Significance
Maternally inherited bacterial endosymbionts in arthropods manipulate host reproduction to increase the number of infected females. Cytoplasmic incompatibility (CI) is one such manipulation, in which infected females can produce offspring by mating with both infected and uninfected males, but uninfected females cannot or seldom produce offspring with infected males. Two bacterial endosymbionts, Wolbachia and Cardinium, are known CI inducers. Here we report a third CI inducer that belongs to a unique clade of Alphaproteobacteria. This bacterial clade was found to cause complete CI between two clades of the coconut beetle, a serious invasive pest of coconut palms. We discuss the potential use of this bacterium as a biological control agent and its effects on speciation of the coconut beetle.
Keywords: biological control, reproductive isolation, speciation, symbiont, Wolbachia
Abstract
Maternally inherited bacterial endosymbionts in arthropods manipulate host reproduction to increase the fitness of infected females. Cytoplasmic incompatibility (CI) is one such manipulation, in which uninfected females produce few or no offspring when they mate with infected males. To date, two bacterial endosymbionts, Wolbachia and Cardinium, have been reported as CI inducers. Only Wolbachia induces complete CI, which causes 100% offspring mortality in incompatible crosses. Here we report a third CI inducer that belongs to a unique clade of Alphaproteobacteria detected within the coconut beetle, Brontispa longissima. This beetle comprises two cryptic species, the Asian clade and the Pacific clade, which show incompatibility in hybrid crosses. Different bacterial endosymbionts, a unique clade of Alphaproteobacteria in the Pacific clade and Wolbachia in the Asian clade, induced bidirectional CI between hosts. The former induced complete CI (100% mortality), whereas the latter induced partial CI (70% mortality). Illumina MiSeq sequencing and denaturing gradient gel electrophoresis patterns showed that the predominant bacterium detected in the Pacific clade of B. longissima was this unique clade of Alphaproteobacteria alone, indicating that this endosymbiont was responsible for the complete CI. Sex distortion did not occur in any of the tested crosses. The 1,160 bp of 16S rRNA gene sequence obtained for this endosymbiont had only 89.3% identity with that of Wolbachia, indicating that it can be recognized as a distinct species. We discuss the potential use of this bacterium as a biological control agent.
Bacterial endosymbionts in arthropods influence host reproduction in various ways. Because bacterial endosymbionts are transmitted mainly through the female host, they manipulate host reproduction to increase the fitness of infected females. Such manipulation was first detected in the early 1970s in Culex pipiens infected with Wolbachia pipientis, a member of the Alphaproteobacteria class (1). W. pipientis induces cytoplasmic incompatibility (CI), in which uninfected females or those infected with a different strain than a male’s Wolbachia produce few or no offspring when they mate with infected males. Several other manipulation phenotypes have been identified in Wolbachia and other bacterial species, including parthenogenesis induction, in which infected females produce daughters without fertilization by males in a haplodiploid system (Wolbachia, Rickettsia sp., Cardinium hertigii); feminization of genetic males, in which genetic male embryos develop phenotypically as females (Wolbachia, C. hertigii); male killing, in which male embryos are killed during development (Wolbachia, Rickettsia sp., Flavobacterium sp., Spiroplasma ixodetis, Spiroplasma poulsonii); and oogenesis, in which uninfected females cannot produce mature oocytes (Wolbachia) (2). Although several bacterial species are known to manipulate host reproduction, only Wolbachia shows all of these phenotypes.
CI is the most common phenotype of Wolbachia. Although the molecular mechanisms are unknown, CI can be explained by the “modification-rescue” system. Wolbachia “modifies” sperm in the testes, and the sperm develops abnormally, resulting in death of the embryo when appropriate Wolbachia is not present in the eggs to “rescue” the embryo from the modification (3). The other known CI inducer, Cardinium (Bacteroidetes), induces only a low level of CI and never causes 100% mortality in incompatible crosses (4–7). Although CI has important effects on host speciation (3, 8), and CI-inducing microbes might be useful for biological control (9), research focusing on such issues has been limited to Wolbachia.
Brontispa longissima (Gestro) (Coleoptera: Chrysomelidae) is a serious pest of coconut palm (Cocos nucifera L.) that is native to Indonesia and New Guinea (10). Around 2000, B. longissima was recorded in southeast and east Asia and has caused serious damage to coconut palms (11). Based on mitochondrial cytochrome oxidase I gene sequences, B. longissima comprises two cryptic species: the Asian clade, distributed over a wide area including Asia and the Pacific region, and is the Pacific clade, distributed in a limited area (12). Recent invasions and outbreaks have been reported only for the Asian clade. The two clades copulate with each other, and sperm are successfully delivered to the spermatheca, but hatchability is much lower than that of intraclade crosses, indicating that CI occurs between them (12); however, what induces this CI remains unknown.
Here we show that different bacterial endosymbionts, a unique clade of Alphaproteobacteria in the Pacific clade and Wolbachia in the Asian clade, induce bidirectional CI between the two B. longissima clades. The former induces complete CI (100% offspring mortality), whereas the latter induces partial CI (70% mortality).
Results
Crossing Experiments.
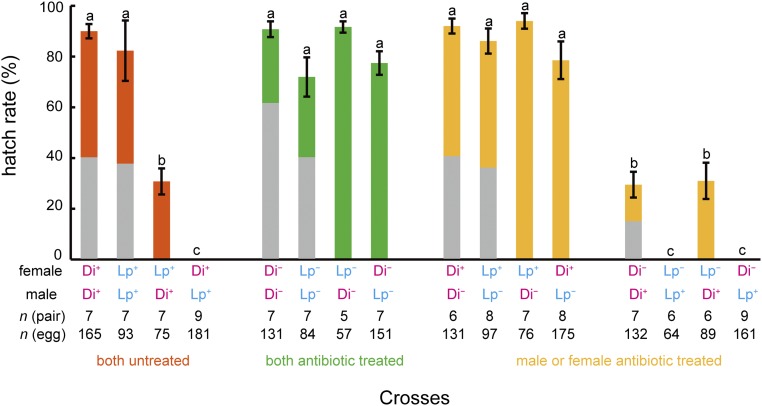
To determine the effect of bacterial endosymbionts on CI between the two clades of B. longissima, we examined egg hatch rates among crosses of antibiotic-treated (indicated by −) or untreated (indicated by +) insects. Two populations were used: B. longissima collected in Lospalos, East Timor, which belongs to the Pacific clade (Lp), and B. longissima collected in Dili, East Timor, which belongs to the Asian clade (Di). Egg hatch rates in the untreated interclade crosses were significantly lower than those in the untreated intraclade crosses (Fig. 1). Hatch rates were significantly lower in the Di+ female × Lp+ male crosses (0%; n = 9 crosses and 181 eggs) compared with the Lp+ female × Di+ male crosses (mean ± SEM, 30.8 ± 5.2%; n = 7 crosses and 75 eggs) (Fig. 1). When bacterial endosymbionts were eliminated by tetracycline treatment in both parental females and males, egg hatch rates in the interclade crosses increased dramatically, reaching levels similar to those of the intraclade crosses (Fig. 1). When parental males were treated with tetracycline but females were untreated, hatch rates also increased significantly in both the intraclade and interclade crosses, whereas when females were treated but males were untreated, hatch rates did not increase (Fig. 1). In the intraclade crosses, hatch rates decreased dramatically when females were treated with tetracycline but males were not. Antibiotic-treated females showed lower hatch rates in Lp (0%; n = 6 crosses and 64 eggs) than in Di (mean ± SEM, 29.5 ± 5.1%; n = 7 crosses and 132 eggs) (Fig. 1). Dissection of females to examine the presence of sperm in the spermatheca showed that 4 of 117 pairs did not copulate during the 1-wk period: one Di+ female × Di− male, one Lp− female × Di− male, one Lp− female × Lp+ male, and one Di+ female × Lp+ male. Data for these pairs were omitted from our analysis. Sex ratios of offspring did not significantly differ from 1:1 in all tested pairs (P > 0.05, binomial test) (Fig. 1).
