Significance
Chronic neuroinflammation and oxidative stress are likely complicit in driving disease progression in Huntington's disease (HD). Here, we describe the mechanism of action of a unique chemical scaffold that is highly selective for activation of NRF2, the master transcriptional regulator of cellular antiinflammatory and antioxidant defense genes. The use of this scaffold revealed that NRF2 activation responses were muted in HD patient-derived neural stem cells, suggesting increased susceptibility of this critical renewable cell population to oxidative stress in HD brain. However, pharmacological activation of NRF2 was able to repress inflammatory responses in mouse microglia and astrocytes, the principal cellular mediators of neuroinflammation, and in blood monocytes from HD patients. Our results suggest multiple protective benefits of NRF2 activation for HD patients.
Keywords: Huntington’s disease, KEAP1/NRF2/ARE signaling, NRF2 inducer, antiinflammatory responses, human neural stem cells
Abstract
The activity of the transcription factor nuclear factor-erythroid 2 p45-derived factor 2 (NRF2) is orchestrated and amplified through enhanced transcription of antioxidant and antiinflammatory target genes. The present study has characterized a triazole-containing inducer of NRF2 and elucidated the mechanism by which this molecule activates NRF2 signaling. In a highly selective manner, the compound covalently modifies a critical stress-sensor cysteine (C151) of the E3 ligase substrate adaptor protein Kelch-like ECH-associated protein 1 (KEAP1), the primary negative regulator of NRF2. We further used this inducer to probe the functional consequences of selective activation of NRF2 signaling in Huntington's disease (HD) mouse and human model systems. Surprisingly, we discovered a muted NRF2 activation response in human HD neural stem cells, which was restored by genetic correction of the disease-causing mutation. In contrast, selective activation of NRF2 signaling potently repressed the release of the proinflammatory cytokine IL-6 in primary mouse HD and WT microglia and astrocytes. Moreover, in primary monocytes from HD patients and healthy subjects, NRF2 induction repressed expression of the proinflammatory cytokines IL-1, IL-6, IL-8, and TNFα. Together, our results demonstrate a multifaceted protective potential of NRF2 signaling in key cell types relevant to HD pathology.
Nuclear factor-erythroid 2 p45-related factor 2 (NRF2)-mediated signaling is a major endogenous cellular defense mechanism against oxidative and xenobiotic stress (1–3). The transcription factor NRF2 is tightly regulated in the cytoplasm by Kelch-like ECH-associated protein 1 (KEAP1), acting as an adaptor between NRF2 and CUL3-based E3 ubiquitin ligase (1, 4–6). In the absence of stress, KEAP1 efficiently mediates the polyubiquitination of NRF2, leading to its degradation by the ubiquitin–proteasome degradation system (UPS) (7–10). During stress conditions, oxidants or electrophiles chemically modify KEAP1 sensor cysteines, causing a conformational change of the protein KEAP1/NRF2/CUL3 complex, and impair NRF2 degradation (11–14). De novo-synthesized NRF2 accumulates to levels overcoming the endogenous sequestration capacity of the remaining free KEAP1, at which point excess NRF2 translocates to the nucleus, binds to antioxidant response elements (AREs) in the promoter/enhancer regions of target genes, and broadly activates transcriptional responses (2, 12, 15). In this manner, NRF2 coordinates the transcriptional regulation of genes encoding phase II enzymes: NAD(P)H:quinone oxidoreductase 1 (NQO1); heme oxygenase (decycling) 1 (HO-1); catalytic (GCLC) and regulatory (GCLM) subunits of glutamate-cysteine ligase, and ∼100 additional downstream targets (2, 16). Additional robust antiinflammatory effects occur through NRF2 activation through molecular mechanisms that are just emerging (17). Nevertheless, there is already a strong body of empirical evidence in vitro and in vivo linking NRF2 activation and suppression of inflammation (18–20).
Oxidative stress and neuroinflammation are among the common pathogenic mechanisms implicated in neurodegenerative disorders, including Huntington's disease (HD) (21, 22). HD, an autosomal-dominant and highly penetrant neurodegenerative disorder, results from the pathological expansion (>39) of a polymorphic trinucleotide repeat sequence (CAG)n within the gene encoding the large, highly conserved protein, huntingtin (HTT) (23). A harmful role for oxidative stress has been described in both HD patients as well as in several experimental models (21, 24), which is potentially due to inherent sensitivity of neurons to damage caused by exposure to an excess of reactive oxygen species (ROS) (25–28). Some antioxidant proteins, such as glutathione peroxidases, catalase, and superoxide dismutase (SOD), but not all canonical ARE gene products, are increased in human HD brain (29), suggesting that NRF2 signaling is not fully engaged as a protective mechanism or may be partially repressed. Pharmacological stimulation of NRF2 in HD mouse models is efficacious and associated with increased expression of antioxidant proteins and reduction of ROS levels in brain (30, 31), which further suggests that protective NRF2 signaling is not fully activated and/or muted.
Sustained brain inflammation contributes significantly to the pathogenesis of age-dependent neurodegenerative disorders (32, 33). The release of proinflammatory cytokines, associated with the harmful effects of activated microglia in brain, creates a cytotoxic environment for neighboring neurons (34–36). In HD, elevated expression of several key inflammatory mediators is observed in blood, striatum, cortex, and cerebellum from postmortem patient tissues (37, 38). Neuroinflammatory responses appear relatively early in HD disease progression, suggesting that mutant HTT promotes the abnormal release of cytokines by activated microglia (39–41). The continual release of proinflammatory mediators by microglia may perpetuate a feedforward cycle, recruiting and activating additional microglia, promoting their proliferation, and leading to additional release of proinflammatory factors with ever-increasing exacerbation of disease pathogenesis (22, 40, 42–45).
It remains uncertain whether NRF2 signaling in human HD is intact or disease-modified and/or differentially impacted in different cell types. Our recent work identified a unique structural scaffold of triazole-containing small molecules that potently induces expression of canonical ARE genes (NQO1, GCLM, GCLC, HMOX1/HO-1) in primary mouse cells and causes a pronounced reduction of ROS (46). Here, we elucidated the molecular mechanism by which the most potent lead compound developed from this scaffold induces NRF2 activation, and used this highly selective probe to examine specific activation responses and potential benefits of NRF2 signaling in mouse and human HD models.
Results
MIND4 Is Neuroprotective in MPTP-Toxicity Mouse Model.
