Significance
In this work we report on the previously uncharacterized clinical and biological role for EGFL7 in acute myeloid leukemia (AML). Patients with increased EGFL7 mRNA expression had lower complete remission rates and shorter overall and event-free survival, demonstrating the clinical relevance of EGFL7 expression in cytogenetically normal AML. Our results show that AML blasts are able to synthesize and secrete EGFL7 protein, promoting autocrine blast cell growth. Inhibition of EGFL7 results in decreased proliferation and induces apoptosis of AML cells. Taken together, our data provide the rationale for targeting EGFL7 using blocking antibodies as a therapy for patients with AML.
Keywords: EGFL7, acute myeloid leukemia, clinical outcome
Abstract
Epithelial growth factor-like 7 (EGFL7) is a protein that is secreted by endothelial cells and plays an important role in angiogenesis. Although EGFL7 is aberrantly overexpressed in solid tumors, its role in leukemia has not been evaluated. Here, we report that levels of both EGFL7 mRNA and EGFL7 protein are increased in blasts of patients with acute myeloid leukemia (AML) compared with normal bone marrow cells. High EGFL7 mRNA expression associates with lower complete remission rates, and shorter event-free and overall survival in older (age ≥60 y) and younger (age <60 y) patients with cytogenetically normal AML. We further show that AML blasts secrete EGFL7 protein and that higher levels of EGFL7 protein are found in the sera from AML patients than in sera from healthy controls. Treatment of patient AML blasts with recombinant EGFL7 in vitro leads to increases in leukemic blast cell growth and levels of phosphorylated AKT. EGFL7 blockade with an anti-EGFL7 antibody reduced the growth potential and viability of AML cells. Our findings demonstrate that increased EGFL7 expression and secretion is an autocrine mechanism supporting growth of leukemic blasts in patients with AML.
Acute myeloid leukemia (AML) is a clonal hematopoietic disease characterized by the proliferation of immature blasts in the bone marrow (BM) and blood (1). Genetic alterations, including chromosomal translocations and deletions and gene mutations leading to aberrant downstream target gene expression, contribute to AML initiation and maintenance. Previously, our group demonstrated that increased miRNA-126-3p (miR-126) expression in patients with cytogenetically normal AML (CN-AML) correlated with shorter overall survival (OS). Furthermore, we found miR-126 to be essential for leukemia stem cell (LSC) homeostasis, and in vivo targeting of miR-126 in a patient-derived xenograft model resulted in prolonged survival in secondary bone marrow transplant (BMT) recipients (2). miR-126 is located within intron 7 of a protein-coding gene known as Epithelial growth factor-like 7 (EGFL7) (3). Although we and others (2, 4, 5) have shown an important role for miR-126 in AML biology, we know of no studies that have been performed to understand the prognostic and functional implications of expression of its host gene, EGFL7, in AML.
EGFL7 is a secreted protein of ∼30 kDa and plays an important physiological role in angiogenesis (6–8). Unlike other angiogenic factors (e.g., VEGF), physiological EGFL7 expression and function has been restricted mainly to the endothelial cells where it regulates survival, migration, and differentiation (6). Aberrant expression of EGFL7 has been shown to be involved in tumor growth and disease progression of several solid tumors, including hepatocellular carcinoma, malignant glioma, and breast, lung, and pancreatic cancers (9), but its role in hematopoietic malignancies is currently unknown. Therefore, we investigated the prognostic and biological function of EGFL7 expression in AML.
We show that EGFL7 mRNA and protein expression is increased in patient AML blasts compared with normal BM mononuclear cells (NBM-MNCs) and that high EGFL7 mRNA expression levels correlate with worse outcome in both younger (age <60 y) and older (age ≥60 y) patients with CN-AML. Furthermore, we demonstrate that AML blasts are capable of secreting EGFL7 protein, leading to enhanced leukemic blast growth. Our data suggest an independent role for EGFL7 in AML but also highlight the importance of this genetic locus in AML via up-regulation of both miR-126 and its host gene EGFL7.
Results
Pretreatment Features and Clinical Outcomes Associated with EGFL7 Expression in Younger Adults with CN-AML.
To evaluate the prognostic significance of EGFL7 mRNA expression in CN-AML, we analyzed one cohort of younger adults (n = 374) and one of older patients (n = 198), for whom EGFL7 expression was measured by RNA-sequencing (RNA-seq) and microarrays, respectively. The median expression value of EGFL7 was used as a cut point to separate the analyzed cohorts into high and low EGFL7 expressers.
Among younger adults, those with high EGFL7 expression (n = 187) were more likely to present with lower platelet (P = 0.002) and WBC (P = 0.001) counts and higher percentages of blood blasts (P < 0.001) than patients with low EGFL7 expression (n = 187). High EGFL7 expressers were also less likely to have leukemic infiltration at extramedullary sites (P = 0.02). With regard to molecular characteristics, patients with high EGFL7 expression more frequently harbored double CEBPA (P < 0.001) and WT1 (P = 0.02) mutations and less frequently harbored DNMT3A (P = 0.004), FLT3-tyrosine kinase domain (FLT3-TKD; P = 0.03), IDH2 (P = 0.01), and NPM1 (P < 0.001) mutations. EGFL7-expresser status associated with significant differences (P = 0.04) in the risk stratification of patients according to the European LeukemiaNet (ELN) guidelines (10). Patients with high EGFL7 expression were more frequently classified in the adverse risk group and less frequently in the favorable risk group than patients with low EGFL7 expression. High EGFL7 expression status associated with high expression of the BAALC (P < 0.001), ERG (P < 0.001), and MN1 (P < 0.001) genes as well as high expression of miR-181a (P < 0.001) and miR-155 (P = 0.008). High EGFL7 expressers were also more likely to express miR-3151 (P < 0.001) (Table S1). Because gene mutations frequently co-occur in CN-AML, we attempted to evaluate whether any mutational combinations are associated with EGFL7 expression. Only the concomitant presence of FLT3-internal tandem duplications (FLT3-ITD) and DNMT3A and NPM1 mutations (FLT3-ITD/DNMT3Amut/NPM1mut) were frequent enough for this analysis. Patients who harbored these three mutations (n = 52) had higher expression of EGFL7 than patients who had WT DNMT3A, NPM1, and FLT3 (n = 82; P = 0.009).
Table S1.
Comparison of clinical and molecular characteristics by EGFL7-expresser status of younger adult patients (age <60 y) with de novo CN-AML
| Characteristic | Low EGFL7 (n = 187) | High EGFL7 (n = 187) | P |
| Age, y | 0.09 | ||
| Median | 47 | 44 | |
| Range | 19–59 | 17–59 | |
| Sex, n (%) | 0.12 | ||
| Male | 88 (47) | 104 (56) | |
| Female | 99 (53) | 83 (44) | |
| Race, n (%) | 0.21 | ||
| White | 170 (93) | 163 (89) | |
| Nonwhite | 13 (7) | 21 (11) | |
| Hemoglobin, g/dL | 0.89 | ||
| Median | 9.3 | 9.2 | |
| Range | 4.6–25.1 | 4.2–14.4 | |
| Platelet count × 109/L | 0.002 | ||
| Median | 67 | 50 | |
| Range | 8–433 | 8–445 | |
| WBC count × 109/L | 0.001 | ||
| Median | 35.6 | 24.3 | |
| Range | 0.6–308.8 | 0.8–475.0 | |
| Blood blasts, % | <0.001 | ||
| Median | 53 | 66 | |
| Range | 0–97 | 0–97 | |
| Bone marrow blasts, % | 0.35 | ||
| Median | 69 | 65 | |
| Range | 10–96 | 19–95 | |
| Extramedullary involvement, n (%) | 0.02 | ||
| Present | 65 (35) | 43 (24) | |
| Absent | 119 (65) | 139 (76) | |
| ASXL1, n (%) | 0.78 | ||
| Mutated | 6 (3) | 7 (4) | |
| WT | 174 (97) | 167 (96) | |
| CEBPA, n (%) | <0.001 | ||
| Double mutated | 5 (3) | 50 (28) | |
| WT | 173 (97) | 129 (72) | |
| DNMT3A, n (%) | 0.004 | ||
| Mutated | 85 (47) | 56 (31) | |
| R882-mutated | 61 | 45 | |
| Non–R882-mutated | 24 | 11 | |
| WT | 97 (53) | 122 (69) | |
| FLT3-ITD, n (%) | 0.20 | ||
| Present | 66 (36) | 78 (43) | |
| Absent | 118 (64) | 105 (57) | |
| FLT3-TKD, n (%) | 0.03 | ||
| Present | 26 (14) | 12 (7) | |
| Absent | 155 (86) | 165 (93) | |
| IDH1, n (%) | 0.85 | ||
| Mutated | 14 (8) | 15 (8) | |
| WT | 168 (92) | 163 (92) | |
| IDH2, n (%) | 0.01 | ||
| Mutated | 25 (14) | 10 (6) | |
| WT | 157 (86) | 168 (94) | |
| NPM1, n (%) | <0.001 | ||
| Mutated | 130 (74) | 76 (43) | |
| WT | 45 (26) | 99 (57) | |
| RUNX1, n (%) | 0.17 | ||
| Mutated | 7 (4) | 13 (7) | |
| WT | 175 (96) | 165 (93) | |
| TET2, n (%) | 0.86 | ||
| Mutated | 20 (11) | 18 (10) | |
| WT | 162 (89) | 160 (90) | |
| WT1, n (%) | 0.02 | ||
| Mutated | 13 (7) | 27 (15) | |
| WT | 169 (93) | 151 (85) | |
| ELN genetic group*, n (%) | 0.04 | ||
| Favorable | 110 (62) | 90 (52) | |
| Intermediate | 48 (27) | 47 (27) | |
| Adverse | 19 (11) | 35 (20) | |
| BAALC†, n (%) | <0.001 | ||
| High | 38 (24) | 135 (73) | |
| Low | 122 (76) | 49 (27) | |
| ERG†, n (%) | <0.001 | ||
| High | 47 (25) | 139 (74) | |
| Low | 138 (75) | 48 (26) | |
| MN1†, n (%) | <0.001 | ||
| High | 53 (30) | 127 (69) | |
| Low | 121 (70) | 58 (31) | |
| miR-181a†, n (%) | <0.001 | ||
| High | 53 (38) | 97 (61) | |
| Low | 87 (62) | 62 (39) | |
| miR-3151, n (%) | <0.001 | ||
| Expressed | 5 (4) | 46 (29) | |
| Not expressed | 135 (96) | 113 (71) | |
| miR-155†, n (%) | 0.008 | ||
| High | 59 (42) | 92 (58) | |
| Low | 81 (58) | 67 (42) |
Within patients with CN-AML, the ELN favorable risk category comprises patients with double-mutated CEBPA and patients with mutated NPM1 without FLT3-ITD or with FLT3-ITDlow. The ELN intermediate risk category includes patients with WT NPM1 without FLT3-ITD, with WT NPM1 and FLT3-ITDlow, or with mutated NPM1 and FLT3-ITDhigh. The ELN adverse risk category comprises patients with NPM1 with FLT3-ITDhigh and/or mutated RUNX1 and/or mutated ASXL1 (if they do not co-occur with a favorable AML subtypes) and/or mutated TP53. FLT3-ITDlow is defined by a FLT3-ITD/FLT3 WT allelic ratio less than 0.5, and FLT3-ITDhigh is defined as by a FLT3-ITD/FLT3 WT allelic ratio of equal to or more than 0.5.
