Significance
As insecticide resistance is rapidly spreading, alternative tools for mosquito control are urgently needed. Beauveria bassiana is equally effective at killing insecticide-resistant and insecticide-susceptible mosquitoes. Better understanding of fungus–mosquito interactions is critical for improvement of its efficacy. Here we discover a contributory role for the gut microbiota in promoting fungal killing of mosquitoes via down-regulation of antimicrobial peptides and dual oxidase in the midgut. Fungal infection results in dysbiosis of mosquito gut microbiota by significantly increasing gut bacterial loads and decreasing bacterial diversity. In particular, fungal infection causes overgrowth and translocation of the opportunistic pathogen Serratia marcescens from the gut to the hemocoel, thus promoting mosquito death. Our study may lead to new strategies for biological control of mosquitoes.
Keywords: Anopheles, gut microbiota, dysbiosis, entomopathogenic fungus, immunity
Abstract
The insect gut microbiota plays crucial roles in modulating the interactions between the host and intestinal pathogens. Unlike viruses, bacteria, and parasites, which need to be ingested to cause disease, entomopathogenic fungi infect insects through the cuticle and proliferate in the hemolymph. However, interactions between the gut microbiota and entomopathogenic fungi are unknown. Here we show that the pathogenic fungus Beauveria bassiana interacts with the gut microbiota to accelerate mosquito death. After topical fungal infection, mosquitoes with gut microbiota die significantly faster than mosquitoes without microbiota. Furthermore, fungal infection causes dysbiosis of mosquito gut microbiota with a significant increase in gut bacterial load and a significant decrease in bacterial diversity. In particular, the opportunistic pathogenic bacterium Serratia marcescens overgrows in the midgut and translocates to the hemocoel, which promotes fungal killing of mosquitoes. We further reveal that fungal infection down-regulates antimicrobial peptide and dual oxidase expression in the midgut. Duox down-regulation in the midgut is mediated by secretion of the toxin oosporein from B. bassiana. Our findings reveal the important contribution of the gut microbiota in B. bassiana-killing activity, providing new insights into the mechanisms of fungal pathogenesis in insects.
Mosquitoes transmit a wide range of pathogens that cause diseases such as malaria, dengue, yellow fever, and Zika, which have a devastating impact on human health (1). Although vector control via insecticides is a major tool for disease control, intensive use of insecticides poses risks to humans and the environment and creates intensive pressure for mosquitoes to develop resistance. Thus, alternative tools for mosquito control are urgently needed (2).
An environmentally friendly alternative to chemical insecticides is offered by entomopathogenic fungi (3, 4). Among them is Beauveria bassiana (Cordycipitaceae), which has been widely used for the biological control of agricultural insect pests (5) and insect vectors of human diseases, including mosquitoes (6). This fungus is equally effective at killing insecticide-resistant and insecticide-susceptible mosquitoes, and is considered a next-generation control agent against mosquitoes (7). However, the relatively slow action of fungal pathogens, compared with chemical insecticides, has hampered their widespread application (8). To develop approaches to accelerate the speed at which a fungal pathogen kills its host, a better understanding of fungus–mosquito interactions is critical.
The mosquito gut is colonized by diverse communities of commensal bacteria, the microbiota, that play important roles in host physiology, particularly in modulation of host immune response and the outcome of pathogen infection (9–11). The gut microbiota has been recognized as a virtual “organ,” which is integrated into the biological system of the host and indispensable to its health (12–14). Coexistence between the insect and its microbiota is mostly harmonious, and in most cases is beneficial to the insect.
The protective roles of the gut microbiota against incoming intestinal pathogens have been studied in mosquitoes. In Anopheles and Aedes, removal of the gut microbiota with antibiotics renders the mosquito more susceptible to infection by the apicomplexan parasite Plasmodium and by the dengue virus (15–17). Gut bacteria protect their host insects against invading pathogens by stimulating the host immune response (16) or by producing antimalarial compounds (18).
However, recent studies have shown that the resident bacteria can also promote or assist the gut infection of incoming pathogens (19). The midgut bacterium Serratia odorifera enhances viral infection in Aedes (20) and Anopheles mosquitoes (21). Furthermore, pathogens can manipulate the microbiota to enhance infection. Abraham et al. have reported that the human pathogenic bacterium Anaplasma phagocytophilum (causative agent of anaplasmosis) appropriates the antibacterial protein of the tick vector, alters the host gut microbiota, and enables the pathogen to more efficiently colonize the tick (22).
Unlike viruses, bacteria and parasites, which need to be ingested to cause disease, pathogenic fungi primarily attack insects by penetrating the host integument and proliferating in the hemolymph (23). The interplay between the gut microbiota and fungal entomopathogens has not been examined. Outstanding issues include: Can gut microbiota protect insects from fungal infection? Do microbiota and fungal pathogens interact, or do they act independently? Can commensal gut bacteria become virulent when a fungal pathogen infects an insect? Understanding the tripartite interactions between mosquito host, resident microbiota, and fungal pathogen may yield new insights into pathogen–insect interactions, and may assist in the development of new insect control strategies and disease interventions.
In the present study, we investigated the interplay between the pathogenic fungus B. bassiana and the gut microbiota of the mosquito Anopheles stephensi. We found that topical infection by B. bassiana down-regulated the mosquito midgut immune responses via production of the toxin oosporein, caused dysbiosis of gut microbiota and translocation of bacteria from the gut to the hemocoel, where they switched from asymptomatic gut symbionts to hemocoelic pathogens, and accelerated fungal killing of mosquitoes.
Results
Effect of the Gut Microbiota on B. bassiana Pathogenesis in Adult Mosquitoes.
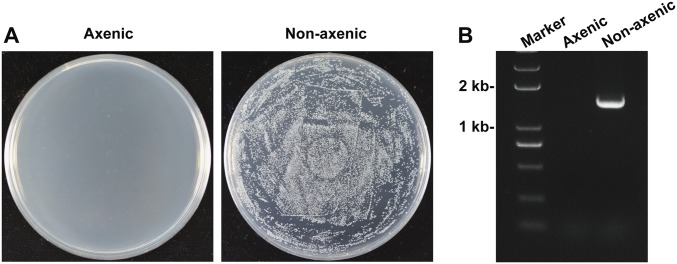
To investigate a possible role of the gut microbiota in fungal pathogenesis in the mosquito, axenic mosquitoes were generated via treatment with oral antibiotics. This treatment did not influence adult mosquito survival (24). The efficacy of elimination of midgut bacteria was confirmed by plating gut homogenates onto Luria–Bertani (LB) agar plates and performing PCR analysis using bacterial 16S ribosomal RNA (rRNA) gene universal primers (Fig. S1 and Table S1).
Fig. S1.
Generation of axenic mosquitoes. The efficacy of elimination of midgut bacteria confirmed by (A) culturing mosquito (n = 10) gut homogenates on LB agar plates and (B) by performing PCR analysis on mosquito (n = 10) gut homogenates using universal 16S rRNA gene primers.
Table S1.
Primers used in this study
| Primer | Sequence, 5′-3′ | Application |
| 16S-27F | AGAGTTTGATCCTGGCTCAG | PCR of eubacterial 16S rRNA gene |
| 16S-1492R | TACGGYTACCTTGTTACGACTT | |
| 16SF | TCCTACGGGAGGCAGCAGT | qPCR of eubacterial 16S rRNA gene |
| 16SR | GGACTACCAGGGTATCTAATCCTGTT | |
| SmF1 | ACGTTCATCAATTGACGTTACTCGCA | qPCR of S. marcescens16S rRNA gene |
| SmR1 | AACCGCCTGCGTGCGCTTTA | |
| 338F | ACTCCTACGGGAGGCAGCA | PCR of V3 and V4 hypervariable regions of eubacterial 16S rRNA gene |
| 806R | GGACTACHVGGGTWTCTAAT | |
| cDNA S7F | AGAACCAGCAGACCACCATC | qPCR of AsS7 as endogenous control for quantification of relative expression of immune genes |
| cDNA S7R | CGACGCACAGTAGCACAAAC | |
| gDNA AsS7F | TCCTGGAGCTGGAAATGAAC | qPCR of AsS7 as endogenous control for quantification of midgut bacteria |
| gDNA AsS7R | GCCGGGTCTGAACCTTCTGG | |
| GAM1F | GTACGTCAGCCGGAAGGGAG | qPCR of Gambicin 1 |
| GAM1R | CGTAATGAACGAGGACGAACAGC | |
| CEC1F | GGAAGCGGGACGCCTGAA | qPCR of Cecropin 1 |
| CEC1R | CCTTGACACCTGCCACCACC | |
| DEF1F | AGTCGTGGTCCTGGCGGCTCT | qPCR of Defensin 1 |
| DEF1R | ACGAGCGATGCAATGCGCGGCA | |
| ATTF | AAAGCCAGAGCGGCAACAC | qPCR of Attacin |
| ATTR | TCAGTAACCGTGCGTGAAAGTC | |
| FBN9F | AACAATCTGACCGCACTGC | qPCR of Fbn9 |
| FBN9R | TGTGACGCATTCCCTGTAG | |
| DuoxF | GGAACTGTTCTCGGCTGTCA | qPCR of Duox |
| DuoxR | TGATGTCCCAGAGCGTGAAC | |
| gpdF | CCCAGAACATCATTCCCAGC | qPCR of fungal gpd for quantification of B. bassiana |
| gpdR | TCAATGCGGGCAGTCAAGTC | |
| dsDuoxF | TAATACGACTCACTATAGGGGCTCGTCGGAAGTTTGTGAAAAA | RNAi of mosquito Duox |
| dsDuoxR | TAATACGACTCACTATAGGGTCGCATCATCTCACTCAGCTCTCC | |
| dsGFPF | TAATACGACTCACTATAGGGAGATGGTGAGCAAGGGCGAGGAGCTGT | RNAi of GFP as negative control |
| dsGFPR | TAATACGACTCACTATAGGGAGTTACTTGTACAGCTCGTCCATGCCG |
F, forward; R, reverse.