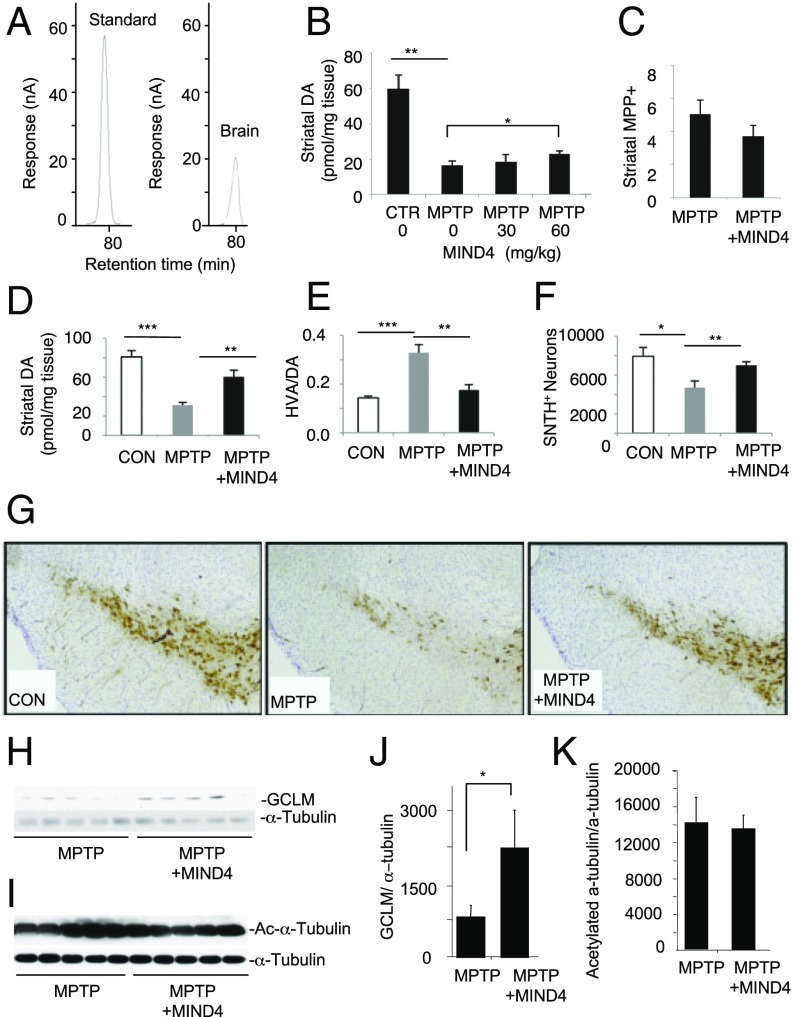
We first sought to evaluate the in vivo neuroprotective potential of the new structural scaffold of NRF2-inducing compounds (46). We tested and established the brain permeability of 5-nitro-8-{[5-(phenoxymethyl)-4-phenyl-4H-1,2,4-triazol-3-yl]thio}quinoline (MIND4), but we were unable to detect the presence in brain of the less hydrophobic analog 5-nitro-2-{[5-(phenoxymethyl)-4-phenyl-4H-1,2,4-triazol-3-yl]thio}pyridine (MIND4-17) (Figs. 1A and 2A, and Fig. S1). Studies published to date have shown that NRF2 activation is protective and associated with a reduction in oxidative damage in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) mouse model (47, 48), which was selected for a short-term efficacy trial with MIND4. In a pilot experiment, MIND4 was tested at two different doses in MPTP-treated mice using a subacute treatment paradigm (49). The higher dose of 60 mg/kg attenuated the loss of dopamine and did not change MPTP metabolism significantly (Fig. 1 B and C). Using the same 60 mg/kg dose and subacute regimen of treatment, we found that MIND4 was protective for dopaminergic neurons in MPTP mice, as demonstrated by an increased level of residual dopamine and a decreased dopamine turnover rate [homovanillic (HVA)/dopamine (DA)] in the drug treatment group compared with the vehicle-treated control (Fig. 1 D–F). Furthermore, stereological analysis demonstrated that the MPTP/MIND4-treated mice had more remaining DA neurons in the substantia nigra than the MPTP/vehicle-treated group (Fig. 1G). Analysis of pharmacodynamic changes showed that treatment at the selected dose was associated with an increase of antioxidant NRF2-responsive GCLM protein levels, albeit not uniform among the treated animals (n = 5) (Fig. 1 H and J). The levels of acetylated α-tubulin, a prototypic substrate of SIRT2 deacetylase, identified as a MIND4 target in vitro, remained largely unchanged (Fig. 1 I and K). The results suggest that NRF2 induction contributes to MIND4 protective activity in MPTP-treated mice.
Fig. 1.
MIND4 is neuroprotective in MPTP-toxicity mouse model. (A) Brain permeability of MIND4, administered by i.p. injections to WT mice. Chromatograms of MIND4 standard (1 µg/mL) and MIND4-treated brain are shown. HPLC analysis detected MIND4 at 0.45 μg/mL concentration in mouse cortical extracts, providing preliminary evidence for compound brain permeability. (B) MIND4 effective dose was established in mice using subacute MPTP treatment paradigm: MPTP-untreated control (CTR) and MPTP-treated mice were administered by i.p. vehicle alone or MIND4 at 30 and 60 mg/kg dose. Vehicle or MIND4 were administered 10 min before and 50 min after each MPTP injection. Levels of dopamine (DA) were determined by HPLC-ECD. Statistical significant increase of DA was observed in mice treated with MIND4 at 60 mg/kg. n = 5–8. *P < 0.05; **P < 0.01. (C) MIND4 at 60 mg/kg before and after administration of 20 mg/kg MPTP does not change the metabolism of neurotoxin in the striatum. MPP+ was examined by HPLC 90 min after the second MIND4 administration. (D–G) Degeneration of dopaminergic neurons was induced in mice using subacute MPTP-treatment paradigm (20 mg/kg i.p. injection once daily for 4 d) and MIND4 administered by i.p. at established effective dose 60 mg/kg) 10 min before and 50 min after each MPTP injection. (D and E) MIND4 attenuated MPTP-induced dopamine (DA) depletion (D) and high DA turnover rate (E) (n = 5–8). (F) Stereological quantification of TH neurons (n = 5–8) (*P < 0.05; **P < 0.01; ***P < 0.001). (G) MIND4 treatment preserves SN dopaminergic neurons in MPTP mice. (H–K) Analysis of pharmacodynamic markers in striatal samples (D–G): NRF2 activation (increase GCLM levels) and SIRT2 inhibition (increase α-tubulin acetylation) in MPTP-challenged brains from MIND4-treated and untreated mice. Levels of GCLM (H) and acetylated α-tubulin (I) and α-tubulin (loading control) were detected by immunoblotting. (J) Densitometry analysis of H. Mean of GCLM signal is statistically significantly greater in MIND4-treated then in untreated samples (n = 5). *P < 0.5. (K) Densitometry analysis of I. Mean of acetylated α-tubulin shows no statistically significant difference in cohorts of MIND4-treated vs. untreated cohorts. Levels of total α-tubuin were used for normalization in J and K.
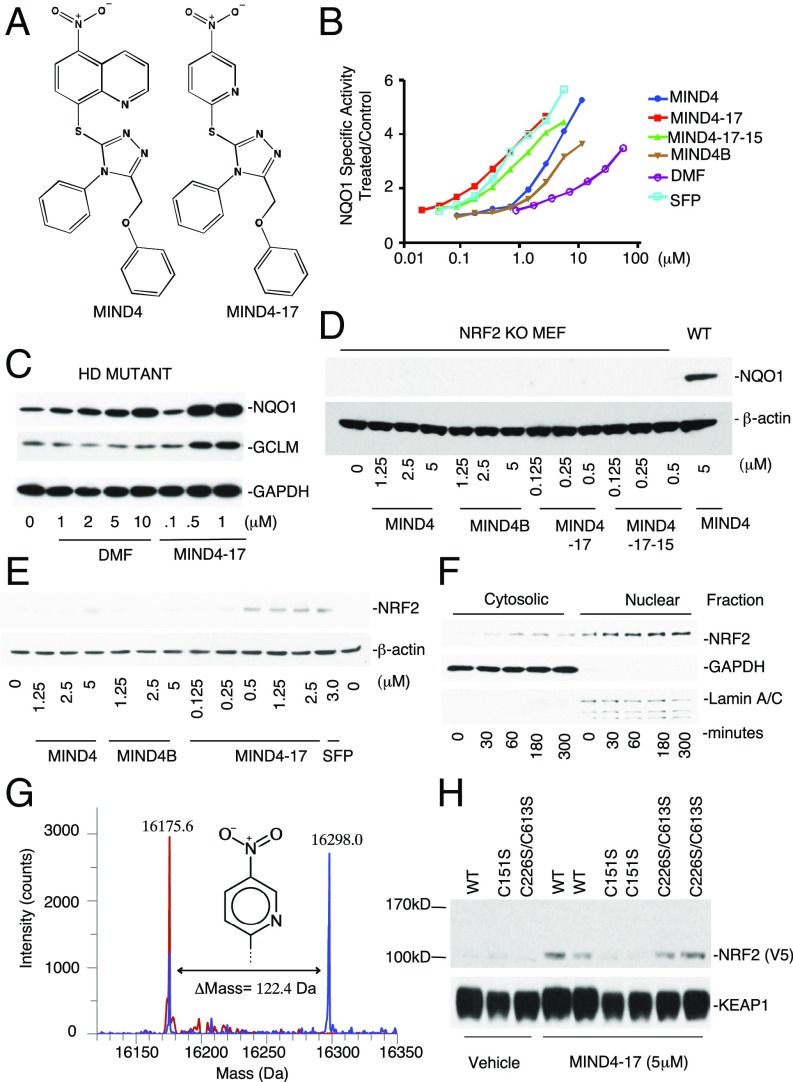
Fig. 2.