Fig. 1.
Crossing tests among two populations in the absence or presence of antibiotic treatment in B. longissima. Lp. B. longissima collected in Lospalos, East Timor (Pacific clade); Di, B. longissima collected in Dili, East Timor (Asian clade); −, antibiotic-treated insects; +, antibiotic-untreated insects. Error bars indicate SEM. Means with the same letters do not differ significantly (Tukey–Kramer test, P > 0.05). Gray parts indicate percentage of females in each cross. In pairs, n = 9 for Di+ Lp+ (female × male, the same in the following), Di− Lp+, and n = 8 for Lp+ Lp−, Di+ Lp−, and n = 6 for Di+ Di−, Lp− Lp+, Lp− Di+, n = 5 for Lp− Di−, and n = 7 for the rest of the crosses. In the number of eggs, n = 165, 93, 75, 181, 131, 84, 57, 151, 131, 97, 76, 175, 132, 64, 89, and 161 for Di+ Di+, Lp+ Lp+, Lp+ Di+, Di+ Lp+, Di− Di−, Lp− Lp−, Lp− Di−, Di− Lp−, Di+ Di−, Lp+ Lp−, Lp+ Di−, Di+ Lp−, Di− Di+, Lp− Lp+, Lp− Di+, and Di− Lp+, respectively.
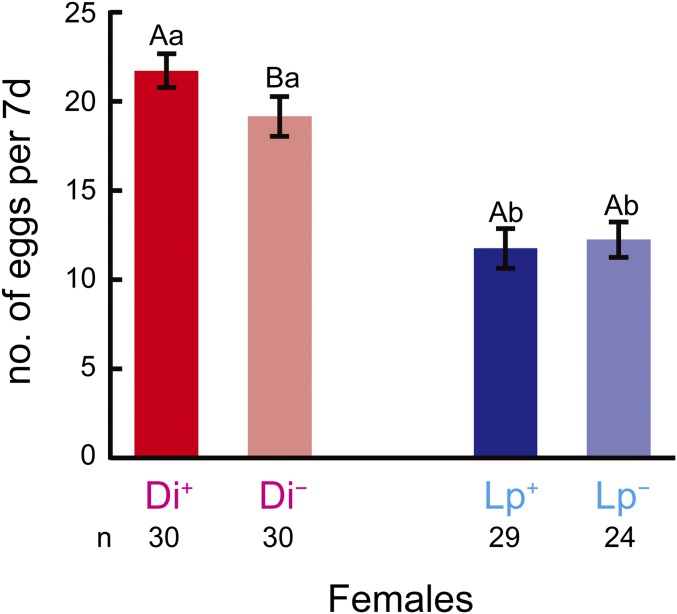
The number of eggs produced per female in 7 d did not differ between antibiotic-treated and untreated Lp females (Mann–Whitney test, U = 390.5, P = 0.451). Antibiotic-treated Di females produced fewer eggs than untreated females (U = 295.5, P = 0.022). Lp females produced fewer eggs than Di females (U = 786.5, P < 0.001 for untreated females vs. U = 598.5, P < 0.001 for antibiotic-treated females) (Fig. 2).
Fig. 2.
Number of eggs produced every 7 d per female of B. longissima. Lp, B. longissima collected in Lospalos, East Timor (Pacific clade); Di, B. longissima collected in Dili, East Timor (Asian clade); −, antibiotic-treated insects; +, antibiotic-untreated insects. Error bars indicate SEM. Means with the same capital letters do not differ significantly between antibiotic-treated and untreated females in the same clade, and means with the same lowercase letters do not differ significantly between Lp and Di in the same treatment (P > 0.05, Mann−Whitney U test). n = 30 for Di+ and Di−, n = 29 for Lp+, and n = 24 for Lp−.
Sequences and Phylogenetic Relationships of Bacterial Endosymbionts in B. longissima.
