Abstract
A common path to the formation of complex 3D structures starts with an epithelial sheet that is patterned by inductive cues that control the spatiotemporal activities of transcription factors. These activities are then interpreted by the cis-regulatory regions of the genes involved in cell differentiation and tissue morphogenesis. Although this general strategy has been documented in multiple developmental contexts, the range of experimental models in which each of the steps can be examined in detail and evaluated in its effect on the final structure remains very limited. Studies of the Drosophila eggshell patterning provide unique insights into the multiscale mechanisms that connect gene regulation and 3D epithelial morphogenesis. Here we review the current understanding of this system, emphasizing how the recent identification of cis-regulatory regions of genes within the eggshell patterning network enables mechanistic analysis of its spatiotemporal dynamics and evolutionary diversification. It appears that cis-regulatory changes can account for only some aspects of the morphological diversity of Drosophila eggshells, such as the prominent differences in the number of the respiratory dorsal appendages. Other changes, such as the appearance of the respiratory eggshell ridges, are caused by changes in the spatial distribution of inductive signals. Both types of mechanisms are at play in this rapidly evolving system, which provides an excellent model of developmental patterning and morphogenesis.
Keywords: enhancer, network, evolution, signal, dynamics
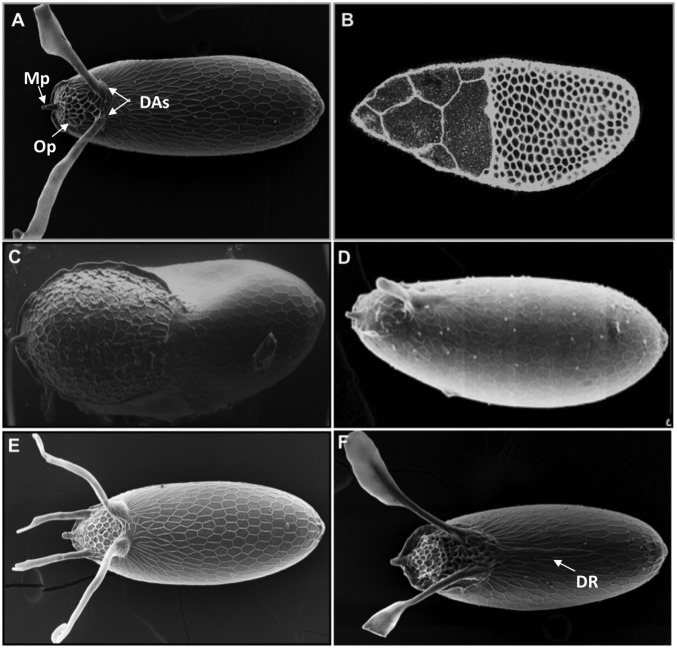
The Drosophila eggshell is a proteinaceous structure that houses the future embryo and mediates its interaction with the environment (Fig. 1A). It provides a point for sperm entry and controls the gas exchange needed for embryo respiration. The eggshell is derived from the follicular epithelium, a cell sheet that envelops the germ line cyst, comprising one oocyte and 15 nurse cells (Fig. 1B). This cell sheet is patterned by the combined activities of several signaling pathways, including the epidermal growth factor receptor (EGFR) and bone morphogenetic protein (BMP) pathways (1–3). The EGFR pathway is activated by Gurken (GRK), a TGFα-like ligand that is secreted from the oocyte and generates a dorsoventral gradient of EGFR signaling in the follicle cells (4–6) (Fig. 2A). The BMP pathway is activated by the BMP2/4 homolog Decapentaplegic (DPP), which is distributed in the anteroposterior gradient (7) (Fig. 2B). The joint activities of the EGFR and DPP pathways induce the formation of several eggshell structures, including the respiratory dorsal appendages and the operculum, the region of the eggshell from which the larva hatches when the embryogenesis is completed. Importantly, the follicle cells do not divide during the patterning of the dorsal eggshell structures (8). Consequently, the patterning and morphogenesis of these structures can be studied without the added complexities associated with changing cell numbers.
Fig. 1.
Drosophila eggshell, a complex structure derived from an epithelial sheet. (A) Scanning electron microscopy (SEM) image of the eggshell of D. melanogaster. The most prominent features are the two dorsal appendages (DAs), the micropyle (Mp), and the operculum (Op). (B) The egg chamber midway through oogenesis stained with phalloidin. (C) SEM image of the eggshell resulting from uniform activation of DPP signaling in the follicle cells. (D) SEM image of the eggshell resulting from reduced EGFR signaling. (E) SEM image of the eggshell from D. virilis, a species with four dorsal appendages. (F) SEM image of the eggshell from D. willistoni, a species with two dorsal appendages and a dorsal ridge (DR). All SEM images present the dorsal views of the eggshell (anterior to the left).
Fig. 2.