Characterization of NRF2 inducer MIND4-17. (A) Structures of parent MIND4 and lead-inducer MIND4-17. (B) Concentration-dependent activity test of MIND4-17, structural analogs MIND4-17-15, MIND4, MIND4B, and control NRF2 inducers SFP and DMF in quantitative NQO1 inducer bioassay. The SD in each data point (n = 8) is <5%. (C–F and H) Levels of NRF2 and NRF2-responsive NQO1 and GCLM proteins, and GADPH, β-actin, LAMIN used as loading controls, were detected by immunoblotting. (C) Induction of NRF2-responsive NQO1 and GCLM proteins in MIND4- and DMF-treated mutant HD ST14A cells. (D) Compound activity test shows no induction of NQO1 in NRF2-KO MEFs treated with MIND4-17 and its analogs. Induced NQO1 level in WT MEFs is shown for comparison. (E) Concentration-dependent stabilization of NRF2 protein in WT MEFs treated with MIND4-17, MIND4, MIND4B, or SFP. (F) Time-dependent accumulation of NRF2 in cytoplasmic and nuclear fractions from WT MEFs treated with 0.5 μM MIND4-17. Cell lysis and biochemical fractionation were performed at the indicated treatment times. (G) MIND4-17 covalently modifies a single cysteine-151 in the BTB domain of KEAP1. Overlay of the deconvoluted intact protein mass spectra obtained from BTB (48-190/S182A) (theoretical mass, 16,175.6 Da) in buffer (red) vs. treated with MIND4-17 (blue). (H) MIND4-17 treatment stabilizes NRF2 in COS1 cells coexpressing NRF2-V5 and KEAP1 WT or double-mutant C226S/C613S KEAP1, but not a single-mutant C151 KEAP1.
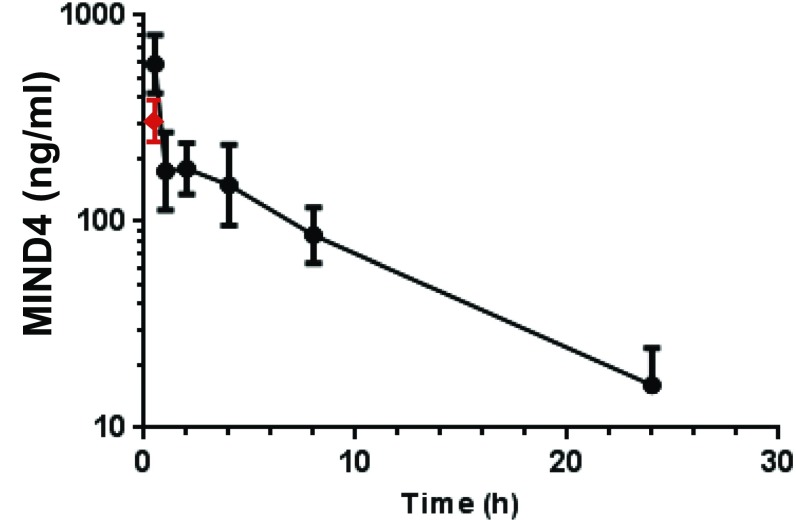
Fig. S1.
MIND4 pharmacokinetics in serum (filled circles) were examined by HPLC analysis in WT male C57BL/6 mice (n = 3) at 0.5, 1.0, 2, 4, 8, and 24 h at compound treatment dose of 50 mg/kg; blood Cmax = 477 ng/mL at 0.5-h time point. Brain levels (red triangle) were analyzed in cortices from mice (n = 3) killed at 0.5 h after i.p. injection; Cbrain = 120 ng/mL and respective brain/serum ratio, 0.25.
Characterization of Triazole-Containing Inducers of NRF2 Activation.
To elucidate the mechanism of NRF2 activation, first we compared the potencies of MIND4, MIND4-17, and a series of structural triazole-containing analogs in a quantitative NQO1 inducer bioassay in murine Hepa1c1c7 cells (Fig. 2 A and B, and Fig. S2A) (50, 51). The concentration that doubles the specific activity (CD value) for NQO1 was used as a measure of inducer potency and compound ranking. Within this lead series, MIND4-17 was confirmed to be the inducer of highest potency (CD = 0.15 μM), comparable to the naturally occurring inducer sulforaphane (SFP) (CD = 0.18 μM) and significantly more potent than the clinically approved NRF2 activator dimethyl fumarate (DMF) (CD = 9 μM) (Fig. 2 A and B) (52–54). A comparative analysis of MIND4-17 and DMF on induction of NRF2-responsive NQO1 and GCLM in mutant HD rat embryonic striatal cells ST14A treated with compounds for 24 h clearly showed higher potency of activation mediated by MIND4-17 (Fig. 2C).
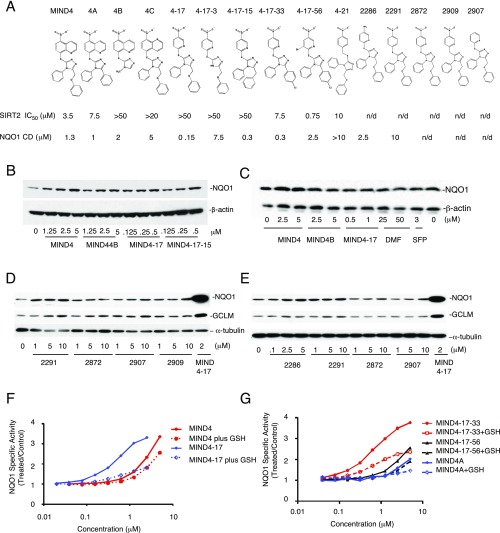
Fig. S2.
Characterization of MIND4-17 and its structural analogs. (A) Structures of MIND4-17 and lead series of structural analogs. Established NQO1 CD and SIRT2 IC50 are shown. (B and C) Dose–response activity tests of MIND4-17 and MIND4-17 analogs in WT mouse embryonic fibroblasts (MEFs): (B) WT and (C) KEAP1-null. Cells were treated with compounds for 24 h. Levels of induced NQO1 protein and β-actin (loading control) were detected by immunoblotting. (D and E) Dose–response activity tests of MIND4-17 analogs in rat embryonic ST14A cells. Levels of NQO1, GCLM, and loading control α-tubulin were detected by immunoblotting. Cells were treated with compounds for 24 h. (F and G) Negative effects of GSH on the NQO1 inducer activity of (F) MIND4-17 and MIND4 and (G) MIND4-17-33 and MIND4-17-56 analogs in Hepa1c1c7 cells. Each compound was incubated in cell culture medium in the presence or absence of 1 mM GSH at 37 °C for 30 min before administering to cells. After a 48-h exposure, the NQO1 enzyme activity was determined in cell lysates. Results are shown as average values of eight replicate wells. The SD in each case was less than 5%.
As the parent compound MIND4 was initially identified as a SIRT2 inhibitor, activities for this lead series were also tested in a biochemical SIRT2 deacetylation assay (55). In agreement with our previous data (46), there was no correlation between the potencies of SIRT2 inhibition and induction of NQO1; MIND4-17 showed no detectable SIRT2 inhibition activity (Fig. S2A).
Next, we established that induction of NQO1 by MIND4-17 and other structural analogs is NRF2 dependent. NQO1 expression was assessed over a series of concentrations in WT, NRF2-null, or KEAP1-null mouse embryonic fibroblasts (MEFs) after 24-h treatment (56–59). In WT MEFs, MIND4-17 exhibited the highest potency, whereas other MIND4 analogs increased the levels of NQO1 protein to varying degrees (Fig. S2B). In contrast, the effects of MIND4-17 and other MIND4 analogs on induction of NQO1 expression were absent in NRF2-null cells (Fig. 2D). In KEAP1-null MEFs, which have constitutively high transcription of NRF2-responsive genes (57, 59), treatment with MIND4-17 and other MIND4 analogs did not up-regulate NQO1 expression further (Fig. S2C). These results demonstrated that induction of NQO1 expression by MIND4-17 and other structural analogs is NRF2 dependent and KEAP1 dependent.
Stabilization of NRF2 is an essential step of activation of the pathway, which has been well-defined for KEAP1-modifying inducers of NRF2 signaling such as SFP (7, 8, 52, 58). Thus, we tested whether the stabilization of NRF2 in cells treated with MIND4-17 and its structural analogs preceded the induction of NQO1. Accumulation of total NRF2 protein was readily detectable in WT MEFs after 5-h exposure to SFP and MIND4-17 and, to a lesser extent, to the lower potency inducers MIND4 and MIND4B (Fig. 2E). In cells treated with MIND4-17, the induction of NQO1 was preceded by nuclear accumulation of NRF2 as early as 30 min after cell exposure to MIND4-17 (0.5 μM) and remained elevated for at least 5 h (Fig. 2F).
NRF2-Inducing Mechanism of MIND4-17.
The chemical requirements for the NRF2-inducing activity of MIND4-17 became evident from a structure–activity relationship (SAR) study (Fig. S3; see SI Materials and Methods for detailed compound synthesis schemes) (60, 61). Analogs of MIND4-17 with O or C substitutions of the S atom were completely inactive, illuminating the essential requirements for the sulfur for inducer activity (Fig. S2 D and E). The dependence of inducer activity on an electron-withdrawing group and electron-deficient aromatic system was also apparent (Fig. S2 D and E). Together, these results suggested that the S group of MIND4-17 could be chemically reactive with cysteine nucleophiles, and that this property is essential for NRF2 activation.
Fig. S3.
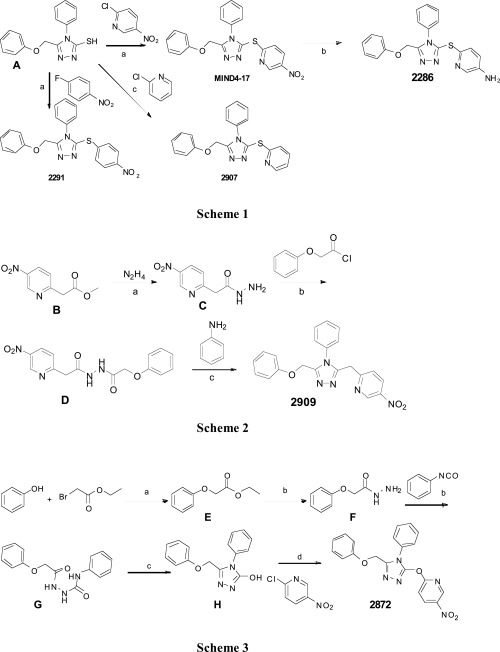
Design and synthesis schemes of MIND4-17 analogs 2872, 2286, 2291, 2907, and 2909 for SAR study. Scheme 1: (a) K2CO3, DMF, 100 °C, 1 h; (b) SnCl2⋅2H2O, EtOH, reflux; (c) Pd2(dba)3, Xanthphos, Hunig's base, Dioxane reflux. Scheme 2: (a) EtOH, reflux; (b) DCM, Et3N, RT; (c) PCl3, 1,2-dichlorobenzene, 190 °C. Scheme 3: (a) acetone, K2CO3 reflux; (b) EtOH, reflux; (c) 2 M KOH reflux; (d) K2CO3, DMF, 100 °C. (A) A starting compound for synthesis of MIND4-17 and its analogs, 5-(phenoxymethyl)-4-phenyl-4H-1,2,4-triazole-3-thiol. (B–H) Intermediates of organic reactions: (B) methyl 2-(5-nitropyridin-2-yl)acetate; (C) 2-(5-nitropyridin-2-yl)acetohydrazide; (D) 2-(5-nitropyridin-2-yl)-N′-(2-phenoxyacetyl)acetohydrazide; (E) ethyl 2-phenoxyacetate; (F) 2-phenoxyacetohydrazide; (G) 2-(2-phenoxyacetyl)-N-phenylhydrazine-1-carboxamide; and (H) 5-(phenoxymethyl)-4-phenyl-4H-1,2,4-triazol-3-ol.
To test this hypothesis, MIND4-17 and other triazole-containing analogs were preincubated with reduced glutathione (GSH) followed by incubation with intact cells and measurement of NRF2 activation responses. Such pretreatment negatively affected the ability of MIND4-17 to induce the NRF2 response (Fig. S2 F and G). Furthermore, preincubation with GSH inhibited NRF2 inducer activity in direct correlation with NRF2 activation potency; that is, MIND4-17 was the most strongly reactive and thus the most profoundly negatively affected. These results indicated that the reactivity of the S group is essential for MIND4-17–mediated activation of NRF2 and involves reaction with cysteine nucleophile(s) of putative target(s).
It is well established that reactive cysteine residues of KEAP1 serve as sensors for electrophiles and oxidants, and that chemical modifications of these cysteines disable the KEAP1/CUL3 complex, ultimately leading to NRF2 activation (7, 11, 12, 53, 62). Located within the Broad complex, Tramtrack, and Bric-à-Brac domain (BTB domain), cysteine-151 (C151) is one of the major sensors in KEAP1, the modification of which is sufficient for robust activation of NRF2 (7, 12, 53, 63–65). It was therefore conceivable that C151 of KEAP1 is targeted by MIND4-17. To investigate this possibility, we purified recombinant BTB domain of KEAP1 protein and examined by mass spectrometry (MS) its covalent modification in the absence or presence of MIND4-17 (66). The mass for the protein incubated with dimethyl sulfoxide (DMSO) (control reaction) was determined to be 16,175.6 Da, in good agreement (within <10 ppm) to its calculated mass of 16,175.7686 Da (Fig. 2G). Incubation of BTB with MIND4-17 resulted in a reduction in the peak for the nonmodified protein, and the emergence of a dominant peak at 16,298.0 Da, indicating covalent modification of the protein. The observed shift of 122.4 Da was as would be predicted for a single cysteine being modified by formation of a 3-nitropyridine adduct with MIND4-17 (calculated mass difference, 123.09 Da).
Next, we validated the putative KEAP1-modyfying mechanism of NRF2 activation by MIND4-17 in live cells (12). COS1 cells were cotransfected with plasmids encoding NRF2-V5 and KEAP1 WT, mutant C151S (cysteine-151 is replaced by serine), or double-mutant C226S/C613S; β-galactosidase was used to monitor transfection efficiencies. In accord with our previous observations, MIND4-17 stabilized NRF2 in cells expressing WT KEAP1 (Fig. 2H). In cells expressing a double-mutant C226S/C613S KEAP1, MIND4-17 was still able to stabilize NRF2 protein to levels similar to those for WT KEAP1 (Fig. 2H). In contrast, MIND4-17 failed to stabilize NRF2 in cells expressing the single C151S mutant KEAP1 (Fig. 2H). These results therefore were consistent with MIND4-17–dependent activation of NRF2 through a highly selective and covalent modification of the KEAP1 sensor cysteine C151.
To determine whether MIND4-17 promiscuously alkylated other reactive cysteines within the proteome, we used a quantitative cysteine reactivity profiling strategy to globally identify cysteine residues that demonstrate significant loss of reactivity upon preincubation with compounds (67, 68). Briefly, a promiscuous cysteine reactive iodoacetamide-alkyne (IA) probe coupled to isotopic linkers was used to monitor changes in cysteine reactivity by quantitative MS (69–71). (For detailed protocol, see SI Materials and Methods.) Of the >300 reactive cysteine-containing peptides identified in our MS studies, only four residues showed a greater than twofold change in cysteine reactivity upon treatment with MIND4-17 (Dataset S1). Within the subset of cysteines with no change in reactivity were hyperreactive cysteine residues such as the active-site nucleophile of GAPDH, attesting to the low reactivity of the MIND4-17 across even highly reactive cysteines within the proteome.
MIND4-17 Causes Conformational Change in the KEAP1:NRF2 Protein Complex in Live Cells.
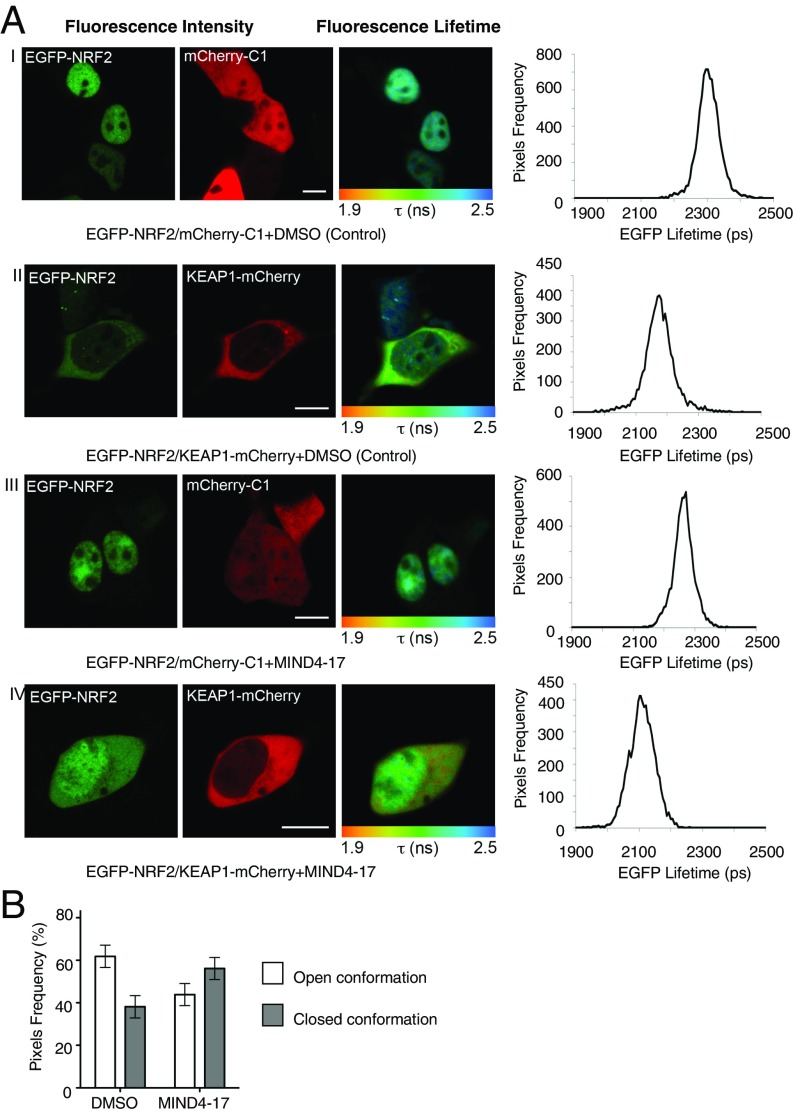
We further validated the MIND4-17 mechanism of NRF2 activation using a recently developed FRET/fluorescence lifetime imaging microscopy (FLIM) methodology (13, 72) and examined whether MIND4-17 treatment arrests the NRF2:KEAP1 complex in the closed conformation, thus permitting de novo NRF2 accumulation and translocation in nuclei to activate ARE-gene transcription. The effects of MIND4-17 on the conformational changes of the KEAP1:NRF2 complex were determined in HEK293 cells ectopically expressing fluorescent EGFP-NRF2 and KEAP1-mCherry fusion proteins, or EGFP-NRF2 and free mCherry as a negative control. As expected, in vehicle-treated cells transfected with EGFP-NRF2 and free mCherry, most of the NRF2 was in the nucleus (Fig. 3A, first row, images), and the lifetime of EGFP fluorescence in the cytoplasm was 2,324 ± 7 ps (n = 6) (Fig. 3A, first row, graph). When KEAP1-mCherry was coexpressed with EGFP-NRF2, NRF2 was largely cytoplasmic (Fig. 3A, second row, images), and the EGFP lifetime was significantly reduced to 2,184 ± 42 ps (n = 6, P = 1.14E-05) (Fig. 3A, second row, graph), indicating the occurrence of FRET between the fluorophores and demonstrating that the two fusion proteins interact. No significant changes were observed in the subcellular localization of EGFP-NRF2 upon addition of MIND4-17 to cells coexpressing EGFP-NRF2 and free mCherry (Fig. 3A, third row, images and graph). In contrast, exposure to 1 μM MIND4-17 for 1 h promoted the nuclear accumulation of EGFP-NRF2 in cells coexpressing EGFP-NRF2 and KEAP1-mCherry (Fig. 3A, fourth row, images), and the EGFP lifetime was significantly reduced further to 2,147 ± 50 ps (n = 17, P = 5.98E-09) (Fig. 3A, fourth row, graph).
Fig. 3.
MIND4-17 arrests KEAP:NRF2 complex in the closed conformation in live cells. (A) Fluorescence lifetime imaging of cells coexpressing either EGFP-NRF2 plus free mCherry or EGFP-NRF2 plus KEAP1-mCherry, and treated with vehicle (DMSO) or MIND4-17 (1 μM) for 1 h. HEK293 cells were transfected with the indicated constructs, and the fluorescence lifetime of EGFP and the FRET efficiency were quantified in the cytoplasm 24 h later. The first column shows the EGFP intensity images from which the lifetime data were derived, whereas the second column shows mCherry intensity images. The third column depicts EGFP fluorescence lifetime where pixel color corresponds to the mean lifetime of EGFP, ranging from 1.9 to 2.5 ns as indicated in the color scale below each image. The fourth column shows the lifetime data histograms for each image, with lifetime on the x axis and pixel frequency on the y axis. (B) Quantification of the open and closed conformations of the KEAP1:NRF2 protein complex based on the FRET efficiency distribution across the cytoplasm. The subpopulation of FRET efficiency between 0% and 15% was assigned the “open” conformation of the complex, whereas the FRET population between 15% and 30% was assigned the “closed” conformation of the complex. Data represent means ± SD from 6 to 17 cells.
Further quantification of the FRET efficiency revealed that, in the cytoplasm of vehicle-treated cells, 62% of the KEAP1:NRF2 complex was in the open conformation, and 38% in the closed conformation. Treatment with MIND4-17 inverted this ratio to 44% and 56% of KEAP1:NRF2 complexes being in the open and the closed conformations, respectively (Fig. 3B). These results obtained by FRET/FLIM analysis in live cells showed that MIND4-17 arrests the NRF2:KEAP1 protein complex in the closed conformation, which is similar to previously reported effects of the electrophilic NRF2 inducers SFP and sulfoxythiocarbamate alkyne (13) but in contrast to the effects of the nonelectrophilic triazole inducer HB229, which directly disrupts the KEAP1:NRF2 protein:protein interaction (73).
Taken together, these data delineate a mechanism of NRF2 activation by MIND4-17 through specific covalent modification of KEAP1 sensor-cysteine C151, subsequently causing accumulation and nuclear translocation of de novo-synthesized NRF2, and activation of NRF2-dependent gene transcription. Thus, MIND4-17 acts as a highly KEAP1-selective stress-mimicking compound (74) and was next used for investigating NRF2 signaling responses in HD model systems.
NRF2 Activation by MIND4-17 Induces Antiinflammatory Effects in Microglia and Macrophages.
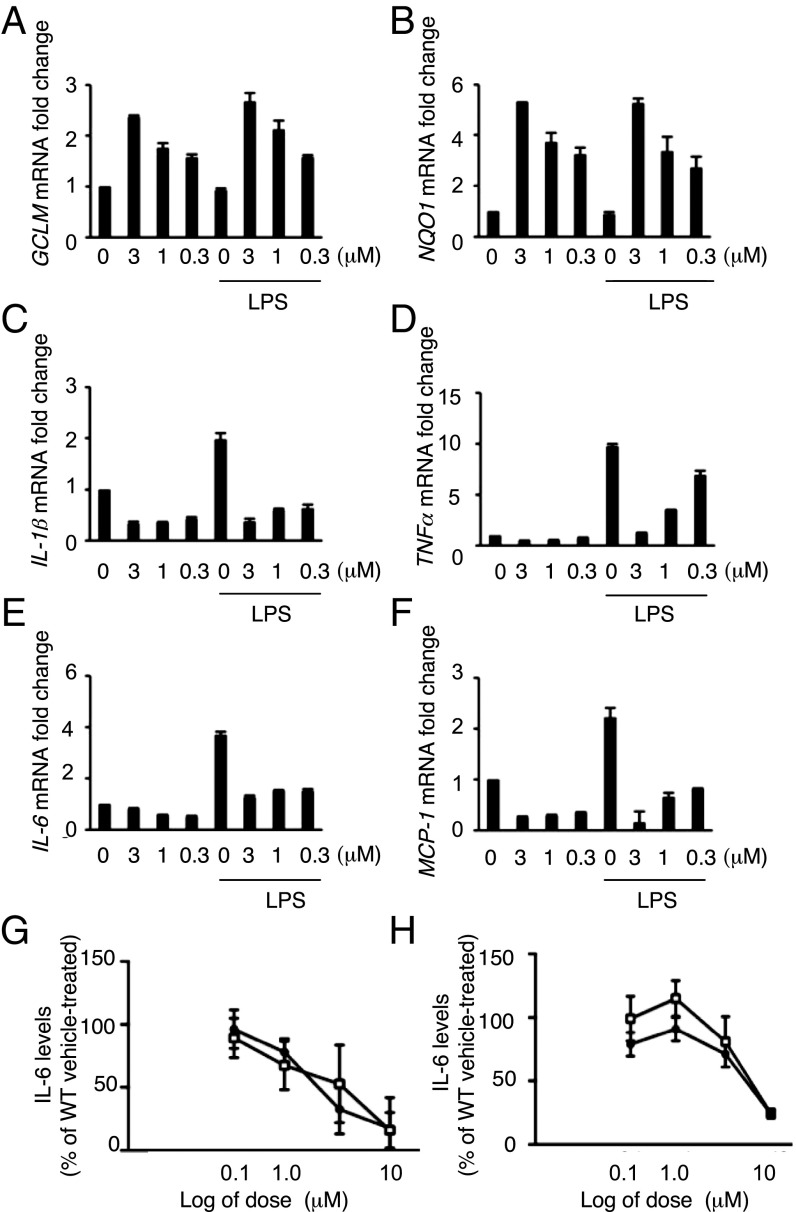
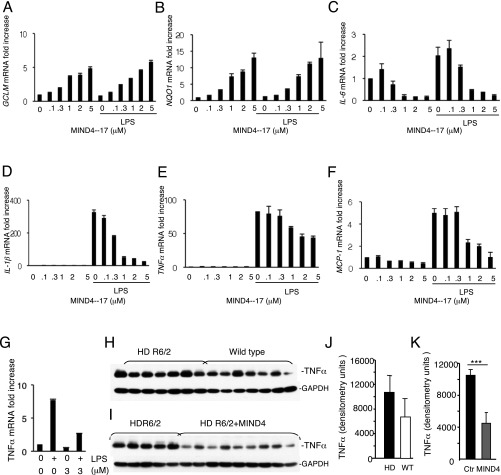
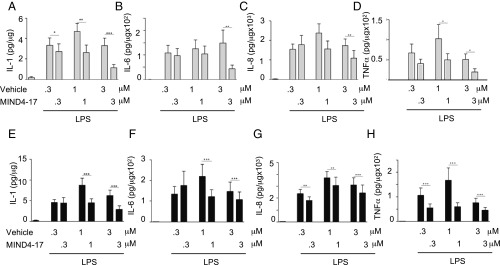
NRF2 activation-dependent antiinflammatory responses have been well described in microglia cells (75, 76). It has been also shown that genetic or pharmacologic NRF2 activation counteracts release of proinflammatory cytokines in the context of exposure to UV radiation (20). Thus, we first validated the effect of MIND4-17 treatment on repression of proinflammatory cytokines in microglial cells and also in peripheral macrophages that are known to infiltrate CNS in late-stage HD. As expected, treatment of mouse microglial BV2 cells with MIND4-17 resulted in a potent and concentration-dependent increases in transcription of the canonical ARE genes GCLM and NQO1 in both resting and lipopolysaccharide (LPS)-activated microglial cells (Fig. 4 A and B). Concomitantly, treatment with MIND4-17 resulted in a concentration-dependent repression of the proinflammatory factors IL-6, IL-1β, TNFα, and MCP-1 (Fig. 4 C–F). MIND4-17 treatment similarly repressed IL-6, IL-1β, TNFα, and MCP-1 and activated NQO1 and GCLM genes in resting and LPS-stimulated bone marrow-derived macrophages (Fig. S4 A–F) (77).
Fig. 4.
NRF2 activation by MIND4-17 has antiinflammatory effects in mouse microglia cells. (A–F) Evaluation of NRF2-specific transcriptional responses in resting and LPS-activated BV2 microglial cells treated with MIND4-17. Concentration-dependent effects of MIND4-17 treatment on mRNA expression of NRF2-responsive antioxidant GCLM (A) and NQO1 (B), and proinflammatory cytokine Il-6 (C), IL-1β (D), TNFα (E), and MCP-1 (F) genes. Transcriptional expression was measured using qRT-PCR with gene-specific primers in duplicates (n = 2). Means (filled bars) and SE bars are shown. (G and H) MIND4-17 concentration-dependent effects on secretion of IL-6 in primary microglia (G) and astrocytes (H) from WT and HD (YAC 128); inflammatory responses were induced with CSE and IFN-γ. Combined graphs of two independent experiments are shown. Filled circles (WT) and open squares (YAC128) represent mean values (n = 2) from compound-treated cells expressed as a percentage from vehicle (DMSO)-treated cells (100%), SE bars are shown.
Fig. S4.
Treatment with MIND4-17 reduces expression of proinflammatory cytokines. (A–F) Evaluation of NRF2-specific transcriptional responses in rested and LPS-activated iBMM macrophages treated with MIND4-17. Concentration-dependent effects of MIND4-17 on mRNA expression of NRF2-responsive genes GCLM (A) and NQO1 (B), and inflammatory factors Il-6 (C), IL-1β (D), TNFα (E), and MCP-1 (F), and effects of MIND4 at 3 μM on TNFα (G). Transcriptional expression was measured using qRT-PCR with gene-specific primers and assessed in duplicates (n = 2). Means (filled bars) and SE bars are shown. (H–K) MIND4 treatment represses expression of inflammatory TNFα protein in brain of HD mouse model R6/2. (H and I) TNFα and GAPDH protein levels were detected by immunoblotting in cortical extracts from 10-wk old WT (n = 7) and R6/2 (n = 7) mice (H) and in R6/2 mice treated by for 2 wk with vehicle (control) (n = 5) or MIND4 (n = 9) at 50 mg/kg, administered by daily i.p. injections (I). GAPDH levels were used as loading control. (J) Quantification analysis of TNFα levels (H) in WT (open bars) and in R6/2 (black bars) brain samples shows no statistical difference. GAPDH protein was used to normalize TNFα levels. (K) Quantification analysis of TNFα levels (I) in vehicle-treated (black bars) and in MIND4-treated (gray bars) R6/2 brain samples. GAPDH protein was used to normalize TNFα levels. MIND4 treatment resulted in statistically significant reduction of TNFα levels in HD cortices. Student’s t test, ***P < 0.001.
Antiinflammatory NRF2 Activation Response in HD and WT Primary Mouse Microglia and Astrocytes.
Having established that MIND4-17 represses cytokine expression, next we probed for antiinflammatory effects of NRF2 activation in brain-resident nonneuronal cells, namely microglia and astrocytes, and specifically examined the effect on release of IL-6, an established marker of inflammation in HD model systems (40, 78).
MIND4-17 was first evaluated for its impact on inflammatory responses in primary microglia derived from WT and HD mutant YAC128 mice (79). The YAC128 mouse model of HD expresses the full-length human transgene with 128 CAG repeats, and replicates key elements of HD phenotypes and selective neurodegeneration. Previous studies in peripheral blood monocytes from YAC128 mice focused on IL-6 as a marker of inflammation in HD (80). Release of IL-6 from WT and YAC128 microglia was induced by stimulation with control standard endotoxin (CSE) and IFN-γ. MIND4-17 reduced the amount of IL-6 secreted from WT and YAC128 primary microglia in a concentration-dependent manner and in highly similar fashion (Fig. 4G). Experiments were next extended to primary astrocytes derived from WT and YAC128 mice. MIND4-17 treatment resulted in a similar concentration-dependent reduction of IL-6 release in CSE-stimulated primary astrocytes derived from YAC128 and WT mice (Fig. 4H).
Taking advantage of the brain permeability of the MIND4 analog, which, similar to MIND4-17, repressed cytokine expression in LPS-induced microglia (Fig. S4G), we next conducted a semiacute 2-wk treatment of symptomatic HD R6/2 mice (81). A statistically significant reduction of cortical TNFα levels was detected (Fig. S4 H–K), consistent with the antiinflammatory effects of the NRF2 activation observed in primary mouse microglia and astrocytes (Fig. 4 G and H). These results suggest that the antiinflammatory NRF2 activation response remains available for induction in symptomatic R6/2 mice with well-progressed HD neurological phenotype. Interestingly, cortical TNFα levels were not significantly different between untreated WT and R6/2 mice (Fig. S4J), suggesting the TNFα itself is unlikely to be the sole driver of neuroinflammation in HD.
Antiinflammatory NRF2 Activation Responses Are Intact in Primary Monocytes from HD Patients.
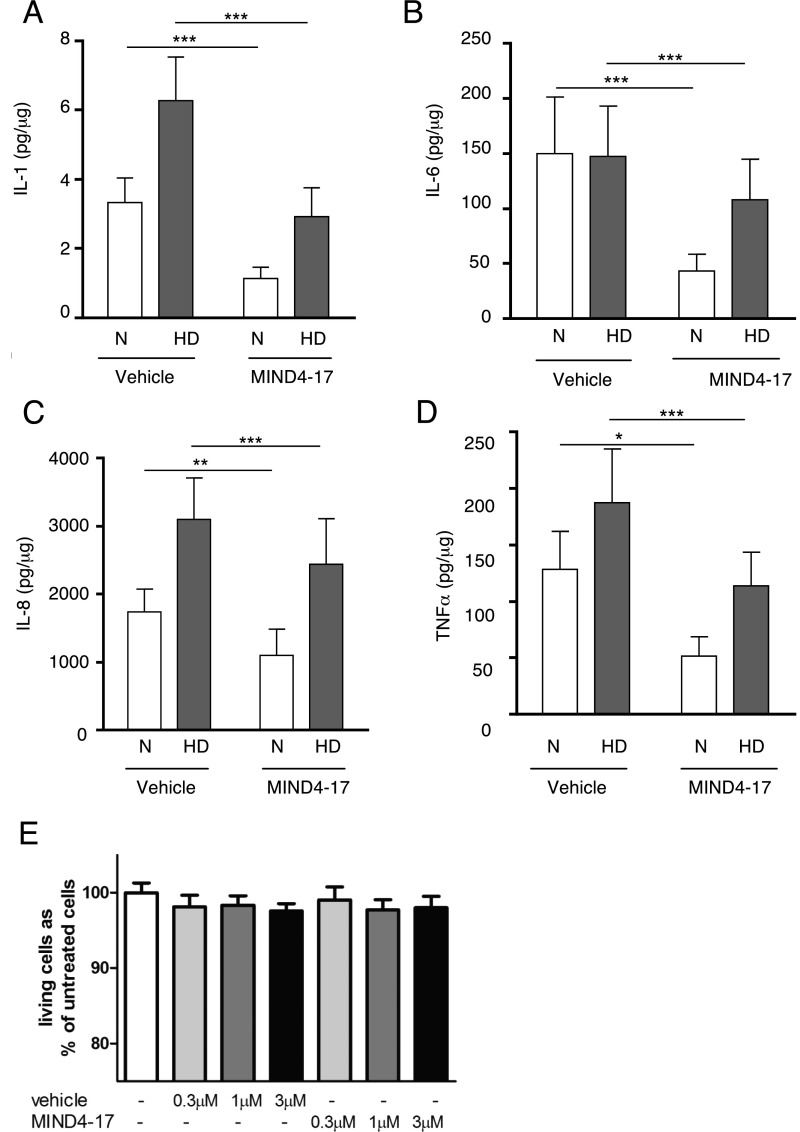
Finally, to validate these HD model results in human HD, the antiinflammatory effects of NRF2 induction by MIND4-17 were examined in primary human monocytes derived from HD patients and nondisease controls. Ex vivo peripheral immune cells from HD patients produce significantly more proinflammatory cytokines in response to LPS and IFN-γ stimulation than cells isolated from control subjects (40, 78). Here, we induced production of the proinflammatory cytokines IL-1, IL-6, IL-8, and TNFα in primary monocytes by stimulation with LPS and IFN-γ. Pretreatment with MIND4-17, before induction with LPS/IFN-γ significantly decreased the levels of all four proinflammatory cytokines in both HD and control monocytes (Fig. 5 and Fig. S5 A–D). In HD monocytes, MIND4-17 significantly repressed the expression of all four cytokines at the 3 and 1 μM concentrations and repressed IL-8 and TNFα expression at the lowest tested concentration of 0.3 μM, demonstrating a highly similar pattern of the compound effects in the disease and nondisease counterparts. Neither vehicle (DMSO) nor MIND4-17 affected the viability of primary human monocytes at any of the concentrations tested (Fig. S5E).
Fig. 5.
Characterization of antiinflammatory NRF2-dependent responses in primary human monocytes treated with MIND4-17. (A–H) MIND4-17 represses expression of induced inflammatory cytokines in primary monocytes from normal subjects (A–D) and HD patients (E–H). IL-1b, IL-6, IL-8, and TNFα, production by LPS- and IFN-γ–stimulated monocytes, measured by multiplex ELISA, was compared in cells treated with different concentrations of MIND4-17 and with corresponding doses of vehicle (DMSO). Linear mixed model on log transformed data; n = 10 controls, n = 13 for HD patients; ±SEM; *P < 0.05; **P < 0.01, ***P < 0.001 (one-way ANOVA, P = 0.61, n = 13).
Fig. S5.
MIND4-17 mediates antiinflammatory NRF2 activation responses in human primary monocytes from HD patients and healthy controls. (A–D) MIND4-17 represses expression of induced inflammatory cytokines in primary monocytes from HD patients and nondisease controls. Expression levels are shown. IL-1 (A), IL-6 (B), IL-8 (C), and TNFα (D) production by IFN-γ− and LPS-stimulated HD patient monocytes. Cytokine expression, measured by multiplexed ELISAs, was compared in cells treated with 3 μM MIND4-17 or vehicle (DMSO). (Linear mixed model on log transformed data; n = 10 nondisease, n = 13 for HD patients; *P < 0.05, **P < 0.01, ***P < 0.001.) (E) MIND4-17 is not toxic for human monocytes. Lack of apparent toxicity of MIND4-17 treatment at tested doses. The 48-h treatment of primary human monocytes with MIND4-17 and vehicle does not show any effect on cell viability measured with LDH assays.
The responses in human monocytes were consistent with the effects observed in the mouse models. Overall, the results suggest that NRF2 activation in nonneuronal primary HD cells is intact, amenable to activation, and capable to mediate antiinflammatory protective responses.
NRF2 Activation Responses Are Muted in Human HD Neural Stem Cells.
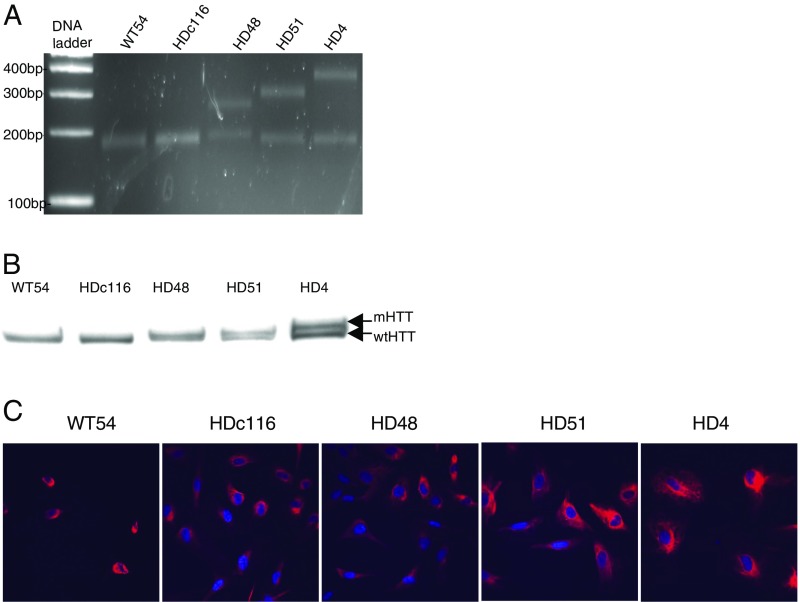
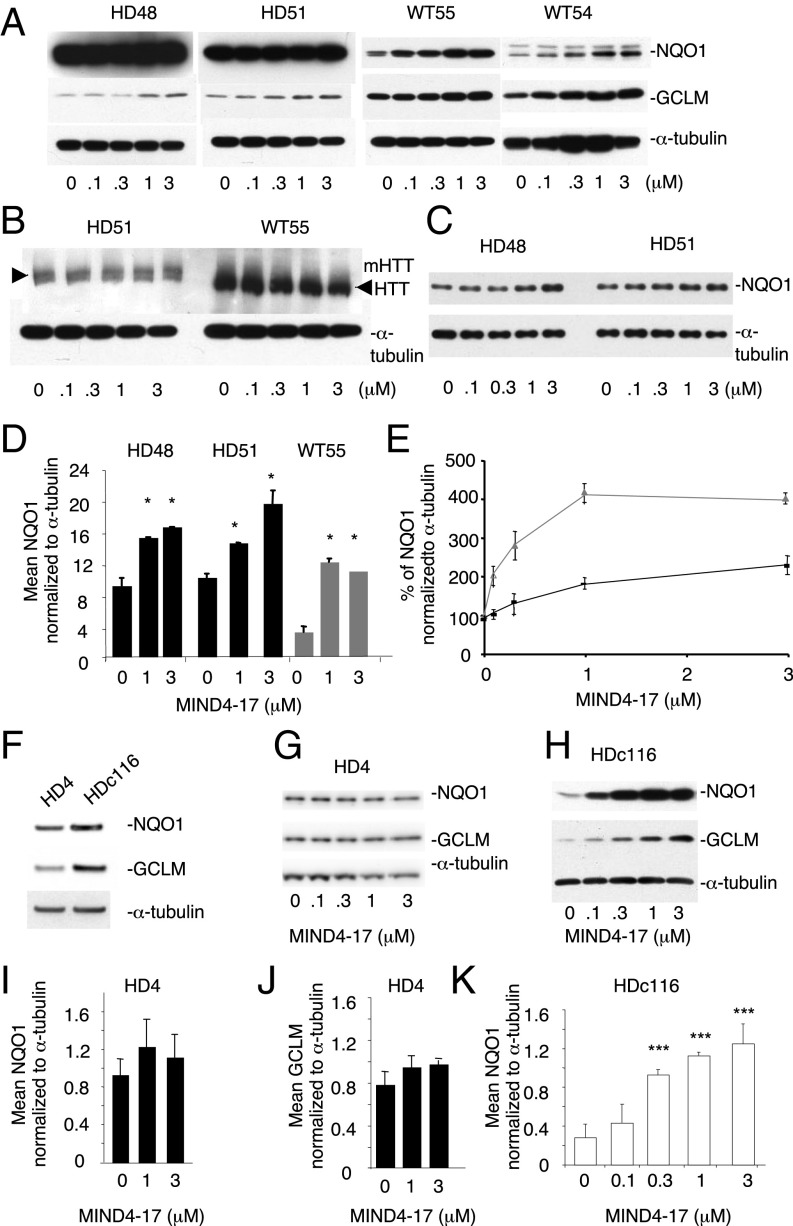
We next evaluated NRF2 activation responses in experimental neural HD models of human origin. Nestin-positive HD48 (mutant 42CAG) and HD51 (mutant 51CAG) and nondisease WT55 and WT54 neural stem cells (NSCs) were differentiated from human induced pluripotent stem cells (iPSCs), based on a protocol described for mouse ES cells (82) (Fig. S6; see SI Materials and Methods for detailed protocols). Cells were treated for 24 h with MIND4-17 at a concentration range of 0.1–3 μM, and levels of the canonical NRF2-responsive proteins NQO1 and GCLM (Fig. 6A) and HTT (Fig. 6B) were examined by immunoblotting. The levels of NQO1 were significantly higher in both HD cell lines, and thus a lower amount of protein per lane was loaded to illuminate the compound effects (Fig. 6C; note multiple NQO1 molecular-weight bands reflecting known protein isoforms). Treatment with MIND4-17 induced a concentration-dependent increase of the NRF2-responsive proteins NQO1 and GCLM in HD and nondisease cells. The MIND4-17–dependent increase of NQO1 and GCLM levels started at 1 μM in both HD and at 0.1 μM in both nondisease cell lines. MIND4-17 treatment had no detectable effects on HTT levels.
Fig. S6.
Characterization of human neuronal HD and nondisease cells, NSCs, and mature neurons. (A) PCR analysis of CAG triplet repeat length on genomic DNA from human NSC WT54 (17CAG); HDc116 (containing corrected to WT 21CAG); HD48 (mutant allele with 42 CAGs); HD51 (mutant allele with 50 CAGs); HD4-parent of HDc116 (containing 72 CAGs). The expected sizes of mutant 42 CAG, 50 CAG, and 72 CAG PCR products in ranges of 180–210, 250–300, and 350–450 bp, respectively, are shown with reference to DNA ladder standard. (B) Western blot analysis of expression WT and mutant HTT (indicated by arrows) of NSC cell lines detected using anti-HTT antibody Ab1 against 1–17 aa of HTT. (C) Immunofluorescence microscopy using anti-Nestin antibody of human NSCs derived from iPSCs. Percentages of cells positive for Nestin staining: WT54, 98.75% (n = 80 cells); HDc116, 99% (n = 301 cells); HD48, 98.92% (n = 556 cells); HD51, 98.25% (n = 228 cells); HD4, 100% (159 cells).
Fig. 6.
Activation of NRF2 signaling in human iPSC-derived NSCs. (A) MIND4-17 induces NQO1 and GCLM protein expression in HD48, HD51 and WT54, WT55 NSCs. Cells were treated with MIND4-17 for 24 h at indicated concentrations. Protein levels were detected by immunoblotting. (B) No effects of MIND4-17 treatment on mutant or WT HTT levels in HD51 and WT55 NSCs in were detected. (C) HD48 and HD51 extracts from A were diluted 1:3 and NQO1 levels reexamined. (D) Densitometric analysis of NQO1 expression in HD48 and HD51 (black bars) and WT54 (gray bars) NSCs. Data are presented as mean ± SD of two independent experiments. *P < 0.05 by t test; n = 4. (E) Relative induction of NQO1 expression in WT54 (gray triangle/line) and HD51 (black triangle/line) NSCs treated with MIND4-17 at indicated concentrations. Each data point represents a mean ± SD of two independent experiments; n = 4. (F–I) Effects of MIND4-17 in mutant HD4 and in genetically corrected isogenic nondisease HD116c. (F) Basal levels of NQO1, GCLM, and α-tubulin in HD4 and HD116c, detected by immunoblotting. (G and H) MIND4-17 induces expression of NQO1 and GCLM in the corrected HD116c (H) but not in the parental HD4 (G) cell lines. NQO1 and GCLM protein expression levels and α-tubulin were detected by immunoblotting. (I and J) Densitometric analysis of NQO1 (I) and GCLM (J) induction in HD4 from G shows no induction of NRF2-responsive proteins by MIND4-17. Levels of α-tubulin served as the loading control and were used for normalization of NQO1 or GCLM signals. (K) Densitometric analysis of NQO1 induction in HD116c. Signals were normalized to the levels of α-tubulin. Data are presented as means ± SD; t test with Holm correction for multiple comparison. ***P < 0.01; n = 4.
To determine whether the magnitude of NRF2 response varied in HD vs. nondisease cells, we compared the maximum fold-induction of NQO1 expression. A maximal approximately twofold response was observed in each HD cell line at 3 μM MIND4-17 (Fig. 6 D and E). Nondisease WT cells showed a maximum approximately fourfold response at 1 μM MIND4-17, which plateaued by 3 μM (Fig. 6 D and E). These data suggest that NRF2 induction could be compromised in HD due to suppressive influence of the expanded CAG repeat mutation on pathway activation; alternatively, a reduced fold change in response to MIND4-17 could be due to a preexisting partial or full activation in response to mHTT-induced homeostatic changes in the HD cells.
To distinguish between these two possibilities, we compared responses in NSC HD4 with a severe 72 CAG mutation and its isogenic-counterpart NSC HD116c, in which the 72 CAG expansion was corrected to a nonpathological 21 CAG repeat length by homologous recombination (83). In HD4 NSCs, the basal levels of NQO1 and GCLM proteins were similar to HD116c (Fig. 6F), indicating the NRF2 pathway was not partially activated. Treatment with MIND4-17 did not induce a significant increase of either NQO1 or GCLM expression in HD4 cells (Fig. 6 G, I, and J). In sharp contrast, MIND4-17 treatment induced a concentration-dependent increase of NQO1 and GCLM expression in HDc116 cells (Fig. 6 H and K), similar to that observed in WT54 and WT55 NSCs (Fig. 6A). The induction of both NRF2-responsive proteins in the corrected HDc116 cells was significant at 0.3 μM concentration of MIND4-17. A maximum fourfold response was observed at concentrations of 1 and 3 μM, which was similar to the response in nondisease NSCs.
Together, these data show that the HD mutation negatively impacts NRF2 signaling and suggest that interference is even greater for the extreme CAG pathological expansions, which are associated with juvenile clinical cases.
SI Materials and Methods
Formulation, Tolerability, and Pharmacokinetics of MIND4.
MIND4 was solubilized at 5 mg/mL in 7.5% Cremophor EL (BASF)/2.375% ethanol in PBS. WT mice C57BL/6 (n = 3) were subjected to compound treatment at escalated dosing from 50 mg/kg up to a final dose of 275 mg/kg administered daily by i.p. injection. No weight loss or sudden death was observed, suggesting lack of acute toxicity of MIND4 in that dose range. Mice were killed 2 h following the last drug administration and cortical samples isolated according to standard procedure and subjected to HPLC analysis as described (96, 97). Pharmacokinetics was performed in male C57BL/6 mice (n = 3) at 0.5, 1.0, 2, 4, 8, and 24 h after i.p. injection of MIND4 at 50 mg/kg and collected serum samples analyzed by HPLC. