will be interesting to determine what aspect of the stress system induces ELS-mediated HDAC1 expression. Causative evidence that increased HDAC1 levels contribute to the pathogenesis of schizophrenia stems from our observation that the HDAC inhibitor MS-275 ameliorates ELS-induced phenotypes and that overexpression of HDAC1 in the prefrontal cortex leads to impaired short-term memory, synaptic plasticity, and PPI. It would be interesting to test if Hdac1 knockout mice are resistant to ELS-induced phenotypes. Moreover, we cannot distinguish if the phenotypes caused by increased HDAC1 expression are exclusively due to its catalytic activity or may also involve processes independent of HDAC1 activity. The view that HDAC1-mediated gene expression plays a role in the development of schizophrenia-like phenotypes is supported by our finding that HDAC levels were increased at the promoter regions of three genes implicated in schizophrenia, namely, Pvalb, Gad1, and Kcnv1. Increased promoter binding of HDAC1 was associated with reduced expression of these genes, which was observed in patients, in mice exposed to ELS, and in mice overexpressing HDAC1. Because HDAC1 levels were specifically increased in neurons in our overexpression model, we suggest that increased expression of neuronal HDAC1 is sufficient to drive schizophrenia-like phenotypes in mice. This finding is in line with the reported predominantly neuronal expression pattern of HDAC1 in the adult brain (14, 23). However, in our experimental approach, we cannot distinguish between different neuronal populations. In the future, it will be important to determine the precise cell types in which HDAC1 is increased in response to ELS and to decipher how HDAC1 impacts the plasticity of the corresponding neuronal circuits. The finding that HDAC1 affects the expression of Gad1 and Pvalb indicates a role in inhibitory neurons, which is in line with a previous study showing that HDAC1 is expressed in GABAergic neurons regulating GAD1 expression (41). Gad1, Pvalb, and Kcnv1 were selected for analysis as examples (Fig. S8). There are certainly additional genes deregulated in response to increased HDAC1 levels, and future genome-wide approaches will be necessary to identify these networks. We observed reduced basal excitatory synaptic transmission and impaired N-methyl-d-aspartate receptor-dependent LTP in the prefrontal cortex of mice overexpressing neuronal HDAC1. We also observed a deficit in basal synaptic transmission, which is in line with an observation that HDAC1 acts as a transcriptional repressor of GluR2 subunit-containing α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors, thereby down-regulating synaptic strength (42). It is therefore likely that increased HDAC1 affects the function of multiple neuronal cell types and cellular pathways. Another interesting observation is that HDAC1 levels are elevated in the mPFC and hippocampus of patients with schizophrenia and mice exposed to ELS. However, only overexpression of HDAC1 in the mPFC, but not in the dorsal (14) or ventral hippocampus (this study), causes schizophrenia-like phenotypes. This finding indicates that HDAC1 in the mPFC, a brain region intimately linked to schizophrenia (43, 44), may control different molecular pathways compared with other brain regions, such as the hippocampus. In line with this observation, the concentration of systemically injected MS-275 leading to increased bulk levels of histone 3 acetylation in the prefrontal cortex of mice is fourfold lower compared with the dose needed to observe the same effect in hippocampal tissue (45). Interestingly, we also observed increased HDAC1 levels in blood samples from mice exposed to ELS and in patients with schizophrenia. Although increased HDAC1 levels in blood are unlikely to contribute to schizophrenia-like phenotypes, this finding is in line with the view that adverse early life events and other risk factors induce similar gene expression changes in various cell types (46).
Taken together, these findings suggest that exposure to ELS leads to increased HDAC1 expression in several tissues and cell types. However, only in selected cells, such as neurons of the mPFC, will increased HDAC1 expression trigger pathological events. It will be interesting to understand the molecular basis for this phenomenon.
It has to be mentioned that a previous study reported reduced HDAC1 level in the prefrontal cortex of mice exposed to ELS, which was also linked to cognitive deficits, however (47). The discrepancy between that study and our data is likely due to the fact that an infant maternal separation protocol and BALB/c mice were used (47). It is well known that BALB/c and C57BL6J mice differ regarding cognitive function and stimulus-induced gene expression (48, 49). Moreover, there is by now substantial evidence from independent studies (refs. 21, 22 and this study) that HDAC1 levels are increased in human patients with schizophrenia, that overexpression of HDAC1 in the prefrontal cortex of mice increases stereotype behavior (24) and induces schizophrenia-like phenotypes (this study), and that the HDAC inhibitor MS-275 alleviates cognitive phenotypes (ref. 50 and this study).
Nevertheless, great care has to be taken when interpreting results from animal experiments in the context of human diseases. This caution is especially true for complex diseases such as schizophrenia. However, the relevance of our findings is substantiated by the observation that HDAC1 levels are increased in blood samples from patients with schizophrenia who had encountered ELS, compared with patients without ELS.
In conclusion, our data strongly support a role of HDAC1 function in the etiology of schizophrenia. Elevated HDAC1 expression is likely due to adverse environmental factors, such as ELS, that occur within a vulnerable postnatal phase. The finding that altered DNAme may play a key role in the up-regulation of HDAC1 also merits further investigation and suggests that combinational therapies or even dietary changes that are known to affect DNAme (51) could be suitable therapeutic strategies. We show that the analysis of HDAC1 levels in blood from patients allows the identification of individuals with ELS experience. Therefore, this biomarker approach might be useful to identify schizophrenic patients who will benefit from therapeutic intervention with HDAC inhibitors. In addition, it should be tested if DNAme of the Hdac1 gene could also be used as a biomarker. It will also be interesting to see if different stressors will affect blood Hdac1 levels and if patients that experienced ELS can be further classified based on the categories of stressors. Although some drugs, such as MS-275/Entinostat, are in clinical testing, other HDAC inhibitors, such as vorinostat or valproate, are already in clinical use. The latter are even used for the treatment of psychosis. Our data therefore point to two immediate translational avenues. First, blood Hdac1 levels should be measured when treating schizophrenic patients with valproate to correlate therapeutic efficacy with Hdac1 levels, and, second, more specific compounds, such as the US Food and Drug Administration-approved HDAC inhibitor vorinostat, should be tested in clinical studies treating schizophrenic patients with ELS experience and/or elevated blood Hdac1 levels. In sum, our data may help to develop better therapeutic avenues to treat schizophrenia.