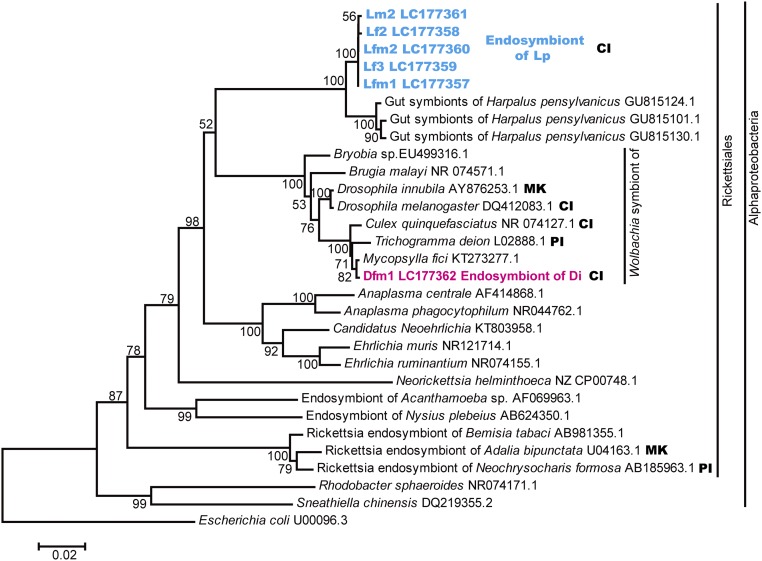
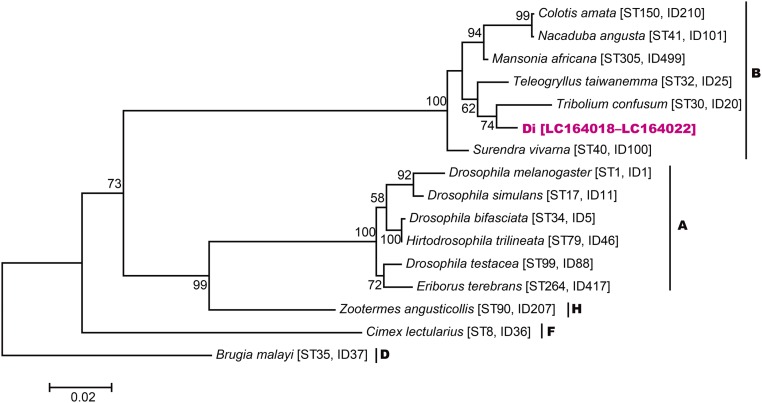
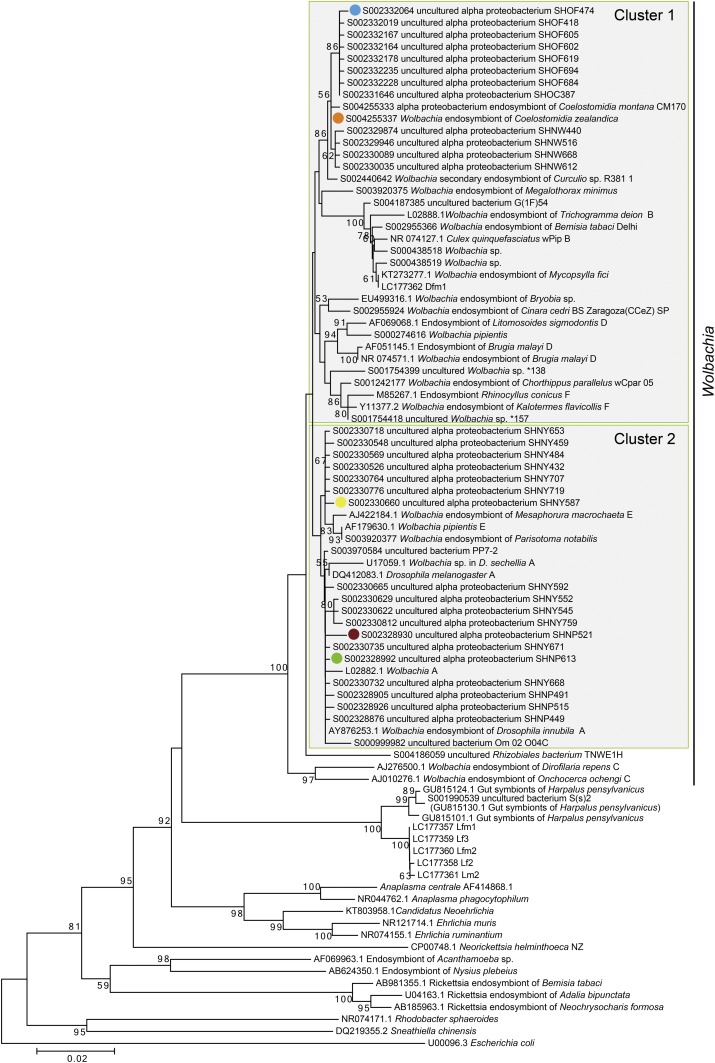
To detect and analyze phylogenetic relationships of bacterial endosymbionts, total genomic DNA was extracted from the testes or ovaries of Di and Lp adults, and bacterial 16S rRNA gene sequences were analyzed using universal bacterial 16S rRNA primers. For Lp, 1,160-bp 16S rRNA gene sequences were obtained from 21 clones derived from three males and three females (LC177357–LC177361). The closest BLASTn match (for LC177357) was with a sequence of unidentified bacterial gut symbionts of a beetle (GU815124.1) with 97.9% (1,129 of 1,153) identity. The closest BLASTn match with an identified sequence was with Wolbachia (EU499316.1), with 89.3% (1,038 of 1,162) identity. For Di, 1,159-bp 16S rRNA gene sequences were obtained from 22 clones derived from three males and three females (LC177362). The closest BLASTn match was with a sequence of Wolbachia (KT273277.1), with 99.7% (1,155 of 1,159) identity. The phylogenetic tree showed that bacterial endosymbionts detected from Lp beetles formed a unique clade in Alphaproteobacteria with 100% bootstrap support, whereas endosymbionts detected from Di beetles belonged to Wolbachia (Fig. 3). A maximum likelihood tree for concatenated multilocus sequence typing (MLST) sequences (gatB, coxA, hcpA, ftsZ, and fbpA) indicated that Di beetles were infected with Wolbachia endosymbionts belong to supergroup B (Fig. S1).
Fig. 3.
Neighbor-joining tree of the unique clade of alphaproteobacterial endosymbionts detected in B. longissima collected in Lospalos (Lp) and Wolbachia sp. detected in B. longissima collected in Dili (Di). Calculations were based on 1,160-bp fragments of 16S rRNA gene sequences. Bootstrap values (>50%) calculated on the basis of 1,000 replications are shown near the branches. CI, cytoplasmic incompatibility; MK, male killing; PI, parthenogenesis induction.
Fig. S1.
Maximum likelihood phylogenetic tree of the concatenated MLST data (2,073 or 2,079 bp). Letters next to the species names indicate the supergroups. The ID codes and the sequence type numbers are obtained from the MLST database. Maximum likelihood bootstrap values based on 1,000 replicates (>50%) are given at the nodes.
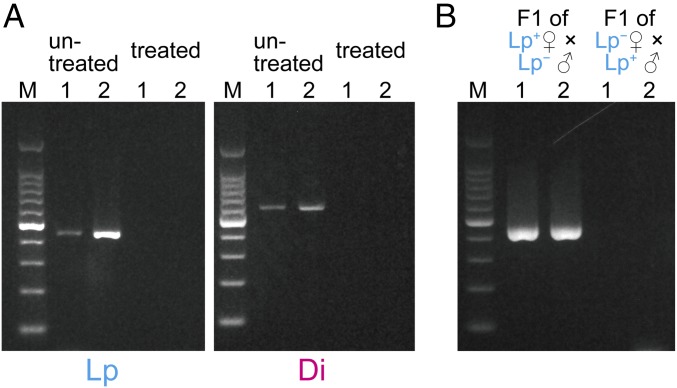
Elimination of the endosymbionts by antibiotic treatment was confirmed for eight adults from each cross using specific primers for Lp (designed in the present study) and primers for Wolbachia (13) for Di (Fig. 4A).
Fig. 4.