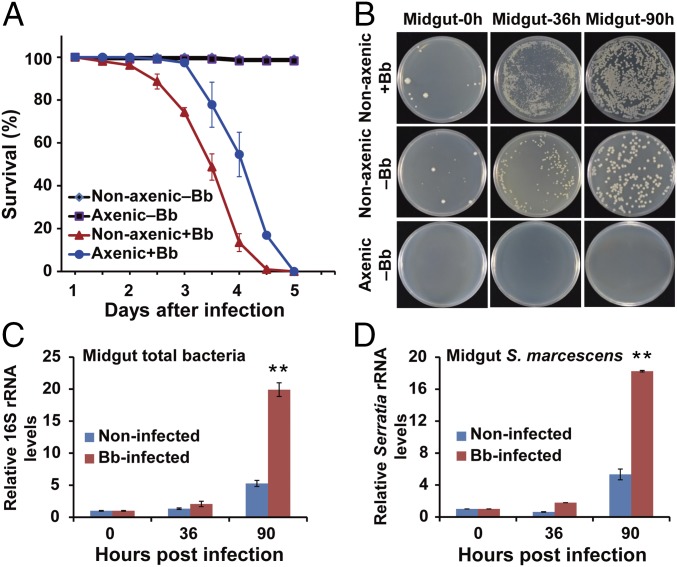
The insect bioassay was conducted using female mosquitoes with and without gut microbiota (nonaxenic and axenic, respectively). After topical inoculation with a B. bassiana conidial suspension, adult nonaxenic mosquitoes died significantly faster (median lethal time, LT50 79.9 ± 3.2 h) than did axenic mosquitoes (LT50 95.2 ± 3.1 h) (P < 0.05, t test) (Fig. 1A). This result suggests that the gut microbiota accelerates killing of mosquitoes by B. bassiana.
Fig. 1.
Effect of gut microbiota on pathogenesis in A. stephensi infected by the fungus B. bassiana (Bb). (A) Survival of axenic (without microbiota) and nonaxenic (with microbiota) mosquitoes (n = 50) following topical infection (+Bb) or no topical infection (−Bb) with B. bassiana. (B) Load of midgut cultivable bacteria from Bb-infected nonaxenic, noninfected nonaxenic, and noninfected axenic mosquitoes (n = 20) at 0, 36, and 90 h post fungal infection. Bacterial load was determined by plating the homogenate of mosquito midguts with 10,000 dilution on LB agar plates. Representative images are shown. (C and D) Number of midgut total bacteria (C) and S. marcescens (D) in Bb-infected and noninfected mosquitoes (n = 15) at 0, 36, and 90 hpi; quantification was by 16S rRNA gene-based qPCR analysis. Three biological replicates were conducted. Error bars indicate SD. Double asterisks represent a significant difference determined by the Student’s t test at P < 0.01.
B. bassiana Infection Causes Dysbiosis of the Gut Microbiota.
We next tested whether fungal infection affected the homeostasis of mosquito gut microbiota by determining the cultivable bacterial loads in the midgut of the mosquitoes at 36 and 90 h post topical fungal infection (hpi). Bacterial load was significantly increased in B. bassiana-infected mosquitoes at 36 and 90 hpi compared with noninfected controls treated with 0.01% Triton X-100 (Fig. 1B). Quantitative PCR (qPCR) showed that total bacterial load in infected mosquitoes at 90 hpi was significantly higher (∼3.7-fold higher) than in noninfected controls (P < 0.001) (Fig. 1C).
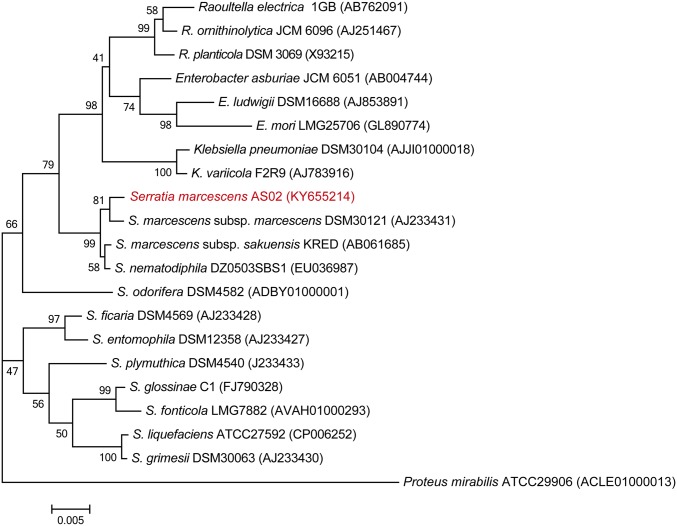
Based on 16S rRNA gene sequence analysis, the predominant cultivable bacterium had high similarity to Serratia marcescens (99% identity) (Fig. S2). We validated the proliferation of S. marcescens in the gut of infected mosquitoes using Serratia-specific 16S rRNA gene PCR primers (Table S1). Consistent with proliferation of the total gut bacteria, S. marcescens numbers significantly increased (by 3.4-fold, P = 0.014) in the infected mosquitoes at 90 hpi compared with noninfected controls (Fig. 1D). S. marcescens is a prevalent midgut bacterium in laboratory-reared and field-collected mosquitoes (25, 26), and can also be an opportunistic pathogen in mosquitoes under certain conditions (20, 27, 28).
Fig. S2.
Phylogenetic analysis of the translocating isolate S. marcescens AS02 with 20 related bacteria. The phylogenetic relationships were inferred from the alignment of 1,508 bp of 16S rRNA gene indicating the positions of S. marcescens AS02 relative to selected Serratia species and other genera within the family Enterobacteriaceae. The evolutionary history was inferred by using the neighbor-joining method conducted in MEGA7 software. A sequence of Proteus mirabilis (a member of the Enterobacteriaceae family) was used as the outgroup. Bootstrap values (percentages of 1,000 tree replications) greater than 40% are displayed at the nodes. Sequence GenBank accession numbers are shown in parentheses. (Bar, 0.005 substitutions per nucleotide position.)
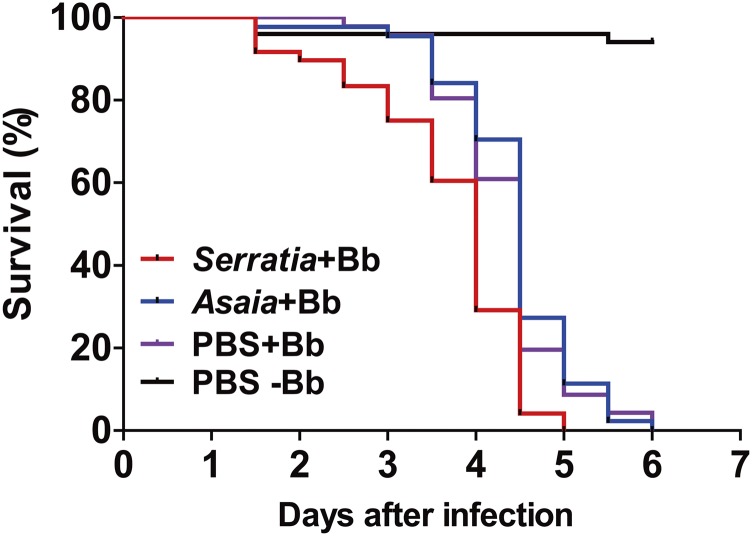
To determine whether the proliferated bacteria contributed to the speed of the kill, we reintroduced the overproliferating S. marcescens recovered from the midgut of B. bassiana-infected mosquitoes into the midgut of axenic mosquitoes via sugar meals. We found that the reintroduction of S. marcescens restored mosquito susceptibility to fungal infection, in comparison with nonvirulent symbiotic bacterium Asaia sp.-treated mosquitoes (29) and PBS controls (Fig. S3).
