Significance
Double plant homeodomain finger 2 (DPF2) is a regulator of myeloid differentiation and implicated in a range of human cancers, including acute myelogenous leukemia. Recruitment of DPF2 to chromatin has been shown to alter the expression of target genes and inhibit myeloid differentiation. Here, we present the crystal structure of the human DPF2 tandem plant homeodomain finger domain and comprehensive structure-guided biochemical and in vivo analyses. Combined, our data delineate the determinants of DPF2’s chromatin recruitment and establish its regulatory role in human hematopoietic stem/progenitor cell differentiation.
Keywords: X-ray crystallography, myeloid differentiation, tandem PHD finger, isothermal titration calorimetry, protein–protein interaction
Abstract
Double plant homeodomain finger 2 (DPF2) is a highly evolutionarily conserved member of the d4 protein family that is ubiquitously expressed in human tissues and was recently shown to inhibit the myeloid differentiation of hematopoietic stem/progenitor and acute myelogenous leukemia cells. Here, we present the crystal structure of the tandem plant homeodomain finger domain of human DPF2 at 1.6-Å resolution. We show that DPF2 interacts with the acetylated tails of both histones 3 and 4 via bipartite binding pockets on the DPF2 surface. Blocking these interactions through targeted mutagenesis of DPF2 abolishes its recruitment to target chromatin regions as well as its ability to prevent myeloid differentiation in vivo. Our findings suggest that the histone binding of DPF2 plays an important regulatory role in the transcriptional program that drives myeloid differentiation.
The founding member of the d4 family of proteins, d4, zinc, and double plant homeodomain (PHD) finger 2 (DPF2, also known as requiem/REQ or ubi-d4), was initially discovered as a factor required for apoptosis in myeloid cells (1). d4 proteins, which in humans also include DPF1 and DPF3b, are characterized by an N-terminal requiem domain, a central C2H2-type zinc finger domain, and a C-terminal tandem PHD finger (2). PHD fingers, which contain two zinc finger motifs, are notable for their ability to read a diverse number of posttranslational modifications, including unmodified, methylated, or acetylated lysines, as well as unmodified arginines (3). Besides such putative binding capabilities of the DPF2 tandem PHD finger domain, relatively little is known about the regulation and function of DPF2 or its remaining individual domains. Previous studies have shown that DPF2 bridges SWI/SNF components and RelB/p52 to affect noncanonical NF-κB signaling (4), acts as a globin switching factor (5), and is a target for Staufen-1–mediated mRNA decay (5). Notably, DPF2 is expressed ubiquitously in human tissues compared with DPF1 and DPF3b (6, 7).
DPF2, along with DPF1 and DPF3b, has been implicated in a range of human cancers, including cervical cancer and acute myelogenous leukemia (AML) (8–13). Runt-related transcription factor 1 (RUNX1, also known as AML1) functions as an AML tumor-suppressor gene, which is frequently inactivated through somatic mutations and chromosomal translocations, including t(8;21), which produces the AML1–ETO fusion protein (14). Recent work has shown that recruitment of DPF2 into a RUNX1-containing repressor complex inhibits the expression of RUNX1 target genes, including the myeloid-specific microRNA miR-223, and inhibits myeloid differentiation (8). DPF2 recruitment appears to depend on arginine methylation events of RUNX1, as it is blocked by mutation of RUNX1 Arg223 or chemical inhibition of the type I arginine methyltransferase PRMT4. Knockdown of either DPF2 or PRMT4 increases miR-223 gene expression and myeloid differentiation. These findings suggest a model in which DPF2 and RUNX1 form a methylation-dependent repressive complex in AML, although it remains unclear whether the two proteins bind each other directly or act concertedly as part of a larger complex.
Here, we present the crystal structure of the human DPF2 tandem PHD finger domain at a 1.6-Å resolution. We demonstrate that the DPF2 tandem PHD finger domain binds acetylated H3 and H4 histone tails, identify the primary determinants of histone recognition, and confirm these interactions in vivo. We further show that a histone-binding–deficient DPF2 mutant fails to inhibit myeloid differentiation of human hematopoietic stem/progenitor cells (HSPCs), demonstrating the importance of this interaction for DPF2 function. Finally, we map the protein–protein interaction network of DPF2 in leukemia cells and find that DPF2 and RUNX1 share many common interaction partners, including components of the SWI/SNF chromatin-remodeling complex. Together, these data support the conclusion that the histone binding of DPF2 plays an important regulatory role in myeloid differentiation.
Results
Crystallization and Structure Determination of Human DPF2.
Based on secondary structure prediction and sequence conservation analyses, we designed a series of N-terminal truncation constructs of Homo sapiens DPF2 for recombinant protein expression in Escherichia coli (Table S1). We identified a fragment corresponding to the tandem PHD finger domain, encompassing residues 270–391, hereafter referred to as DPF2PHD. DPF2PHD was prone to oxidation as observed by mass spectrometry and required immediate use in any experiments following purification. Crystals of DPF2PHD were obtained in the tetragonal space group I422 and diffracted to 1.6-Å resolution. The structure was solved by single-wavelength anomalous dispersion using anomalous X-ray diffraction data, taking advantage of the four endogenously bound Zn2+ ions per DPF2PHD molecule. The final model contains residues 270–386, has excellent stereochemical properties, and was refined to Rfree and Rwork values of 18.7% and 14.6%, respectively. For further details of the data collection and refinement statistics, see Table S2.