Materials/Subjects and Methods
Animals and Behavioral Experiments.
Mice (C57BL/6J) were housed under standard conditions with free access to food and water. All experiments were carried out in accordance with the animal protection law and were approved by the District Government of Germany (Animal Care protocol 10/0186).
Maternal separation (ELS) was performed according to a recent protocol published by Niwa et al. (28). In brief, the mothers were removed from the cage and the pups were subsequently placed in a Petri dish filled with bedding. The dish was then placed for 6 h each day in a different room (32 °C) before the pups were returned to their mothers. Control groups were handled identically, but not removed from their mothers. At day 22, mice were weaned and then housed in isolation.
The open field test was performed to assay explorative behavior and locomotion. Each mouse was placed in the center of an open arena (length of 1 m, width of 1 m, side walls were 20 cm high), and behavioral activity was recorded for 5 min using the VideoMot2 tracking system (TSE Systems).
The elevated plus maze test was used to test basal anxiety. Animals were placed individually in a uniformly gray plastic arena consisting of two nonwalled (open) and two walled (closed) arms for 5 min (10 × 40 cm each, walls were 40 cm high). Time spent in open versus closed arms was measured using a camera (VideoMot2 system).
The novel object recognition test was used to measure short-term memory in mice. Mice were habituated to an open arena (length of 1 m, width of 1 m, side walls were 20 cm high) for 5 min on two consecutive days. Twenty-four hours after this habituation period, mice were exposed for 5 min to the familiar arena with two identical objects placed at an equal distance (18 cm from the side walls). During a short retention period in the home cage for 5 min, one of the two identical objects was replaced with a new object. During the testing phase, mice were exposed to this situation and short-term memory was assessed by scoring exploration of the new object on the basis of direct contact with the object using the VideoMot2 tracking system. For the assessment of long-term memory, the test was performed as described above but mice were exposed to the novel object not 5 min but 24 h after the initial exposure to the two familiar objects. In case of the experiments described in Fig. 2E, the objects used from training at PND120 and after vehicle or MS-275 treatment were different. The index for object preference was calculated as the percentage of time spent with the novel object using the following formula: (time spent with novel object/time spent with both objects) * 100%.
PPI was used to measure sensorimotor gating function. The experiments were conducted as previously described using an apparatus from TSA Systems. In brief, mice were placed individually in a small cylindrical cage with an integrated stainless floor grid (80 × 40 × 45 cm) that was placed on a sensitive transducer platform in a sound-attenuating cabinet. Acoustic stimuli were delivered through loudspeakers above the cage and startle response signals presented by TSA startle response software. During one test session, mice were habituated first for 3 min to 65-dB background noise, followed by a 2-min baseline recording. After the baseline recording, mice were exposed to six pulse-alone trials, with each consisting of 120-dB startle stimuli intensity for a duration of 40 ms to decrease the influence of within-session habituation and to scale down the initial startle response to a stable plateau. The startle reaction to acoustic stimuli was recorded with the presentation of the startle stimuli for a time window of 100 ms. PPI of startle activity was conducted by trials presenting startle stimuli at 120 dB for 40 ms alone or preceding nonstartling prepulses of 5, 10, 15, 20, and 25 dB above the 65-dB background noise (i.e., 70, 75, 80, 85, 90 dB). An interval of 100 ms with background nose was introduced between each prepulse and pulse-alone stimulus. Each trial (startle pulse alone; pulse preceded by 70, 75, or 80 dB; or no stimulus) was presented in a pseudorandom order, with intertrial intervals ranging from 8 to 22 s. The startle response amplitude was defined as the average of the maximum force detected during a reaction to a 120-dB startle stimulus. The percentage of PPI was calculated using the following formula: (%) = 100 − [(startle amplitude after prepulse and pulse)/(startle amplitude after pulse only) * 100].
Patients with Schizophrenia.
Postmortem tissue from patients with schizophrenia and controls was obtained with ethical approval and upon informed consent from the Harvard Brain Tissue Resource Center (Boston), Brainnet Europe II (Munich), and the brain bank of Hospital Sant Joan de Deu (Spain). Samples were matched for age and postmortem delay.
For analysis of blood samples, the patients were recruited from a large network of clinical centers across Germany, comprising both academic and nonacademic facilities. These centers are clinical collaborators of an ongoing study on genotype–phenotype relationships and the neurobiology of the longitudinal course of major psychiatric disorders (www.kfo241.de and www.PsyCourse.de). This study combines comprehensive longitudinal deep phenotyping, longitudinal sampling of various types of biomaterial, and state-of-the art proband management and biobanking, as well as advanced data protection, governed by stringent standard operating procedures for a multicenter study framework like ours (52–55). Blood was collected in Pax gene tubes and incubated for 2 h at room temperature. Subsequently, tubes were stored at −20 °C for 72 h before being transferred to −80 °C for long-term storage. RNA was then isolated according to a user’s manual. All patients included in this study were suffering from paranoid schizophrenia [DSM-IV diagnosis code 295.30 (35)]. They were diagnosed using a comprehensive phenotyping inventory, including a structured diagnostic interview (56), clinical rating scales, questionnaires, and additional information systematically gleaned from medical records (whenever available). Structured diagnostic interviews and rating scales were administered by psychologists, graduate students with at least a bachelor’s degree in psychology, board-certified psychiatrists, or psychiatric residents who underwent training in structured diagnostic assessments. For the assessment of ELS, we asked patients to complete the childhood trauma screener (CTS) (31). The CTS is a five-item questionnaire measuring physical, sexual, and emotional abuse, as well as physical and emotional neglect, in childhood and adolescence. Based on validated threshold values (32), patients were classified as either having experienced ELS (n = 38) or not (n = 39). For human samples, written informed consent was obtained from all study participants before inclusion to the study from the local ethical committees. The experiments were approved by the ethical committee of the Georg-August University Göttingen, and the ethical committee of the Ludwig-Maximilians University, Munich.