Fig. S3.
Survival of mosquitoes (n = 50) fed S. marcescens or Asaia sp. after topical infection with a 5 × 108 conidia per mL suspension of B. bassiana. Control mosquitoes were fed PBS. Bacteria were introduced into adult mosquito midgut via a sugar meal; female adults were allowed to feed for 24 h on 5% sucrose containing bacteria at a final concentration of 108 cells per mL. Introduction of S. marcescens (Serratia+Bb) significantly increased mosquito susceptibility to fungal infection compared with the PBS+Bb treatment [log-rank (Mantel–Cox) test, P < 0.001]. The experiments were performed in three biological replicates. The log-rank test was used to assess the significance of differences between two survival curves using GraphPad Prism software.
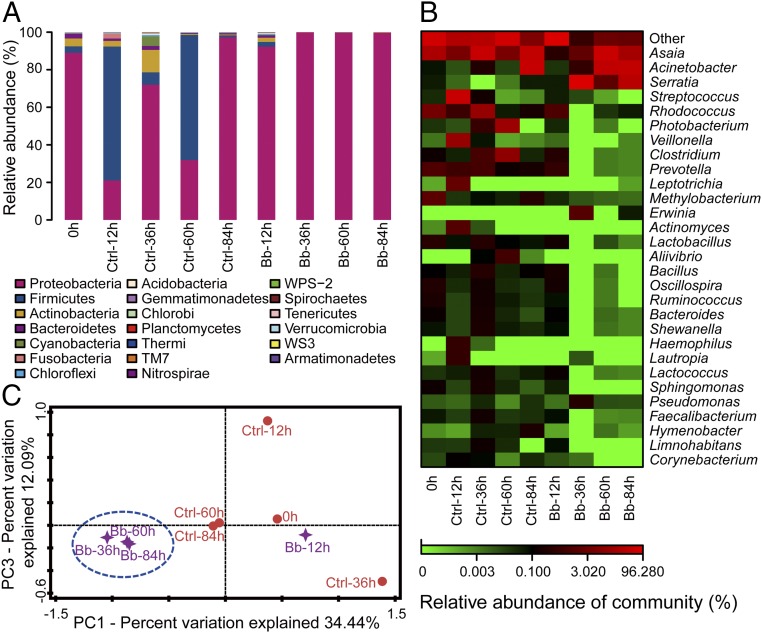
We further assessed the dynamic composition and diversity of the midgut bacteria in the noninfected mosquitoes and the fungus-infected mosquitoes at 0, 12, 36, 60, and 84 hpi by deep sequencing of 16S rRNA genes (Table S2). In the noninfected mosquitoes, the midgut bacteria were diverse and dominated by bacteria of six phyla: Proteobacteria, Firmicutes, Actinobacteria, Bacterioidetes, Fusobacteria, and Cyanobacteria (Fig. 2A). The abundance of Proteobacteria and Firmicutes changed dynamically over time, possibly because of changes in mosquito physiology. The Proteobacteria Acinetobacter, Photobacterium, and Asaia, the Firmicutes Streptococcus, and the Actinobacteria Rhodococcus increased in abundance over time in the noninfected mosquitoes (Fig. 2B).
Table S2.
Number of analyzed 16S rRNA gene sequences for gut bacteria of noninfected or Bb-infected mosquitoes at five time points after fungal topical inoculation in deep sequencing
| Ctrl | Bb | ||||||||
| 0 h | 12 h | 36 h | 60 h | 84 h | 12 h | 36 h | 60 h | 84 h | |
| Sequences | 85,506 | 75,521 | 65,040 | 78,164 | 72,895 | 81,394 | 17,344 | 75,314 | 64,836 |
Ctrl, noninfected.
Fig. 2.
Fungal infection alters the composition of gut microbiota in mosquitoes. (A) Histogram showing temporal changes, at the phylum level, in noninfected (Ctrl; Triton treatment as control) and Bb-infected mosquitoes (n = 40) over 84 h. (B) Heat map showing temporal changes, at the genus level, in Ctrl and Bb-infected mosquitoes. (C) Principal component analysis of unweighted jack-knifed UniFrac distances of microbial communities from Ctrl and Bb-infected mosquitoes.
Fungal infection decreased the bacterial diversity in comparison with noninfected mosquitoes (Table S3). Starting at 36 hpi, the single phylum of Proteobacteria predominated in B. bassiana-infected mosquitoes (Fig. 2A). The composition and diversity of the midgut bacterial population changed markedly in mosquitoes after topical infection by B. bassiana, resulting in almost exclusive colonization by three genera of Proteobacteria: Acinetobacter, Serratia, and Asaia. Serratia overgrew in the fungus-infected mosquitoes, yet was not dominant in noninfected mosquitoes (Fig. 2B).
Table S3.
Three diversity indexes analyzed upon deep sequencing for each sample of gut microbiota of noninfected or Bb-infected mosquitoes at five time points after fungal topical inoculation
| Ctrl | Bb | |||||||||
| 0 h | 12 h | 36 h | 60 h | 84 h | 12 h | 36 h | 60 h | 84 h | ||
| Chao1 | Average | 773 | 1140.92 | 901.46 | 1445.73 | 548.93 | 948.11 | 36.93 | 133.79 | 163.08 |
| SD | 52.16 | 93.19 | 21.86 | 120.16 | 54.27 | 48.57 | 1.92 | 17.1 | 24.11 | |
| Shannon | Average | 2.5 | 3.53 | 3.71 | 3.89 | 1.91 | 1.35 | 0.42 | 1.58 | 1.79 |
| SD | 0.03 | 0.02 | 0.03 | 0.02 | 0.02 | 0.02 | 0 | 0.01 | 0.01 | |
| Observed | Average | 499.7 | 563.6 | 707.7 | 688.5 | 293.2 | 601.5 | 27.9 | 69.2 | 68.5 |
| SD | 9.89 | 15.14 | 11.37 | 18.72 | 10.03 | 11.51 | 0.32 | 4.08 | 3.34 | |
Ctrl, noninfected.
Principal coordinate analysis (PCA) of unweighted jack-knifed UniFrac distances of microbial communities showed that the first and second principal coordinates, which explained 34.4 and 12.1% of the variance in the data, respectively, separated infected mosquitoes from noninfected mosquitoes starting at 36 hpi (Fig. 2C). These results suggest that fungal infection can alter bacterial composition, reduce bacterial diversity, and result in dysbiosis of the gut microbiota.
Fungal Infection Promotes Translocation of Opportunistic Pathogenic Midgut Bacteria.
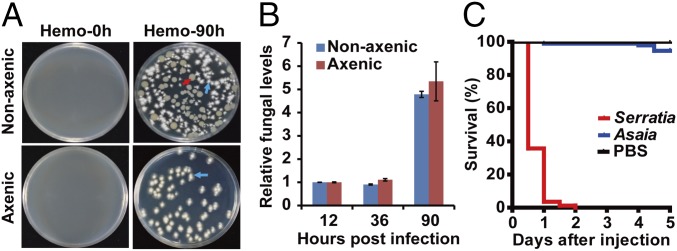
Previous studies have shown that translocation of overgrowing bacteria from the gut to the hemocoel promotes infection and death (19, 30, 31). To determine whether fungal infection results in bacterial translocation from the gut to the hemocoel, the hemolymph from infected nonaxenic and axenic mosquitoes was collected and plated onto LB plates. In nonaxenic mosquitoes, 90 h after infection by B. bassiana, both B. bassiana and bacterial colonies were present in the hemolymph (Fig. 3A). In contrast, in noninfected nonaxenic mosquitoes and in infected axenic mosquitoes, no bacterial colonies were found in the hemolymph (Fig. 3A). The 16S rRNA gene sequence identified the predominant bacterium isolated from the hemolymph of B. bassiana-infected mosquitoes as S. marcescens, which overgrew in the mosquito midgut after fungal infection (Fig. 1D). These results indicated that fungal infection leads to translocation of S. marcescens from the midgut to the hemocoel at 90 hpi.
Fig. 3.
Translocation of gut bacteria to mosquito hemocoel after topical infection with B. bassiana. (A) Growth of bacteria and fungi from the hemolymph of nonaxenic and axenic mosquitoes at 0 and 90 hpi with B. bassiana. The red arrow indicates bacterial colonies. The blue arrows indicate fungal colonies. (B) qPCR-based quantification of fungal load in nonaxenic and axenic mosquitoes (n = 15) at 12, 36, and 90 hpi. Fungal levels are expressed as that of fungal gpd mRNA relative to A. stephensi ribosomal protein S7 (AsS7) mRNA. (C) Survival of mosquitoes (n = 100) following injection of 100 CFUs of S. marcescens, 100 CFUs of Asaia sp., or PBS (control) into the hemolymph. Experiments were performed in three replicates with similar results. Error bars indicate SD.