Crystal Structure of DPF2PHD Reveals a Characteristic Tandem PHD Finger Domain.
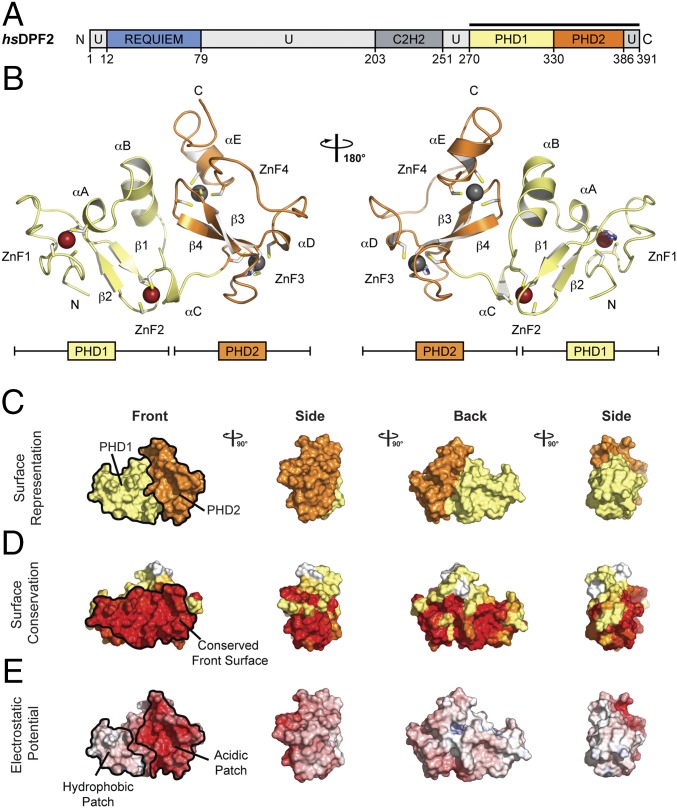
DPF2PHD contains two individual PHD finger motifs, the first comprised of residues 270–329 and the second of residues 330–386 (hereafter referred to as PHD1 and PHD2, respectively) (Fig. 1A). Each PHD finger contains the canonical architecture first described for the Williams syndrome transcription factor (Fig. 1 and Fig. S1) (15). This includes a two-stranded antiparallel β-sheet (β1–2 in PHD1 and β3–4 in PHD2) and two zinc atoms coordinated by a His-Cys3 motif followed by a Cys4 motif in a cross-brace topology (Cys273, Cys276, His303, Cys306 and Cys295, Cys298, Cys324, Cys327 for PHD1; and Cys330, Cys333, His353, Cys356 and Cys345, Cys348, Cys371, Cys374 for PHD2) (Fig. S1A). PHD1 and PHD2 adopt a similar conformation, with a RMSD of ∼1.9 Å over 45 Cα atoms (Fig. S1B). The two PHD fingers mainly differ in the presence of an additional short α-helix in PHD1 (αB, residues 311–319), resulting in three flanking helices in PHD1 (αA, αB, and αC) compared with two in PHD2 (αD and αE) (Fig. 1B).
Fig. 1.
Structure of the human DPF2 tandem PHD finger domain. (A) Domain structure. Light gray, predicted unstructured regions (U); blue, requiem domain (REQUIEM); dark gray, C2H2 zinc finger domain (C2H2); yellow, PHD finger 1 (PHD1); orange, PHD finger 2 (PHD2). The bar above the domain structure marks the crystallized fragment. (B) Structure of the DPF2 tandem PHD finger domain in ribbon representation, colored as in A. Zn2+ atoms are depicted as red (PHD1) and gray (PHD2) spheres. (C) Surface representation of DPF2PHD colored as in A (yellow, PHD1; orange, PHD2). (D) Surface representation of DPF2PHD colored according to a multispecies sequence alignment (Fig. S2). (E) Surface representation of DPF2PHD colored according to electrostatic potential. The electrostatic potential is plotted onto the surface and colored in a gradient from −10 kBT/e (red) to +10 kBT/e (blue).
Evolutionary Conserved DPF2PHD Front Surface Binds Histones in a Bipartite Manner.
A multispecies sequence alignment shows DPF2PHD to be highly evolutionary conserved from H. sapiens to Nematostella vectensis, with ∼50% sequence identity among all species tested (Fig. S2). Whereas the surface of DPF2PHD maintains the high level of conservation observed for the entire domain, the front surface appears to be the most invariant region of the protein (Fig. 1D). This surface contains a large hydrophobic patch on its N-terminal half and a large negatively charged patch on its C-terminal half, corresponding to PHD1 and PHD2, respectively (Fig. 1 C and E). Additionally, among human d4 family members, DPF2PHD shows an even greater level of conservation, with ∼80% sequence identity (Fig. S3A). Despite being closely related in sequence, a structural comparison of the DPF3bPHD solution NMR structure in complex with various histone peptides and our DPF2PHD crystal structure uncovered a movement in the relative position of the individual PHD domains, corresponding to a rotation by ∼25° (Fig. 2A) (16). Because of the high sequence identity between the two proteins, the absence of obvious differences in the PHD1–PHD2 interface, and the presence of a predicted hinge region between the two PHD finger domains, the observed conformational changes appear to be a result of ligand binding (17).