(A) Detection of bacterial endosymbionts in the testes or ovaries of B. longissima. Beetles were treated with tetracycline or untreated. Lp, B. longissima collected in Lospalos, East Timor (Pacific clade); Di, B. longissima collected in Dili, East Timor (Asian clade); 1, male; 2, female. (B) Inheritance of previously unidentified alphaproteobacterial endosymbionts of B. longissima.
Inheritance of Lp endosymbionts was examined by PCR with the specific primers for offspring of intraclade crosses. A strong band was detected from offspring larvae of the Lp+ female × Lp− male crosses, whereas no band was detected in the offspring eggs from the Lp− female × Lp+ male crosses (Fig. 4B).
Analysis of Microbial Composition.
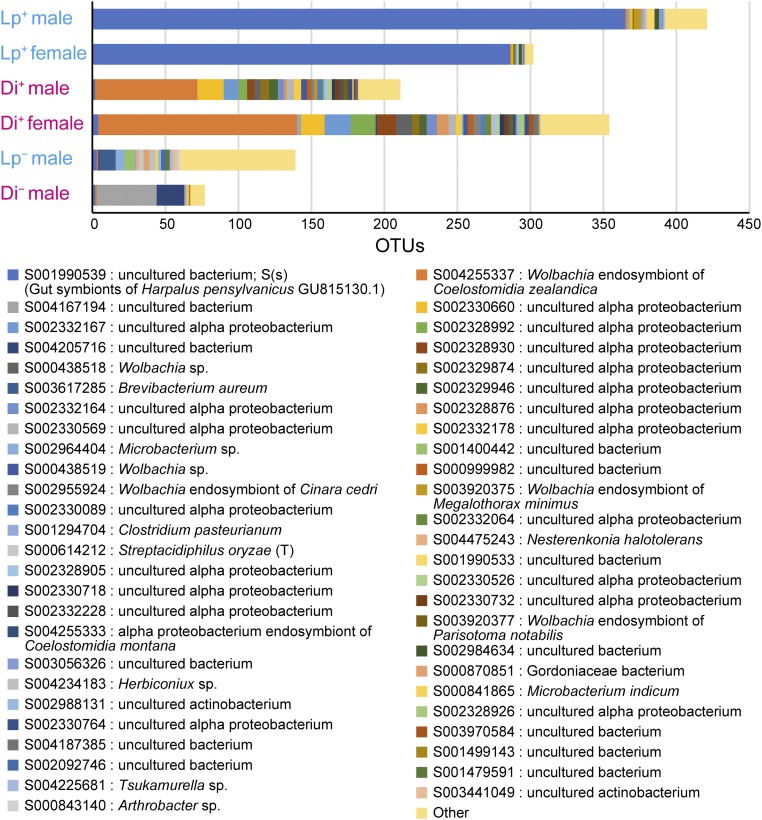
We performed Illumina MiSeq sequencing to check for the possible presence of other bacteria not detected by the foregoing cloning procedure. We obtained a total of 215,058 (mean ± SEM, 35,843 ± 5,393 reads per sample) high-quality pair-end 16S rRNA reads from the testes or ovaries of B. longissima. Operational taxonomic units (OTUs) were generated using USEARCH (14) with identity ≥97%. After excluding the OTUs containing 18S rRNA genes of the host beetles, a total of 1,504 OTUs were generated. The predominant OTU in both female and male Lp+ was S001990539, uncultured bacterium alone (Fig. 5). Although one predominant OTU, S004255337, a Wolbachia endosymbiont of Coelostomidia zealandica, was detected in Di+, other Alphaproteobacteria were found as well (Fig. 5). Phylogenetic analysis revealed that major Alphaproteobacteria detected in Di+ belonged to different clusters within Wolbachia (Fig. S2). The number of detected OTUs was rather small in antibiotic-treated insects (Fig. 5).
Fig. 5.
Microbial composition of the testes or ovaries of B. longissimia. Lp, B. longissima collected in Lospalos, East Timor (Pacific clade); Di, B. longissima collected in Dili, East Timor (Asian clade); −, antibiotic-treated insects; +, antibiotic-untreated insects. The horizontal axis indicates the number of OTUs assigned to Ribosomal Database Project release 11 (RDP) bacterial 16S rRNA V3 region.
Fig. S2.
Phylogenetic distance tree of Anaplasma obtained from OTUs of male and female Brontispa longissima collected in Dili, East Timor (Asian clade). Circle colors indicate the top five numbers of OTUs in Fig. 5. The number at each branch indicates the bootstrap values (>50%) (100 replications). The phylogenetic tree was constructed in MEGA6 using neighbor-joining method, and based on the full length 16S rRNA sequences of the mapped OTUs that belonged to Anaplasma in Fig. 5.
PCR and Denaturing Gradient Gel Electrophoresis.
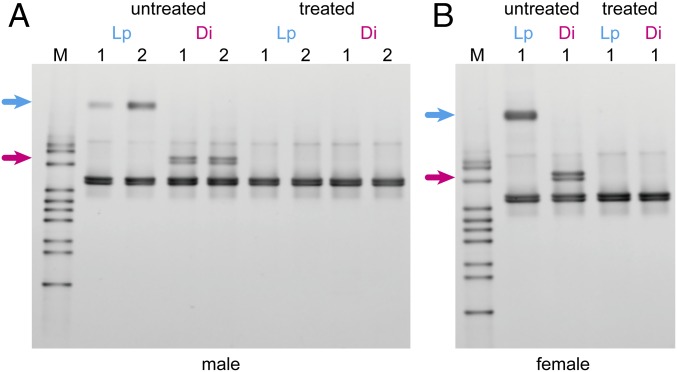
In addition to the Illumina MiSeq sequencing, we performed PCR and denaturing gradient gel electrophoresis (DGGE) analyses to check for the possible presence of other bacteria. DGGE patterns of the variable V3 region of 16S rRNA genes showed that one band disappeared due to the antibiotic treatment in both males and a female of Lp, and two closely spaced bands disappeared in Di (Fig. 6).
Fig. 6.
Denaturing gradient gel electrophoresis profiles of bacterial 16S rRNA genes in the testes or ovaries of B. longissima. Male (A) and female (B) beetles were treated with tetracycline or untreated. Lp, B. longissima collected in Lospalos, East Timor (Pacific clade); Di, B. longissima collected in Dili, East Timor (Asian clade). Upper arrows indicate the band that disappeared after antibiotic treatment of Lp. Lower arrows indicate the bands that disappeared after antibiotic treatment of Di.
Discussion
In this study, we used crossing tests between different clades of B. longissima (Di and Lp) and analyses of the 16S rRNA gene sequences to demonstrate that bidirectional CI between the two clades is induced by different endosymbionts. We found that the complete CI (100% mortality) between Di females and Lp males was induced by a unique clade of Alphaproteobacteria, and that the partial CI (70% mortality) between Lp females and Di males was induced by Wolbachia (Figs. 1, 3, and 4A). With antibiotic treatment, the hatch rates in interclade crosses became as high as those in intraclade crosses (Fig. 1).
Several factors may cause reproductive isolation among sympatric populations, including a host shift (15), a change in breeding time (16), and infection with CI inducers (3, 8). Nasonia parasitoid wasps have three sibling species (Nasonia vitripennis, Nasonia longicornis, and Nasonia giraulti) and show bidirectional CI among the species, caused by different strains of Wolbachia (17, 18). In this case, Wolbachia may play a causal role in speciation (3, 19). Our findings regarding B. longissima suggest that this may be another case in which bidirectional CI causes speciation. Although we used B. longissima obtained from East Timor alone in this study, CI between the two clades occurs in coconut beetles from different locations; individuals from Papua New Guinea belonging to the Pacific clade show complete CI when crossed with those from Japan belonging to the Asian clade (12). In the Papua New Guinea beetles, we detected the same endosymbionts as in the beetles from Lp (Fig. S3). We also detected the same endosymbionts in beetles of the Pacific clade collected from different places, including Australia and Indonesia (Sumba) (Fig. S3).
Fig. S3.
Detection of the previously unidentified bacterial endosymbiont in B. longissima from different locations: 1–3, Australia; 4–6, Papua New Guinea; 7–9, Indonesia (Sumba): 10 and 11, East Timor (Puno); 12–14, East Timor (Home); 15–17, East Timor (Raca); 18 and 19, East Timor (Dili). Specimens in 1–17 belong to the Pacific clade; specimens in 18 and 19, to the Asian clade.
No effects of this bacterium on host reproduction other than CI were found in the present study. Sex ratio of the two clades was maintained at ∼50% in all crosses (Fig. 1), indicating that neither feminization nor male killing was caused by this bacterium in B. longissima. Because the same Wolbachia species may induce different phenotypes in different host species (20, 21), further research on the phenotypes of this Alphaproteobacterium in other insect species is warranted.
Our phylogenetic tree indicates that bacterial symbionts in Lp form a clear monophyletic group in class Alphaproteobacteria, order Rickettsiales (Fig. 3). In bacteriology, similarities of ≥97% (22) or >98.7% (23) in 16S rRNA gene sequences are required for individuals to be classified as the same species. Sequences of bacterial symbionts of Lp have only 89.3% identity with a sequence of Wolbachia (EU499316.1), the closest BLASTn match of identified bacteria. Although further studies on morphologic or anatomic characteristics are needed to determine the taxonomic status of this Alphaproteobacterium, we can recognize it as a distinct species based on the sequences of 16S rRNA genes. This distinct species may elucidate the origin and relationships among supergroups of Wolbachia owing to its similarity. So far, the outgroups used for this purpose are too divergent, and this distinct species could serve as a suitable outgroup in a phylogenetic study of Wolbachia (24, 25).
Detection of bacterial symbionts using universal bacterial 16S rRNA primers for PCR and cloning identified only two endosymbionts, a unique clade of Alphaproteobacteria in Lp and Wolbachia in Di. Our findings indicate that only these bacterial endosymbionts are responsible for CI in the two clades, which is consistent with Illumina MiSeq sequencing and DGGE patterns showing only one predominant bacterium at the strain level and only one band in Lp (Figs. 5 and 6). For Di, however, Illumina MiSeq sequencing showed that major OTUs belong to different clusters within Wolbachia (Fig. S2). DGGE patterns also showed two closely spaced bands that disappeared with antibiotic treatment. Thus, we infer that Di is infected by more than one Wolbachia strain. Although this finding is inconsistent with the Di Wolbachia cloning and MLST sequence results (gatB, coxA, hcpA, ftsZ, and fbpA), the limited number of analyses (i.e., 22 clones from six beetles in the cloning and direct sequencing from two beetles in MLST) may explain the detection of only a single strain.
The use of the bacterial symbiont Wolbachia as a biological control agent against insect pests has been attempted. A simple technique is to release males infected with CI-inducing Wolbachia into target uninfected populations to suppress population growth of the target, as in the sterile insect technique (SIT) (9). Guaranteeing the sterility of males is essential in SIT, and strong CI expression can effectively suppress a target population. Various intensities of CI expression have been reported in the CI inducers Wolbachia and Cardinium. In Wolbachia, 0–100% CI has been reported in different host species (26–28). In Cardinium, 70% hatchability has been reported in incompatible crosses in spider mites (5), and incompatible crosses in the parasitoid wasp Encarsia pergandiella produced only 7% and 38% of the number of offspring in compatible crosses and showed an 8% pupation rate (6, 7). Therefore, the previously unidentified alphaproteobacterial endosymbiont, which induces complete CI, is a strong candidate agent for biological control of the coconut beetle.
Different effects from CI caused by Wolbachia may contribute to pest control. Infection by Wolbachia decreases the fecundity of host insects, and releasing transinfected insects with low fecundity into the field may suppress the pest population (29–31). In B. longissima, egg production is higher in the Asian clade than in the Pacific clade (32). In this study, elimination of the previously unidentified bacterial symbiont did not influence the short-term (7 d) egg production of this beetle (Fig. 2). To assess the potential of this endosymbiont as a biological control agent, we intend to investigate the effects of such elimination on total egg production and longevity, as well as the effects of transinfection with these endosymbionts on fecundity. Finally, Wolbachia infection may suppress the transmission of pathogens to humans by mosquitoes, including dengue, malaria, and zika, along with plant diseases transmitted by planthoppers (33). Further investigation may reveal that the endosymbiont of B. longissima has such effects on pathogen transmission as well.
Materials and Methods
Insects.
Two colonies of B. longissima maintained in our laboratory were used for this study. One colony was initiated from insects collected in Dili, East Timor in July 2014 (referred to as Di), and the other was initiated from insects collected in Lospalos, East Timor in July 2014 (referred to as Lp). Di belongs to the Asian clade, and Lp belongs to the Pacific clade (34). Insects were reared on fresh leaves of Phoenix canariensis Chabaud as described previously (12).
Antibiotic Treatment.
To eliminate bacterial endosymbionts, newly hatched larvae were reared on an artificial diet, Diet C, as described previously (35) as a control or Diet C with tetracycline. After the artificial diet was autoclaved, 0.05% (wt/wt) tetracycline was added to it. Because beetles do not lay fertile eggs when reared on the artificial diet, emerged adults were reared on fresh leaves of P. canariensis brushed with 0.05% tetracycline solution (in distilled water) or distilled water alone as a control.
Crossing Experiments.