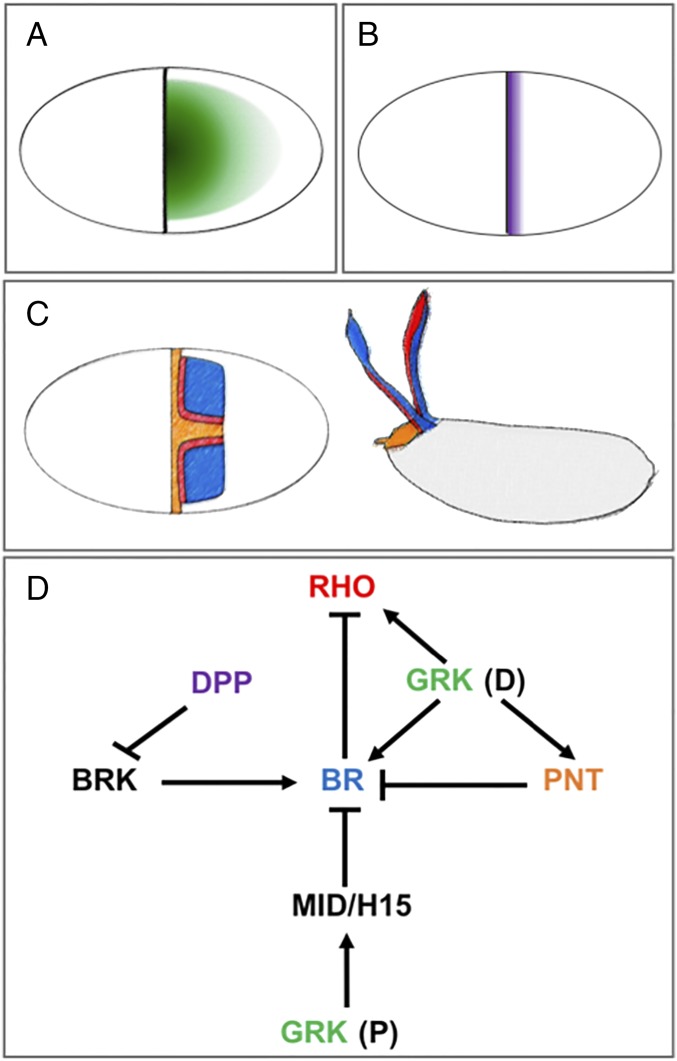
Inductive signals, target genes, and genetic interactions involved in eggshell patterning. (A) The dorsoventral pattern of EGFR signaling. (B) The anteroposterior pattern of DPP signaling. (C) The fate map for the formation of the respiratory dorsal appendages and the operculum. Each of the appendages is derived from a primordium comprising a 2D domain of cells expressing Broad (BR, blue) and an adjacent line of cells expressing Rhomboid (RHO, red). Cells between the two primordia express the transcription factor Pointed (PNT, orange) and contribute to the formation of the operculum. (D) The network of some of the key interactions involved in eggshell patterning. GRK controls br at two different time points: first, when it is distributed in a posterior-to-anterior gradient (P), and then when it is distributed in a dorsoventral (D) gradient (see text for details).
Changes in the EGFR and DPP signaling profiles can cause dramatic alterations of eggshell morphology. For example, uniform activation of the DPP pathway eliminates the appendages and results in eggshells with greatly expanded operculum (Fig. 1C). At the same time, decreasing the levels of EGFR signaling can result in eggshells with one appendage (Fig. 1D). These and many other important observations were made two decades ago, when eggshell morphology was used as a sensitive readout in genetic screens that discovered the genes involved in the early steps of body axis specification (9, 10). Genetic studies of eggshell patterning identified numerous molecular components that interpret the GRK and DPP signals in the follicle cells (Fig. 2). Most importantly, it was established that each of the two dorsal appendages is derived from a 2D primordium comprising a patch of cells expressing the Zn-finger transcription factor Broad (BR) and an adjacent L-shaped stripe of cells that express rhomboid (RHO), which encodes a ligand-processing enzyme in the Drosophila EGFR pathway (11). Cells that express BR and RHO form the top and bottom parts of the future dorsal appendages, respectively (Fig. 2C).
Work over the past decade has identified the transcriptional factors that control these genes and has started to connect them in a regulatory network (2, 12–18). Based on this network, we can predict how the expression patterns of multiple genes will respond to a range of genetic perturbations, including the quantitative changes in the distribution and levels of inductive signals (Fig. 2D). In addition, this network provides a starting point for exploring the evolution of eggshell morphology, which varies greatly across drosophilids. For instance, the eggshell of Drosophila virilis has four dorsal appendages (Fig. 1E), whereas the eggshell of Drosophila willistoni has a new dorsal structure, the dorsal ridge, that extends from the base of the dorsal appendages toward the posterior of the eggshell (Fig. 1F). What caused these changes in eggshell morphologies? Can they be attributed to changes in the inductive signals or in the regulatory network that interprets these inputs in the follicle cells? Here we review the current knowledge of the eggshell patterning network and the ongoing studies of its evolutionary diversification.