Fig. 2.
Structural comparison of the DPF2 and DPF3b tandem PHD finger domains. (A) A ribbon representation of DPF2PHD (Left, colored as in Fig. 1A), DPF3bPHD•H3K14Ac (Center, colored in gray, PDB ID code 2KWJ) (16), and a superposition of the two, based on alignment of PHD2s (Right). The hinge between PHD1 and PHD2 is indicated by an arrow. (B) Surface representations of the DPF3bPHD•H3K14Ac (Upper, PDB ID code 2KWJ) and the DPF3bPHD•H4K16Ac (Lower, PDB ID code 2KWN) solution NMR structures (16). Boxes show a cartoon representation of the indicated binding sites superposed with our DPF2PHD crystal structure. Residues constituting the bipartite histone-recognition motif are shown in stick representation.
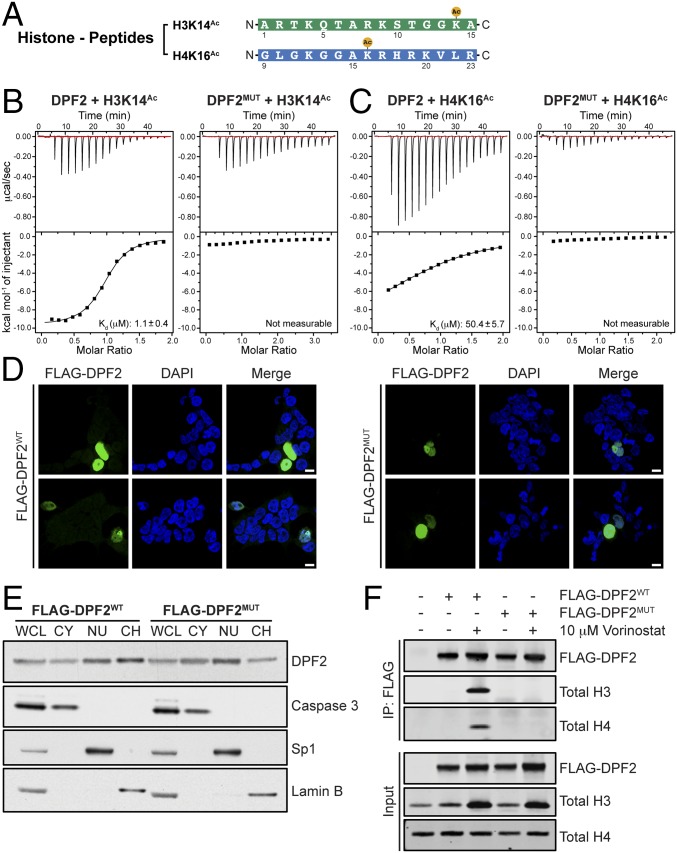
These results led us to explore the possibility that DPF2PHD binds histone tails, possibly with a preference for posttranslational modifications that differs from DPF3b. Indeed, isothermal titration calorimetry (ITC) measurements revealed that DPF2PHD binds H3 or H4 peptides with dissociation constants (Kd) of ∼1 and ∼50 μM, respectively, but independent of their acetylation status in physiological buffer conditions (Fig. 3 A–C, Table 1, and Fig. S4 A and B). As previously described for DPF3bPHD and consistent with the high conservation of d4 family tandem PHD finger domains, DPF2PHD preferentially binds to H3 histone peptides (Fig. 3 B and C and Table 1) (16). To investigate whether histone tail binding to DPF2 is governed by electrostatic interactions, we determined the dissociation constants in reduced salt conditions. In line with the electrostatic properties of the DPF2 peptide binding surface, we observed substantially tighter binding to all histone peptides in low-salt conditions (Table S3). In contrast to the histone tail interactions, we were unable to detect DPF2PHD binding to monomethyl- and asymmetric dimethyl-arginine RUNX1 peptides, even under no-salt conditions (Table 1 and Fig. S4C). These data suggest that the previously described methylation-dependent interaction between DPF2 and RUNX1 occurs indirectly (8).
Fig. 3.
DPF2 binds histone tails in vitro and in vivo. (A) Sequences and modifications of histone-tail peptides used in biochemical interaction studies. (B) Representative ITC data for interactions between an acetylated H3K14Ac (Left) and DPF2 wild-type (DPF2WT), and H3K14Ac and a DPF2 triple mutant (DPF2MUT; F275A, R300A, and D346A) (Right) at 100 mM salt concentration. Upper parts of each box show raw data and Lower parts show integrated heat changes corrected for heat from dilution. (C) Representative ITC data for interactions between an acetylated H4K16Ac (Left) and DPF2WT, and H4K16Ac, and DPF2MUT (Right) at 100 mM salt concentration. Upper parts of each box show raw data and Lower parts show integrated heat changes corrected for heat from dilution. (D) Immunofluorescence localization analysis of FLAG-DPF2. 293T cells were transfected with FLAG-DPF2 or FLAG-DPF2MUT and costained for anti-FLAG and DAPI to visualize nuclei. Upper and Lower panels represent different cells from the same experiment. (Scale bars, 10 μm.) (E) Subcellular DPF2 localization analysis. Whole-cell lysate (WCL), cytoplasmic (CY), nuclear (NU), and chromatin bound (CH) fractions of FLAG-DPF2– or FLAG-DPF2MUT–transfected 293T cells were isolated and resolved by SDS-PAGE and probed for antibodies against FLAG and controls for each fraction. (F) In vivo analysis of the DPF2-histone interaction. 293T cells were transfected with FLAG-DPF2 or FLAG-DPF2MUT constructs followed by a 24-h incubation with the histone deacetylase (HDAC) inhibitor Vorinostat or DMSO. FLAG constructs were immunoprecipitated using anti-FLAG magnetic beads and lysates were probed for total histone H3 and H4. Input (10%) is shown in the Lower panel and probed with antibodies to total H3, total H4, and FLAG.