Injection of MS-725.
For i.p. injections of MS-275, a protocol previously described by Engmann et al. (50) was applied. In brief, MS-275 stock solution (100 μg/μL) was diluted 1:80 in DMSO and PBS to obtain a 1.25-μg/μL working solution. Mice were then injected i.p. with 12.5 mg/kg of MS-275 for 10 d. Control mice received a PBS-DMSO mixture.
Stereotaxic Injection of AAV Particles.
For stereotaxic injections of AAV particles in the prefrontal cortex and ventral hippocampus of mice, animals were anesthetized and placed in a stereotaxic frame, and the heads of mice were cleaned and disinfected with 70% ethanol. After removal of the skin and connective tissue on the skull, two holes were drilled according to the desired coordinates. In the case of the prefrontal cortex, the two holes were drilled according to the following coordinates: anterior/posterior, +1.40 mm relative to bregma; medial/lateral ±0.45 mm. In the case of the ventral hippocampus, the coordinates were as follows: anterior/posterior, −3.00 mm relative to bregma; medial/lateral ±3.00 mm. Glass capillaries filled with mineral oil on the top, a small air bubble in the middle, and respective AAV particles in the bottom were placed on a Nanoliter 2000 microinjector. The microinjector was connected to an ultra-microsyringe pump to control the speed and volume of injection. For injections with HDAC1-GFP-AAV or GFP-AAV, 1 μL with 1.0 * 108 transducing units was injected per hemisphere.
Tissue Dissection and RNA Isolation.
Mice were killed by cervical dislocation, and the brain tissue was quickly isolated on ice. Unless otherwise indicated, tissue was harvested immediately after the end of the experiment. Tissue was snap-frozen in liquid nitrogen and stored at −80 °C until processing. For RNA extraction, tissue was homogenized in TRI Reagent (Sigma–Aldrich) and RNA was extracted according to the manufacturer’s instructions. RNA was treated with DNaseI (Invitrogen) before sequencing. Briefly, RNA was resuspended in water and treated with 2 units of DNaseI for 20 min at 37 °C. DNA-free RNA was then repurified with phenol/chloroform and resuspended in diethyl pyrocarbonate water according to standard protocols. RNA was stored at −80 °C (57).
qPCR.
Total RNA was extracted using the TRI Reagent according to the manufacturer’s recommendations, and the concentration of RNA was determined using a Nano-Drop ND-1000 spectrophotometer (Thermo Scientific) and a bioanalyzer (Stratagene). RNA samples were reverse-transcribed into first-strand cDNA with reverse transcriptase (Roche). Real-time PCR detection of a desired sequence was carried out using an LC480 LightCycler (Roche Applied Science). Primers for gene expression analysis were obtained according to the Universal Probe Library (Roche). Data were normalized to the housekeeping gene Gapdh. Primers used for ChIP and MeDIP were custom-made (a list of primers is provided in Table S1).
Table S1.
Primer sequences
| Primer | Assay | Sequence (5′-3′) forward | Sequence (5′-3′) reverse |
| Pvalbmouse | UPL | ggcaagattggggttgaag | agcagtcagcgccacttag |
| Gad1mouse | UPL | ttccgggacatgagcagt | atacaacctttggctggctgcatgt |
| Kcnv1mouse | UPL | gtctggtccaggtcttgagg | ccaagtgagcgaatcct |
| Pvalbhuman | UPL | caaaattggggttgacgaa | ggggcagtcagtgcttctta |
| Gad1human | UPL | tcaagtaaagatggtgatgggata | gccatgatgctgtacatgttg |
| Kcnv1human | UPL | aaagctgggcagacattcc | aaatatagagattcccacggatagaa |
| Hdac1human | UPL | ccaagtaccacagcgatgac | tggacagtcctcaccaacg |
| Hdac2human | UPL | cagatcgtgtaatgacggtatca | ccttttccagcaccaatatcc |
| Hdac3human | UPL | gacctatgacaggactgatgagg | gaactcattgggtgcctctg |
| Hdac8human | UPL | ccaacaaatcctcaactacatca | aacacgctgtcttcattataggc |
| Gapdhhuman | UPL | agccacatcgctcagacac | agccacatcgctcagacac |
| Hdac1mouse | UPL | tggtctctaccgaaaaatggag | tcatcactgtggtacttggtca |
| Hdac2mouse | UPL | ctccacgggtggttcagt | cccaattgacagccatatca |
| Hdac3mouse | UPL | ttcaacgtgggtgatgactg | ttagctgtgttgctccttgc |
| Hdac8mouse | UPL | gcagctggcaactctgatt | gtcaagtatgtccagcaacgag |
| Gapdhmouse | UPL | aagagggatgctgcccttac | ccattttgtctacgggacga |
| Kcnv1mouse | ChIP | tctttcagttccagcggagt | ctgccttgctctgcctctat |
| Pvalbmouse | ChIP | gtttcaaatgtggtcaggt | ggttgttggatatcagagc |
| Gad1mouse | ChIP | gcctcctagacttgagagtcg | gagacagaagggagaccaa |
| GR site in Hdac1mouse | MeDIP | tggggaccaaataagagaacc | aggctggtctctaacccaca |
| Non-GR site in Hdac1mouse | MeDIP | tgtctagctcccccttcaga | tagggcctgcagttgctagt |
| Pvalbmouse | MeDIP | tatgagcagaaaggatgaggag | aggctcctatcgccttcttgatg |
| Kcnv1mouse | MeDIP | catcaagctggtggaaagagtagg | gcactgggaagttgggaag |
| Gad1mouse | MeDIP | gccatactggctcagtagagcgctc | ccagaatgcactcagcacgctctg |
UPL, Universal Probe Library (Roche).