Patterning Network in D. melanogaster
Most of our understanding of tissue patterning by inductive signals is derived from studies of patterns in one dimension, such as the stripes of gene expression in the Drosophila blastoderm (19). However, the majority of experimentally observed patterns, including those in the Drosophila blastoderm, have a strong 2D component, reflecting their joint control by several inductive signals. Eggshell patterning by the EGFR and DPP pathways provides an excellent model for studying the dynamic emergence of 2D gene expression patterns and features several generally applicable regulatory principles (Fig. 2 A and B). These principles include the combined effects of two signaling gradients that act through a network of transcription factors, converging on the enhancers of their target genes (Fig. 2D). Another important aspect of this system, which has been revealed only recently, is that different features of the emerging 2D patterns are established by processes that are separated in time. In this section, we discuss the network of the inductive cues and transcription factors that govern the 2D pattern of BR, which acts as a master regulator of the dorsal appendage formation.
The dorsoventral gradient of EGFR signaling controls br through a feedforward loop, a network motif in which EGFR activates both br and its repressor (17, 18, 20, 21). The induction threshold for the repressor is higher than that for br, which explains why it is expressed only in cells exposed to intermediate levels of GRK. The activating part of the feedforward loop is mediated by the Iroquois transcription factor Mirror (MIRR), and the repressive part relies on the ETS-domain factor Pointed (PNT) (12, 13, 15, 22). Given the dorsoventral pattern of the EGFR activation by GRK (Fig. 2A), this circuit predicts that the br expression domain should look like a horseshoe and extend all the way to the anterior border of the oocyte-associated follicle cells. The real pattern is different, however; the horseshoe is broken at the posterior and pushed away from the anterior border (Fig. 2C). These effects are mediated by two distinct repression events, which are separated both in time and in space. The anterior repression depends on the anterior gradient of DPP signaling (2, 17, 23), which controls br through a two-tier cascade that involves repression of BRK, a direct target of DPP in multiple stages of Drosophila development (24–26). The posterior repression of br relies on the earlier phase of EGFR signaling, when the oocyte nucleus is located at the posterior of the oocyte. At this point of oogenesis, the EGFR pathway is activated in a posterior-to-anterior gradient and represses br in the posterior half of the follicular epithelium through the T-box transcription factors Midline (MID) and H15 (Fig. 2D) (16, 27, 28).
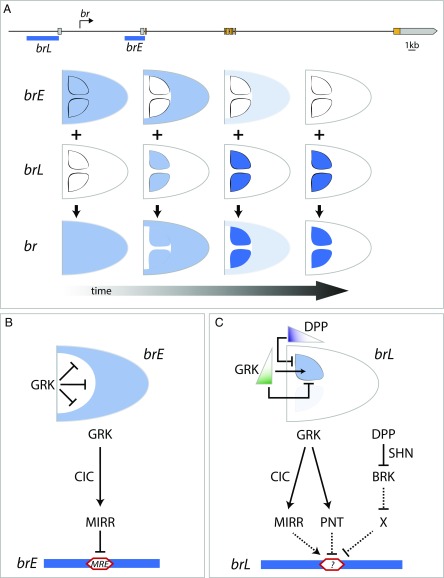
The effects of these inductive signals and transcription factors were discovered through their effects on the eggshell morphology and the patterns of BR protein, without direct analysis of the transcriptional regulation of the br gene. Important aspects of this regulation have been revealed only recently. Specifically, dissection of the genomic region of br revealed the existence of two distinct enhancers, early and late, that combine their activities over time to generate the dynamic expression of br in the follicle cells (Fig. 3A) (29). An early enhancer (brE) drives uniform expression before stage 10 of oogenesis. At stage 10, this enhancer is repressed in a wide dorsal domain and starts to subside in the rest of the follicular epithelium. At the same time, the late enhancer (brL) is activated in two lateral domains within the brE-free zone, foreshadowing the formation of the two dorsal appendage primordia (Figs. 2C and 3A).
Fig. 3.
Summary of the cis-regulatory network controlling the spatiotemporal pattern of br. (A) Schematic of the genomic locus of br with exons indicated in color and positions of identified enhancers shown as blue horizontal bars. For simplicity, a single isoform, br-RA, is shown. The dynamic pattern of br expression emerges from the time-dependent activities of two distinct enhancers, the early (brE) and the late (brL). Dorsal views of the egg chambers are shown in all panels. (B) brE, initially active in a uniform pattern, is repressed in dorsal cells by the EGFR signaling. This repression depends on MIRR, an Iroquois-family transcription factor that is induced by GRK and binds directly to an MIRR response element (MRE) within brE. GRK-dependent activation of mirr is mediated by the transcription factor Capicua (CIC). (C) brL integrates both repressive and activatory inputs from EGFR signaling and repressive input from DPP signaling. MIRR activates brL by unknown mechanisms in the same domain that it represses brE. High levels of EGFR signaling activate PNT, which represses brL in the dorsal midline. DPP signaling represses brL in the anteriormost follicle cells. This involves a chain of repressive steps. DPP first directly represses the expression of transcriptional repressor BRK in the anteriormost follicle cells. In the rest of the tissue, BRK promotes the activity of brL by antagonizing an as-yet unknown repressor.