Table 1.
Isothermal titratrion calorimetry measurements
| Protein | Peptide | Kd (μM) | ΔH (kcal/mol) | ΔS (cal/mol/deg) |
| DPF2WT | H3 | 1.0 ± 0.3 | −6.2 ± 0.2 | 6.8 |
| H3K14Ac | 1.1 ± 0.4 | −9.7 ± 0.2 | −5.3 | |
| H4 | 30.2 ± 13.2 | −6.3 ± 0.7 | −0.2 | |
| H4K16Ac | 50.4 ± 5.7 | −9.1 ± 0.5 | −10.5 | |
| DPF2MUT | H3 | Not measurable | Not measurable | Not measurable |
| H3K14Ac | Not measurable | Not measurable | Not measurable | |
| H4 | Not measurable | Not measurable | Not measurable | |
| H4K16Ac | Not measurable | Not measurable | Not measurable | |
| DPF3b* | H3 | 2.3 ± 0.7 | −4.4 ± 0.2 | 11.1 |
| H3K14Ac | 0.5 ± 0.1 | −6.6 ± 0.1 | 6.7 | |
| H4 | 28.8 ± 7.8 | −3.3 ± 0.4 | 9.7 | |
| H4K16Ac | 46.9 ± 10.0 | −4.6 ± 0.6 | 4.4 | |
| DPF2WT | RUNX1R223Me | Not measurable | Not measurable | Not measurable |
| RUNX1R223ADMA | Not measurable | Not measurable | Not measurable |
Data reported in Zeng et al. (16).
Together, our results establish that histone tail binding constitutes a common property shared among different members of the d4 family.
Identification of Essential Residues for DPF2 Histone Tail Binding.
Despite extensive efforts, in all DPF2PHD crystals we obtained, crystal packing was mediated by the same surface involved in histone tail binding, thus interfering with the determination of the complex structure. To identify individual DPF2 residues involved in histone binding, we therefore modeled the DPF2PHD interactions with H3K14Ac and H4K16Ac using the NMR structures of DPF3bPHD in complex with histone peptides as references (16). The resulting model suggests that histone peptides interact in a bipartite fashion with two distinct binding sites on the highly conserved front surface of DPF2 (Fig. 2B). The first binding site engages the acetyl-lysine residue with a hydrophobic pocket on the surface of PHD1, which is primarily composed of Phe275, Leu307, and Trp322. The second binding site is located on the negatively charged PHD2 surface and includes DPF2 residues Glu326 and Asp346, which make electrostatic interactions with Lys4 and Arg2 residues of the histone H3 tail, respectively.
Using the structural models of the DPF2PHD•H3K14Ac and DPF2PHD•H4K16Ac complexes as guides, we attempted to identify specific DPF2 residues critical for histone tail binding through site-directed mutagenesis followed by ITC. Alanine mutation of DPF2 residues Phe275 and Arg300 in the first binding site and Asp346 in the second binding site, hereafter referred to as DPF2MUT, dramatically reduced the interaction with histone tail peptides to levels that were not measurable by ITC in physiological buffer conditions (Fig. 3 B and C and Table 1). The observed loss of histone tail binding was not a result of improper DPF2MUT protein folding, as confirmed by circular dichroism (CD) spectroscopy (Fig. S5).
Together, these results pinpoint essential binding pockets in the DPF2PHD surface that directly mediate its interactions with histone tails.
DPF2 Binds Acetylated Histones H3 and H4 in Vivo.
We used our structure-guided mutational analysis as a guide to validate the interaction of DPF2 with H3 and H4 histones in vivo. First, we tested the cellular localization of FLAG-tagged DPF2 and DPF2MUT and confirmed that both proteins show identical localization patterns by cell fractionation or immunofluorescence microscopy and are predominantly localized to the nucleus (Fig. 3 D and E, Table S4). In line with our biochemical analysis, wild-type DPF2 but not DPF2MUT is able to bind H3 or H4 in vivo (Fig. 3F). However, this interaction was dependent on acetylation of H3 and H4, suggesting that additional determinants modulate the DPF2–histone interaction in an acetylation sensitive fashion in vivo. Together, these results demonstrate that DPF2 residues Phe275, Arg300, and Asp346 within the tandem PHD finger domain are essential for its recruitment to histones.
Repression of Myeloid Differentiation Is Dependent on DPF2 Chromatin Recruitment.