The median expression value was used as the cut point.
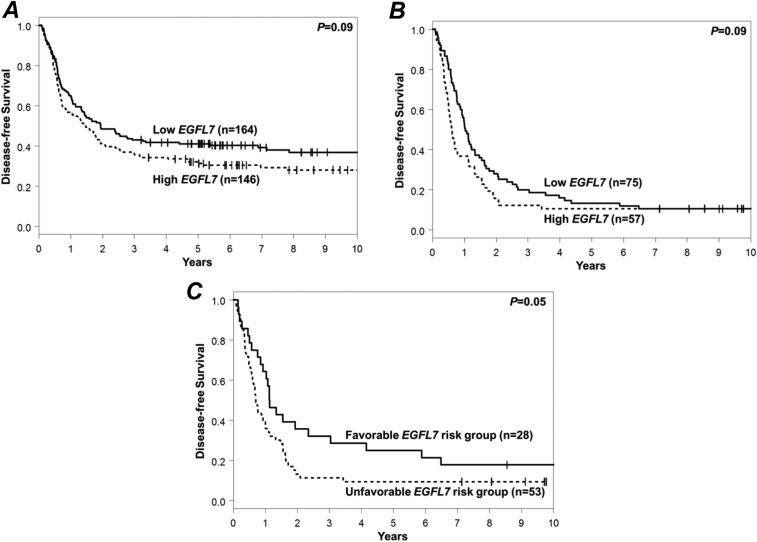
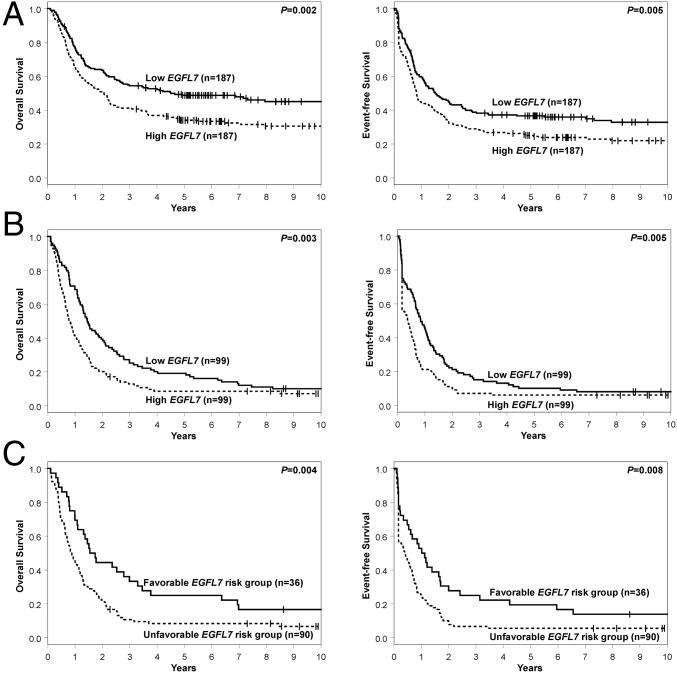
With regard to the clinical outcome, high EGFL7 expression status associated with a lower complete remission (CR) rate (78 vs. 88%, P = 0.01). Patients with high EGFL7 expression showed a trend for shorter disease-free survival (DFS) (P = 0.09, 5-y rates, 31 vs. 41%) (Fig. S1A) and had shorter OS (P = 0.002, 5-y rates, 34 vs. 49%) (Fig. 1A) than patients with low EGFL7 expression. High EGFL7 expressers also had shorter event-free survival (EFS) (P = 0.005, 5-y rates, 25 vs. 37%) (Fig. 1A and Table S2).
Fig. S1.
Prognostic significance of EGFL7 in younger and older CN-AML patients. (A and B) Impact of EGFL7 expression levels on DFS of younger (age <60 y) (A) and older (age ≥60 y) (B) adult patients. (C) DFS according to EGFL7 risk group in older CN-AML patients. The favorable risk group comprised patients with EGFL7 low expression/high methylation; the unfavorable risk group comprised the remaining patients (high expression/low methylation, high expression/high methylation, low expression/low methylation). The median values of EGFL7 expression and EGFL7 promoter methylation were used as cutoffs.
Fig. 1.
Prognostic significance of EGFL7 in younger and older CN-AML patients. (A and B) The association of EGFL7 expression levels with OS and EFS of younger adult patients (age <60 y) (A) and older patients (age ≥60 y) (B). (C) OS and EFS according to EGFL7 risk group in older CN-AML patients. The favorable risk group was comprised of patients with EGFL7 low expression/high promoter methylation; the unfavorable risk group included the remaining patients (high expression/low promoter methylation, high expression/high promoter methylation, low expression/low promoter methylation). The median values of EGFL7 expression and EGFL7 promoter methylation were used as the high/low cut points.
Table S2.
Treatment outcomes according to EGFL7 expression in 374 younger adult patients (age <60 y) with de novo CN-AML
| End point | Low EGFL7* (n = 187) | High EGFL7* (n = 187) | P† |
| Complete response, n (%) | 165 (88) | 146 (78) | 0.01 |
| DFS | 0.09 | ||
| Median, y | 1.9 | 1.5 | |
| % disease-free at 5 y (95% CI) | 41 (34–49) | 31 (24–39) | |
| OS‡ | 0.002 | ||
| Median, y | 4.5 | 2.1 | |
| % alive at 5 y (95% CI) | 49 (41–56) | 34 (27–41) | |
| EFS§ | 0.005 | ||
| Median, y | 1.5 | 0.8 | |
| % event-free at 5 y (95% CI) | 37 (30–43) | 25 (19–31) |
The median was the cut point used to dichotomize the variable.
P values for categorical variables are from Fisher’s exact test; P values for the time-to-event variables are from the log-rank test.
The median follow-up for those alive is 8.2 y, range: 0.6–18.1 y (n = 143).
The median follow-up for those who have not had an event is 7.6 y, range: 0.6–18.1 y (n = 106).
Pretreatment Features and Clinical Outcomes Associated with EGFL7 Expression in Older Adults with CN-AML.
Among older patients, those with high EGFL7 expression more frequently harbored double CEBPA mutations (P = 0.01), FLT3-ITD (P < 0.001) and RUNX1 mutations (P < 0.001), and less frequently harbored NPM1 (P < 0.001) and TET2 (P = 0.001) mutations. They were also less frequently classified in the favorable and more frequently in the intermediate or adverse risk group of the ELN classification than patients with low EGFL7 expression (P < 0.001). High EGFL7 expressers were more likely to have high expression of the BAALC (P < 0.001), ERG (P < 0.001), and MN1 (P < 0.001) genes as well as miR-181a (P = 0.02) and miR-155 (P = 0.05) (Table S3).
Table S3.
Comparison of clinical and molecular characteristics by EGFL7-expresser status of older patients (age ≥60 y) with de novo CN-AML
| Characteristic | Low EGFL7 (n = 99) | High EGFL7 (n = 99) | P |
| Age, y | 0.21 | ||
| Median | 68 | 70 | |
| Range | 60–83 | 60–81 | |
| Sex, n (%) | 0.09 | ||
| Male | 45 (45) | 58 (59) | |
| Female | 54 (55) | 41 (41) | |
| Race, n (%) | 0.81 | ||
| White | 89 (90) | 90 (92) | |
| Nonwhite | 10 (10) | 8 (8) | |
| Hemoglobin (g/dL) | 0.70 | ||
| Median | 9.4 | 9.4 | |
| Range | 5.4–15.0 | 6.0–13.1 | |
| Platelet count × 109/L | 0.26 | ||
| Median | 79 | 60 | |
| Range | 4–271 | 11–850 | |
| WBC count ×109/L | 0.14 | ||
| Median | 28.4 | 21.1 | |
| Range | 1.0–450.0 | 1.0–434.1 | |
| Blood blasts, % | 0.22 | ||
| Median | 37 | 57 | |
| Range | 0–97 | 0–99 | |
| Bone marrow blasts, % | 0.85 | ||
| Median | 68 | 66 | |
| Range | 4–97 | 17–96 | |
| Extramedullary involvement, n (%) | 0.87 | ||
| Present | 24 (25) | 22 (23) | |
| Absent | 73 (75) | 74 (77) | |
| ASXL1, n (%) | 0.68 | ||
| Mutated | 12 (12) | 15 (15) | |
| WT | 86 (88) | 84 (85) | |
| CEBPA, n (%) | 0.01 | ||
| Double mutated | 0 (0) | 7 (7) | |
| WT | 99 (100) | 92 (93) | |
| DNMT3A, n (%) | 0.45 | ||
| Mutated | 28 (29) | 34 (34) | |
| R882-mutated | 17 | 19 | |
| Non–R882-mutated | 11 | 15 | |
| WT | 68 (71) | 65 (66) | |
| FLT3-ITD, n (%) | <0.001 | ||
| Present | 20 (20) | 43 (43) | |
| Absent | 79 (80) | 56 (57) | |
| FLT3-TKD, n (%) | 0.67 | ||
| Present | 11 (11) | 14 (14) | |
| Absent | 87 (89) | 85 (86) | |
| IDH1, n (%) | 0.29 | ||
| Mutated | 16 (16) | 10 (10) | |
| WT | 83 (84) | 89 (90) | |
| IDH2, n (%) | 1.00 | ||
| Mutated | 22 (22) | 21 (21) | |
| WT | 77 (78) | 78 (79) | |
| NPM1, n (%) | <0.001 | ||
| Mutated | 72 (73) | 43 (43) | |
| WT | 27 (27) | 56 (57) | |
| RUNX1, n (%) | <0.001 | ||
| Mutated | 7 (7) | 26 (26) | |
| WT | 89 (93) | 73 (74) | |
| TET2, n (%) | 0.001 | ||
| Mutated | 35 (36) | 15 (15) | |
| WT | 63 (64) | 84 (85) | |
| WT1, n (%) | 1.00 | ||
| Mutated | 4 (4) | 4 (4) | |
| WT | 95 (96) | 95 (96) | |
| ELN genetic group*, n (%) | <0.001 | ||
| Favorable | 64 (65) | 28 (28) | |
| Intermediate | 20 (20) | 37 (37) | |
| Adverse | 14 (14) | 34 (34) | |
| BAALC†, n (%) | <0.001 | ||
| High | 29 (29) | 69 (70) | |
| Low | 70 (71) | 30 (30) | |
| ERG†, n (%) | <0.001 | ||
| High | 25 (25) | 74 (75) | |
| Low | 74 (75) | 25 (25) | |
| MN1†, n (%) | <0.001 | ||
| High | 24 (37) | 48 (70) | |
| Low | 41 (63) | 21 (30) | |
| miR-181a†, n (%) | 0.02 | ||
| High | 35 (42) | 53 (60) | |
| Low | 48 (58) | 35 (40) | |
| miR-3151†, n (%) | 0.15 | ||
| High | 31 (40) | 40 (53) | |
| Low | 46 (60) | 36 (47) | |
| miR-155†, n (%) | 0.05 | ||
| High | 34 (40) | 49 (55) | |
| Low | 52 (60) | 40 (45) |
In patients with CN-AML, the ELN favorable risk category comprises patients with double-mutated CEBPA and patients with mutated NPM1 without FLT3-ITD or with FLT3-ITDlow. The ELN intermediate risk category includes patients with WT NPM1 without FLT3-ITD, WT NPM1 and FLT3-ITDlow, or mutated NPM1 and FLT3-ITDhigh. The ELN adverse risk category comprises patients with WT NPM1 with FLT3-ITDhigh and/or mutated RUNX1 and/or mutated ASXL1 (if they do not co-occur with a favorable AML subtype) and/or mutated TP53. FLT3-ITDlow is defined by a FLT3-ITD/FLT3 WT allelic ratio less than 0.5, and FLT3-ITDhigh is defined as by a FLT3-ITD/FLT3 WT allelic ratio equal to or more than 0.5.