MeDIP/ChIP.
MeDIP.
Sample preparation for MeDIP and sequencing was conducted as described previously (58). We isolated the mPFC from five mice at 3 mo of age and performed MeDIP sequencing for all five samples. Briefly, 200 ng of sheared genomic DNA was end-repaired and adaptor-ligated using a NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (E6240 kit; New England Biolabs), followed by the immunoprecipitation of methylated regions using 5-mC monoclonal antibody (BI-MECY-0100; Eurogentech). Immunoprecipitated as well as input DNA was PCR-amplified using 2× Phusion High-Fidelity PCR Master Mix (ThermoFisher Scientific) as per the kit’s protocol. Sequencing for MeDIP and input DNA libraries was performed on a HiSeq 2000 instrument (Illumina) using the standard Illumina protocol at Fasteris SA. Sequenced reads were mapped using Bowtie 2 (59), and further analysis for methylated regions was done with the MEDIPS package (60) using input as a control. A false discovery rate of 0.1 was taken as the cutoff value for significantly methylated regions compared with input samples. Methylated regions were annotated using the R package ChIPseeker. BAM files for all replicates were merged, sorted, and indexed using SAMtools. A WIG file was produced via the MEDIPS package and was used for visualization in the Integrative Genome Browser. The program ngsplot was used to plot the DNAme across the high-, medium-, and low-expressed genes. Genes ware classified into high-, medium-, and low-expressed according to expression level as mentioned earlier (58). Briefly, we calculated the fragments per kilobase of exon per million reads (FPKM) values for the genes of the RNA-sequencing data from the same region from wild-type mice (n = 3) using cufflinks. Genes with an average FPKM between 1 and 5 were considered as low-expressed genes, genes with 5–30 FPKM were considered as medium-expressed genes, and genes with an FPKM higher than 30 were considered as high-expressed genes. An aggregate gene plot was created using ngsplot with the parameters −MW 5 (smoothing of average profiles), −RB 0.01 (trimmed mean to remove extreme values), and −F protein_coding. For ELS samples, immunoprecipitation was carried out directly using the sheared genomic DNA without any end-repair and adapter-ligation steps. qPCR was done from immunoprecipitated and input DNA for the glucocorticoid receptor-binding site and control region in the Hdac1 gene based on the wild-type MeDIP-sequencing data. For identification of the GR-binding site and control region, the enriched region sequences from the wild-type mice MeDIP-sequencing data were taken. Further analysis was performed using the JASPER database for the identification of GR-binding sites using the default parameters.
ChIP.
All procedures were carried out in DNA low-binding tubes from Eppendorf (or Diagenode, where indicated). Tissue was lysed in radioimmunoprecipitation assay-SDS [140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 10 mM Tris⋅HCl (pH 8), 1% SDS] buffer for 10 min at 4 °C in a rotating wheel in sonication tubes (Diagenode). Samples were then sonicated for 20 min in a Bioruptor plus NGS device (Diagenode) at the high setting and spun down every four cycles to ensure homogeneous sonication. Chromatin was centrifuged at 18,000 × g for 5 min at 4 °C, and the supernatant was snap-frozen as sheared chromatin. A small sample was checked for efficient sonication in a Bioanalyzer (Agilent) and used to estimate chromatin concentration in the sample after reversion of cross-linking, which was achieved by treating samples with 0.1 μg/μL RNaseA (Qiagen) and then digesting protein with proteinase K (PK; Roth). Chromatin was precleared by diluting it to 10-fold the initial volume in immunoprecipitation buffer [150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 50 mM Tris⋅HCl (pH 8), 20 mM EDTA, 0.1% SDS], adding 20 μL of protein A-coded magnetic Dynabeads (Life Technologies), and incubating the mixture at 4 °C for at least 4 h in a rotating wheel. For HDAC1 ChIP, 4 μL of antibody (Diagenode) was incubated with 500 μg of precleared chromatin overnight at 4 °C in a rotating wheel. Immunoprecipitated chromatin was then recovered by adding 15 μL of magnetic Dynabeds, with further incubation for 2 h at 4 °C in a rotating wheel. Samples were then washed twice with immunoprecipitation buffer, three times with wash buffer [100 mM Tris⋅HCl (pH 8), 500 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 20 mM EDTA], twice with immunoprecipitation buffer, and twice with TE buffer (10 mM Tris⋅HCl, 1 mM EDTA). Samples were kept on ice at all times. Beads were collected with a magnetic stand (Invitrogen). After the last wash, immunoprecipitation and input samples (input samples constituted 10% of the material used for ChIP) were collected and eluted in 20 μL of EB buffer [10 mM Tris⋅HCl (pH 8)] containing 0.1 μg/μL RNaseA and then incubated at 37 °C for 30 min under gentle agitation in a Thermomix (Eppendorf). Samples were then further diluted 1:2 with WB buffer [100 mM Tris⋅HCl (pH 8), 20 mM EDTA, 2% SDS], and 1 μL of PK (20 mg/mL) was added. Samples were then incubated overnight at 65 °C under agitation in a Thermomix. DNA was recovered on the magnetic stand and reeluted with EB buffer for 10 min at 65 °C. Inputs were carried out in parallel after the immunoprecipitation. DNA was precipitated with SureClean (Bioline) and linear polyacrylamide (Ambion), washed twice with 70% EtOH, and quantified using a Qubit Fluorometer (Life Technologies).