Having previously shown that DPF2 is recruited to the pre-miR-223 promoter region (8), we investigated whether this recruitment is mediated by the interaction of DPF2 with histones. To reduce the influence of endogenous DPF2, we overexpressed wild-type DPF2 and the histone-binding–deficient mutant DPF2MUT in MOLM-13 cells, which have low endogenous DPF2 levels compared with other AML cell lines (Fig. 4 A and B). Next, we performed ChIP assays using an anti-DPF2 antibody and primer pairs that amplify the RUNX1 binding site within the pre-miR-223 promoter region. Whereas wild-type DPF2 is primarily located at region 4 of the pre-miR-223 promoter region in MOLM-13 cells (Fig. 4 C–E), DPF2MUT displayed substantially reduced recruitment to this promoter region (P < 0.005), comparable to a non-DPF2 responsive promoter (Fig. 4D) (8). Consistently, there was no significant difference between DPF2WT and DPF2MUT at the nonresponsive promoter (P > 0.1) (Fig. 4D).
Fig. 4.
DPF2 recruitment to histones inhibits myeloid differentiation. (A) Expression of DPF2 in a panel of acute leukemia cell lines compared with normal CD34+ CB cell control. DPF2 was detected with an anti-DPF2 antibody at a concentration of 1:1,000 (sc-101943, Santa Cruz). (B) Overexpression of DPF2 wild-type (DPF2WT) or DPF2 triple mutant (DPF2MUT; F275A, R300A, and D346A) protein in MOLM-13 cells as assessed by Western blot analysis. (C) Occupancy of the miR-223 promoter regions in MOLM-13 cells, overexpressing DPF2WT or DPF2MUT protein. Blue and yellow bars represent regions 4 and 5 of the miR-223 promoter region, respectively. Data represent the mean ± SD from three independent experiments. (D) Occupancy of a non-DPF2 responsive promoter (albumin) region in MOLM-13 cells, overexpressing DPF2WT or DPF2MUT protein. Data represent the mean ± SD from three independent experiments. (E) Schematic representation of the miR-223 promoter region. Two black lines represent the RUNX1 binding site in the miR-223 promoter region. (F) Overexpression of DPF2WT or DPF2MUT protein in human CB CD34+ cells as assessed by Western blot analysis. (G) DPF2MUT blocks the repressive effect of DPF2 on myeloid differentiation of HSPCs. CD34+ cells expressing DPF2WT, DPF2MUT, or a vector control were cultured in myeloid differentiation-promoting cytokines for 7 d. Myeloid differentiation was determined by FACS analysis for expression of CD11b. A representative experiment is shown. (H) Data represent the mean ± SD of percent CD11b+ cells from three independent experiments. *P < 0.01 by Student’s t test.
Next, we examined the effect of DPF2 chromatin recruitment on the myeloid differentiation of human cord blood (CB) CD34+ cells by using lentiviral expression of either wild-type DPF2 or DPF2MUT (Fig. 4F). After 7 d in myeloid differentiation-promoting cultures, the proportion of CD11b+ cells originating from wild-type DPF2-overexpressing CD34+ cells was markedly reduced, from 87.5 to 44.1%, compared with control cells. In contrast, DPF2MUT overexpression did not reduce the number of CD11b+ cells (Fig. 4 G and H). Thus, DPF2 negatively regulates myeloid differentiation and this activity is dependent on its histone binding ability.
To further understand how DPF2 regulates HSPC biology, we performed RNA sequencing of the CD34+ CB cells transduced with either DPF2 knockdown (KD) or scrambled control shRNAs (Fig. S6A). Experiments were conducted in triplicate. We found 438 genes that were differentially expressed between the DPF2 KD and the control shRNA transfected cells, of which 133 and 305 were down- and up-regulated, respectively. Functional annotation of the RNA sequencing results by gene-set enrichment analysis (GSEA) using the Molecular Signature Database revealed that only a single gene set, Negative Regulation of Transcription-DNA Dependent, was enriched in DPF2 KD CD34+ cells. In contrast, three gene sets, Mitosis, Cell Cycle Regulation, and M-Phase, were down-regulated in the DPF2 KD CD34+ cells (Fig. S6 B and C). Additionally, we carried out an independent classification of the enriched molecular functions using the Gene Ontology Consortium (GO) database, which identified regulation of transcription, RNA processing, and cell-cycle regulation, consistent with the Molecular Signature Database findings (Tables S5–S7).
Thus, DPF2 plays an active role in the regulation of the transcriptional program alterations that are associated with myeloid differentiation.
Common Binding Partners Bridge DPF2 and Methylated RUNX1.