Virgin beetles (age 30–40 d) were paired in a Petri dish (60-mm diameter, 25-mm height) with fresh leaves of P. canariensis for 7 d for copulation. Eggs were collected and kept in a Petri dish containing a paper towel saturated with distilled water. The hatch rate was monitored for 5 d after oviposition. When the hatch rate was zero, the females were dissected to check for the presence of sperm in the spermatheca. Females without sperm were considered unmated and omitted from the data. Between five and nine pairs were investigated in each cross (Fig. 1). Sex ratios of F1 offspring were examined for one to three pairs in antibiotic-treated and untreated intraclade crosses. Antibiotic treatment and crossing were conducted at 25 ± 1 °C and 70 ± 5% relative humidity with a 12-h:12-h light:dark photoperiod.
Differences in hatch rates among crosses were analyzed by one-way ANOVA and the Tukey–Kramer test after arcsine transformation. The number of eggs produced per 7 d was compared between antibiotic-treated and untreated females in the same clade, and between Lp and Di in the same treatment, using the Mann–Whitney U test. The sex ratio of offspring was analyzed using the binomial test. These analyses were conducted with R version 3.3.1 (36).
DNA Extraction, PCR, Cloning, Sequencing, and Phylogenetic Analysis of 16S rRNA.
Total genomic DNA was extracted from the testes or ovaries of Di and Lp adults obtained from the colonies (three males and three females from each clade). Beetles were dissected in distilled water to remove the testes or ovaries, which were placed on a clean bench under UV lighting for 5 min to eliminate possible surface contamination from other organs. DNA was extracted using the Qiagen DNeasy Blood and Tissue Kit. To amplify 16S rRNA genes, PCR was conducted with universal bacterial 16S rRNA primers (forward, 5′-GCT TAA CAC ATG CAA G-3′; reverse, 5′-CCA TTG TAG CAC GTG T-3′) (37), under the following conditions: an initial cycle at 94 °C for 1 min, followed by 30 cycles at 98 °C for 10 s, 55 °C for 15 s, and 68 °C for 90 s. The 20-µL PCR mixture contained 5× PrimeSTAR GXL Buffer (TaKaRa Bio), 0.2 mM dNTP mixture, 0.5 µM each primer, 0.5 U of TaKaRa PrimeSTAR GXL DNA polymerase, and 2.0 µL of DNA. PCR products were purified with NucleoSpin Gel and PCR Clean-up (Macherey-Nagel) and were inserted into the TA cloning vector (pMD20-T; TaKaRa Bio), which was used to transform Escherichia coli DH5α-competent cells (Competent Quick; Toyobo). The blue-white selection system with ampicillin and X-Gal was used. At least four white colonies from each individual were subjected to colony PCR with primers pUC/M13 (forward, 5′-CGC CAG GGT TTT CCC AGT CAC GAC-3′; reverse, 5′-TCA CAC AGG AAA CAG CTA TGA C-3′). PCR products of the expected length (∼1.4 kb) were sent to FASMAC (Atsugi, Japan) for sequencing. The obtained 16S rRNA gene sequences were aligned using CLUSTAL X (38).
The phylogenetic tree was constructed by the neighbor-joining method with MEGA6 software (39); sequences from representative Alphaproteobacteria were included in the analysis. The evolutionary distances were computed by the Kimura two-parameter method, which accounts for differing rates of transitions vs. transversions (40). Nodal support was evaluated with 1,000 bootstrap resamplings (41). The sequence data were deposited in the DDBJ/EMBL/GenBank database (accession nos. LC177357–LC177362).
MLST Analysis.
MLST analysis was conducted to determine the supergroup of Wolbachia infecting Di. Five ubiquitous genes (gatB, coxA, hcpA, ftsZ, and fbpA) were analyzed in one male and one female (Fig. S1) (13). The sequence data were deposited in the DDBJ/EMBL/GenBank database (accession nos. LC164018–LC164022).
Confirmation of the Elimination of Bacterial Endosymbionts by the Antibiotic Treatment.
Bacterial endosymbionts were detected using primers designed in the present study for Lp (forward L355F, 5′-GCT ATG CCG CGT GAG TGA TT-3′; reverse L749R, 5′-ACA CAG AAA TAA AAA TTC CTA C-3′) and primers (13) for Di (forward wsp_F1, 5′-GTC CAA TAR STG ATG ARG AAA C-3′; reverse wsp_R1, 5′- CYG CAC CAA YAG YRC TRT AAA-3′), with annealing temperatures of 58 °C. After crossing tests, the elimination was examined in eight antibiotic-treated adults of each clade. DNA extraction was conducted as described above.
In addition, inheritance of a unique clade of Alphaproteobacteria was examined in the offspring of crosses of Lp+ females × Lp− males and Lp− females × Lp+ males. DNA was extracted from the whole body of first instar larvae in the former crosses and from whole eggs in the latter crosses, because no larvae hatched in the latter crosses. Ten larvae or eggs were analyzed for each cross.
Analysis of Microbial Composition.
To check for the possible presence of other bacteria not detected by the foregoing cloning procedure, microbial communities in testes and ovaries were analyzed by Illumina MiSeq 16S rRNA gene sequencing. One male and one female reared on the artificial diet without tetracycline, and one male reared on the artificial diet with tetracycline were used for each clade. To amplify the variable V3 region of 16S rRNA gene, PCR was conducted with primers (forward 357F, 5′-CC TAC GGG AGG CAG CAG-3′; reverse 518R, 5′-ATT ACC GCG GCT GG-3′) (42) under the following conditions: an initial cycle at 95 °C for 2 min, followed by 35 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min. The 12.5-µL PCR mixture contained 2× GoTaq Green Master Mix (Promega), each primer, and 1.5 µL of DNA. PCR products were used for library production using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs Japan). The nucleotide sequence of the PCR products was determined using the Illumina MiSeq Reagent Kit v3.
We developed a custom pipeline for analysis of the 16S rRNA gene V3 region sequences. First, we excluded 16S reads that did not have primer sequences. Second, we trimmed the reads to maintain high quality of the first 100 bases using the FASTX-Toolkit (hannonlab.cshl.edu/fastx_toolkit/). Third, we merged and filtered the pair-end reads using MeFiT (https://github.com/nisheth/MeFiT) (43), with an average quality value ≥25. We used all merged reads of the 16S rRNA gene V3 region to generate OTUs using the USEARCH program (www.drive5.com) (14) with ≥97% pairwise identity cutoff. The OTUs including 18S rRNA genes of the host beetles were detected by BLASTn match against the National Center for Biotechnology Information nucleotide database, and were excluded from the analysis. We constructed our own 16S rRNA reference database from the publicly available Ribosomal Database Project release 11 (RDP) with ≥1200 bases. Each OTU was assigned to bacterial 16S rRNA by a BLASTn search with coverage ≥90% and pairwise identity ≥90%. All sequences of the 16S rRNA V3 region used in this study were deposited in the DNA Data Bank of Japan with accession numbers DRA005752.
PCR and DGGE.