The identification of these enhancers made it possible to determine whether the effects of the key transcription factors controlling br are direct and whether they affect the early or the late phases of br expression (29). For instance, it was established that MIRR controls both enhancers, but with opposite signs: repressing brE and activating brL (Fig. 3 B and C). On the other hand, PNT represses brL, but does not affect brE. The DPP pathway controls only the late phase of br expression within the appendage primordia. The net effect of DPP is repressive and is fully dependent on BRK (23). Specifically, the DPP pathway directly represses brk in the anterior follicle cells, by acting through a well-characterized direct mechanism (Fig. 3C) (26). In the rest of the follicular epithelium, BRK positively regulates brL, likely by repressing an unknown repressor (23).
Established under the joint action of the EGFR and DPP signals, BR represses rho, confining its expression to the anterior border of the BR domain. As mentioned above, RHO is a ligand-processing protease within the Drosophila EGFR pathway. Remarkably, although neither RHO nor its target (Spitz, SPI) is essential for proper eggshell patterning (15, 18), the BR-dependent repression of rho is important. Specifically, low levels of the BR protein resulting from the activity of the brE enhancer “protect” the brL enhancer from the PNT-mediated repression. In the absence of the early br expression, EGFR activation induces premature expression of rho, which is normally expressed only after the brE enhancer is repressed by GRK. Precocious expression of RHO results in ectopic secretion of SPI and leads to ectopic high levels of EGFR activation. This in turn leads to the PNT-mediated repression of the brL enhancer and results in loss of the dorsal appendages (30). Thus, the 2D pattern of BR is shaped by the dynamic interplay of at least two enhancers with very different spatiotemporal activities.
Some of the cis and trans components in the network establishing the dorsal appendage primordia are shown in Fig. 3 B and C. Based on this network, we can explain the observations made in numerous genetic experiments. For instance, uniform activation of DPP signaling leads to eggshells with no appendages and a large, dorsally located operculum (Fig. 1C). We now understand that this effect is caused by direct repression of BRK, which leads, through a short cascade, to repression of the brL enhancer. As another example, uniform activation of the EGFR pathway also results in eggshells with no appendages, but the operculum is now formed around the entire dorsoventral axis (31). This effect is caused by the PNT-dependent repression of brL. In both examples, brL is repressed, but the mechanisms of repression are different. Note that because all of the effects of the patterning cues ultimately converge on the brL enhancer, future studies of the eggshell patterning network should focus on a more complete understanding of this regulatory element. Along with identifying the missing transacting elements, such as a missing repressor that is antagonized by BRK, this requires identifying the cis-regulatory sequences that respond to the transcription factors controlled by the EGFR and DPP pathways.
From D. melanogaster to Other Species
Explaining the morphological diversity of eggshells in drosophilids motivated much of the mechanistic studies of the eggshell patterning network (32–38). For example, in contrast to D. melanogaster, in which each of the two dorsal appendages is formed from a separate primordium, the four appendages in D. virilis arise not from four, but from two separate primordia (Fig. 4A). Similar to D. melanogaster, each primordium comprises a 2D domain of br bordered by a line of rho-expressing cells. Thus, it appears that the same genes are involved in patterning of appendages, but their expression patterns are different (39). What mediates these changes in gene expression? Is it possible to trace them to changes in the inductive signals and the mechanisms by which they are interpreted by the follicle cells? Although work addressing these questions has just begun, it is already clear that multiple mechanisms are at play in this rapidly evolving system.
Fig. 4.
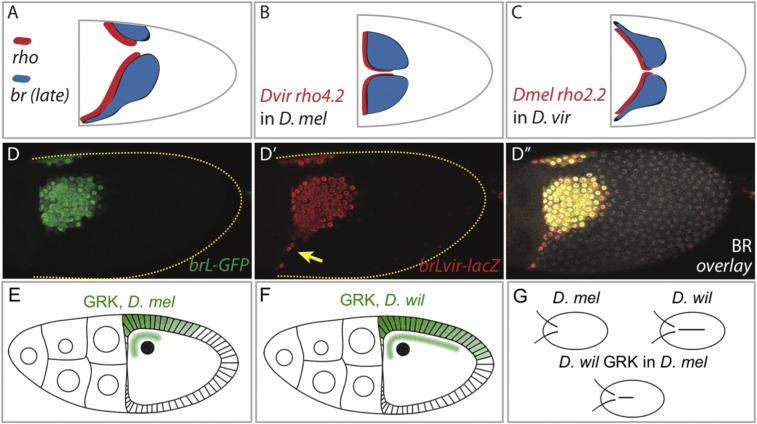
Two different mechanisms for the evolution of br patterning: changes in the regulatory region of br and changes in the inductive signals. (A) Schematic representation of the expression domains of br and rho in D. virilis. (B) Summary of the activity of the rho enhancer from D. virilis in D. melanogaster. (C) Summary of the activity of the rho enhancer from D. melanogaster in D. virilis. (D–D′′′) Comparison of the activities of the brL enhancers from D. melanogaster (D; brL-GFP in green) and D. virilis (D′; brLvir-lacZ, red), analyzed in D. melanogaster. (D′′) Merged image including immunostaining for endogenous Br (white nuclear staining) for orientation. The yellow arrow indicates the lateral expansion of D. virilis brL. (E) Schematic of GRK localization in D. melanogaster (lateral view). (F) Schematic of GRK localization in D. willistoni (lateral view). (G) Schematics of eggshells in D. melanogaster, D. willistoni, and D. melanogaster patterned by GRK from D. willistoni.