The median expression value was used as the cut point.
With regard to mutational combinations, again only the FLT3-ITD/DNMT3Amut/NPM1mut mutational combination was frequent enough for analysis of its potential association with EGFL7 expression in older CN-AML patients. However, in contrast to younger patients, EGFL7 expression in older CN-AML patients with FLT3-ITD/DNMT3Amut/NPM1mut (n = 21) did not differ significantly from that in patients who had WT DNMT3A, NPM1, and FLT3 (n = 51; P = 0.79).
Concerning clinical outcome, older CN-AML patients with high EGFL7 expression were less likely to achieve a CR (58 vs. 76%, P = 0.01). High EGFL7 expression status associated with shorter OS (P = 0.003, 5-y rates, 9 vs. 19%) (Fig. 1B) and EFS (P = 0.005, 5-y rates, 6 vs. 10%) (Fig. 1B), and in these patients there was a trend for the association of high EGFL7 expression with shorter DFS (P = 0.09, 5-y rates, 11 vs. 13%) (Fig. S1B and Table S4).
Table S4.
Treatment outcomes according to EGFL7 expression in 198 older patients (age ≥60 y) with de novo CN-AML
| End point | Low EGFL7* (n = 99) | High EGFL7* (n = 99) | P† |
| Complete response, n (%) | 75 (76) | 57 (58) | 0.01 |
| DFS | 0.09 | ||
| Median, y | 1.0 | 0.6 | |
| % disease-free at 5 y (95% CI) | 13 (7-22) | 11 (4-20) | |
| OS‡ | 0.003 | ||
| Median, y | 1.4 | 0.8 | |
| % alive at 5 y (95% CI) | 19 (12–27) | 9 (4–15) | |
| EFS§ | 0.005 | ||
| Median, y | 0.8 | 0.4 | |
| % event-free at 5 y (95% CI) | 10 (5–17) | 6 (2–12) |
From Affymetrix microarray platform HG-U133 Plus 2.0. The median was the cut point used to dichotomize the variable.
P values for categorical variables are from Fisher’s exact test; P values for time-to-event variables are from the log-rank test.
The median follow-up for those alive is 9.2 y, range: 2.3–13.9 y (n = 15).
The median follow-up for those who have not had an event is 9.2 y, range: 7.3–13.9 y (n = 10).
Association of EGFL7 Expression with Clinical Outcome in the Context of miR-126 Expression.
As previously mentioned, EGFL7 is the host gene for miR-126, which has been shown to associate independently with prognosis of CN-AML patients (2). Because EGFL7 expression was also found to be prognostic, we decided to evaluate whether one of these two transcripts (i.e., EGFL7 or miR-126) had a stronger association with clinical outcome than the other. We therefore studied 300 younger and 171 older CN-AML patients with available EGFL7 and miR-126 expression data. In both age groups, we detected high correlation in the expression levels of EGFL7 and miR-126 (r = 0.84 in younger CN-AML patients and r = 0.64 in older CN-AML patients). This high degree of correlation did not allow us to evaluate the association of EGFL7 and miR-126 expression with prognosis in multivariable Cox-regression models, which include both covariates. Nevertheless, univariable analyses indicated a stronger association of EGFL7 expression with outcome than for miR-126 for both younger and older CN-AML patients (Table S5).
Table S5.
Univariable models for expression of EGFL7 and miR-126 and associations with outcome in 300 younger adults and 171 older patients with CN-AML
| Models* | DFS | OS | EFS | |||
| HR (CI) | P value | HR (CI) | P value | HR (CI) | P value | |
| Univariable model: EGFL7 | 1.40 (1.02–1.91) | 0.03 | 1.68 (1.02–2.28) | <0.001 | 1.53 (1.17–2.02) | 0.002 |
| Univariable model: miR-126 | 1.15 (0.84–1.57) | 0.38 | 1.37 (1.02–1.84) | 0.04 | 1.34 (1.02–1.76) | 0.04 |
| Univariable model: EGFL7 | 1.33 (0.90–1.98) | 0.15 | 1.52 (1.11–2.08) | 0.01 | 1.45 (1.06–1.99) | 0.02 |
| Univariable model: miR-126 | 1.21 (0.81–1.78) | 0.35 | 1.29 (0.94–1.77) | 0.11 | 1.37 (1.01–1.87) | 0.05 |
In all models, the median was the cut point used to dichotomize the variable.
Prognostic Significance of EGFL7 Promoter Methylation Status.
We hypothesized that the epigenetic regulation of EGFL7 could also provide prognostic information, and thus we evaluated the EGFL7 promoter methylation status in a subgroup of older CN-AML patients (n = 126) using a methylated DNA-capture technique, followed by next-generation sequencing (MethylCap-Seq), as described previously (11). We used the median value of EGFL7 promoter methylation to dissect our cohort and found that patients with high EGFL7 promoter methylation showed a trend toward higher CR rates (73 vs. 56%, P = 0.06) and had longer OS (P = 0.05) than patients with low EGFL7 promoter methylation. There also was a trend for longer EFS in patients with high EGFL7 promoter methylation (P = 0.09) but no significant association of DFS with EGFL7 promoter methylation status.
Prognostic Significance of Integrated EGFL7 mRNA Expression and EGFL7 Promoter Methylation Status.
The combination of high EGFL7 promoter methylation with low EGFL7 expression identified a subset of older CN-AML patients with better outcome (n = 36, hereafter referred to as the “EGFL7 favorable risk group”) compared with the remaining patients of the cohort (n = 90, the “EGFL7 unfavorable risk group”). Patients in the EGFL7 favorable risk group showed a trend for higher CR rates (78 vs. 59%, P = 0.06) and had longer DFS (P = 0.05) (Fig. S1C). Five years after diagnosis, 25% of these patients remained alive and leukemia-free, in contrast to only 9% of patients in the EGFL7 unfavorable risk group. Patients in the EGFL7 favorable risk group also had longer OS (P = 0.004, 5-y rates, 25 vs. 4%) and EFS (P = 0.008, 5-y rates, 19 vs. 6%) than those in the EGFL7 unfavorable risk group (Fig. 1C and Table S6).
Table S6.
Treatment outcomes according to EGFL7 risk group in 126 older patients (age ≥60 y) with de novo CN-AML
| End point | Unfavorable EGFL7 risk group* (n = 90) | Favorable EGFL7 risk group* (n = 36) | P† |
| CR, n (%) | 53 (59) | 28 (78) | 0.06 |
| DFS | 0.05 | ||
| Median, y | 0.7 | 1.1 | |
| % disease-free at 5 y (95% CI) | 9 (3–19) | 25 (11–42) | |
| OS‡ | 0.004 | ||
| Median, y | 0.9 | 1.6 | |
| % alive at 5 y (95% CI) | 8 (4–15) | 25 (12–40) | |
| EFS§ | 0.008 | ||
| Median, y | 0.4 | 1.1 | |
| % event-free at 5 y (95% CI) | 6 (2–12) | 19 (9–34) |
The favorable risk group consists of patients with low EGFL7 AFFY expression and high EGFL7 methylation; the unfavorable risk group consists of patients with high EGFL7 AFFY expression and/or low EGFL7 methylation. AFFY data are from the Affymetrix microarray platform HG-U133 Plus 2.0.
P values for categorical variables are from Fisher’s exact test; P values for time to event variables are from the log-rank test.
The median follow-up for those alive is 9.2 y, range: 2.3–13.9 y (n = 15).
The median follow-up for those who have not had an event is 9.2 y, range: 7.3–13.9 y (n = 10).
In multivariable analyses of older CN-AML patients, the EGFL7 favorable risk group was shown to be an independent marker of longer DFS (P = 0.03) after adjusting for extramedullary involvement (P = 0.02) and of longer OS (P < 0.001) after adjusting for miR-155 expression status (P < 0.001) and platelet counts (P < 0.001) (Table 1). Because of the relatively small number of patients in the EGFL7 favorable risk group, a final model could not be constructed for EFS. We could, however, generate three separate three-variable models, in which the association of favorable EGFL7 status with longer EFS remained significant after adjusting for other covariables (P = 0.002, P < 0.001, and P < 0.001) (Table S7). With regard to CR, EGFL7 risk group status was not significant in multivariable analysis.
Table 1.
Multivariable analyses of outcome according to the EGFL7 risk group in 126 older patients (age ≥60 y) with de novo CN-AML
| Variables in final models | DFS | OS | ||
| HR (95% CI) | P | HR (95% CI) | P | |
| EGFL7 risk group, favorable vs. unfavorable | 0.57 (0.34–0.95) | 0.03 | 0.45 (0.29–0.71) | <0.001 |
| Extramedullary involvement, present vs. absent | 1.99 (1.13–3.51) | 0.02 | — | — |
| miR-155, high vs. low* | — | — | 2.47 (1.65–3.70) | <0.001 |
| Platelets, continuous | — | — | 1.22 (1.09–1.36) | <0.001 |
Hazard ratios (HR) greater than (or less than) 1.0 indicate higher (or lower) risk for relapse or death (in DFS) or for death (in OS) for the higher value of the continuous variables and for the first category listed in the categorical variables.
The median expression value was used as the cut point.
Table S7.
Multivariable analyses of event-free survival according to the EGFL7 risk group in 126 older patients (age ≥60 y) with de novo CN-AML
| Variables considered | EFS | |||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| EGFL7 risk group, favorable vs. unfavorable | 0.48 (0.31–0.77) | 0.002 | 0.44 (0.28–0.70) | <0.001 | 0.46 (0.29–0.72) | <0.001 |
| WBC count, continuous | 1.22 (1.05–1.40) | 0.007 | 1.20 (1.04–1.39) | 0.01 | 1.17 (1.01–1.36) | 0.04 |
| miR-155, high vs. low* | 1.68 (1.14–2.47) | 0.008 | — | — | — | — |
| Platelets, continuous | — | — | 1.16 (1.04–1.28) | 0.006 | — | — |
| Extramedullary involvement, present vs. absent | — | — | — | — | 1.63 (1.05–2.53) | 0.03 |
HRs greater than (or less than) 1.0 indicate higher (or lower) failure to achieve complete remission, relapse, or death (EFS) for the higher value of the continuous variables and the first category listed for the categorical variables.
The median expression value was used as the cut point.
EGFL7 Expression in AML.