All buffers were supplemented with a proteinase inhibitor mixture (reference no. 04 693 132 001; Roche).
Immunoblotting and Immunohistochemistry.
Brain tissue was homogenized in TX buffer (50 mM Tris, 150 mM NaCl, 2 M EDTA, 1% Triton-X, protease inhibitors), incubated for 15 min at 4 °C, and centrifuged for 10 min (9,391 × g). The supernatant was used for immunoblotting. Immunoblots were performed using fluorescent secondary antibodies, and data were quantified using an Odyssey Imager (Licor). Antibodies were diluted either in 0.5% milk phosphate buffered saline or 0.5% milk TRIS buffered saline, respectively. Immunostaining was performed as described previously (14, 61) and analyzed using a Leica SP2 confocal microscope. The following antibodies were commercially purchased and used at the cited concentrations: GAPDH (1:5,000; Chemicon), HDAC1 (1:1,000; Diagenode), GFP, Cy3-labeled (goat anti-rabbit, 1:500; Jackson ImmunoResearch), and Alexa 488-labeled (donkey anti-mouse, 1:500; Invitrogen). Immunostaining was performed as described previously (57, 62) and analyzed using a Leica SP2 confocal microscope. The following antibodies were commercially purchased and used at the cited concentrations: HDAC1 (1:1,000, H-51; Santa Cruz Biotechnology) and Pvalb (1:1,000; Swant). Secondary antibodies were as follows: Alexa 633-labeled (1:2,000, goat anti-rabbit; Jackson ImmunoResearch), Cy3-labeled (1:500, goat anti-rabbit; Jackson ImmunoResearch), and Alexa 488-labeled (1:500, donkey anti-mouse; Invitrogen).
Electrophysiology.
Mice overexpressing HDAC1-GFP or GFP in the mPFC were killed by cervical dislocation. After quick decapitation and removal of the brain, 400-μm-thick coronal slices containing the prefrontal area (1.9–1.1 mm anterior to bregma) were cut using a VT1200M vibratome (Leica) in ice-cold artificial cerebrospinal fluid (ACSF) equilibrated with 95% O2/5% CO2 and containing the following: 250 mM sucrose, 24 mM NaHCO3, 25 mM glucose, 2.5 mM KCl, 1.25 mM NaH2PO4, 2 mM CaCl2, and 1.5 mM MgCl2 (pH 7.4). The slices were then kept for at least 2 h before the start of recordings in ACSF that was continuously gassed with 95% O2/5% CO2 and contained 120 mM NaCl instead of sucrose (63). Experiments were performed in the same solution at a flow rate of 4 mL⋅min−1 in a submerged chamber (2 mL) and at room temperature (22–24 °C). Recordings of fEPSPs were performed with glass pipettes filled with ACSF in the mPFC, including prelimbic and cingulate cortex. A stimulation electrode (0.3–0.5 MΩ) was placed in layers II–III of the prefrontal cortex to stimulate electrically the input fibers of pyramidal cells. Evoked fEPSPs were recorded with a glass electrode (2–2.5 MΩ) in layer V, which is the location of the dendrites and cell bodies of pyramidal neurons (64). Brain slices were viewed using an Olympus microscope fitted with fluorescence and infrared differential interference contrast videomicroscopy. Fluorescent mPFC neurons were identified by epifluorescence, and only slices with obvious GFP expression in the prefrontal area were selected for experiments.
Basal synaptic transmission and paired-pulse facilitation (PPF) were measured before inducing LTP. Stimulus–response curves at stimulus intensities of 40–85 μA were constructed to measure levels of excitatory synaptic transmission. To study short-term plasticity, PPF was assessed by recording the synaptic response to a pair of stimuli at 0.033 Hz with an interpulse interval of 50 ms. For PPF experiments, the stimulation intensity was set to elicit fEPSPs with a magnitude of ∼30% of the maximum (defined as a response with a detectable population spike). PPF was estimated as the ratio between the slopes of the second and first fEPSPs (percentage). Basal synaptic transmission was monitored at 0.05 Hz for a minimum of 10 min. To induce LTP in the mPFC, five trains of TBS with an inter–theta-train interval of 20 s were applied (65). Each train consisted of eight bursts delivered at 5 Hz. Each burst consisted of four pulses delivered at 100 Hz. The duration of pulses was 0.2 ms, and the stimulation intensity was set to provide fEPSPs with amplitude of ∼50% from the value corresponding to a sweep for which a population spike is first detectable. In all LTP experiments, the level of LTP represents the average fEPSP slope measured during 50–60 min after TBS normalized to the baseline fEPSP slope. The level of STP was determined as the maximal potentiation within the first 1 min after TBS. The data were acquired using an EPC10 amplifier at a sampling rate of 10 kHz, low-pass-filtered at 3 kHz, analyzed using PatchMaster software (Heka Elektronik), and presented using SigmaPlot (Systat Software, Inc.). Stimulus artifacts on figures were blanked to facilitate the perception of fEPSPs.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Research Group KFO241/PsyCourse Fi981-4 and Fi981 11-1 (A.F.), DFG Project 179/1-1/2013 (A.F.), and an European Research Council (ERC, 648898) consolidator grant (to A.F.). P.F. was supported by the KFO241/PsyCourse Project FA 241/16-1. F.S. was supported by the DFG Grant SA1050/2-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data from sequencing experiments reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE90150).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613842114/-/DCSupplemental.