There was no obvious antagonism between the translocating S. marcescens and B. bassiana grown on LB plates (Fig. 3A). Next, we examined in vivo whether the translocating bacteria affected the fungal colonization of the mosquito hemocoel. qPCR with fungus-specific GPD gene primers showed no significant differences in fungal load between nonaxenic and axenic mosquitoes (Fig. 3B), suggesting that translocation of bacteria into the hemocoel does not affect fungal proliferation.
Further, we tested whether S. marcescens isolated from the fungus-infected mosquito hemolymph could cause systemic infection. Injection of 100 CFUs of the isolated S. marcescens directly into the mosquito hemocoel caused up to 94.5% mortality within 1 d. In contrast, injection of 100 CFUs of the avirulent symbiont Asaia sp. into the hemocoel caused only 3.6% mortality within 5 d (Fig. 3C). These data show that fungal infection results in translocation of opportunistic bacteria such as S. marcescens from the midgut to the hemocoel, where they switch from asymptomatic gut symbionts to hemocoelic pathogens and facilitate fungal killing of mosquitoes.
Fungal Infection Down-Regulates Immune Gene Expression in the Midgut.
Given the dysbiosis of gut microbiota in fungus-infected mosquitoes, we reasoned that decreased immune responses might account, in part, for the bacterial overproliferation. To test this hypothesis, we used qPCR to assess changes in the expression profiles of five effector genes encoding antimicrobial peptides (AMPs), chosen based on their roles in mosquito midgut immunity and controlling bacterial proliferation (32).
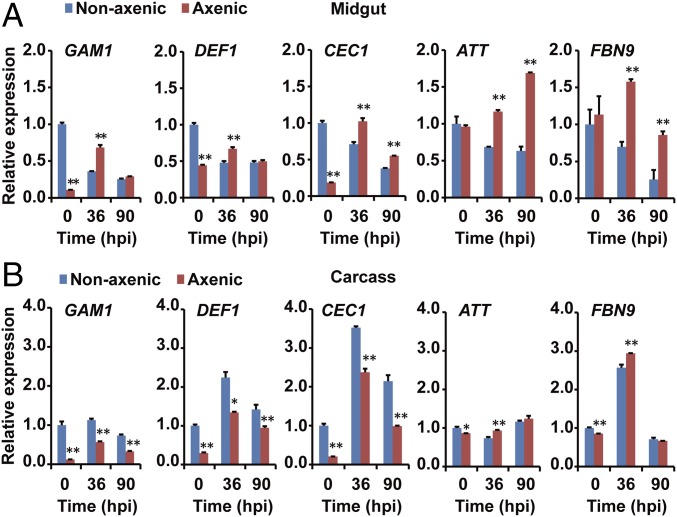
In the midgut, in the absence of fungal infection, effector genes encoding Gambicin 1 (GAM1), Defensin 1 (DEF1), Cecropin 1 (CEC1), Attacin (ATT), and FBN9 (Fibrinogen-related protein family) were expressed at high levels (Fig. 4A), a likely reflection of enhanced basal immunity induced by the resident bacteria. After 36 and 90 hpi with B. bassiana, all five effector genes in the midgut were significantly down-regulated by approximately twofold (Fig. 4A). Conversely, in the midgut of axenic mosquitoes and in the absence of fungal infection, all AMP genes were expressed at low levels, and were significantly up-regulated at 36 h after fungal infection.
Fig. 4.
Expression of five AMPs in the midgut is down-regulated after topical infection with B. bassiana. qPCR analysis of expression levels of AMPs in the (A) midgut and (B) carcass of nonaxenic and axenic mosquitoes (n = 20) at 0, 36, and 90 hpi. Gene expression of each sample was normalized to that of nonaxenic mosquitoes at time 0 (taken as 1). Three biological replicates were conducted. Error bars indicate SD. Single and double asterisks represent a significant difference determined by the Student’s t test at P < 0.05 and P < 0.01, respectively.
In the carcass, the expression pattern of the effector genes was similar in nonaxenic and axenic mosquitoes (Fig. 4B). All of the effector genes were significantly up-regulated at 36 hpi and then declined at 90 hpi, even though the gut microbiota such as S. marcescens had translocated to the hemocoel of the nonaxenic mosquitoes (Fig. 3A). These observations suggest that immune responses in the carcass of the mosquito are not influenced by midgut bacteria during fungal infection. Taken together, these data demonstrate that fungal infection strongly suppressed the expression of immune effector genes in nonaxenic mosquito midguts, which might cause dysbiosis of the midgut microbiota.
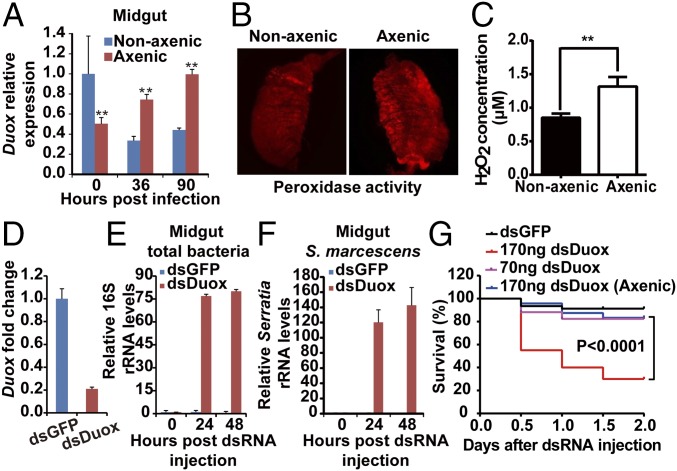
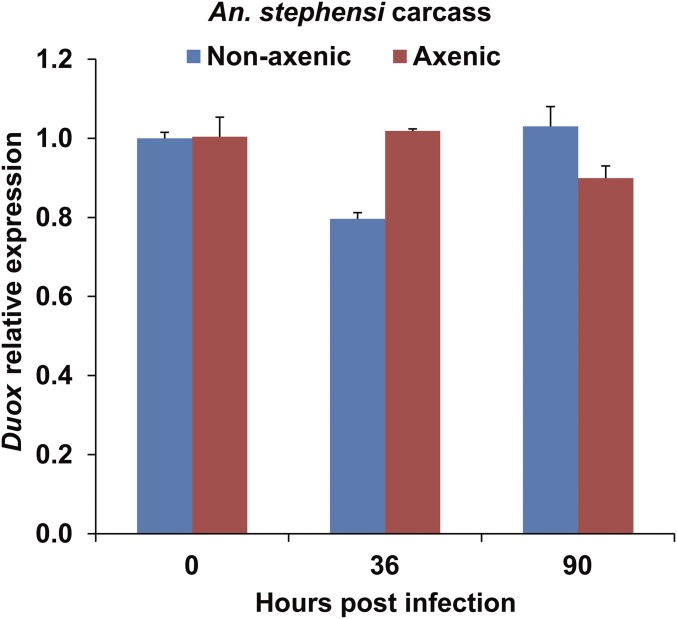
The number of bacteria in the midgut is also controlled by reactive oxygen species (ROS) produced by dual oxidase (Duox) in the insect midgut epithelium (24). To explore this aspect in our experimental system, we further tested the expression patterns of Duox in the midgut of mosquitoes infected with B. bassiana. Expression of Duox was significantly higher in the midguts of nonaxenic mosquitoes than in axenic mosquitoes (Fig. 5A). However, following fungal infection, Duox expression was significantly down-regulated at 36 hpi in the nonaxenic mosquitoes and significantly up-regulated in the midgut of axenic mosquitoes (Fig. 5A). In contrast, after fungal infection, there was no significant difference in the expression of Duox in the carcasses of axenic and nonaxenic mosquitoes (Fig. S4).
Fig. 5.
Expression of mosquito Duox following fungal infection and effect of Duox silencing on midgut bacterial load and host survival. (A) Duox mRNA levels in the midgut of nonaxenic and axenic mosquitoes (n = 20) at 0, 36, and 90 hpi with B. bassiana. (B) Fluorescence staining for peroxidase activity. (C) H2O2 concentration in the midgut of nonaxenic and axenic mosquitoes (n = 5) infected by B. bassiana at 60 h. (D) Midgut Duox silencing efficiency in mosquitoes (n = 20) injected with 70 ng of dsGFP or dsDuox. (E and F) Effect of Duox silencing on midgut total bacterial load (E) (n = 15) and S. marcescens (F) at 0, 24, and 48 h post dsRNA injection (n = 15); levels are relative to readings at time 0 taken as 1. (G) Survival of Duox-silenced nonaxenic and axenic mosquitoes (n = 50) injected with different amounts of dsDuox. Experiments were performed in three biological replicates. Error bars indicate SD. Double asterisks represent a significant difference determined by the Student’s t test at P < 0.01.
Fig. S4.
Expression of Duox in the carcass of nonaxenic and axenic mosquitoes following fungal infection. Quantitative RT-PCR analysis of Duox mRNA levels in the carcass of nonaxenic and axenic mosquitoes (n = 20) at 0, 36, and 90 h post topical infection with B. bassiana. Error bars indicate SD. Three biological replicates were conducted.