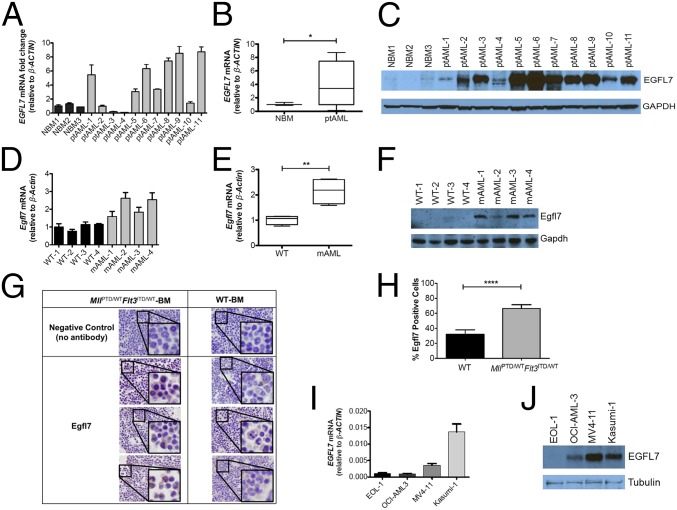
To validate the RNA-seq and microarray data further, we measured EGFL7 mRNA and protein in patient AML blasts and NBM-MNCs using real-time RT-PCR and Western blotting, respectively (Table S8 for cytogenetic and molecular characteristics of the patients). As shown in Fig. 2 A–C, higher levels of EGFL7 mRNA and protein were observed in AML blasts from the majority of patients analyzed than in NBM-MNCs. We next investigated the Egfl7 expression in our MllPTD/WTFlt3ITD/WT mouse model of CN-AML (12) and compared it with Egfl7 expression in WT murine controls. We found significant increases in Egfl7 mRNA and Egfl7 protein in the BM of MllPTD/WTFlt3ITD/WT leukemic blasts compared with WT NBM-MNCs (Fig. 2 D–F). To evaluate the expression of Egfl7 protein in AML further, we compared immunohistochemistry for Egfl7 in our MllPTD/WTFlt3ITD/WT AML and in WT murine BM. We found substantial increases in Egfl7 in samples of MllPTD/WTFlt3ITD/WT leukemic BM compared with normal controls (Fig. 2 G and H and Fig. S2). We also measured EGFL7 mRNA and EGFL7 protein expression in a panel of four human AML cell lines (EOL1, OCI-AML3, MV4-11, and Kasumi-1). We found Kasumi-1 cells to have the highest expression of EGFL7 mRNA and MV4-11 cells to have the highest expression of EGLF7 protein (Fig. 2 I and J).
Table S8.
Cytogenetic and molecular characteristics of the leukapheresis samples from AML patients that were profiled for EGFL7 expression and were used in functional studies
| Sample ID | Karyotype | FLT3-ITD | FLT3-TKD | NPM1 | CEBPA | BCR-ABL | PML-RARA |
| ptAML1 | 46,XX[20] | Negative | Negative | WT | WT | Negative | Negative |
| ptAML2 | 46,XX[20] | Negative | Negative | Mutated | WT | Negative | Negative |
| ptAML3 | 47,XY,+13[cp2]/48,sl,+4,del(4)(p12p14), del(4)(p14p16)[cp8]/46,XY[10] | Negative | Negative | Mutated | WT | Negative | Negative |
| ptAML4 | 46,XY[28] | Negative | Negative | WT | WT | Negative | Negative |
| ptAML5 | N/A | Negative | Negative | WT | WT | Negative | Negative |
| ptAML6 | 46,XY,del(11)(p13p15)[7]/46,XY[13] | Negative | Negative | Mutated | WT | Negative | Negative |
| ptAML7 | 42–46,XY,add(2)(p13),del(5)(q13q31), del(9)(q13q22),der(11)t(11;11)(p15;q13), add(11)(q25),dup(11)(q14q23),del(12)(p13), -13,ins(14;?)(q24;?),-17,add(19)(p13.3), add(21)(p11.2)[cp8]/41–46,sl,+mar[cp12] .ish del(5)(EGR1),der(11)t(11;11),add(11)(MLL++),dup(11)(MLL++) | Negative | Negative | WT | WT | Negative | Negative |
| ptAML8 | 46,XX[20] | Negative | Negative | Mutated | WT | Negative | Negative |
| ptAML9 | 46,XX[20] | Negative | Negative | WT | WT | Negative | Negative |
| ptAML10 | 46,XX,inv(16)(p13q22)[20] | Negative | Negative | WT | WT | Negative | Negative |
| ptAML11 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
Fig. 2.
EGFL7 is up-regulated in human and mouse AML cells. (A) NBM samples from healthy donors (n = 3) were compared with leukapheresis samples of AML patients (ptAML, n = 11). EGFL7 levels were measured in AML samples by real-time RT-PCR, and the results were normalized to β-ACTIN RNA levels. (B) Mean ± SD of EGFL7 mRNA expression levels between NBM and AML in aggregate. *P < 0.05. (C) EGFL7 protein levels in human NBM and leukapheresis samples of AML patients were determined by immunoblotting with GAPDH as loading control. (D) Normal BM from WT mice (n = 4) was compared with murine AML blasts of the MllPTD/WTFlt3ITD/WT mouse model (mAML, n = 4) for the detection of mouse Egfl7 mRNA by RT-PCR with β-Actin as internal control. (E) Mean ± SD of Egfl7 mRNA between murine NBM and murine AML blasts, in aggregate. **P < 0.01. (F) Mouse Egfl7 protein levels in WT murine controls (n = 4) and murine AML blasts from the MllPTD/WTFlt3ITD/WT mouse model (mAML) (n = 4) were assessed by immunoblotting using Gapdh as loading control. (G) Immunohistochemistry of Egfl7 in NBM of WT (n = 3) control mice vs. BM from MllPTD/WTFlt3ITD/WT leukemic mice (n = 3) using an Egfl7-specific antibody or no antibody controls. (Original magnification: 100×; Insets with same areas across the samples are magnified at same strength.) (H) Percent of Egfl7+ cells in NBM of WT mice vs. BM from MllPTD/WTFlt3ITD/WT leukemic mice. ****P < 0.0001. (I and J) EGFL7 mRNA (I) and EGFL7 protein (J) expression levels in four human AML cell lines (EOL1, OCI-AML3, MV4-11, and Kasumi-1). The relative expression of EGFL7 mRNA was measured with real-time RT-PCR normalized to β-ACTIN. For immunoblotting, β-Tubulin was used as loading control.
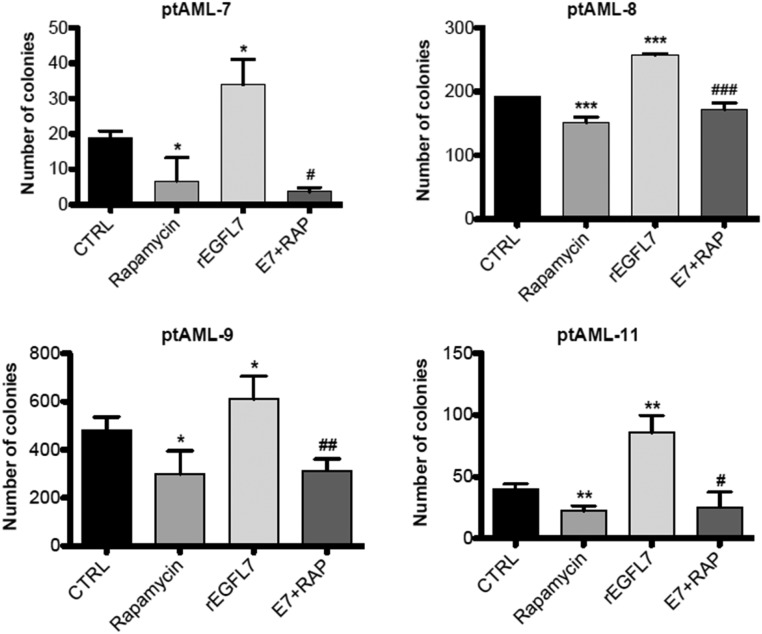
Fig. S2.
Primary blasts of AML patients (n = 4) were plated in triplicate in methylcellulose medium supplemented with rapamycin (E7+RAP; 10 nM), rhEGFL7 (250 nM) alone, or in combination. Colonies with more than 50 cells were enumerated after 10 d of culture, *P < 0.05, **P < 0.01, and ***P < 0.001 vs. CTRL; #P < 0.05, ##P < 0.01, ###P < 0.001, rEGFL7 vs. E7+RAP. There was no statistical difference between rapamycin and E7+RAP in all tested patient samples. CTRL, control; E7, rEGFL7; RAP, rapamycin.
EGFL7 Expression and Secretion by AML Blasts.
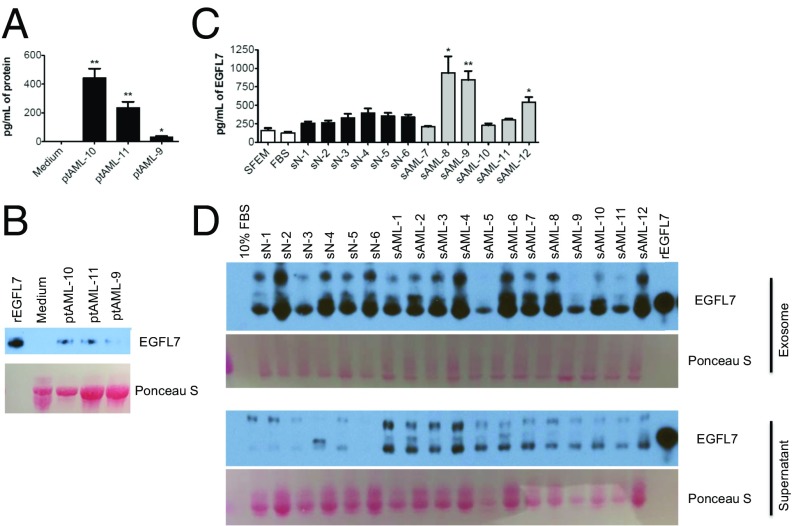
EGFL7 is normally expressed and secreted by endothelial cells to promote angiogenesis. We therefore sought to determine if AML blasts also acquired the ability to synthesize and secrete EGFL7 protein. Blasts from three patients with AML were cultured for 24 h in medium with 10% FBS and cytokines. EGFL7 levels were measured in the medium using an ELISA. As shown in Fig. 3A, we found significantly increased levels of EGFL7 protein in the medium from the AML blasts compared with the medium from wells without blasts. These results were confirmed using Western blotting (Fig. 3B). Next, we measured EGFL7 protein in the serum from healthy donors (n = 6) and AML patients (n = 6) using an ELISA. Significant increases in the level of EGFL7 were found in three of the six samples from the AML patients compared with normal controls (Fig. 3C). In our effort to validate these results using whole serum and Western blotting, we found that the serum was highly saturated with protein and interfered with resolution of the blot. When we separated the serum into the exosome-containing and supernatant fractions, we found variable high levels of EGFL7 in the exosomes in both normal and AML serum; however, there was substantially more “free” nonexosomal EGFL7 in the supernatant fraction from the AML patients than in the fraction from normal controls (Fig. 3D).
Fig. 3.