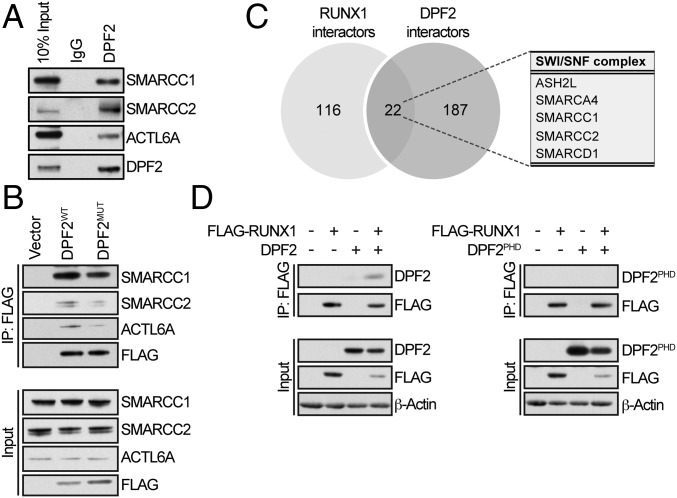
To decipher the regulatory role of DPF2 in myeloid differentiation, we analyzed its protein–protein interaction network, performing a comprehensive immunoprecipitation and mass spectrometry analysis of endogenous DPF2 from two different t(8;21) leukemia cell lines, SKNO-1 and Kasumi-1. This analysis revealed that DPF2 interacts with members of the SWI/SNF complex and several other epigenetic regulatory protein assemblies (Fig. S7 and Table S8). The interaction of endogenous DPF2 with the three SWI/SNF components BAF155/SMARCC1, BAF179/SMARCC2, and BAF53/ACTL6A, was confirmed by Western blot in Kasumi-1 cells (Fig. 5A). Next, we tested whether the interaction of DPF2 with the SWI/SNF complex members depends on the histone-binding ability of DPF2PHD. Indeed, when FLAG-tagged DPF2MUT was immunoprecipitated from 293T cells, we observed a marked reduction in its interaction with these three SWI/SNF components compared with wild-type DPF2 (Fig. 5B). Thus, the histone-binding ability of DPF2PHD is required for the interaction of DPF2 with the SWI/SNF chromatin-remodeling complex.
Fig. 5.
Protein–protein interaction network of DPF2. (A) Confirmation of SWI/SNF interacting proteins identified by the mass spectrometry analysis of proteins precipitated with endogenous DPF2. Whole-cell lysates from Kasumi-1 cells were immunoprecipitated with DPF2-conjugated beads and interacting proteins were analyzed by Western blot. (B) Confirmation of SWI/SNF interacting proteins identified by the mass spectrometry analysis using FLAG-DPF2 constructs. Wild-type FLAG-DPF2 (DPF2WT) and FLAG-DPF2MUT proteins were immunopreciptated from 293T cells using anti-FLAG magnetic beads and analyzed by Western blot. (C) Overlapping DPF2 and RUNX1 interaction partners. Comparison of mass spectrometry analysis of the proteins interacting with endogenous DPF2 and proteins interacting with the dimethyl-Arg223-RUNX1 peptide. DPF2 interacting proteins are normalized to an IgG control and methyl-RUNX1 interacting proteins are normalized to a nonmethylated peptide control. There were 22 protein interaction partners shared between the two datasets. (D) In vivo analysis of the DPF2–RUNX1 interaction. Coimmunoprecipitation of FLAG-RUNX1 with DPF2 and the isolated DPF2PHD domain. FLAG-RUNX1 and DPF2 or DPF2PHD were overexpressed in HEL cells by lentiviral transduction. Cells were lysed after 72 h, immunoprecipitated with anti-FLAG magnetic beads, and analyzed by Western blot. Input is 10% of the nuclear extract.
A comparison of previously determined RUNX1 interaction partners that specifically interact with a dimethylated Arg223 RUNX1 peptide and the newly established DPF2 interaction network identified 22 common interaction partners (Fig. 5C and Tables S8 and S9). Notably, these interaction partners include five members of the SWI/SNF complex. Although previous studies showed that DPF2 interacts with RUNX1 in a methylation-dependent manner (8), RUNX1 was not identified as a DPF2 interaction partner in our immunoprecipitation/mass spectrometry experiments. However, DPF2 coimmunoprecipitated with FLAG-tagged RUNX1 in HEL cells and this interaction was dependent on the presence of the N-terminal region of DPF2, which encompasses the requiem and central C2H2 zinc finger domains (Fig. 5D). Together, these data suggest that the DPF2–RUNX1 interaction is mediated by chromatin modification complexes.
Discussion
We have conducted a comprehensive biochemical, structural, and functional analysis of the d4 family member DPF2, identifying its important role in blocking myeloid differentiation. We determined the crystal structure of the DPF2 tandem PHD finger domain and demonstrated its binding to histone tails. The histone-binding ability of DPF2 is essential for its in vivo regulatory role in the myeloid differentiation of HSPCs, and for its recruitment to specific chromatin sites, including the pre-miR-223 promoter.
The recognition of histone acetylation and other epigenetic modifications by reader domains, including PHD domains, is a major mechanism of epigenetic regulation and a source of dysregulation in hematopoietic malignancies. For example, chromosomal translocations that fuse the NUP98 gene with genes that encode PHD domain-containing proteins, such as JARID1A or PHF23, have been identified in patients with AML (18–21). In both cases, the PHD domains of JARID1A or PHF23, which function as methyl-lysine recognition domains, are conserved in the fusion protein and shown to be critical for Hox gene activation and leukemia induction in vivo (22).
The tandem PHD domains of the MOZ and MORF histone acetyltransferases represent another example of histone reader domains that regulate hematopoiesis. The MOZ and MORF PHD domains recognize acetylated, crotonylated, and butyrylated H3K14, and acetylated H3K9 in vitro (23). Both acetyltransferase genes are involved in chromosomal translocations in leukemia that result in their fusion to CBP, EP300, or TIF2 (24–26).