Along with the Illumina MiSeq sequencing, PCR and DGGE were performed to examine the possible presence of other bacteria. Two males and one female reared on the artificial diet with or without tetracycline were used for each clade. To amplify the variable V3 region of 16S rRNA gene, PCR was conducted with primers (forward 357FGC, 5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG-3′; reverse 518R) (42) under the following conditions: an initial cycle at 94 °C for 1 min; 35 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; and a final cycle of 72 °C for 7 min. The 20-µL PCR mixture contained 10× ExTaq Buffer (TaKaRa Bio), 0.2 mM dNTP mixture, 0.5 µM each primer, 0.5 U of TaKaRa ExTaq polymerase, and 2.0 µL of DNA. PCR products were purified with NucleoSpin Gel and PCR Clean-up (Macherey-Nagel).
DGGE was performed with the DCode Universal Mutation Detection System (Bio-Rad) in polyacrylamide gels; the gradient was from 6% acrylamide with 30% denaturant to 12% acrylamide with 60% denaturant [100% denaturant = 7 M urea and 40% (vol/vol) formamide]. Gels were poured with the aid of a gradient delivery system (Bio-Rad). PCR products (∼100 ng) were electrophoresed in 1× TAE buffer (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA) at 60 °C for 5 h at 200 V. Gels were stained with SYBR Green I (TaKaRa Bio) for 30 min and then photographed under UV light using the Gel Doc XR+ system (Bio-Rad).
SI Materials and Methods
Multilocus Sequence Typing Analysis.
To determine the supergroup of Wolbachia infecting Di, multilocus sequence typing (MLST) analysis was conducted (13). DNA was extracted from one male and one female as described in the analysis of 16S rRNA gene. Five ubiquitous genes (gatB, coxA, hcpA, ftsZ, and fbpA) were amplified by using specific primers (13) under the following conditions: an initial cycle of 94 °C for 2 min; 37 cycles of 94 °C for 30 s, 54 °C (59 °C for ftsZ) for 45 s, and 72 °C for 1 min 30 s; and a final cycle of 72 °C for 10 min. The 20-µL PCR mixture contained 10× ExTaq Buffer (TaKaRa Bio), 0.2 mM dNTP mixture, 0.5 µM each primer, 0.5 U of TaKaRa ExTaq polymerase, and 2.0 µL of DNA. PCR products were purified with NucleoSpin Gel and PCR Clean-up (Macherey-Nagel) and were sent to FASMAC (Atsugi, Japan) for sequencing. Obtained sequences were aligned by using CLUSTAL X (37). The phylogenetic tree was constructed by the maximum likelihood method for concatenated MLST sequences by using MEGA6 software (38); sequences from other Wolbachia representatives were included. The nucleotide substitution model GTR + G was selected by using jModelTest 2.1.10. Nodal support was evaluated with 1,000 bootstrap resamplings (40). The sequence data were deposited in the DDBJ/EMBL/GenBank database (accession nos. LC164018–LC164022).
Phylogenetic Tree Construction.
The full-length 16S rRNA gene sequences of the OTUs that belonged to Anaplasma in Fig. 5 were aligned using the multiple sequence alignment program MUSCLE in MEGA6 (38, 44). The phylogenetic tree was constructed using neighbor-joining method with 100 bootstrap replications. Wolbachia supergroups B, D, and F were distributed within Cluster 1. Supergroups A and E were distributed within cluster 2. The major species, S004255337 (Wolbachia endosymbiont of C. zealandica) in male and female B. longissima, belonged to cluster 1 (Fig. S2).
Presence of the Previously Unidentified Alphaproteobacterial Endosymbiont in B. longissima from Different Locations.
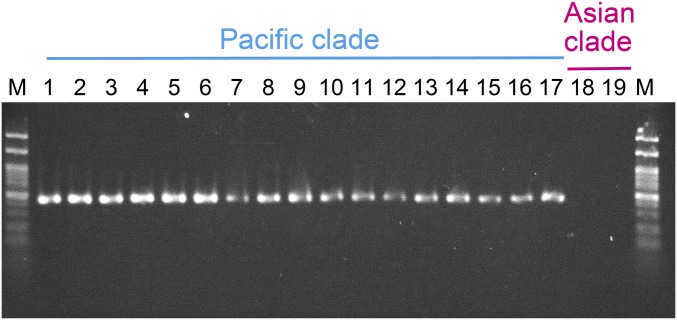
Presence of the bacterial endosymbiont was tested as described in the methods section under “Confirmation of the elimination of bacterial endosymbionts by the antibiotic treatment” for the specimens from the following locations: Australia, Papua New Guinea, Indonesia (Sumba), and East Timor. Details of the specimens have been described previously (12). A total of 29 specimens that belong to the Pacific clade and two specimens that belong to the Asian clade were analyzed. DNA extracted from the thorax muscle was used (12) and a single band was detected in all specimens belonging to the Pacific clade (Fig. S3), indicating that all were infected with the previously unidentified alphaproteobacterial endosymbiont.
Acknowledgments
We thank Kazuki Miura, Natsuko Kinoshita, and Mika Murata for their suggestions on the study design; Katsuya Fukami and the Materials Management Center of Kyushu University for arranging transfer of the materials used in this study; Brendan Trewin for helpful comments on the manuscript; and Shun-ichiro Iwase and Katsuto Fukuda for their technical assistance. This research was supported by Japan Society for the Promotion of Science KAKENHI Grant JP26850031 and JP25430194.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences have been deposited in the DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank database (accession nos. LC177357–LC177362, LC164018–LC164022, DRA005752).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618094114/-/DCSupplemental.