EGFL7 is a secreted protein and is increased in the serum of some AML patients. (A) Blasts of AML patients (ptAML, n = 3) were cultured in SFEM medium + 10% FBS supplemented with cytokines for 24 h. The EGFL7 protein level in the cell-culture supernatant was detected by ELISA and was compared with that in medium from wells without cultured cells. **P < 0.01, *P < 0.05. (B) Blasts of AML patients (n = 3) were cultured in SFEM medium + 10% FBS supplemented with cytokines for 24 h. The EGFL7 protein in the cell-culture supernatant was assessed by immunoblotting with rEGFL7 as a positive control and medium from wells without cultured cells as a negative control. Ponceau S staining shows the loading control for protein. (C) EGFL7 protein levels in sera from normal healthy controls (sN, n = 6) and AML patients (sAML, n = 6) were determined by the EGFL7 ELISA kit. SFEM medium alone and 10% FBS serve as blank controls. *P < 0.05, **P < 0.01. (D) An equal volume of serum from AML patients (sAML, n = 12) or normal healthy donors (sN, n = 6) was subjected to the separation of exosomal vs. nonexosomal eluant using the ExoQuick kit (System Biosciences). EGFL7 protein levels in both isolated exosomes and the supernatant were determined by immunoblotting. SFEM containing 10% FBS served as a negative control, and rEGFL7 was used as a positive control. Ponceau S staining shows the loading of proteins.
EGFL7 Stimulation Leads to Enhanced AML Blast Cell Growth.
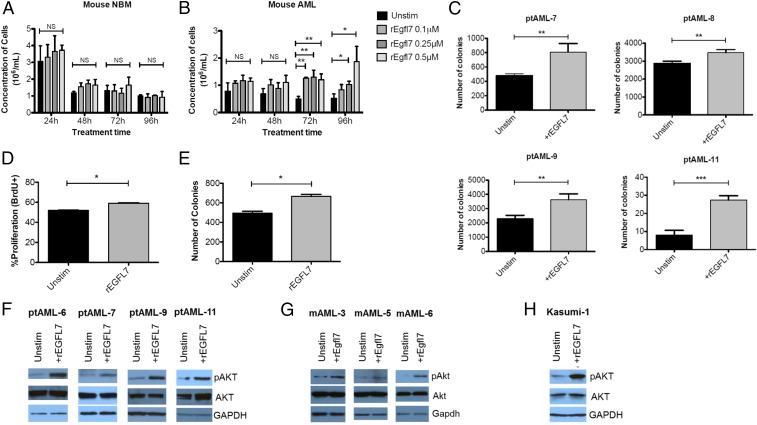
To compare the effect of Egfl7 protein on murine NBM-MNCs with its effect on murine AML blasts, we cultured blasts in minimal medium [Serum-Free Expansion Medium (SFEM; STEMCELL Technologies) + 2% BSA] and then stimulated them with recombinant Egfl7 (rEgfl7). Although the normal mouse BM did not expand at any time point (Fig. 4A), we found significant expansion of the murine AML blasts cells at 72 and 96 h poststimulation with rEgfl7 (Fig. 4B). We further validated the stimulatory effect of EGFL7 on growth of patient human AML blasts using CFU assays. We found an increase in the number of CFUs when rEGFL7 was added to the methylcellulose (Fig. 4C). In addition, we treated the Kasumi-1 AML cell line with rEGFL7, which we found, in concordance with previous reports (13), to express high levels of EGFL7 mRNA. rEGFL7 treatment of these cells led to an increase in the fraction of proliferating cells, as measured by BrdU incorporation (percent of BrdU+ cells in control vs. rEGFL7 treatment: 52 vs. 60, P = 0.02) (Fig. 4D) and by the number of colonies formed in CFU assays (Fig. S2C). In agreement with the increased cell growth in response to EGFL7, we found consistent increases in phosphorylated AKT (pAKT) in human (Fig. 4E) and mouse (Fig. 4F) AML cells, as well as in Kasumi-1 cells (Fig. 4G). These findings are in agreement with previous reports of increased pAKT levels in response to EGFL7 stimulation (14, 15).
Fig. 4.
EGFL7 stimulates the proliferation of human and mouse AML cells. (A and B) BM cells from WT (A) and MllPTD/WTFlt3ITD/WT (B) mice were treated without (Unstim) or with 0.1, 0.25, or 0.5 μM rEgfl7 in Iscove’s Modified Dulbecco’s Medium (IMDM) + 2% BSA for 24, 48, 72, or 96 h. At the indicated time points, the number of viable cells was determined by Trypan blue dye exclusion assay. Each condition was repeated in triplicate. *P < 0.05, **P < 0.01; NS, not significant. (C) Blasts of the indicated AML patients (20,000 cells) were mixed with methylcellulose medium in the absence or presence of 0.25 μM rEGFL7 and were plated onto 2-cm dishes for 10 d. Colonies with more than 50 cells were enumerated using a light microscope. Each condition for each patient (n = 4) was plated in triplicate; **P < 0.01, ***P < 0.001. (D) Kasumi-1 cells were stimulated with 100 nM rEGFL7 for 4 h in RPMI1640 with 10% FBS. Cell proliferation was assessed using APC-BrdU/7AAD staining coupled with flow cytometry; *P < 0.05. (E) Kasumi-1 cells (2,500 cells) were mixed with methylcellulose medium in the absence or presence of 100 nM recombinant human EGFL7 and were scored after 10 d. Each condition was plated in triplicate in three independent experiments. *P < 0.05. (F) Blasts from AML patients (n = 4) were cultured in SFEM + 2% BSA in the absence or presence of 0.25 μM rEGFL7 for 20 min. Total proteins were extracted for immunoblotting of pAKT-S473 and total AKT. GAPDH was used as loading control. (G) AML blasts from MllPTD/WTFlt3ITD/WT mice (n = 3) were cultured in IMDM medium + 2% BSA in the absence or presence of 0.25 μM rEgfl7 for 20 min. Total proteins were extracted for immunoblotting of pAkt-S473 and total Akt. Gapdh was used as loading control. (H) Exponentially growing Kasumi-1 cells were starved in serum-free RPMI1640 medium for 1 h, followed by the addition of 100 nM recombinant EGFL7 for 5 min. Total proteins were extracted for immunoblotting of pAKT-S473, total AKT, and GAPDH.
To validate the role of the pAKT pathway in EGFL7-induced blast growth further, we tested whether inhibition of downstream targets of pAKT, such as the mTOR pathway, would abrogate the increased blast cell growth phenotype of rEGFL7. We found that although treatment with rEGFL7 consistently increased the number of AML primary blast colonies, this effect was abolished when the blasts were pretreated with the mTOR inhibitor rapamycin (Fig. S2), suggesting a role for AKT signaling in EGFL7-stimulated blast growth.
Differential Effect of EGFL7 Silencing on AML Blasts and Normal Hematopoietic Cells.
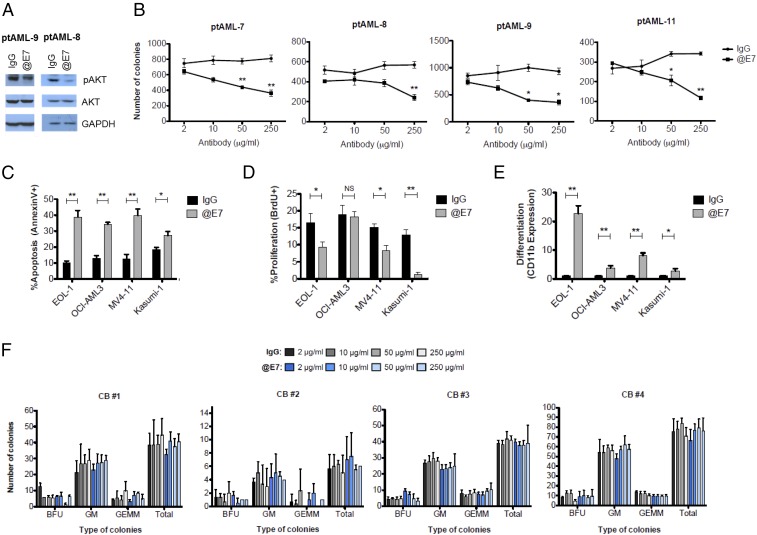
To evaluate the effects and therapeutic potential of inhibiting EGFL7 in AML, we performed experiments with a commercially available antibody that binds to the EGFL7 protein and inhibits its downstream effects (16). Immunoblotting performed after treatment of patient AML blasts with the anti-EGFL7 antibody showed a decrease in the levels of pAKT (Fig. 5A). More importantly, treatment of human AML blasts with increasing concentrations of the anti-EGFL7 blocking antibody led to significant decreases in cell growth, assessed by the numbers of colonies in CFU assays, compared with normal IgG controls (Fig. 5B). In human AML cell lines (EOL-1, OCI-AML3, MV4-11, and Kasumi-1), EGFL7 inhibition led to significant decreases in the viability of leukemic cells in all cell lines (Fig. 5C), along with concomitant decreases in the fraction of proliferating cells in three of the four cell lines tested (i.e., EOL-1, MV4-11, and Kasumi-1) (Fig. 5D). We also evaluated the effect of anti-EGFL7 treatment on differentiation of AML cells by measuring CD11b expression. CD11b is a surface marker reported to be up-regulated upon differentiation of leukemic cell lines into macrophage-like cells (17, 18). We found that EGFL7 inhibition led to significant increases in CD11b expression in all four cell lines that were treated with the anti-EGFL7 antibody compared with treatment with human IgG control (Fig. 5E).
Fig. 5.
EGFL7 inhibition results in decreased human AML cell growth without affecting normal hematopoietic cells. (A) Blasts from AML patients were cultured in SFEM with 10% FBS in the presence of 50 μg/mL of normal human IgG or anti-EGFL7 (@E7) antibody for 1 h. Total proteins were extracted for immunoblotting of pAKT-S473 and total AKT. GAPDH was used as loading control. (B) Human primary blasts (400,000) from AML patients (n = 4) were treated with 2, 10, 50, or 250 μg/mL of IgG control or anti-EGFL7 antibody in SFEM containing 10% FBS and cytokines for 2 h. Twenty thousand cells were plated in triplicate in methylcellulose medium and scored after 14 d of culture for mean ± SD colony numbers. *P < 0.05, **P < 0.01. (C) Forty-eight hours after IgG control vs. anti-EGFL7 antibody treatment (50 μg/mL) of AML cell lines, apoptosis was evaluated by Annexin V/7AAD staining, (D and E) Cell proliferation was measured using BrdU incorporation (D), and differentiation analysis was evaluated by CD11b expression (E). CD11b is depicted as the ratio of the CD11b expression value of the examined sample to the CD11b expression value of the corresponding IgG-treated control. *P < 0.05, **P < 0.01. (F) CD34+ CB cells from four different donors were plated in methylcellulose medium in the presence of increasing concentrations (2, 10, 50, or 250 μg/mL) of human IgG or anti-EGFL7 and were scored after 14 d of culture. Along with total number of colonies, colony types [erythroid burst-forming units (BFU), granulocyte/monocyte (GM), or granulocyte/erythrocyte/monocyte/megakaryocyte colonies (GEMM)] were enumerated also.
We hypothesized that dependency on EGFL7 signaling could be a feature that distinguishes leukemic blasts from healthy hematopoietic cells. To test this hypothesis, we treated CD34-selected umbilical cord blood cells (CD34+ CBCs) from four separate donors with a range of anti-EGFL7 antibody concentrations and performed CFU assays. In contrast to the effect of anti-EGFL7 treatment on leukemic blasts, EGFL7 inhibition did not impede the proliferation/survival of colonies (measured by number of CFUs) or alter myeloid differentiation (measured by CFU types) of normal hematopoietic cells (Fig. 5F).