Collectively, these studies indicate that loss of the normal recognition of histone modifications can have widespread effects on myeloid transcription and HSPC differentiation and self-renewal. The findings also suggest that targeting oncogenic epigenetic readers, such as PHD domains, could be a therapeutic strategy for treating myeloid malignancies. In one example of such an approach, chemical inhibitors that target the bromodomain of the BET family member, BRD4, have shown efficacy in treating mixed lineage leukemia gene-rearranged leukemia, as well as multiple myeloma and acute lymphoblastic leukemia (ALL) (27–29).
DPF2 is mutated in several human cancers including AML, lymphoma, and ALL (30), with mutation hot spots occurring within the N-terminal region as well as the PHD domains. Several of these N-terminal residues are predicted to be sites of posttranslational modifications, including multiple arginines that represent potential targets of the arginine methyltransferase enzymes PRMT1, PRMT4, and PRMT5 (31, 32). We, and others, have shown that myeloid differentiation is regulated by specific arginine methylation events, and PRMT4 modifies several proteins in the larger DPF2 interactome, including RUNX1 and the SWI/SNF subunit BAF155 (8, 33). Thus, the recognition of histone modifications by the PHD domains of DPF2 potentially integrates multiple epigenetic inputs and likely enables the targeting of DPF2 to specific chromatin sites.
Our data highlight the important role of DPF2 in AML; DPF2 is a potential new therapeutic target that can possibly trigger the myeloid differentiation or apoptosis of leukemia cells (Fig. 6). Although DPF2 is ubiquitously expressed, we show that it is overexpressed in AML cell lines, and in AML patient samples, where it represents a poor prognosis indicator (Fig. S8). Overexpression or mutation of DPF2 is expected to alter its histone-binding properties. However, further work is needed to elucidate the mechanism by which histone modifications modulate DPF2’s role in controlling myeloid differentiation, especially in light of studies implicating several histone acetyltransferases in hematopoietic differentiation and self-renewal. Nonetheless, the results presented here provide a solid foundation for further studies aiming to trigger the differentiation of AML cells through targeted manipulation of DPF2.
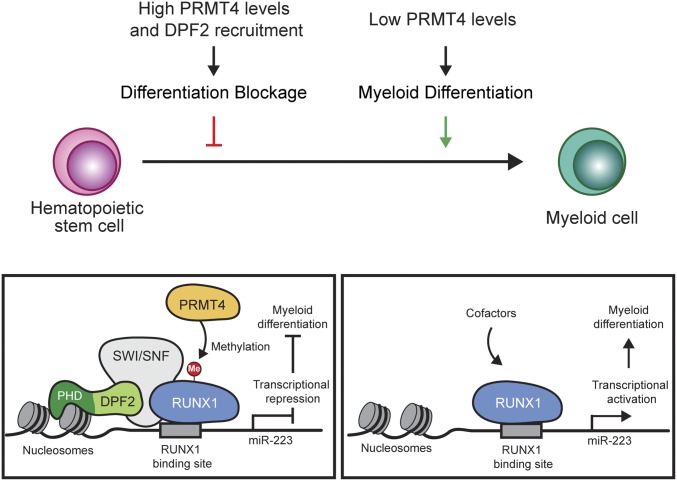
Fig. 6.
Model for the inhibitory role of DPF2 in myeloid differentiation. Histone-binding of DPF2 along with PRMT4 mediated methylation of RUNX1 residue Arg223 trigger the formation of a DPF2 containing repressive complex that is recruited to the miR-223 promoter. The interaction between DPF2 and methylated RUNX1 is either mediated by the DPF2 N-terminal region or bridged by components of a larger complex, such as the SWI/SNF chromatin remodeling complex. Formation of the repressive DPF2 containing RUNX1 complex on the miR-223 promoter region results in the repression of miR-223 transcription and a subsequent block of myeloid differentiation. As hematopoietic stem cells differentiate, the reduction in PRMT4 expression leads to disassembly and replacement of the repressive transcriptional complex by an activating complex that up-regulates transcription of miR-223, among other genes.
Methods
Details for X-ray diffraction data collection and structure refinement are described in SI Methods and Table S2. Further details of molecular cloning, protein expression, purification, crystallization, CD spectroscopy, ITC measurements, cell culture, cell fractionation, mass spectrometry, immunofluorescence microscopy, immunoprecipitation, generation of lentivirus, immunoblot analysis, flow cytometry, RNA sequencing, and ChIP assays are described in SI Methods and Tables S1 and S3–S9.
Supplementary Material
Acknowledgments
We thank Alina Patke for critical reading of the manuscript; David King for mass spectrometry analysis; and Jens Kaiser and the scientific staff of Stanford Synchrotron Radiation Lightsource (SSRL) Beamline 12-2 for their support with X-ray diffraction measurements. The operations at SSRL are supported by the Department of Energy and the National Institutes of Health. We acknowledge the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program for their support of the Molecular Observatory at the California Institute of Technology and the Beckmann Institute Laser Resource Center for access to their Circular Dichroism Spectrometer. The DPF2 interaction analysis was carried out at the Taplin Mass Spectrometry Facility at Harvard Medical School. F.M.H. was supported by a PhD fellowship of the Boehringer Ingelheim Fonds. A.M.D. was supported by a National Institutes of Health Research Service Award 5 T32 GM07616. S.D.N. was supported by National Institutes of Health, National Cancer Institute Grant R01 CA166835. A.H. is a Faculty Scholar of the Howard Hughes Medical Institute, an Investigator of the Heritage Medical Research Institute, and was supported by the Albert Wyrick V Scholar Award of the V Foundation for Cancer Research, a Kimmel Scholar Award of the Sidney Kimmel Foundation for Cancer Research, and a Teacher-Scholar Award of the Camille and Henry Dreyfus Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5VDC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700328114/-/DCSupplemental.