Early models of eggshell patterning suggested that quantitative changes in the distribution of the GRK gradient can change the number of dorsal appendages (33, 40, 41). This possibility was conclusively ruled out by an elegant experiment that relied on chimera egg chambers established by pole cell transplantation (42). In this case, egg chambers contained somatic cells from D. melanogaster and the germ line cells derived from D. virilis. As a result, the follicular epithelium of D. melanogaster was patterned by GRK from D. virilis. The eggshells formed by these mosaic egg chambers were indistinguishable from the wild-type eggshells of D. melanogaster. These results show that, at least in this case, changes in eggshell morphology cannot be explained by changes in the inductive signal alone, pointing to the need to consider alterations in the network that interprets the inductive signal.
As a first step in this direction, Nakamura et al. (42) focused on the regulatory region of rho and identified the orthologous enhancer in D. virilis. This enhancer recapitulates the endogenous pattern of rho expression in D. virilis and drives expression in a broken V-shaped pattern. However, when introduced in D. melanogaster, this enhancer drives expression in an L-shaped pattern that is very close to the endogenous pattern of rho in D. melanogaster (Fig. 4B). Thus, changes in the pattern of rho expression are caused by changes in trans to the regulatory region of rho. Consistent with this scenario, the L-shaped activity rho enhancer from D. melanogaster is altered when this enhancer is introduced to D. virilis. As expected, this enhancer is now active in a broken V-shaped pattern (Fig. 4C).
As mentioned in the previous section, one of the main regulators of rho is BR, which represses rho in cells that form the upper part of the future dorsal appendages (11, 43). At this point of oogenesis, br is regulated by the brL enhancer. Thus, perhaps one way to alter the expression of rho is to alter the activity of the brL enhancer. Indeed, the wild- type expression of rho in both species can be predicted by the expression pattern of br. In both species, rho is expressed at the anterior border of the br domain (11, 21, 30, 43). If this is correct, then the regulatory region of brL should be changed in a way that alters the lateral extent of its activity. In particular, the expression of brL should be expanded in the lateral direction, to generate the “handle” characteristic of the wild-type pattern of BR in D. virilis. This handle is responsible for defining the anteriormost pair of appendages (39, 44).
To test this possibility, we have begun to identify the regulatory region of brL in D. virilis and assay its activity in D. melanogaster. Our preliminary results demonstrate that the brL enhancer from D. virilis indeed drives the expression more broadly than the brL enhancer from D. melanogaster (Fig. 4D′). The most noticeable difference is the expression in a lateral group of cells, resulting in a pattern that begins to resemble the endogenous pattern in D. virilis (Fig. 4D–D′′). Based on this, we propose that the evolution of the brL enhancer is responsible for the diversification of eggshell patterning and morphogenesis. To test this hypothesis, we will have to identify which sequence changes within the brL result in the expanded expression of br in D. virilis. The identification of these changes is ongoing.
Although changes in the spatial distribution of GRK could not explain the differences between the eggshell structures of D. virilis and D. melanogaster, they are responsible for the diversification of other aspects of eggshell morphogenesis. A clear example of the functional capabilities of changes of inductive signals is provided by studies of the dorsal ridge, a lumen-like structure along the dorsalmost side of the eggshells of several Drosophila species, including D. willistoni (Fig. 1F) (32, 36) In contrast to D. melanogaster, which lacks the dorsal ridge and is patterned by GRK localized around the oocyte nucleus (Fig. 4E), D. willistoni is patterned by the GRK profile that is significantly extended toward the posterior end of the follicular epithelium (45) (Fig. 4F). The origin of this dramatic change in the distribution of GRK requires further study, but it has been already established that GRK from D. willistoni is both necessary for dorsal ridge formation and sufficient for generating a partial dorsal ridge in eggshells of D. melanogaster (Fig. 4G). Thus, diversification of eggshell morphogenesis can be caused by the combined effects of changes at very top (GRK) and very bottom (BR) of the patterning network.
Summary and Outlook
Eggshell morphogenesis in drosophilids provides an excellent opportunity for the detailed analysis of multiple steps that connect inductive signals and transcription factors to 2D patterns of gene expression and 3D structures. The experimental studies of this system have come a long way from the identification of the key components of signaling pathways through their effects on eggshell morphologies to the detailed analysis of gene regulatory sequences and computational models that capture multiple aspects of signaling, transcription, and morphogenesis (30, 46, 47). Much work remains to be done to characterize the regulatory sequences and their control by transcription factors. Furthermore, some of the important players, most notably the activators, are yet to be identified and placed within the existing network.