After fungal infection, the diminished Duox expression was consistent with the weaker ROS signal (Fig. 5B) and lower hydrogen peroxide (H2O2) production (Fig. 5C) in the midgut of nonaxenic mosquitoes. To validate the role of ROS in the control of gut bacteria, we silenced Duox by systemic injection of Duox double-stranded RNA (dsDuox). The dsDuox silencing reduced midgut Duox mRNA levels by 79% (Fig. 5D) and markedly promoted proliferation of midgut bacteria (Fig. 5E), including S. marcescens (Fig. 5F). A high dose of dsDuox (170 ng) significantly reduced the survival of nonaxenic mosquitoes but did not impact survival of axenic mosquitoes (Fig. 5G). These data suggest that fungal infection causes dysregulation of AMPs and Duox in the midgut, which in turn might result in dysbiosis of the midgut microbiota.
B. bassiana Produces the Toxin Oosporein, Which Down-Regulates Duox Expression in the Midgut.
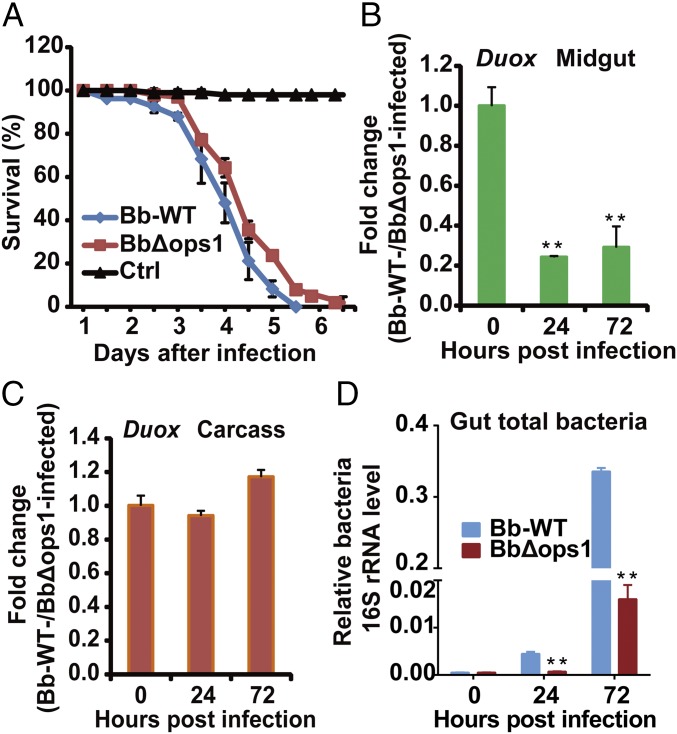
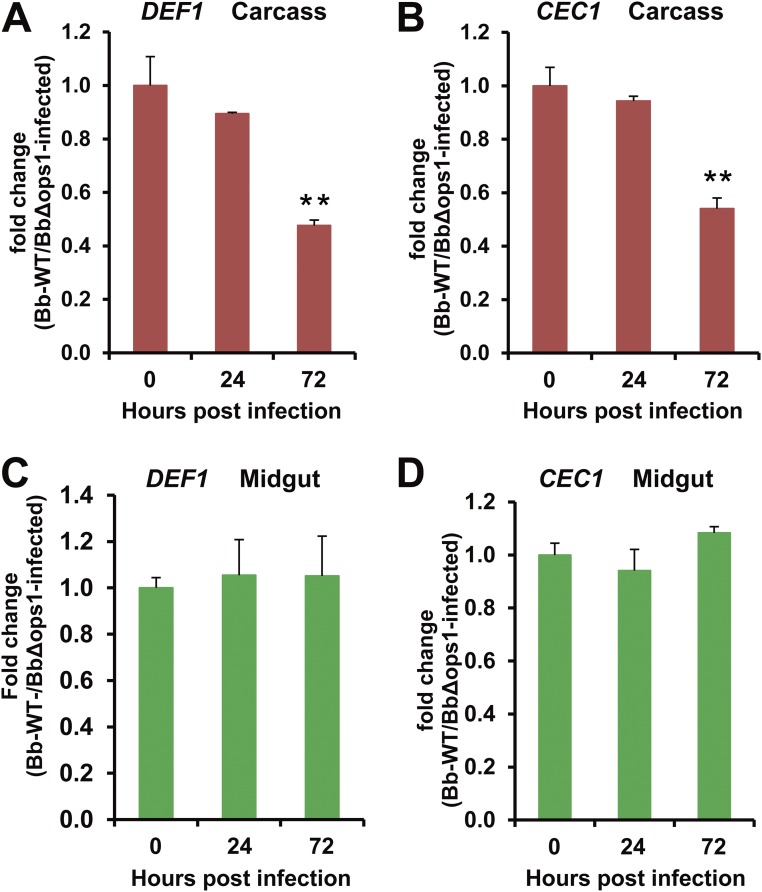
The fungal pathogen B. bassiana produces oosporein, which can down-regulate expression of antifungal peptide genes in the insect fat body (33). To test whether oosporein can inhibit midgut immune responses, we generated an oosporein-nonproducing mutant strain, BbΔops, by disrupting the oosporein synthase 1 gene (Bbops1; required for oosporein biosynthesis) in B. bassiana. We found that deletion of Bbops1 resulted in a significant decrease in fungal virulence against adult female mosquitoes (P < 0.05) (Fig. 6A). qPCR analysis showed that expression of the AMP genes def1 and cec1 was not significantly different in the midgut of mosquitoes infected by B. bassiana WT and by the BbΔops mutant (Fig. S5). However, in the carcass, at 72 hpi, expression of def1 and cec1 was significantly lower in mosquitoes infected with B. bassiana WT than in those infected by the mutant BbΔops.
Fig. 6.
Effect of oosporein on fungal virulence, mosquito Duox expression, and midgut bacterial growth. (A) Survival of adult female A. stephensi (n = 50) following topical infection with B. bassiana WT or BbΔops1; control mosquitoes were not infected. (B and C) Effect of Bbops1 disruption on mRNA levels of Duox in the midgut (B) (n = 20) and carcass (C) of mosquitoes (n = 20) following topical infection with B. bassiana WT and BbΔops1. (D) Effect of Bbops1 disruption on midgut total bacteria determined by 16S rRNA gene-based qPCR analysis (n = 15). Experiments were conducted in three biological replicates. Error bars indicate SD. Double asterisks represent a significant difference determined by the Student’s t test at P < 0.01.
Fig. S5.
Effect of Bbops1 disruption on mRNA expression of DEF1 and CEC1. mRNA levels of DEF1 in the (A) carcass and (C) midgut, and mRNA levels of CEC1 in the (B) carcass and (D) midgut of mosquitoes (n = 20) following topical infection with B. bassiana WT and BbΔops. Error bars indicate SD. Double asterisks represent a significant difference determined by the Student’s t test at P < 0.01. Three biological replicates were performed.
Duox expression was significantly lower in the midgut of mosquitoes infected by B. bassiana WT than in those infected by BbΔops (Fig. 6B). In contrast, Duox expression in the carcass was not significantly different in mosquitoes infected by B. bassiana WT or by the BbΔops mutant (Fig. 6C). Accordingly, midgut bacterial load at 24 and 72 hpi was significantly higher in mosquitoes infected by B. bassiana WT than in those infected by BbΔops (Fig. 6D). Taken together, our data suggest that B. bassiana produces the toxin oosporein to mediate interactions with the mosquitoes’ immune systems. It appears that oosporein is involved in the down-regulation of Duox in the midgut and suppression of AMP genes in the carcass, which might result in dysbiosis of the midgut bacteria and promotion of fungal killing of mosquitoes.
Discussion
Entomopathogenic fungi gain access to the hemocoel cavity through the external cuticle, where they take up nutrients, produce toxins, destroy host cells, and eventually kill their hosts (23). Historically, fungal infections have primarily been studied as interactions between the fungus and the host insect, without consideration of interactions with the gut microbiota. To address this gap in knowledge, we have investigated the role of the gut microbiota in the interactions of the pathogenic fungus B. bassiana with its mosquito hosts. We now report that the fungus interacts with the gut microbiota to promote mosquito death. Reintroduction of gut bacteria into axenic mosquitoes enhanced the susceptibility of the mosquitoes to fungal infection.