References
- 1.Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraepelin E. Dementia Praecox and Paraphrenia. Livingstone; Edinburgh, Scotland: 1919. [Google Scholar]
- 3.Heilbronner U, Samara M, Leucht S, Falkai P, Schulze TG. The longitudinal course of schizophrenia across the lifespan: Clinical, cognitive, and neurobiological Aspects. Harv Rev Psychiatry. 2016;24:118–128. doi: 10.1097/HRP.0000000000000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyegbe C, Campbell D, Butler A, Ajnakina O, Sham P. The emerging molecular architecture of schizophrenia, polygenic risk scores and the clinical implications for GxE research. Soc Psychiatry Psychiatr Epidemiol. 2014;49:169–182. doi: 10.1007/s00127-014-0823-2. [DOI] [PubMed] [Google Scholar]
- 6.Stepniak B, et al. Accumulated common variants in the broader fragile X gene family modulate autistic phenotypes. EMBO Mol Med. 2015;7:1565–1579. doi: 10.15252/emmm.201505696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinz A, Deserno L, Reininghaus U. Urbanicity, social adversity and psychosis. World Psychiatry. 2013;12:187–197. doi: 10.1002/wps.20056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown AS. Epidemiologic studies of exposure to prenatal infection and risk of schizophrenia and autism. Dev Neurobiol. 2012;72:1272–1276. doi: 10.1002/dneu.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt A, Malchow B, Hasan A, Falkai P. The impact of environmental factors in severe psychiatric disorders. Front Neurosci. 2014;8:19. doi: 10.3389/fnins.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akdeniz C, Tost H, Meyer-Lindenberg A. The neurobiology of social environmental risk for schizophrenia: An evolving research field. Soc Psychiatry Psychiatr Epidemiol. 2014;49:507–517. doi: 10.1007/s00127-014-0858-4. [DOI] [PubMed] [Google Scholar]
- 11.Fischer A. Epigenetic memory: The Lamarckian brain. EMBO J. 2014;33:945–967. doi: 10.1002/embj.201387637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer A. Targeting histone-modifications in Alzheimer’s disease. What is the evidence that this is a promising therapeutic avenue? Neuropharmacology. 2014;80:95–102. doi: 10.1016/j.neuropharm.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 13.Nestler EJ, Peña CJ, Kundakovic M, Mitchell A, Akbarian S. Epigenetic basis of mental illness. Neuroscientist. 2016;22:447–463. doi: 10.1177/1073858415608147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahari-Javan S, et al. HDAC1 regulates fear extinction in mice. J Neurosci. 2012;32:5062–5073. doi: 10.1523/JNEUROSCI.0079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koseki T, et al. Exposure to enriched environments during adolescence prevents abnormal behaviours associated with histone deacetylation in phencyclidine-treated mice. Int J Neuropsychopharmacol. 2012;15:1489–1501. doi: 10.1017/S1461145711001672. [DOI] [PubMed] [Google Scholar]
- 16.Hyman SE. Target practice: HDAC inhibitors for schizophrenia. Nat Neurosci. 2012;15:1180–1181. doi: 10.1038/nn.3200. [DOI] [PubMed] [Google Scholar]
- 17.Kurita M, et al. HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat Neurosci. 2012;15:1245–1254. doi: 10.1038/nn.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong E, Guidotti A, Grayson DR, Costa E. Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc Natl Acad Sci USA. 2007;104:4676–4681. doi: 10.1073/pnas.0700529104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki T, et al. Augmentation of atypical antipsychotics with valproic acid. An open-label study for most difficult patients with schizophrenia. Hum Psychopharmacol. 2009;24:628–638. doi: 10.1002/hup.1073. [DOI] [PubMed] [Google Scholar]
- 20.Haddad PM, Das A, Ashfaq M, Wieck A. A review of valproate in psychiatric practice. Expert Opin Drug Metab Toxicol. 2009;5:539–551. doi: 10.1517/17425250902911455. [DOI] [PubMed] [Google Scholar]
- 21.Benes FM, et al. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci USA. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma RP, Grayson DR, Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: analysis of the National Brain Databank microarray collection. Schizophr Res. 2008;98:111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakovcevski M, et al. Prefrontal cortical dysfunction after overexpression of histone deacetylase 1. Biol Psychiatry. 2013;74:696–705. doi: 10.1016/j.biopsych.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aas M, et al. A systematic review of cognitive function in first-episode psychosis, including a discussion on childhood trauma, stress, and inflammation. Front Psychiatry. 2014;4:182. doi: 10.3389/fpsyt.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Kloet ER, Sibug RM, Helmerhorst FM, Schmidt MV. Stress, genes and the mechanism of programming the brain for later life. Neurosci Biobehav Rev. 2005;29:271–281. doi: 10.1016/j.neubiorev.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Ellenbroek BA, van den Kroonenberg PT, Cools AR. The effects of an early stressful life event on sensorimotor gating in adult rats. Schizophr Res. 1998;30:251–260. doi: 10.1016/s0920-9964(97)00149-7. [DOI] [PubMed] [Google Scholar]
- 28.Niwa M, Matsumoto Y, Mouri A, Ozaki N, Nabeshima T. Vulnerability in early life to changes in the rearing environment plays a crucial role in the aetiopathology of psychiatric disorders. Int J Neuropsychopharmacol. 2011;14:459–477. doi: 10.1017/S1461145710001239. [DOI] [PubMed] [Google Scholar]
- 29.Powell SB, Zhou X, Geyer MA. Prepulse inhibition and genetic mouse models of schizophrenia. Behav Brain Res. 2009;204:282–294. doi: 10.1016/j.bbr.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maccari S, Krugers HJ, Morley-Fletcher S, Szyf M, Brunton PJ. The consequences of early-life adversity: Neurobiological, behavioural and epigenetic adaptations. J Neuroendocrinol. 2014;26:707–723. doi: 10.1111/jne.12175. [DOI] [PubMed] [Google Scholar]