References
- 1.Yen JH, Barr AR. The etiological agent of cytoplasmic incompatibility in Culex pipiens. J Invertebr Pathol. 1973;22:242–250. doi: 10.1016/0022-2011(73)90141-9. [DOI] [PubMed] [Google Scholar]
- 2.Duron O, et al. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008;6:27. doi: 10.1186/1741-7007-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werren JH. Biology of Wolbachia. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 4.Hunter MS, Perlman SJ, Kelly SE. A bacterial symbiont in the Bacteroidetes induces cytoplasmic incompatibility in the parasitoid wasp Encarsia pergandiella. Proc Biol Sci. 2003;270:2185–2190. doi: 10.1098/rspb.2003.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotoh T, Noda H, Ito S. Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity (Edinb) 2007;98:13–20. doi: 10.1038/sj.hdy.6800881. [DOI] [PubMed] [Google Scholar]
- 6.Perlman SJ, Kelly SE, Hunter MS. Population biology of cytoplasmic incompatibility: Maintenance and spread of Cardinium symbionts in a parasitic wasp. Genetics. 2008;178:1003–1011. doi: 10.1534/genetics.107.083071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris LR, Kelly SE, Hunter MS, Perlman SJ. Population dynamics and rapid spread of Cardinium, a bacterial endosymbiont causing cytoplasmic incompatibility in Encarsia pergandiella (Hymenoptera: Aphelinidae) Heredity (Edinb) 2010;104:239–246. doi: 10.1038/hdy.2009.130. [DOI] [PubMed] [Google Scholar]
- 8.Brucker RM, Bordenstein SR. Speciation by symbiosis. Trends Ecol Evol. 2012;27:443–451. doi: 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Zabalou S, et al. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc Natl Acad Sci USA. 2004;101:15042–15045. doi: 10.1073/pnas.0403853101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waterhouse DF. Brontispa longissima (Gestro) In: Pressley M, editor. Biological Control: Pacific Prospects. Inkata Press; Melbourne, Australia: 1987. pp. 134–141. [Google Scholar]
- 11.Nakamura S, Konishi K, Takasu K. Invasion of the coconut hispine beetle, Brontispa longissima: Current situation and control measures in Southeast Asia. In: Ku TY, Chiang MY, editors. Proceedings of an International Workshop on Development of a Database (APASD) for Biological Invasion. Taiwan Agricultural Chemicals and Toxic Substance Research Institute, Taichung, Taiwan and Food and Fertilizer Technology Center for the Asia and Pacific Region, Taipei; Taiwan: 2006. pp. 1–9. [Google Scholar]
- 12.Takano S, et al. Two cryptic species in Brontispa longissima (Coleoptera: Chrysomelidae): Evidence from mitochondrial DNA analysis and crosses between the two nominal species. Ann Entomol Soc Am. 2011;104:121–131. [Google Scholar]
- 13.Baldo L, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 15.Berlocher SH, Feder JL. Sympatric speciation in phytophagous insects: Moving beyond controversy? Annu Rev Entomol. 2002;47:773–815. doi: 10.1146/annurev.ento.47.091201.145312. [DOI] [PubMed] [Google Scholar]
- 16.Marshall DC, Cooley JR. Reproductive character displacement and speciation in periodical cicadas, with description of new species, 13-year Magicicada neotredecem. Evolution. 2000;54:1313–1325. doi: 10.1111/j.0014-3820.2000.tb00564.x. [DOI] [PubMed] [Google Scholar]
- 17.Breeuwer JAJ, Werren JH. Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature. 1990;346:558–560. doi: 10.1038/346558a0. [DOI] [PubMed] [Google Scholar]
- 18.Bordenstein SR, Werren JH. Bidirectional incompatibility among divergent Wolbachia and incompatibility level differences among closely related Wolbachia in Nasonia. Heredity (Edinb) 2007;99:278–287. doi: 10.1038/sj.hdy.6800994. [DOI] [PubMed] [Google Scholar]
- 19.Stouthamer R, Breeuwer JAJ, Hurst GDD. Wolbachia pipientis: Microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki T, Kubo T, Ishikawa H. Interspecific transfer of Wolbachia between two lepidopteran insects expressing cytoplasmic incompatibility: A Wolbachia variant naturally infecting Cadra cautella causes male killing in Ephestia kuehniella. Genetics. 2002;162:1313–1319. doi: 10.1093/genetics/162.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaenike J. Spontaneous emergence of a new Wolbachia phenotype. Evolution. 2007;61:2244–2252. doi: 10.1111/j.1558-5646.2007.00180.x. [DOI] [PubMed] [Google Scholar]
- 22.Stackebrandt E, Goebel BM. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 23.Stackebrnadt E, Ebers J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 24.Werren JH, Baldo L, Clark ME. Wolbachia: Master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 25.Casiraghi M, et al. Phylogeny of Wolbachia pipientis based on gltA, groEL, and ftsZ gene sequences: Clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology. 2005;151:4015–4022. doi: 10.1099/mic.0.28313-0. [DOI] [PubMed] [Google Scholar]
- 26.Wade MJ, Stevens L. Microorganism-mediated reproductive isolation in flour beetles (genus Tribolium) Science. 1985;227:527–528. doi: 10.1126/science.3966160. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki T, Ishikawa H. Wolbachia infections and cytoplasmic incompatibility in the almond moth and the Mediterranean flour moth. Zoolog Sci. 1999;16:739–744. [Google Scholar]
- 28.Merçot H, Charlat S. Wolbachia infections in Drosophila melanogaster and D. simulans: Polymorphism and levels of cytoplasmic incompatibility. Genetica. 2004;120:51–59. doi: 10.1023/b:gene.0000017629.31383.8f. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann AA, Turelli M, Harshman LG. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics. 1990;126:933–948. doi: 10.1093/genetics/126.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stouthamer R, Luck RF. Influence of microbe-associated parthenogenesis on the fecundity of Trichogramma deion and T. pretiosum. Entomol Exp Appl. 1993;67:183–192. [Google Scholar]
- 31.Calvitti M, Moretti R, Lampazzi E, Bellini R, Dobson SL. Characterization of a new Aedes albopictus (Diptera: Culicidae)-Wolbachia pipientis (Rickettsiales: Rickettsiaceae) symbiotic association generated by artificial transfer of the wPip strain from Culex pipiens (Diptera: Culicidae) J Med Entomol. 2010;47:179–187. doi: 10.1603/me09140. [DOI] [PubMed] [Google Scholar]
- 32.Takano S, Takasu K, Murata M, Nguyen TH, Nakamura S. Comparative developmental and reproductive biology of geographical populations from two cryptic species in Brontispa longissima (Coleoptera: Chrysomelidae) Entomol Sci. 2013b;16:335–340. [Google Scholar]
- 33.Hoffmann AA, Ross PA, Rašić G. Wolbachia strains for disease control: Ecological and evolutionary considerations. Evol Appl. 2015;8:751–768. doi: 10.1111/eva.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takano S, et al. Rapid discrimination of two cryptic species within Brontispa longissima (Gestro) (Coleoptera: Chrysomelidae) by PCR-RFLP. J Pest Sci. 2013a;86:151–155. [Google Scholar]
- 35.Murata M, et al. A comparison of artificial diets for mass rearing Brontispa longissima (Coleoptera: Chrysomelidae) as hosts for the larval and the pupal parasitoids. J Econ Entomol. 2012;105:802–809. doi: 10.1603/ec11373. [DOI] [PubMed] [Google Scholar]
- 36.R Core Team 2015 R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna). Available at https://www.R-project.org/. Accessed May 31, 2016.
- 37.O’Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 41.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 42.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parikh HI, Koparde VN, Bradley SP, Buck GA, Sheth NU. MeFiT: Merging and filtering tool for illumina paired-end reads for 16S rRNA amplicon sequencing. BMC Bioinformatics. 2016;17:491. doi: 10.1186/s12859-016-1358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]