In the present study, fungal infection caused the opportunistic bacterial pathogen S. marcescens to outgrow others and translocate from the gut to the hemocoel. Whereas S. marcescens persists in the mosquito midgut without causing apparent illness, injection of S. marcescens into the hemocoel leads to rapid death (Fig. 3C), indicating that S. marcescens switches from being an asymptomatic gut symbiont to a hemocoelic pathogen following fungal topical infection. Additionally, oral ingestion of a large amount of S. marcescens induced an elevated rate of mosquito mortality following fungal topical inoculation. S. marcescens is also a pathogenic bacterium to Drosophila both by oral infection to the gut and injection into the hemocoel (28). It was previously shown that the commensal midgut microbiota contributes to lepidopteran mortality induced by the pathogenic bacterium Bacillus thuringiensis (30, 31). Moreover, Caccia et al. reported that mortality of the cotton leafworm Spodoptera littoralis was increased by the gut bacteria Serratia and Clostridia species invading the body cavity through toxin-induced epithelial lesions (31). In Anopheles, the symbiotic bacterium Asaia is responsible for inhibiting Wolbachia transmission but antibiotic microbiome perturbation enables Wolbachia transmission (34). Examples of the contributory role of gut microbiota to invading intestinal pathogens are also found among vertebrates. For example, the gut commensal microbiota promotes viral infection directly, by activating the immunosuppressive cytokine, or indirectly, by stimulating the proliferation of target cells (35–37).
Bacterial overproliferation is limited by the delicate balance between the commensal gut microbiota and the immune system of the mosquito host. Our data show that mosquito midgut commensal bacteria trigger a basal level of immunity that enhances the expression of AMPs, which in other studies has been shown to be mainly through the IMD pathway (27) and to control the proliferation of the bacterial population (16, 35, 38). Constitutive activation of the gut immune response is detrimental to insect health. Thus, regulatory mechanisms that dampen the basal immune response are required to avoid unhealthy excesses and prevent chronic lethal reactions (24), but the immune system must remain responsive to acute infectious challenges (39). Our results show that the mosquito’s systemic immune response is significantly induced but that the midgut immune response in nonaxenic mosquitoes is significantly down-regulated at 36 hpi. These data suggest that mosquitoes might regulate their immune response to prioritize the fight against acute fungal infection. Such prioritizing modulation of the immune response might be a factor that causes dysregulation of the midgut immune response following topical fungal infection. A recent study by Barreaux et al. showed that the immune response of the Anopheles gambiae mosquito becomes dramatically induced by a small number of injected Sephadex beads, and that melanization is prioritized for one bead rather than distributed over all beads (40). However, dysbiosis of midgut microbiota might also be caused by other factors. Damage to the gut by B. bassiana could change gut physiology, which leads to dysbiosis and translocation of gut microbes to the hemocoel, as indicated in the case of the pathogenic bacterium B. thuringiensis (31).
The suppression of immune responses by invading fungi has been attributed to the combined activity of enzymes and immunosuppressive toxins (41). Entomopathogenic fungi produce a large array of secondary metabolites that are toxic to insects, such as bassianolide, beauvericin, beauverolides, cordycepin, destruxins, and oosporein. Most of these are required for full fungal virulence, via weakening the host immune defenses or damaging the muscular system (23, 42). The destruxins produced by Metarhizium robertsii induce flaccid paralysis and visceral muscle contraction by targeting the Ca2+ channel in insects (43–45). A recent study showed that the toxin oosporein produced by B. bassiana promotes fungal infection by inhibiting polyphenol oxidase activity and down-regulating expression of antifungal peptide genes in the insect fat body (33). Our study reveals that the toxin oosporein specifically mediates down-regulation of Duox expression in the midgut, which reduces midgut ROS production. Duox-dependent ROS generation plays a major role in gut immunity and the control of gut-associated bacteria (24, 46). In Drosophila, opportunistic pathogenic bacteria can be discriminated and controlled by triggering the Duox-dependent gut immunity (47, 48). Down-regulation of Duox in the midgut may cause the opportunistic pathogen S. marcescens to outgrow other commensal bacteria.
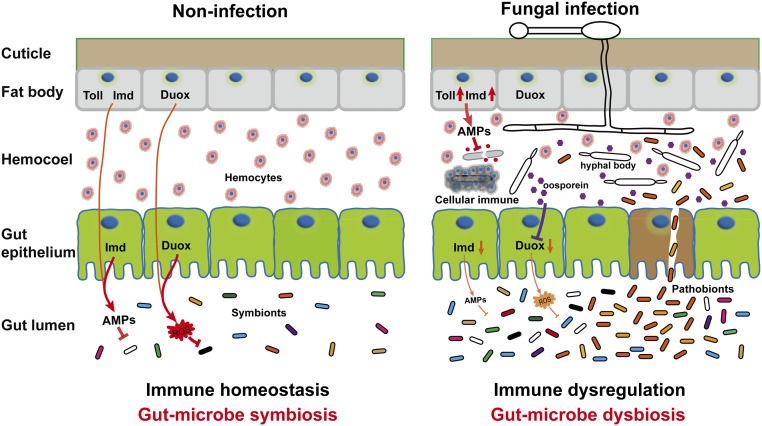
In conclusion, we have discovered a contributory role for the gut microbiota in promoting fungal killing of mosquitoes. We propose a model in which a fungal pathogen interacts with the midgut microbiota to accelerate mosquito death via down-regulation of antimicrobial peptides and dual oxidase in the midgut (Fig. S6). The down-regulated midgut immune responses might account, in part, for microbiota dysbiosis, and bacterial translocation from the gut to the hemocoel results in the acceleration of mosquito death by B. bassiana. These findings provide new insights into the mechanisms of fungal pathogenesis in insects. Understanding of fungus–insect–microbiota interactions may lead to new strategies for biological control of mosquitoes, and consequently the prevention of vector-borne disease transmission.
Fig. S6.
Model of the proposed mechanism of B. bassiana interactions with the gut microbiota to accelerate mosquito death. When the mosquitoes are topically infected by B. bassiana, expression of antimicrobial peptides (AMPs) and Duox in the midgut is down-regulated. Duox down-regulation in the midgut is mediated by the toxin oosporein secreted from B. bassiana. The immune dysregulation in the midgut might result in dysbiosis of gut microbiota and translocation of the opportunistic pathogenic bacterium S. marcescens from the gut to the hemocoel, where switching from asymptomatic gut symbiont to hemocoelic pathogen accelerates the killing of mosquitoes by the fungus.
Materials and Methods
Mosquito Rearing and Antibiotic Treatment.
A. stephensi (Dutch strain) mosquitoes were maintained as previously described (49). Axenic mosquitoes were generated via oral antibiotic treatment as previously described (24). Experimental details can be found in SI Materials and Methods.
Fungal Infection.
To conduct fungal infection, adult female A. stephensi were sprayed with fungal conidia suspension (5 × 108 conidia per mL). Mosquitoes sprayed with sterile 0.01% Triton X-100 were used as control. Experimental details can be found in SI Materials and Methods.
Deep Sequencing.
At each of five time points (0, 12, 36, 60, and 84 h) after fungal infection, mosquitoes were dissected to collect midguts. The bacterial DNA was purified using Gentra Puregene Yeast/Bact. Kit B (Qiagen). The V3 and V4 variable regions of the 16S rRNA gene were amplified and sequenced on the Illumina MiSeq platform. Experimental details can be found in SI Materials and Methods.
Quantitative Real-Time PCR Analysis.
To quantify gene expression, qRT-PCR was performed using SYBR dye technology. Experimental details can be found in SI Materials and Methods. Primers are shown in Table S1.
In Vivo Detection of Reactive Oxygen Species.
ROS production in intact midguts was measured using the intracellular ROS-sensitive fluorescent dye dihydroethidium. Experimental details can be found in SI Materials and Methods.
dsRNA-Mediated Gene Silencing.
To conduct RNAi-mediated gene silencing, mosquitoes were injected with 70 or 140 ng of dsDuox. Control mosquitoes were injected with dsGFP. Experimental details can be found in SI Materials and Methods.
B. bassiana OpS1 Gene Disruption.
The Bbops1 gene was disrupted in B. bassiana Bb252 by homologous recombination using Agrobacterium tumefaciens-mediated transformation (50). Experimental details can be found in SI Materials and Methods.
SI Materials and Methods
Mosquito Rearing and Antibiotic Treatment.
Anopheles stephensi (Dutch strain) mosquitoes were maintained on 10% sucrose at 26 ± 1 °C and 80 ± 5% relative humidity, with a 12-h/12-h day–night cycle. Larvae were fed cat food pellets and ground fish food supplement (49). Axenic female mosquitoes were reared with 10% sucrose solution containing penicillin (10 unit/mL), streptomycin (10 μg/mL), and gentamicin (15 μg/mL) from the first day of emergence. At day 5 post emergence, the antibiotic solution was replaced by sterile water, and mosquitoes were starved overnight before topical fungal infection or oral bacterial infection. Subsequently, the mosquitoes were fed a sugar cube and sterile water that was replaced twice a day.
Fungal Culture and Infection Bioassays.