- 31.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 32.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verwer RW, Meijer RJ, Van Uum HF, Witter MP. Collateral projections from the rat hippocampal formation to the lateral and medial prefrontal cortex. Hippocampus. 1997;7:397–402. doi: 10.1002/(SICI)1098-1063(1997)7:4<397::AID-HIPO5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 34.Parent MA, Wang L, Su J, Netoff T, Yuan LL. Identification of the hippocampal input to medial prefrontal cortex in vitro. Cereb Cortex. 2010;20:393–403. doi: 10.1093/cercor/bhp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. 4th Ed American Psychiatric Association Publishing; Arlington, VA: 1994. [Google Scholar]
- 36.Grabe HJ, et al. Ein Screeninginstrument für Missbrauch und Vernachlässigung in der Kindheit: der Childhood Trauma Screener (CTS) Psychiatr Prax. 2012;39:109–115. German. doi: 10.1055/s-0031-1298984. [DOI] [PubMed] [Google Scholar]
- 37.Glaesmer H, et al. [The childhood trauma screener (CTS) - development and validation of cut-off-scores for classificatory diagnostics] Psychiatr Prax. 2013;40:220–226. German. doi: 10.1055/s-0033-1343116. [DOI] [PubMed] [Google Scholar]
- 38.van den Buuse M. Modeling the positive symptoms of schizophrenia in genetically modified mice: Pharmacology and methodology aspects. Schizophr Bull. 2010;36:246–270. doi: 10.1093/schbul/sbp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guidotti A, Dong E, Tueting P, Grayson DR. Modeling the molecular epigenetic profile of psychosis in prenatally stressed mice. Prog Mol Biol Transl Sci. 2014;128:89–101. doi: 10.1016/B978-0-12-800977-2.00004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jawahar MC, Murgatroyd C, Harrison EL, Baune BT. Epigenetic alterations following early postnatal stress: A review on novel aetiological mechanisms of common psychiatric disorders. Clin Epigenetics. 2015;7:122–134. doi: 10.1186/s13148-015-0156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subburaju S, Coleman AJ, Ruzicka WB, Benes FM. Toward dissecting the etiology of schizophrenia: HDAC1 and DAXX regulate GAD67 expression in an in vitro hippocampal GABA neuron model. Transl Psychiatry. 2016;6:e723. doi: 10.1038/tp.2015.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu Z, et al. The Rett syndrome protein MeCP2 regulates synaptic scaling. J Neurosci. 2012;32:989–994. doi: 10.1523/JNEUROSCI.0175-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Zhuo C, Qin W, Zhu J, Yu C. Altered spontaneous brain activity in schizophrenia: a meta-analysis and a large-sample study. BioMed Res Int. 2015;2015:204628. doi: 10.1155/2015/204628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert D, Martens GJ, Kolk SM. Molecular underpinnings of prefrontal cortex development in rodents provide insights into the etiology of neurodevelopmental disorders. Mol Psychiatry. 2015;20:795–809. doi: 10.1038/mp.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simonini MV, et al. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc Natl Acad Sci USA. 2006;103:1587–1592. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt A, et al. Members of the WFSBP Task Force on Biological Markers Consensus paper of the WFSBP Task Force on Biological Markers: Criteria for biomarkers and endophenotypes of schizophrenia, part III: Molecular mechanisms. World J Biol Psychiatry. 2016;26:1–27. doi: 10.1080/15622975.2016.1224929. [DOI] [PubMed] [Google Scholar]
- 47.Adler SM, Schmauss C. Cognitive deficits triggered by early life stress: The role of histone deacetylase 1. Neurobiol Dis. 2016;94:1–9. doi: 10.1016/j.nbd.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer A, Sananbenesi F, Schrick C, Spiess J, Radulovic J. Regulation of contextual fear conditioning by baseline and inducible septo-hippocampal cyclin-dependent kinase 5. Neuropharmacology. 2003;44:1089–1099. doi: 10.1016/s0028-3908(03)00102-3. [DOI] [PubMed] [Google Scholar]
- 49.Lalonde R, Strazielle C. Relations between open-field, elevated plus-maze, and emergence tests as displayed by C57/BL6J and BALB/c mice. J Neurosci Methods. 2008;171:48–52. doi: 10.1016/j.jneumeth.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Engmann O, et al. Schizophrenia is associated with dysregulation of a Cdk5 activator that regulates synaptic protein expression and cognition. Brain. 2011;134:2408–2421. doi: 10.1093/brain/awr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGarel C, Pentieva K, Strain JJ, McNulty H. Emerging roles for folate and related B-vitamins in brain health across the lifecycle. Proc Nutr Soc. 2015;74:46–55. doi: 10.1017/S0029665114001554. [DOI] [PubMed] [Google Scholar]
- 52.Demiroglu SY, et al. Managing sensitive phenotypic data and biomaterial in large-scale collaborative psychiatric genetic research projects: Practical considerations. Mol Psychiatry. 2012;17:1180–1185. doi: 10.1038/mp.2012.11. [DOI] [PubMed] [Google Scholar]
- 53.Anderson-Schmidt H, et al. The “DGPPN-Cohort”: A national collaboration initiative by the German Association for Psychiatry and Psychotherapy (DGPPN) for establishing a large-scale cohort of psychiatric patients. Eur Arch Psychiatry Clin Neurosci. 2013;263:695–701. doi: 10.1007/s00406-013-0401-8. [DOI] [PubMed] [Google Scholar]
- 54.Schwanke J, Rienhoff O, Schulze TG, Nussbeck SY. Suitability of customer relationship management systems for the management of study participants in biomedical research. Methods Inf Med. 2013;52:340–350. doi: 10.3414/ME12-02-0012. [DOI] [PubMed] [Google Scholar]
- 55.Nussbeck SY, Skrowny D, O’Donoghue S, Schulze TG, Helbing K. How to design biospecimen identifiers and integrate relevant functionalities into your biospecimen management system. Biopreserv Biobank. 2014;12:199–205. doi: 10.1089/bio.2013.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittchen HU, Fydrich T. 1997. Strukturiertes Klinisches Interview für DSM-IV (Hogrefe, Goettingen, Germany). German.