Discussion
EGFL7 is a secreted protein with a well-characterized role in the physiology of angiogenesis and the pathology of certain solid tumors (6–9). We and others have previously reported on the significance of miR-126 (2, 4, 5), which is located in intron 7 of the EGFL7 gene, in AML, but an independent role of EGFL7 in this disease has not been described to date. Here we analyzed a set of younger adults and a set of older patients with CN-AML to evaluate the prognostic and biologic significance of EGFL7 expression. We found that high EGFL7 expression associates with worse outcome in both studied cohorts. We also found that in the older patients the combination of high EGFL7 promoter methylation and low EGFL7 expression identified a subset of patients with favorable prognosis, independently of other prognostic covariables. In our effort to evaluate the functional role of EGFL7, we screened blasts from a number of AML patients and AML cell lines. In these samples, we found some discordance between the mRNA and the protein levels of EGFL7, indicating that additional, posttranscriptional or posttranslational mechanisms could be involved in the regulation of EGFL7.
Although it seems reasonable that the expression levels of miR-126 and EGFL7 are regulated by the same mechanisms, including methylation, because they stem from the same transcript, mechanistic studies indicate that these genes have different effector functions. EGFL7 is a protein that is secreted by the AML blasts and is capable of directly inducing enhanced cell growth. Treatment of patient AML samples as well as cell lines with recombinant EGFL7 protein led to markedly increased levels of pAKT, a key regulator of cell proliferation. Concordantly, EGFL7 treatment increased the proliferating fraction as well as the number of colonies formed by AML cells. In contrast to this result, we observed no effects on cell proliferation when miR-126 expression was modulated in AML bulk blasts. Instead, we found miR-126 to be essential for LSC homeostasis (2). Whether EGFL7 also has a role in LSCs, dependent on or independent from miR-126, has not yet been determined.
Understanding the individual as well as combinatorial roles of EGFL7 and miR-126 in leukemogenesis could contribute significantly to more efficient therapeutic approaches in patients with aberrant activation of the EGFL7 locus. Our data indicate that EGFL7 could represent a therapeutic target in AML. We found that treatment with an EGFL7-blocking antibody reduced pAKT levels and decreased AML blast growth. In addition, EGFL7 inhibition increased apoptosis, decreased proliferation, and induced differentiation in the majority of the AML cell lines that were tested, independently of their endogenous EGFL7 expression levels. In striking contrast, blocking EGFL7 did not affect growth or differentiation of normal CD34+ umbilical CB cells. Thus, EGFL7-blocking antibodies may have a therapeutic effect, in particular in AML patients with increased EGFL7 expression, while preserving normal BM populations. In patients with concomitant aberrant overexpression of miR-126, EGFL7 blockade could be combined with therapeutic interventions to down-regulate miR-126, to target the LSC compartment additionally. We have previously shown the feasibility of therapeutic manipulation of miR-126 with nanoparticle-conjugated oligonucleotides (NP-antagomiR-126) in a preclinical model (2). In this sense, combining EGFL7-inhibition with NP-antagomiR-126 therapies may improve the treatment of AML patients, because the blocking the growth-promoting functions of EGFL7 on bulk blasts would be combined with the targeting of the therapy-resistant LSCs by the NP-antagomiR-126.
In conclusion, our results demonstrate the clinical and biological relevance of EGFL7 expression in AML. We found that expression levels of EGFL7 are prognostic in CN-AML patients and that patient AML blasts are able to secrete EGFL7 protein and promote the leukemic cell growth in an autocrine fashion. Furthermore, our data indicate that targeting EGFL7 with a monoclonal blocking antibody could provide a therapeutic option for the treatment of AML.
Patients and Methods
All patients provided written informed consent, and all study protocols were in accordance with the Declaration of Helsinki and approved by the institutional review boards at each center. For details concerning the treatment protocols and the molecular profiling of the analyzed AML patients, as well as for details concerning the experimental procedures, see SI Patients and Methods.
SI Patients and Methods
Participating Institutions.
The following Cancer and Leukemia Group B (CALGB)/Alliance for Clinical Trials in Oncology (Alliance) institutions participated in this study and contributed at least three patients. For each of these institutions, the current or last principal investigator is listed as follows: Wake Forest University School of Medicine, Winston-Salem, NC: Heidi Klepin; The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield; North Shore University Hospital, Manhasset, NY: Daniel R. Budman; University of Iowa Hospitals, Iowa City, IA: Daniel A. Vaena; Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine; Washington University School of Medicine, St. Louis: Nancy L. Bartlett; Dana Farber Cancer Institute, Boston: Harold J. Burstein; University of Chicago Medical Center, Chicago: Hedy L. Kindler; Duke University Medical Center, Durham, NC: Jeffrey Crawford; Ft. Wayne Medical Oncology/Hematology, Ft. Wayne, IN: Sreenivasa Nattam; University of North Carolina, Chapel Hill, NC: Thomas C. Shea; University of Maryland Cancer Center, Baltimore: Martin J. Edelman; Dartmouth Medical School, Lebanon, NH: Konstantin Dragnev; University of Vermont Cancer Center, Burlington, VT: Claire Verschraegen; Christiana Care Health Services, Inc., Newark, DE: Gregory Masters; Eastern Maine Medical Center, Bangor, ME: Thomas H. Openshaw; Mount Sinai School of Medicine, New York: Lewis R. Silverman; Weill Medical College of Cornell University, New York: Scott Tagawa; SUNY Upstate Medical University, Syracuse, NY: Stephen L. Graziano; University of Tennessee Cancer Center, Memphis, TN: Harvey B. Niell; Western Pennsylvania Hospital, Pittsburgh: John Lister; University of Massachusetts Medical Center, Worcester, MA: William V. Walsh; University of Puerto Rico, San Juan, Puerto Rico: Eileen I. Pacheco; Rhode Island Hospital, Providence, RI: Howard Safran; Long Island Jewish Medical Center, Lake Success, NY: Daniel R. Budman; Moores University of California San Diego Cancer Center, San Diego: Barbara A. Parker; University of California at San Francisco, San Francisco: Charalambos Andreadis; Walter Reed National Military Medical Center, Bethesda: Mary Kwok; Massachusetts General Hospital, Boston: David Ryan; University of Illinois, Chicago: Arkadiusz Z. Dudek; University of Alabama at Birmingham, Birmingham, AL: Robert Diasio; University of Minnesota, Minneapolis: Bruce A. Peterson; University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Clint Kingsley; Virginia Commonwealth University, Richmond, VA: Steven Grossman; and University of Nebraska Medical Center, Omaha, NE: Apar Ganti.
Treatment Protocols.
All patients included in our study were treated on CALGB/Alliance first-line protocols for patients with AML and received cytarabine/daunorubicin-based induction therapy (19). Per protocol, all patients were to receive at least one induction cycle. For patients with residual leukemia present in a BM biopsy after one induction cycle, a second cycle of induction was administered. None of the protocols included allogeneic stem cell transplantation (SCT) in first CR. Patients enrolled on the treatment protocols also provided written informed consent to their participation in the companion protocols CALGB 20202 (molecular studies in AML), CALGB 8461 (prospective cytogenetic companion), and CALGB 9665 (leukemia tissue bank), which involved collection of pretreatment BM aspirates and blood samples.
Younger (<60 y) adult patients were enrolled in the following treatment protocols: CALGB 19808, CALGB 10503, CALGB 9621, CALGB 10603, CALGB 9222, CALGB 8525, CALGB 9022, CALGB 8721, CALGB 8821, and CALGB 9120. Patients enrolled in CALGB 19808 (n = 113) were randomly assigned to receive induction chemotherapy with cytarabine/daunorubicin, and etoposide with or without PSC-833 (valspodar), a multidrug resistance protein inhibitor (19). Upon attainment of CR, patients were assigned to intensification with high-dose cytarabine and etoposide for stem-cell mobilization followed by myeloablative treatment with busulfan and etoposide supported by autologous peripheral blood SCT. Patients included in CALGB 10503 (n = 112) received cytarabine/daunorubicin-based induction chemotherapy, and those who achieved CR received a further two-step consolidation with chemo-mobilization and autologous SCT, if eligible, or high-dose cytarabine-based consolidation if not. Maintenance with decitabine began as soon as possible after recovery from consolidation (20). Patients enrolled in CALGB 9621 (n = 60) were treated similarly to those enrolled in CALGB 19808, as previously reported (21). Patients in CALGB 10603 (n = 40) were stratified by FLT3 mutation subtype [FLT3-TKD vs. FLT3-ITD-high allelic ratio (>0.70) vs. low allelic ratio (0.05–0.70)] and were randomized to receive cytarabine/daunorubicin-based induction chemotherapy and high-dose cytarabine consolidation in combination with either the multikinase inhibitor midostaurin or placebo. One-year midostaurin or placebo maintenance was administered after the last cycle of consolidation therapy (22). Patients enrolled in CALGB 9222 (n = 27) received cytarabine/daunorubicin-based induction chemotherapy, and those who achieved CR received either three cycles of high-dose cytarabine or three cycles of a non–cross-resistant regimen (the first cycle of this regimen was high-dose cytarabine, the second was cyclophosphamide plus etoposide, and the third was mitoxantrone plus diaziquone) (23). Patients enrolled in CALGB 8525 (n = 17) who achieved CR after cytarabine/daunorubicin-based induction chemotherapy were randomly assigned to consolidation with different doses of cytarabine followed by maintenance treatment (24). Patients who participated in CALGB 9022 (n = 2) and achieved CR after cytarabine/daunorubicin-based induction chemotherapy received one course of high-dose cytarabine consolidation, followed by one course of cyclophosphamide and etoposide, followed by one course of mitoxantrone and diaziquone (25).
Older patients (age ≥60 y) were enrolled in CALGB/Alliance protocols 9720, 10201, 8525, 8923, or 9420. Patients in CALGB 9720 (n = 95) (26) and 9420 (n = 5) (27) received induction chemotherapy consisting of cytarabine in combination with daunorubicin and etoposide, with (in CALGB 9420) or with/without (in CALGB 9720) the multidrug resistance protein modulator PSC-833. The PSC-833 arm of CALGB 9720 was closed after random assignment of 120 patients because of excessive early deaths, and enrollment continued in the chemotherapy-only control arm. After a single consolidation dose, patients in CALGB 9720 were randomly assigned to low-dose recombinant interleukin-2 maintenance therapy or none (28). Interleukin-2 maintenance was not associated with a difference in patient outcome. Patients in CALGB 10201 (n = 65) received induction chemotherapy consisting of cytarabine and daunorubicin, with or without the BCL2 antisense oblimersen sodium. Preliminary results showed no impact of the antisense therapy on outcome (29). Patients in CALGB 8525 (n = 18) were treated with induction chemotherapy consisting of cytarabine in combination with daunorubicin and were randomly assigned to consolidation with different doses of cytarabine followed by maintenance treatment (24). Patients in CALGB 8923 (n = 15) received induction chemotherapy consisting of cytarabine and daunorubicin and were randomly assigned to receive postremission therapy with cytarabine alone or in combination with mitoxantrone (30).
Patient Set.
For survival analyses, pretreatment BM or blood samples were obtained from 572 adult patients (374 younger patients, age 17–59 y, and 198 older patients, age 60–83 y) with de novo CN-AML, who received intensive cytarabine/anthracycline-based first-line therapy on CALGB/Alliance trials, and were alive 30 d after initiation of treatment. Per protocol, no patient received allogeneic SCT in first CR. All patients provided written informed consent, and all study protocols were in accordance with the Declaration of Helsinki and approved by the institutional review boards at each center.