Beauveria bassiana strain ARSEF 252 (Bb252) were grown and maintained on Sabouraud dextrose agar plus yeast extract (SDAY; BD Difco) at 26 °C. Conidial suspensions in 0.01% (vol/vol) Triton X-100 were prepared from 12-d-old cultures. To conduct fungal infection, adult female A. stephensi were sprayed with fungal conidial suspension (5 × 108 conidia per mL). They were subsequently maintained at 26 ± 1 °C and 80 ± 5% relative humidity, with a 12-h/12-h day–night cycle, until they died or were killed for sample collection at the indicated time points. Mosquitoes sprayed with sterile 0.01% Triton X-100 were used as controls. Each treatment was replicated three times with 50 mosquitoes per replicate, and the infection bioassays were repeated three times. Mortality was recorded every 12 h.
Quantification of Culturable Bacteria in the Mosquito Midgut.
The culturable bacteria of mosquito midgut were quantified by colony-forming unit (CFU) assay. To isolate bacteria from the midgut, mosquitoes were surface-sterilized in 75% ethanol for 3 min and then rinsed three times in sterile PBS (pH 7.4). The midguts (pools of 20 mosquitoes) were dissected aseptically in a laminar hood, transferred to 100 μL of sterile PBS, and homogenized. The homogenates of the midguts were serially diluted 1,000 times with PBS and plated onto Luria–Bertani medium (LB) agar plates. The plates were incubated at 27 °C for 2 to 3 d, and CFUs per plate were counted.
Quantification of Midgut Bacteria by Quantitative PCR.
Genomic DNA from A. stephensi midguts was extracted using DNeasy Kits (QIAGEN) according to the manufacturer’s instructions. Bacterial quantitation by qPCR was performed on genomic DNA using universal eubacteria primers to amplify 16S ribosomal RNA (rRNA) fragments. qPCR analysis was performed with the PikoReal 96 (Thermo) using AceQ qPCR SYBR Green Master Mix (Vazyme). The mosquito A. stephensi housekeeping ribosomal protein S7 gene (AsS7) was used as an endogenous control. The primers are shown in Table S1.
Isolation of Bacteria from the Mosquito Hemolymph.
To isolate bacteria from the hemolymph, 10 mosquitoes were surface-sterilized and the hemolymph was collected as previously described using an anticoagulant solution of 60% (vol/vol) Schneider’s insect medium, 10% (vol/vol) FBS, and 30% (vol/vol) citrate buffer (98 mM NaOH, 186 mM NaCl, 1.7 mM EDTA, and 41 mM citric acid buffer, pH 4.5) (51). In brief, mosquitoes were injected in the thorax with the anticoagulant solution, and the perfusion was collected (∼10 μL) from a small perforation in the abdomen using Sigmacote (Sigma)-treated pipet tips and a Pipetman. The hemolymph of each mosquito was mixed with 100 μL of sterile PBS and spread onto the LB agar plates. All of the plates were incubated at 27 °C.
DNA Extraction and Bacterial Identification.
A single-colony isolation technique was used for each of the morphologically distinct colonies visible on the LB agar plates. The bacterial species were identified by 16S rRNA gene sequence comparisons. Briefly, genomic DNAs were isolated from the different bacterial isolates by the phenol extraction method, and a stretch of a 1,508-bp 16S rRNA gene region was PCR-amplified using universal 16S rRNA gene-specific primers (Table S1). The purified PCR products were sequenced using the ABI Prism 3730 sequencer (Applied Biosystems). The obtained 16S rRNA gene sequences were aligned with the closest relatives matching the 16S rRNA gene sequences found in the GenBank database by BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The phylogenetic tree was constructed using the neighbor-joining method with MEGA7 software (52).
DNA Sample Preparation and Deep Sequencing.
At each of five time points (0, 12, 36, 60, and 84 h) after fungal infection, mosquitoes topically inoculated with B. bassiana conidial suspension or sprayed with 0.01% (vol/vol) Triton X-100 (control) were dissected to collect midguts. Each treatment had two replicates, and 40 mosquitoes from two replicates (20 mosquitoes per replicate) were mixed to obtain enough samples. The bacterial DNA was purified using Gentra Puregene Yeast/Bact. Kit B (Qiagen), following the manufacturer’s instructions. A region encompassing the V3 and V4 hypervariable regions of the 16S rRNA gene was amplified using the primers 338F and 806R shown in Table S1. The 16S amplicon sequencing was performed on the Illumina MiSeq platform by Shanghai Biotechnology. The resulting FASTA files were filtered to a minimum read length of 250 bp using Galaxy (53), and blasted against the National Center for Biotechnology Information 16S rRNA bacterial database using BLAST+ and prfectBLAST (54), with standard blastn algorithm settings and 10 maximum target sequences. Further analysis was performed using MEGAN4 (55). Numbers of analyzed sequences per sample are shown in Table S2.
Bacterial Introduction into the Mosquito Midgut via Sugar Meals.
Serratia marcescens and Asaia sp. were cultured in LB broth at 30 °C for 16 h. The bacterial culture was then pelleted by centrifugation (3,000 × g, 5 min), washed twice in sterile PBS, and resuspended in 5% sterile sucrose solution to obtain 108 cells per mL. The bacterial suspension was added to a sterile cotton pad and provided to 2-d-old mosquitoes for 24 h, after which the bacterial cotton pads were replaced with new sterile cotton pads containing 5% sterile sucrose solutions.
Quantitative Real-Time PCR Analysis.
At three time points (0, 36, and 90 h) after fungal infection, 20 nonaxenic or axenic mosquitoes were surface-sterilized by immersion in 75% ethanol for 3 min, and then rinsed three times in sterile PBS and dissected to separate the midgut and the remainder of the body (carcass). Total RNA from mosquito midguts and carcasses was extracted after homogenization with a grinding rod in an RNAiso Plus (Takara). First-strand cDNA was synthesized from total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara) according to the manufacturer’s instructions. A quantitative real-time PCR analysis was performed with the PikoReal 96 (Thermo) using the AceQ qPCR SYBR Green Master Mix (Vazyme). The housekeeping AsS7 gene was used as an endogenous control. The primers are shown in Table S1.
In Vivo Detection of Reactive Oxygen Species.
At 36 h post fungal infection, the midguts of mosquitoes were dissected in PBS and incubated in RPMI 1640 medium supplemented with 10% FBS containing 50 μM the intracellular ROS-sensitive fluorescent dye dihydroethidium (hydroethidine) (Invitrogen) for 10 min at room temperature in the dark. Then, the midguts were washed twice with fresh dye-free medium and the tissues were immediately transferred to a glass slide in a drop of PBS for epifluorescence examination.
Measurement of Hydrogen Peroxide Production.
The production of H2O2 was determined using the Hydrogen Peroxide Assay Kit (Beyotime Biotech) according to the manufacturer’s instructions. In this assay, H2O2 converts Fe2+ to Fe3+, which then complexes with xylenol orange dye to yield a purple product having an absorbance maximum at 560 nm, which could be detected by a spectrometer.
Mosquito midgut epithelia were dissected in ice-cold PBS and opened longitudinally in PBS to remove the gut contents. Briefly, 10 midguts were homogenized in 200 μL lysis buffer and centrifuged at 12,000 × g at 4 °C for 3 to 5 min, and the supernatant was collected. Aliquots of 50 μL of supernatants and 100 μL of test solutions from the Hydrogen Peroxide Assay Kit were incubated at room temperature for 20 min and measured immediately with a spectrometer at a wavelength of 560 nm. The concentration of H2O2 released was calculated according to a hydrogen peroxide standard curve. The measurement was repeated three times.
dsRNA-Mediated Gene Silencing in Adult Female Mosquitoes.
To produce the double-stranded RNA of the Duox gene (dsDuox), the coding region of the Duox gene was amplified from A. stephensi cDNA with forward and reverse primers containing the T7 promoter sequence at their 5′ ends (5′-TAATACGACTCACTATAGGG-3′). The sequences of the primers are given in Table S1. The PCR product was used as a template to synthesize dsRNA in vitro using the MEGAscript RNAi Kit (Ambion). The dsRNA was further purified using the purification column supplied with the kit, eluted with nuclease-free water, and concentrated to 3 μg/μL using a Microcon YM-100 filter (Millipore). As a negative control, we synthesized enhanced green fluorescent protein dsRNA.
To conduct RNAi-mediated gene silencing, cold-anesthetized 4-d-old female mosquitoes were injected with 70 or 140 ng of dsDuox solution into the hemocoel using a Nanoject II microinjector (Drummond). Control mosquitoes were injected with dsGFP. Injected mosquitoes were allowed to recover for 2 to 3 d before dissection of the midgut and quantification of midgut bacteria. The efficiency of dsRNA-mediated gene silencing was determined by qRT-PCR at 2 to 5 d after injection.
Construction of B. bassiana OpS1 Mutant.
For targeted disruption of Bbops1 (an oosporein synthase 1, required for oosporein biosynthesis), the 5′ and 3′ flanking regions of the gene ORF were amplified by PCR from B. bassiana Bb252 genomic DNA and then subcloned into the XbaI and EcoRV sites of the binary vector pBarGFP, respectively (50). The gene disruption construct (pBarGFP-Bbops1) was then transformed into Agrobacterium tumefaciens AGL-1 for targeted gene disruption by homologous recombination. Replacement-specific PCR amplifications of the gene locus were performed with specific primer pairs (Table S1) that amplified either the WT or the mutant gene locus.