Recent studies with the enhancer of br and data from cross-species analysis strongly suggest that the late enhancer of br is one of the main loci for the diversification of eggshell patterning. At this point, the identified regulatory regions for brL in both species are still quite large. Shortening these enhancers will enable more efficient exploration of the cis-regulatory changes involved in the diversification of eggshell patterning. Once these changes are identified and the activity of minimal enhancers is examined in both species, similar to what has been done for the rho enhancer, their functional effects on eggshell patterning can be tested. This can be done using the recently developed genome editing techniques, by swapping the brL regions between the two species. In particular, it will be very interesting to see whether a change in a single enhancer is sufficient to change both the expression pattern of BR and the number of dorsal appendages.
In addition to changes in the cis-regulatory region of br, which appears to be the key node in the appendage patterning network, it is important to keep in mind the mechanisms that rely on the intracellular modulation of inductive signals. In particular, GRK induces several negative feedback loops that modulate the signals sensed by the enhancers of the genes within the patterning network, changing the 2D patterns of br expression and affecting eggshell morphology (15, 48–52). Changing the induction thresholds of these negative feedback regulators provides another degree of flexibility for the evolution of eggshell patterning (18, 28).
One of the most exciting directions for future studies is related to the mechanistic analysis and experimental validation of the computational models that can explain the remarkable morphological diversity of eggshell structures. Current models of eggshell patterning can account for the dynamics of inductive inputs and some of the most important features of the transcriptional network that regulate BR and RHO (20, 21, 28, 30, 37, 53). These models can predict how the spatiotemporal activity of the brL enhancer responds to changes in the inductive signals and changes within the transcriptional network, providing a compact summary for a large number of genetic perturbation experiments. At this point, models of eggshell patterning do not directly use the sequence-specific information, but these capabilities can be added as we acquire more knowledge about the connections between transcription factors and the brL sequence (54, 55). In the future, we envision a unified model that accounts for multiple processes, from inductive signals to enhancers, and can generate all of the observed eggshell morphologies by variations of model parameters and sequence variations.
Acknowledgments
We thank Celeste Berg and Miriam Osterfield for helpful discussions and Matt Niepielko, Vikrant Singh, Nicole Revaitis, Cody Stevens, David Lemon, and Jasmin Imran Alsous for help in preparing the figures. S.Y.S. was supported by National Institute of General Medical Sciences (NIGMS) Grant R01 GM107103. G.P. was supported by the Excellence Initiative of the German Federal and State Governments (EXC294). N.Y. was supported by NIGMS Grant 1R15GM101597-01A1 and National Science Foundation CAREER Award IOS-1149144.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Regulatory Networks and Network Models in Development and Evolution,” held April 12–14, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Gene_Regulatory_Networks.
This article is a PNAS Direct Submission.
References
- 1.Berg CA. The Drosophila shell game: Patterning genes and morphological change. Trends Genet. 2005;21(6):346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124(22):4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- 3.Peri F, Roth S. Combined activities of Gurken and decapentaplegic specify dorsal chorion structures of the Drosophila egg. Development. 2000;127(4):841–850. doi: 10.1242/dev.127.4.841. [DOI] [PubMed] [Google Scholar]
- 4.Cheung LS, Schüpbach T, Shvartsman SY. Pattern formation by receptor tyrosine kinases: Analysis of the Gurken gradient in Drosophila oogenesis. Curr Opin Genet Dev. 2011;21(6):719–725. doi: 10.1016/j.gde.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schüpbach T. Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell. 1987;49(5):699–707. doi: 10.1016/0092-8674(87)90546-0. [DOI] [PubMed] [Google Scholar]
- 6.Goentoro LA, et al. Quantifying the Gurken morphogen gradient in Drosophila oogenesis. Dev Cell. 2006;11(2):263–272. doi: 10.1016/j.devcel.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twombly V, et al. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development. 1996;122(5):1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- 8.Spradling AC. 1993. Developmental genetics of oogenesis. The Development of Drosophila melanogaster (Cold Spring Harbor Lab Press, Cold Spring Harbor, NY), pp 1–70.