- 57.Benito E, et al. HDAC inhibitor-dependent transcriptome and memory reinstatement in cognitive decline models. J Clin Invest. 2015;125:3572–3584. doi: 10.1172/JCI79942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Halder R, et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci. 2016;19:102–110. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- 59.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: Genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30:284–286. doi: 10.1093/bioinformatics/btt650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 62.Stilling RM, et al. K-Lysine acetyltransferase 2a regulates a hippocampal gene expression network linked to memory formation. EMBO J. 2014;33:1912–1927. doi: 10.15252/embj.201487870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eckhardt M, et al. Mice deficient in the polysialyltransferase ST8SiaIV/PST-1 allow discrimination of the roles of neural cell adhesion molecule protein and polysialic acid in neural development and synaptic plasticity. J Neurosci. 2000;20:5234–5244. doi: 10.1523/JNEUROSCI.20-14-05234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang YY, Pittenger C, Kandel ER. A form of long-lasting, learning-related synaptic plasticity in the hippocampus induced by heterosynaptic low-frequency pairing. Proc Natl Acad Sci USA. 2004;101:859–864. doi: 10.1073/pnas.2237201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brennaman LH, et al. Transgenic mice overexpressing the extracellular domain of NCAM are impaired in working memory and cortical plasticity. Neurobiol Dis. 2011;43:372–378. doi: 10.1016/j.nbd.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tse MT, Piantadosi PT, Floresco SB. Prefrontal cortical gamma-aminobutyric acid transmission and cognitive function: Drawing links to schizophrenia from preclinical research. Biol Psychiatry. 2015;77:929–939. doi: 10.1016/j.biopsych.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 67.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Colantuoni C, et al. Age-related changes in the expression of schizophrenia susceptibility genes in the human prefrontal cortex. Brain Struct Funct. 2008;213:255–271. doi: 10.1007/s00429-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 69.Hyde TM, et al. Expression of GABA signaling molecules KCC2, NKCC1, and GAD1 in cortical development and schizophrenia. J Neurosci. 2011;31:11088–11095. doi: 10.1523/JNEUROSCI.1234-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitchell AC, Jiang Y, Peter C, Akbarian S. Transcriptional regulation of GAD1 GABA synthesis gene in the prefrontal cortex of subjects with schizophrenia. Schizophr Res. 2015;167:28–34. doi: 10.1016/j.schres.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subburaju S, Benes FM. Induction of the GABA cell phenotype: An in vitro model for studying neurodevelopmental disorders. PLoS One. 2012;7:e33352. doi: 10.1371/journal.pone.0033352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong E, Tueting P, Matrisciano F, Grayson DR, Guidotti A. Behavioral and molecular neuroepigenetic alterations in prenatally stressed mice: Relevance for the study of chromatin remodeling properties of antipsychotic drugs. Transl Psychiatry. 2016;6:e711. doi: 10.1038/tp.2015.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mellios N, et al. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 2009;65:1006–1014. doi: 10.1016/j.biopsych.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 74.Fung SJ, et al. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 2010;167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- 75.Koh DX, Sng JC. HDAC1 negatively regulates Bdnf and Pvalb required for parvalbumin interneuron maturation in an experience-dependent manner. J Neurochem. 2016;139:369–380. doi: 10.1111/jnc.13773. [DOI] [PubMed] [Google Scholar]
- 76.Vukadinovic Z, Rosenzweig I. Abnormalities in thalamic neurophysiology in schizophrenia: Could psychosis be a result of potassium channel dysfunction? Neurosci Biobehav Rev. 2012;36:960–968. doi: 10.1016/j.neubiorev.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 77.Norkett R, Modi S, Kittler JT. Mitochondrial roles of the psychiatric disease risk factor DISC1. Schizophr Res. January 10, 2017 doi: 10.1016/j.schres.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 78.Lipska BK, et al. Functional genomics in postmortem human brain: Abnormalities in a DISC1 molecular pathway in schizophrenia. Dialogues Clin Neurosci. 2006;8:353–357. doi: 10.31887/DCNS.2006.8.3/blipska. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang B, Jia H, Kast RJ, Thomas EA. Epigenetic changes at gene promoters in response to immune activation in utero. Brain Behav Immun. 2013;30:168–175. doi: 10.1016/j.bbi.2013.01.086. [DOI] [PubMed] [Google Scholar]
- 80.Walss-Bass C, et al. Methionine sulfoxide reductase: A novel schizophrenia candidate gene. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:219–225. doi: 10.1002/ajmg.b.30791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Regland B. Schizophrenia and single-carbon metabolism. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:1124–1132. doi: 10.1016/j.pnpbp.2005.06.023. [DOI] [PubMed] [Google Scholar]