Statistical Analysis.
The statistical significance of the survival data from fungal infection bioassays and dsRNA-mediated gene silencing assays was analyzed with a log-rank (Mantel–Cox) test. Other statistical significance was calculated using the Student’s t test for unpaired comparisons between nonaxenic and axenic mosquitoes, between fungus-infected and Triton-treated mosquitoes, and between B. bassiana WT-infected and BbΔops-infected mosquitoes. A value of P < 0.05 was considered to be statistically significant. All statistics were performed using GraphPad Prism version 5.00 for Windows.
Acknowledgments
We thank Prof. Marcelo Jacobs-Lorena at the Johns Hopkins University School of Public Health for critical comments on the manuscript. We also thank Dr. Sarah Poynton at the Johns Hopkins Editing Referral Service, William H. Welch Medical Library, Johns Hopkins University School of Medicine for editorial assistance. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB11010500) and One Hundred Talents Program of the Chinese Academy of Sciences (Grant 2013OHTP01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The entire 16S rRNA gene sequence dataset reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. PRJNA371598).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703546114/-/DCSupplemental.
References
- 1.Medlock JM, et al. A review of the invasive mosquitoes in Europe: Ecology, public health risks, and control options. Vector Borne Zoonotic Dis. 2012;12:435–447. doi: 10.1089/vbz.2011.0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, Jacobs-Lorena M. Genetic approaches to interfere with malaria transmission by vector mosquitoes. Trends Biotechnol. 2013;31:185–193. doi: 10.1016/j.tibtech.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y, Borovsky D, Hawkings C, Ortiz-Urquiza A, Keyhani NO. Exploiting host molecules to augment mycoinsecticide virulence. Nat Biotechnol. 2012;30:35–37. doi: 10.1038/nbt.2080. [DOI] [PubMed] [Google Scholar]
- 4.Kanzok SM, Jacobs-Lorena M. Entomopathogenic fungi as biological insecticides to control malaria. Trends Parasitol. 2006;22:49–51. doi: 10.1016/j.pt.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Lord JC. From Metchnikoff to Monsanto and beyond: The path of microbial control. J Invertebr Pathol. 2005;89:19–29. doi: 10.1016/j.jip.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Blanford S, et al. Fungal pathogen reduces potential for malaria transmission. Science. 2005;308:1638–1641. doi: 10.1126/science.1108423. [DOI] [PubMed] [Google Scholar]
- 7.Knols BGJ, Bukhari T, Farenhorst M. Entomopathogenic fungi as the next-generation control agents against malaria mosquitoes. Future Microbiol. 2010;5:339–341. doi: 10.2217/fmb.10.11. [DOI] [PubMed] [Google Scholar]
- 8.Lacey LA, Frutos R, Kaya HK, Vail P. Insect pathogens as biological control agents: Do they have a future? Biol Control. 2001;21:230–248. [Google Scholar]
- 9.Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 10.Coon KL, Vogel KJ, Brown MR, Strand MR. Mosquitoes rely on their gut microbiota for development. Mol Ecol. 2014;23:2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors. 2013;6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 13.Gravitz L. Microbiome: The critters within. Nature. 2012;485:S12–S13. doi: 10.1038/485s12a. [DOI] [PubMed] [Google Scholar]
- 14.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 15.Beier MS, Pumpuni CB, Beier JC, Davis JR. Effects of para-aminobenzoic acid, insulin, and gentamicin on Plasmodium falciparum development in anopheline mosquitoes (Diptera: Culicidae) J Med Entomol. 1994;31:561–565. doi: 10.1093/jmedent/31.4.561. [DOI] [PubMed] [Google Scholar]
- 16.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirimotich CM, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narasimhan S, et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe. 2014;15:58–71. doi: 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apte-Deshpande A, Paingankar M, Gokhale MD, Deobagkar DN. Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One. 2012;7:e40401. doi: 10.1371/journal.pone.0040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carissimo G, et al. Antiviral immunity of Anopheles gambiae is highly compartmentalized, with distinct roles for RNA interference and gut microbiota. Proc Natl Acad Sci USA. 2015;112:E176–E185. doi: 10.1073/pnas.1412984112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abraham NM, et al. Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc Natl Acad Sci USA. 2017;114:E781–E790. doi: 10.1073/pnas.1613422114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Wang S. Insect pathogenic fungi: Genomics, molecular interactions, and genetic improvements. Annu Rev Entomol. 2017;62:73–90. doi: 10.1146/annurev-ento-031616-035509. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science. 2010;327:1644–1648. doi: 10.1126/science.1184008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boissière A, et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8:e1002742. doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bando H, et al. Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. Sci Rep. 2013;3:1641. doi: 10.1038/srep01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stathopoulos S, Neafsey DE, Lawniczak MK, Muskavitch MA, Christophides GK. Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens. PLoS Pathog. 2014;10:e1003897. doi: 10.1371/journal.ppat.1003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nehme NT, et al. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 2007;3:e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favia G, et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci USA. 2007;104:9047–9051. doi: 10.1073/pnas.0610451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broderick NA, Raffa KF, Handelsman J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc Natl Acad Sci USA. 2006;103:15196–15199. doi: 10.1073/pnas.0604865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caccia S, et al. Midgut microbiota and host immunocompetence underlie Bacillus thuringiensis killing mechanism. Proc Natl Acad Sci USA. 2016;113:9486–9491. doi: 10.1073/pnas.1521741113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clayton AM, Cirimotich CM, Dong Y, Dimopoulos G. Caudal is a negative regulator of the Anopheles IMD pathway that controls resistance to Plasmodium falciparum infection. Dev Comp Immunol. 2013;39:323–332. doi: 10.1016/j.dci.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng P, Shang Y, Cen K, Wang C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc Natl Acad Sci USA. 2015;112:11365–11370. doi: 10.1073/pnas.1503200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes GL, et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci USA. 2014;111:12498–12503. doi: 10.1073/pnas.1408888111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilks J, Golovkina T. Influence of microbiota on viral infections. PLoS Pathog. 2012;8:e1002681. doi: 10.1371/journal.ppat.1002681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kane M, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuss SK, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cirimotich CM, Ramirez JL, Dimopoulos G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 2011;10:307–310. doi: 10.1016/j.chom.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barreaux AMG, Barreaux P, Koella JC. Overloading the immunity of the mosquito Anopheles gambiae with multiple immune challenges. Parasit Vectors. 2016;9:210. doi: 10.1186/s13071-016-1491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilcinskas A, Gotz P. Parasitic fungi and their interactions with the insect immune system. Adv Parasitol. 1999;43:267–313. [Google Scholar]
- 42.Schrank A, Vainstein MH. Metarhizium anisopliae enzymes and toxins. Toxicon. 2010;56:1267–1274. doi: 10.1016/j.toxicon.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Wang B, Kang Q, Lu Y, Bai L, Wang C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc Natl Acad Sci USA. 2012;109:1287–1292. doi: 10.1073/pnas.1115983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedras MSC, Zaharia LI, Ward DE. The destruxins: Synthesis, biosynthesis, biotransformation, and biological activity. Phytochemistry. 2002;59:579–596. doi: 10.1016/s0031-9422(02)00016-x. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Sanchez E, Lange AB, Orchard I. Effects of the mycotoxin destruxin A on Locusta migratoria visceral muscles. Toxicon. 2010;56:1043–1051. doi: 10.1016/j.toxicon.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Oliveira JHM, et al. Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog. 2011;7:e1001320. doi: 10.1371/journal.ppat.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee KA, et al. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell. 2013;153:797–811. doi: 10.1016/j.cell.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 48.Bae YS, Choi MK, Lee WJ. Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 2010;31:278–287. doi: 10.1016/j.it.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Wang S, et al. Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc Natl Acad Sci USA. 2012;109:12734–12739. doi: 10.1073/pnas.1204158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Fang W, Wang C, St Leger RJ. Insertion of an esterase gene into a specific locust pathogen (Metarhizium acridum) enables it to infect caterpillars. PLoS Pathog. 2011;7:e1002097. doi: 10.1371/journal.ppat.1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith RC, Barillas-Mury C, Jacobs-Lorena M. Hemocyte differentiation mediates the mosquito late-phase immune response against Plasmodium in Anopheles gambiae. Proc Natl Acad Sci USA. 2015;112:E3412–E3420. doi: 10.1073/pnas.1420078112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goecks J, Nekrutenko A, Taylor J. Galaxy Team Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santiago-Sotelo P, Ramirez-Prado JH. prfectBLAST: A platform-independent portable front end for the command terminal BLAST+ stand-alone suite. Biotechniques. 2012;53:299–300. doi: 10.2144/000113953. [DOI] [PubMed] [Google Scholar]
- 55.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]