References
- 1.Gabig TG, Mantel PL, Rosli R, Crean CD. Requiem: A novel zinc finger gene essential for apoptosis in myeloid cells. J Biol Chem. 1994;269:29515–29519. [PubMed] [Google Scholar]
- 2.Chestkov AV, Baka ID, Kost MV, Georgiev GP, Buchman VL. The d4 gene family in the human genome. Genomics. 1996;36:174–177. doi: 10.1006/geno.1996.0440. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez R, Zhou MM. The PHD finger: A versatile epigenome reader. Trends Biochem Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tando T, et al. Requiem protein links RelB/p52 and the Brm-type SWI/SNF complex in a noncanonical NF-kappaB pathway. J Biol Chem. 2010;285:21951–21960. doi: 10.1074/jbc.M109.087783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim MY, et al. Staufen1-mediated mRNA decay induces Requiem mRNA decay through binding of Staufen1 to the Requiem 3'UTR. Nucleic Acids Res. 2014;42:6999–7011. doi: 10.1093/nar/gku388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulikova DA, Mertsalov IB, Simonova OB. [D4 family genes in vertebrates: Genomic organization and expression] Ontogenez. 2013;44:3–9. Russian. doi: 10.7868/s0475145013010047. [DOI] [PubMed] [Google Scholar]
- 7.Gabig TG, et al. Expression and chromosomal localization of the Requiem gene. Mamm Genome. 1998;9:660–665. doi: 10.1007/s003359900840. [DOI] [PubMed] [Google Scholar]
- 8.Vu LP, et al. PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Reports. 2013;5:1625–1638. doi: 10.1016/j.celrep.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theodorou M, et al. Identification of a STAT5 target gene, Dpf3, provides novel insights in chronic lymphocytic leukemia. PLoS One. 2013;8:e76155. doi: 10.1371/journal.pone.0076155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loi S, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 11.Lo KC, et al. Candidate glioblastoma development gene identification using concordance between copy number abnormalities and gene expression level changes. Genes Chromosomes Cancer. 2007;46:875–894. doi: 10.1002/gcc.20474. [DOI] [PubMed] [Google Scholar]
- 12.Hoyal CR, et al. Genetic polymorphisms in DPF3 associated with risk of breast cancer and lymph node metastases. J Carcinog. 2005;4:13. doi: 10.1186/1477-3163-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi YP, Kang S, Hong S, Xie X, Cho NH. Proteomic analysis of progressive factors in uterine cervical cancer. Proteomics. 2005;5:1481–1493. doi: 10.1002/pmic.200401021. [DOI] [PubMed] [Google Scholar]
- 14.Müller AM, Duque J, Shizuru JA, Lübbert M. Complementing mutations in core binding factor leukemias: From mouse models to clinical applications. Oncogene. 2008;27:5759–5773. doi: 10.1038/onc.2008.196. [DOI] [PubMed] [Google Scholar]
- 15.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J Mol Biol. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 16.Zeng L, et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: Automated prediction of hinges in protein structures. Proteins. 2008;70:1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- 18.van Zutven LJ, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45:437–446. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- 19.Reader JC, Meekins JS, Gojo I, Ning Y. A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17)(p15;p13) in acute myeloid leukemia. Leukemia. 2007;21:842–844. doi: 10.1038/sj.leu.2404579. [DOI] [PubMed] [Google Scholar]
- 20.Gough SM, et al. NUP98-PHF23 is a chromatin-modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD histone reader function. Cancer Discov. 2014;4:564–577. doi: 10.1158/2159-8290.CD-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Rooij JD, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013;27:2280–2288. doi: 10.1038/leu.2013.87. [DOI] [PubMed] [Google Scholar]
- 22.Wang GG, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong X, et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat Chem Biol. 2016;12:1111–1118. doi: 10.1038/nchembio.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu Y, et al. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 2012;26:1376–1391. doi: 10.1101/gad.188359.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali M, et al. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J Mol Biol. 2012;424:328–338. doi: 10.1016/j.jmb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein BJ, Lalonde ME, Côté J, Yang XJ, Kutateladze TG. Crosstalk between epigenetic readers regulates the MOZ/MORF HAT complexes. Epigenetics. 2014;9:186–193. doi: 10.4161/epi.26792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ott CJ, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen SC, et al. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci Signal. 2016;9:rs9. doi: 10.1126/scisignal.aaf7329. [DOI] [PubMed] [Google Scholar]
- 32.Geoghegan V, Guo A, Trudgian D, Thomas B, Acuto O. Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat Commun. 2015;6:6758. doi: 10.1038/ncomms7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014;25:21–36. doi: 10.1016/j.ccr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.