Cytogenetic and Molecular Analyses.
Cytogenetic analyses were performed in CALGB/Alliance-approved institutional laboratories, and results were confirmed by central karyotype review (31). The diagnosis of a normal karyotype was based on ≥20 metaphase cells analyzed in BM specimens subjected to short-term (24- or 48-h) unstimulated cultures.
In the group of younger (n = 374) and in a subset of older (n = 154) CN-AML patients, targeted amplicon sequencing using the MiSeq platform (Illumina) was applied to analyze DNA samples for the presence of gene mutations that are established prognosticators in CN-AML [i.e., mutations in the ASXL1, DNMT3A (R882 and non-R882), IDH1, IDH2 (R140 and R172), NPM1, RUNX1, TET2, or WT1 genes and FLT3-TKD mutations], as described previously (32, 33). A variant allele frequency of ≥10% was used as the cut point to distinguish between mutated vs. WT alleles of these genes. The presence of mutations in the CEBPA gene and FLT3-ITD were evaluated using Sanger sequencing (34) and fragment analysis (35), as described previously. For the remaining older CN-AML patients (n = 44), mutations in all the aforementioned genes were evaluated with Sanger sequencing or fragment analysis, as described previously (34–43). Because only double CEBPA mutations are favorable prognostic markers in CN-AML (44, 45), we considered this genotype as mutated and grouped patients with single-mutated CEBPA and those with WT CEBPA together.
RNA Extraction and RNA Expression Quantification.
RNA, cDNA, and real-time PCR were performed using previously published methods (2). All primers/probes were purchased from Applied Biosystems.
Transcriptome Analysis.
In the cohort of younger adults with CN-AML (n = 374) transcriptome analysis was performed with total RNA sequencing. In brief, extracted total RNA was assessed for quality on an Agilent 2100 Bioanalyzer (BioA) using the RNA 6000 Nano chip and for quantity on a Qubit 2.0 Fluorometer (Agilent Technologies) using the RNA HS Assay Kit. Samples with a RNA integrity number (RIN) greater than four, with no visible sign of genomic DNA (gDNA) contamination and a concentration of >40 ng/μL were used for total RNA library generation. RNA-seq libraries were prepared using the Illumina TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero Gold (no. RS1222201) according to the manufacturer's instructions. Sequencing was performed with the Illumina HiSeq. 2500 system using the HiSeq version 3 sequencing reagents to an approximate cluster density of 800,000/mm2. Image analysis, base calling, error estimation, and quality thresholds were performed using HiSeq Controller Software (version 2.2.38) and the Real Time Analyzer (RTA) software (version 1.18.64).
For miRNA profiling a subset of patients with RNA samples of sufficient quality (n = 300) was analyzed with small RNA sequencing (smRNA-seq). smRNA-seq libraries were generated using the NEBNext Multiplex Small RNA Library Prep Set (catalog no. E7300L; New England Biolabs, Inc.). Library-generation steps were performed as described by the manufacturer. The input RNA criteria for smRNA-seq were a Qubit RNA concentration of >50 ng/μL and a BioA RNA RIN value >7. Generation of barcodes and enrichment of fragments with three and five adaptors for smRNA libraries were accomplished by 12 cycles of PCR amplification. Before pooling smRNA-seq libraries for enriching smRNA species, libraries generated from each sample were assessed for the relative amount of smRNA fragments migrating between 140–160 bp using the Agilent Bioanalyzer HS DNA assay. Size selection/enrichment for smRNAs was accomplished using the Sage Science Pippin Prep with 3% precast agarose gel. The profile of the resultant size-selected libraries was ascertained using the Agilent Bioanalyzer HS DNA assay. Each pool of the smRNA-seq libraries was sequenced with other samples with compatible barcodes on an Illumina HiSeq. 0250 V3 single-read 50-bp lane to achieve 5–8 million passed filter reads per sample.
Cutadapt and FastQ were used to apply quality control and adapter trimming to FastQ files. Spliced Transcripts Alignment to a Reference (STAR) software (46) was used to align the short reads to the human genome (GENECODE ver22) (47), and the HTSeq script (48) was used to quantify and annotate long noncoding RNAs (lncRNAs). Raw data were transformed into reads per million (RPM) before statistical analysis. To minimize noise, mRNAs were evaluated in each sample only when at least nine reads were present in a total of 40 million reads.
In the cohort of older CN-AML patients (n = 198) RNA samples studied were analyzed for genome-wide gene expression using Affymetrix U133 plus 2.0 GeneChip (Affymetrix). Double-stranded cDNA was prepared (Invitrogen) from total RNA using T7-Oligo(dT) primer (Affymetrix). In vitro transcription was performed with the BIOARRAY HIGHYIELD RNA Transcript Labeling Kit (T7) (Enzo Life Science). Fragmented, biotinylated RNA samples were hybridized to the U133 plus 2.0 GeneChip for 16 h at 45 °C.
For the gene-expression analysis, summary measures of the expression levels were computed for each probe set using the robust multichip average method, which incorporates quantile normalization of arrays. Expression values were logged (base 2) before analysis. A filtering step was performed to remove probe sets that did not display significant variation in expression across arrays. In this procedure, a χ2 test was used to determine whether the observed variance in the expression of a probe set was significantly larger than the median observed variance in the expression for all probe sets using α = 0.01 as the significance level.
miRNA profiling in the cohort of older CN-AML patients was performed with the Ohio State University (OSU) Comprehensive Cancer Center custom microarrays and analyzed as previously reported (49).
Definition of Clinical Endpoints.
Clinical endpoints were defined according to generally accepted criteria (50). CR required a BM aspirate with cellularity >20% with maturation of all cell lines, <5% blasts, and undetectable Auer rods; in peripheral blood, an absolute neutrophil count of ≥1.5 × 109/L, platelet count of >100 × 109/L, and leukemic blasts absent; and no evidence of extramedullary leukemia, all of which had to persist for ≥4 wk. Relapse was defined by the presence of ≥5% BM blasts, or circulating leukemic blasts, or the development of extramedullary leukemia. DFS was measured from the date of CR until the date of relapse or death (from any cause); patients alive and in continuous first CR were censored at last follow-up. OS was measured from the date of study entry until the date of death (from any cause); patients alive at last follow-up were censored. EFS was measured from the date of study entry until the date of failure to achieve CR, relapse, or death. Patients alive and in CR at last follow-up were censored.
Statistical Analyses for Survival End-Points.
Baseline demographic, clinical, and molecular features were compared between patients with low and high EFGL7 expression using the Wilcoxon rank sum and Fisher’s exact tests for continuous and categorical variables, respectively (51). The estimated probabilities of DFS, OS, and EFS were calculated using the Kaplan–Meier method, and the log-rank test evaluated differences between survival distributions (52). Cox proportional hazard models were used to calculate HRs for DFS, OS, and EFS (53). For the time-to-event endpoints, the proportional hazards assumption was checked for each variable individually. All statistical analyses were performed by the Alliance Statistics and Data Center on a database locked on April 4, 2016 using SAS 9.4 and TIBCO Spotfire S+ 8.2.
Multivariable proportional hazards models were constructed for DFS and OS using a limited forward elimination procedure. Because of the relatively small number of patients in the EGFL7 favorable risk group, a final model could not be constructed for EFS. However, three separate three-variable models could be generated. Variables considered for model inclusion were lncRNA score status (favorable vs. unfavorable), age (as a continuous variable, in 10-y increments), sex (male vs. female), race (white vs. nonwhite), WBC count (as a continuous variable, in 50-unit increments), hemoglobin (as a continuous variable, in one-unit increments), platelet count (as a continuous variable, in 50-unit increments), extramedullary involvement (present vs. absent), ASXL1 mutations (mutated vs. WT), CEBPA mutations (double-mutated vs. single-mutated or WT), DNMT3A mutations (mutated vs. WT), FLT3-ITD (present vs. absent), FLT3-TKD (present vs. absent), IDH1 mutations (mutated vs. WT), IDH2 mutations (mutated vs. WT), NPM1 mutations (mutated vs. WT), TET2 mutations (mutated vs. WT), RUNX1 mutations (mutated vs. WT), WT1 mutations (mutated vs. WT), ERG expression levels (high vs. low), BAALC expression levels (high vs. low), MN1 expression levels (high vs. low), miR-181a expression levels (high vs. low), miR-3151 (expressed vs. not expressed), and miR-155 expression levels (high vs. low). For ERG, BAALC, MN1, miR-181a, and miR-155, the median expression value was used as the cut point to divide patients into high and low expressers. Variables significant at α = 0.20 from the univariable analyses were considered for multivariable analyses. For the time-to-event endpoints, the proportional hazards assumption was checked for each variable individually.
Western Blot Analysis.
Equal numbers of bulk blast cells from AML patients and human NBM-MNCs (AllCells) or murine WT BM cells and MllPTD/WTFlt3ITD/WT leukemic cells were lysed in 100 μL of Pierce RIPA buffer (catalog no. 8990; Thermo Scientific) supplemented with protease and phosphatase inhibitors (Roche) on ice for 20 min. Protein lysates were separated by SDS/PAGE and transferred to PVDF membranes. Membranes were incubated with antibodies for EGFL7 (LS-C153302 and LS-C40134; LifeSpan BioSciences), goat anti-EGFL7 (sc-34116) and rabbit anti-GAPDH antibodies (Santa Cruz Biotechnology), and pAKT-Ser473 and AKT antibodies (Cell Signaling Technology). Proteins were visualized with HRP-conjugated sheep anti-mouse, goat anti-rat, donkey anti-rabbit, and donkey anti-goat secondary antibodies (GE Healthcare) and were detected using ECL Select Western Blotting Detection Reagent (GE Healthcare).
Immunohistochemistry.
Formalin-fixed humeri were washed with distilled water and placed in decalcifying EDTA pH 7.2–7.6 (catalog no. 2516; Poly Scientific R&D Corporation). Samples were placed on a rotating mixer at 37 °C until bones were soft and pliable (∼7 d). Decalcifying EDTA was changed every 48 h until decalcification was complete. Bones then were rinsed with 1× PBS three times for 5 min each, with distilled water three times for 5 min each, with 50% ethanol for 5 min, and with 70% ethanol for 5 min and then were stored in 70% ethanol. Bones were embedded in paraffin blocks and sectioned onto glass slides. Egfl7 immunohistochemistry was performed with goat polyclonal anti-mouse Egfl7 (R-12) antibody (catalog no. sc-34416; Santa Cruz Biotechnology) at 1:50 dilution using the avidin–biotin complex method. Tissue embedding, sectioning, and immunohistochemistry were performed by The OSU Comparative Pathology and Mouse Phenotyping Core. For quantification of Eglf7 BM cells, five separate areas of each slide were evaluated. Cells were counted if a distinct cell membrane and nucleus were seen in the same plane of focus. The evaluator was blinded with regard to the specimen genotype (WT vs. MllPTD/WTFlt3ITD/WT) being evaluated.
ELISA.
Serum of AML patients or normal donors (OSU Leukemia Tissue Bank and Discovery Life Sciences) was diluted in PBS in a 1:100 ratio. One hundred microliters of EGF7 protein standards (provided by the manufacturer) or diluted samples were added to replicate wells in a 96-well precoated plate, and the assay was performed according to the manufacturer’s protocol (Cloud-Clone Corp). Standard curves were generated from the kit standards and used to extrapolate EGFL7 concentrations in blood plasma/serum.
CD34 Selection of Umbilical Cord Blood Cells.
CD34 selection of umbilical cord blood cells was performed as previously published (54).
CFU Assay.
For the CFU assay, blasts of AML patients (20,000 cells) or CD34+ CBCs (3,000 cells) were mixed with MethoCult HM4434 (STEMCELL Technologies) and then were plated on 2-cm dishes for 10–14 d. Colonies with more than 50 cells were counted using a light microscope. Each condition was repeated in triplicate, n = 3 independent experiments.
Cell Proliferation, Apoptosis, and Differentiation.
For the evaluation of apoptosis, cells were stained with Annexin V and 7AAD as previously described (2, 54). For the evaluation of cell proliferation, AML cell lines were incubated with BrdU, fixed, permeabilized, and stained with APC-conjugated anti-BrdU antibody, according to the instructions of the manufacturer (BD Biosciences) and as previously described (2, 54). The proliferating fraction of the cells (i.e., the BrdU+ cells) was evaluated with flow cytometry (LSRII; BD Biosciences). For cell differentiation analysis, cells were stained with a human anti-CD11b antibody (BD Biosciences).
Anti-EGFL7 Antibody.
A commercially available humanized murine monoclonal antibody that binds to the EGFL7 protein and inhibits its function (parsatuzumab) was purchased from Creative Biolabs, Inc. Experiments were performed according to the manufacturer’s instructions. Human IgG was used as control.
Statistical Methods.
For laboratory in vitro experiments, two-tailed Student’s t tests were performed using GraphPad Prism version 5.0a (GraphPad Software). P values <0.05 were considered significant.
Acknowledgments
We thank Donna Bucci and Wacharaphon Vongchucherd of The Alliance National Cancer Institute-Clinical Trials Network Biorepository and Biospecimen Resource for sample processing and storage services, Dr. David M. Lucas and the Leukemia Tissue Bank of The Ohio State University for apheresis sample storage, Karl Kroll for technical support, and Lisa J. Sterling and Christine Finks of The Ohio State University Comprehensive Cancer Center for data management. This work was supported by National Institutes of Health National Cancer Institute Grants CA180821 and CA180882 (to the Alliance for Clinical Trials in Oncology), CA077658, CA180850, CA180861, CA196171, CA140158, CA16058, CA197734, CA135332, and CA102031, the Leukemia Clinical Research Foundation, and the D. Warren Brown Foundation. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703142114/-/DCSupplemental.
References
- 1.Short NJ, Ravandi F. Acute myeloid leukemia: Past, present, and prospects for the future. Clin Lymphoma Myeloma Leuk. 2016;16(Suppl):S25–S29. doi: 10.1016/j.clml.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Dorrance AM, et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia. 2015;29:2143–2153. doi: 10.1038/leu.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikolic I, Plate K-H, Schmidt MHH. EGFL7 meets miRNA-126: An angiogenesis alliance. J Angiogenes Res. 2010;2:9. doi: 10.1186/2040-2384-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Leeuw DC, et al. Attenuation of microRNA-126 expression that drives CD34+38- stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 2014;74:2094–2105. doi: 10.1158/0008-5472.CAN-13-1733. [DOI] [PubMed] [Google Scholar]
- 5.Lechman ER, et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29:602–606. doi: 10.1016/j.ccell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nichol D, Stuhlmann H. EGFL7: A unique angiogenic signaling factor in vascular development and disease. Blood. 2012;119:1345–1352. doi: 10.1182/blood-2011-10-322446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh J, et al. High expression of epidermal growth factor-like domain 7 is correlated with poor differentiation and poor prognosis in patients with epithelial ovarian cancer. J Gynecol Oncol. 2014;25:334–341. doi: 10.3802/jgo.2014.25.4.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bambino K, Lacko LA, Hajjar KA, Stuhlmann H. Epidermal growth factor-like domain 7 is a marker of the endothelial lineage and active angiogenesis. Genesis. 2014;52:657–670. doi: 10.1002/dvg.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan C, et al. The expression of Egfl7 in human normal tissues and epithelial tumors. Int J Biol Markers. 2013;28:71–83. doi: 10.5301/JBM.2013.10568. [DOI] [PubMed] [Google Scholar]
- 10.Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan P, et al. Genome-wide methylation profiling in decitabine-treated patients with acute myeloid leukemia. Blood. 2012;120:2466–2474. doi: 10.1182/blood-2012-05-429175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zorko NA, et al. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood. 2012;120:1130–1136. doi: 10.1182/blood-2012-03-415067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi K, et al. EGF-like-domain-7 is required for VEGF-induced Akt/ERK activation and vascular tube formation in an ex vivo angiogenesis assay. PLoS One. 2014;9:e91849. doi: 10.1371/journal.pone.0091849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolić I, et al. EGFL7 ligates αvβ3 integrin to enhance vessel formation. Blood. 2013;121:3041–3050. doi: 10.1182/blood-2011-11-394882. [DOI] [PubMed] [Google Scholar]
- 16.Johnson L, et al. Anti-EGFL7 antibodies enhance stress-induced endothelial cell death and anti-VEGF efficacy. J Clin Invest. 2013;123:3997–4009. doi: 10.1172/JCI67892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong CK, Ho CY, Lam CWK, Zhang JP, Hjelm NM. Differentiation of a human eosinophilic leukemic cell line, EoL-1: Characterization by the expression of cytokine receptors, adhesion molecules, CD95 and eosinophilic cationic protein (ECP) Immunol Lett. 1999;68:317–323. doi: 10.1016/s0165-2478(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 18.Asou H, et al. Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood. 1991;77:2031–2036. [PubMed] [Google Scholar]
- 19.Kolitz JE, et al. Cancer and Leukemia Group B P-glycoprotein inhibition using valspodar (PSC-833) does not improve outcomes for patients younger than age 60 years with newly diagnosed acute myeloid leukemia: Cancer and Leukemia Group B study 19808. Blood. 2010;116:1413–1421. doi: 10.1182/blood-2009-07-229492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blum W, et al. Alliance for Clinical Trials in Oncology Maintenance therapy with decitabine in younger adults with acute myeloid leukemia in first remission: A phase 2 Cancer and Leukemia Group B Study (CALGB 10503) Leukemia. 2017;31:34–39. doi: 10.1038/leu.2016.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolitz JE, et al. Cancer and Leukemia Group B Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: Final induction results of Cancer and Leukemia Group B Study 9621. J Clin Oncol. 2004;22:4290–4301. doi: 10.1200/JCO.2004.11.106. [DOI] [PubMed] [Google Scholar]
- 22.Stone R, et al. The multi-kinase inhibitor midostaurin (M) prolongs survival compared with placebo (P) in combination with daunorubicin (D)/cytarabine (C) induction (ind), high-dose C consolidation (consol), and as maintenance (maint) therapy in newly diagnosed acute myeloid leukemia (AML) patients (pts) age 18-60 with FLT3 mutations (muts): An international prospective randomized (rand) P-controlled double-blind trial (CALGB 10603/RATIFY [Alliance]) Blood. 2015;126:6. [Google Scholar]
- 23.Moore JO, et al. Sequential multiagent chemotherapy is not superior to high-dose cytarabine alone as postremission intensification therapy for acute myeloid leukemia in adults under 60 years of age: Cancer and Leukemia Group B Study 9222. Blood. 2005;105:3420–3427. doi: 10.1182/blood-2004-08-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer RJ, et al. Cancer and Leukemia Group B Intensive postremission chemotherapy in adults with acute myeloid leukemia. N Engl J Med. 1994;331:896–903. doi: 10.1056/NEJM199410063311402. [DOI] [PubMed] [Google Scholar]
- 25.Moore JO, et al. Granulocyte-colony stimulating factor (filgrastim) accelerates granulocyte recovery after intensive postremission chemotherapy for acute myeloid leukemia with aziridinyl benzoquinone and mitoxantrone: Cancer and Leukemia Group B study 9022. Blood. 1997;89:780–788. [PubMed] [Google Scholar]
- 26.Baer MR, et al. Phase 3 study of the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age and older with acute myeloid leukemia: Cancer and Leukemia Group B Study 9720. Blood. 2002;100:1224–1232. [PubMed] [Google Scholar]
- 27.Lee EJ, et al. Parallel phase I studies of daunorubicin given with cytarabine and etoposide with or without the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age or older with acute myeloid leukemia: Results of Cancer and Leukemia Group B study 9420. J Clin Oncol. 1999;17:2831–2839. doi: 10.1200/JCO.1999.17.9.2831. [DOI] [PubMed] [Google Scholar]
- 28.Baer MR, et al. Low-dose interleukin-2 immunotherapy does not improve outcome of patients age 60 years and older with acute myeloid leukemia in first complete remission: Cancer and Leukemia Group B Study 9720. J Clin Oncol. 2008;26:4934–4939. doi: 10.1200/JCO.2008.17.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcucci G, et al. A phase III randomized trial of intensive induction and consolidation chemotherapy ± oblimersen, a pro-apoptotic Bcl-2 antisense oligonucleotide in untreated acute myeloid leukemia patients >60 years old. J Clin Oncol. 2007;25:7012. [Google Scholar]
- 30.Stone RM, et al. Postremission therapy in older patients with de novo acute myeloid leukemia: A randomized trial comparing mitoxantrone and intermediate-dose cytarabine with standard-dose cytarabine. Blood. 2001;98:548–553. doi: 10.1182/blood.v98.3.548. [DOI] [PubMed] [Google Scholar]
- 31.Mrózek K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- 32.Garzon R, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci USA. 2014;111:18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroll KW, et al. MuCor: Mutation aggregation and correlation. Bioinformatics. 2016;32:1557–1558. doi: 10.1093/bioinformatics/btw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcucci G, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitman SP, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 36.Metzeler KH, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118:6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcucci G, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcucci G, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitman SP, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker H, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendler JH, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and microRNA expression signatures. J Clin Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzeler KH, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paschka P, et al. Wilms’ tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wouters BJ, et al. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113:3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dufour A, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol. 2010;28:570–577. doi: 10.1200/JCO.2008.21.6010. [DOI] [PubMed] [Google Scholar]
- 46.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcucci G, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 50.Cheson BD, et al. Report of the National Cancer Institute-sponsored workshop on definitions of diagnosis and response in acute myeloid leukemia. J Clin Oncol. 1990;8:813–819. doi: 10.1200/JCO.1990.8.5.813. [DOI] [PubMed] [Google Scholar]
- 51.Vittinghoff E, et al. Regression Methods in Biostatistics: Linear, Logistic, Survival and Repeated Measures Models. Springer; New York: 2005. Basic statistical methods; pp. 26–67. [Google Scholar]
- 52.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;282:457–481. [Google Scholar]
- 53.Vittinghoff E, et al. Regression Methods in Biostatistics: Linear, Logistic, Survival and Repeated Measures Models. Springer; New York: 2005. Logistic regression; pp. 139–202. [Google Scholar]
- 54.Dorrance AM, et al. The Rac GTPase effector p21-activated kinase is essential for hematopoietic stem/progenitor cell migration and engraftment. Blood. 2013;121:2474–2482. doi: 10.1182/blood-2012-10-460709. [DOI] [PMC free article] [PubMed] [Google Scholar]