- 9.Nilson LA, Schüpbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol. 1999;44:203–243. doi: 10.1016/s0070-2153(08)60471-8. [DOI] [PubMed] [Google Scholar]
- 10.Pai LM, Barcelo G, Schüpbach T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103(1):51–61. doi: 10.1016/s0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- 11.Ward EJ, Berg CA. Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech Dev. 2005;122(2):241–255. doi: 10.1016/j.mod.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Atkey MR, Lachance JF, Walczak M, Rebello T, Nilson LA. Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror. Development. 2006;133(11):2115–2123. doi: 10.1242/dev.02369. [DOI] [PubMed] [Google Scholar]
- 13.Morimoto AM, et al. Pointed, an ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Development. 1996;122(12):3745–3754. doi: 10.1242/dev.122.12.3745. [DOI] [PubMed] [Google Scholar]
- 14.Goff DJ, Nilson LA, Morisato D. Establishment of dorsal-ventral polarity of the Drosophila egg requires capicua action in ovarian follicle cells. Development. 2001;128(22):4553–4562. doi: 10.1242/dev.128.22.4553. [DOI] [PubMed] [Google Scholar]
- 15.Boisclair Lachance JF, Fregoso Lomas M, Eleiche A, Bouchard Kerr P, Nilson LA. Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development. 2009;136(17):2893–2902. doi: 10.1242/dev.036103. [DOI] [PubMed] [Google Scholar]
- 16.Fregoso Lomas M, Hails F, Lachance JF, Nilson LA. Response to the dorsal anterior gradient of EGFR signaling in Drosophila oogenesis is prepatterned by earlier posterior EGFR activation. Cell Reports. 2013;4(4):791–802. doi: 10.1016/j.celrep.2013.07.038. [DOI] [PubMed] [Google Scholar]
- 17.Yakoby N, Lembong J, Schüpbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008;135(2):343–351. doi: 10.1242/dev.008920. [DOI] [PubMed] [Google Scholar]
- 18.Zartman JJ, Kanodia JS, Cheung LS, Shvartsman SY. Feedback control of the EGFR signaling gradient: Superposition of domain-splitting events in Drosophila oogenesis. Development. 2009;136(17):2903–2911. doi: 10.1242/dev.039545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briscoe J, Small S. Morphogen rules: Design principles of gradient-mediated embryo patterning. Development. 2015;142(23):3996–4009. doi: 10.1242/dev.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lembong J, Yakoby N, Shvartsman SY. Pattern formation by dynamically interacting network motifs. Proc Natl Acad Sci USA. 2009;106(9):3213–3218. doi: 10.1073/pnas.0810728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simakov DS, Cheung LS, Pismen LM, Shvartsman SY. EGFR-dependent network interactions that pattern Drosophila eggshell appendages. Development. 2012;139(15):2814–2820. doi: 10.1242/dev.077669. [DOI] [PubMed] [Google Scholar]
- 22.Boisclair Lachance JF, et al. A comparative study of Pointed and Yan expression reveals new complexity to the transcriptional networks downstream of receptor tyrosine kinase signaling. Dev Biol. 2014;385(2):263–278. doi: 10.1016/j.ydbio.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charbonnier E, et al. BMP-dependent gene repression cascade in Drosophila eggshell patterning. Dev Biol. 2015;400(2):258–265. doi: 10.1016/j.ydbio.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Schüpbach T. The role of brinker in eggshell patterning. Mech Dev. 2006;123(5):395–406. doi: 10.1016/j.mod.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Shravage BV, Altmann G, Technau M, Roth S. The role of Dpp and its inhibitors during eggshell patterning in Drosophila. Development. 2007;134(12):2261–2271. doi: 10.1242/dev.02856. [DOI] [PubMed] [Google Scholar]
- 26.Hamaratoglu F, Affolter M, Pyrowolakis G. Dpp/BMP signaling in flies: From molecules to biology. Semin Cell Dev Biol. 2014;32:128–136. doi: 10.1016/j.semcdb.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 27.Fregoso Lomas M, De Vito S, Boisclair Lachance JF, Houde J, Nilson LA. Determination of EGFR signaling output by opposing gradients of BMP and JAK/STAT activity. Curr Biol. 2016;26(19):2572–2582. doi: 10.1016/j.cub.2016.07.073. [DOI] [PubMed] [Google Scholar]
- 28.Zartman JJ, et al. Pattern formation by a moving morphogen source. Phys Biol. 2011;8(4):045003. doi: 10.1088/1478-3975/8/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs A, Cheung LS, Charbonnier E, Shvartsman SY, Pyrowolakis G. Transcriptional interpretation of the EGF receptor signaling gradient. Proc Natl Acad Sci USA. 2012;109(5):1572–1577. doi: 10.1073/pnas.1115190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung LS, Simakov DS, Fuchs A, Pyrowolakis G, Shvartsman SY. Dynamic model for the coordination of two enhancers of broad by EGFR signaling. Proc Natl Acad Sci USA. 2013;110(44):17939–17944. doi: 10.1073/pnas.1304753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Queenan AM, Ghabrial A, Schüpbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124(19):3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- 32.Hinton HE. Biology of Insect Eggs. Pergamon Press; Oxford, UK: 1981. [Google Scholar]
- 33.Shvartsman SY, Muratov CB, Lauffenburger DA. Modeling and computational analysis of EGF receptor-mediated cell communication in Drosophila oogenesis. Development. 2002;129(11):2577–2589. doi: 10.1242/dev.129.11.2577. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura Y, Matsuno K. Species-specific activation of EGF receptor signaling underlies evolutionary diversity in the dorsal appendage number of the genus Drosophila eggshells. Mech Dev. 2003;120(8):897–907. doi: 10.1016/s0925-4773(03)00164-3. [DOI] [PubMed] [Google Scholar]
- 35.Vreede BM, Lynch JA, Roth S, Sucena E. Co-option of a coordinate system defined by the EGFr and Dpp pathways in the evolution of a morphological novelty. Evodevo. 2013;4(1):7. doi: 10.1186/2041-9139-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niepielko MG, et al. Chorion patterning: A window into gene regulation and Drosophila species’ relatedness. Mol Biol Evol. 2014;31(1):154–164. doi: 10.1093/molbev/mst186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niepielko MG, Ip K, Kanodia JS, Lun DS, Yakoby N. Evolution of BMP signaling in Drosophila oogenesis: A receptor-based mechanism. Biophys J. 2012;102(8):1722–1730. doi: 10.1016/j.bpj.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niepielko MG, Hernáiz-Hernández Y, Yakoby N. BMP signaling dynamics in the follicle cells of multiple Drosophila species. Dev Biol. 2011;354(1):151–159. doi: 10.1016/j.ydbio.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 39.James KE, Berg CA. Temporal comparison of Broad-Complex expression during eggshell-appendage patterning and morphogenesis in two Drosophila species with different eggshell-appendage numbers. Gene Expr Patterns. 2003;3(5):629–634. doi: 10.1016/s1567-133x(03)00136-4. [DOI] [PubMed] [Google Scholar]
- 40.Muratov CB, Shvartsman SY. An asymptotic study of the inductive pattern formation mechanism in Drosophila egg development. Physica D. 2003;186(1-2):93–108. [Google Scholar]
- 41.Pribyl M, Muratov CB, Shvartsman SY. Transitions in the model of epithelial patterning. Dev Dyn. 2003;226(1):155–159. doi: 10.1002/dvdy.10218. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura Y, et al. Soma-dependent modulations contribute to divergence of rhomboid expression during evolution of Drosophila eggshell morphology. Development. 2007;134(8):1529–1537. doi: 10.1242/dev.001578. [DOI] [PubMed] [Google Scholar]
- 43.Ward EJ, Zhou X, Riddiford LM, Berg CA, Ruohola-Baker H. Border of Notch activity establishes a boundary between the two dorsal appendage tube cell types. Dev Biol. 2006;297(2):461–470. doi: 10.1016/j.ydbio.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 44.Osterfield M, Schüpbach T, Wieschaus E, Shvartsman SY. Diversity of epithelial morphogenesis during eggshell formation in drosophilids. Development. 2015;142(11):1971–1977. doi: 10.1242/dev.119404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niepielko MG, Yakoby N. Evolutionary changes in TGFα distribution underlie morphological diversity in eggshells from Drosophila species. Development. 2014;141(24):4710–4715. doi: 10.1242/dev.111898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osterfield M, Du X, Schüpbach T, Wieschaus E, Shvartsman SY. Three-dimensional epithelial morphogenesis in the developing Drosophila egg. Dev Cell. 2013;24(4):400–410. doi: 10.1016/j.devcel.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fauré A, Vreede BM, Sucena E, Chaouiya C. A discrete model of Drosophila eggshell patterning reveals cell-autonomous and juxtacrine effects. PLOS Comput Biol. 2014;10(3):e1003527. doi: 10.1371/journal.pcbi.1003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wasserman JD, Freeman M. An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell. 1998;95(3):355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- 49.Sapir A, Schweitzer R, Shilo BZ. Sequential activation of the EGF receptor pathway during Drosophila oogenesis establishes the dorsoventral axis. Development. 1998;125(2):191–200. doi: 10.1242/dev.125.2.191. [DOI] [PubMed] [Google Scholar]
- 50.Reich A, Sapir A, Shilo B. Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development. 1999;126(18):4139–4147. doi: 10.1242/dev.126.18.4139. [DOI] [PubMed] [Google Scholar]
- 51.Ghiglione C, et al. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell. 1999;96(6):847–856. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 52.Peri F, Bökel C, Roth S. Local Gurken signaling and dynamic MAPK activation during Drosophila oogenesis. Mech Dev. 1999;81(1-2):75–88. doi: 10.1016/s0925-4773(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 53.Lembong J, Yakoby N, Shvartsman SY. Spatial regulation of BMP signaling by patterned receptor expression. Tissue Eng Part A. 2008;14(9):1469–1477. doi: 10.1089/ten.tea.2008.0098. [DOI] [PubMed] [Google Scholar]
- 54.Samee MA, et al. A systematic ensemble approach to thermodynamic modeling of gene expression from sequence data. Cell Syst. 2015;1(6):396–407. doi: 10.1016/j.cels.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samee AH, Sinha S. Evaluating thermodynamic models of enhancer activity on cellular resolution gene expression data. Methods. 2013;62(1):79–90. doi: 10.1016/j.ymeth.2013.03.005. [DOI] [PubMed] [Google Scholar]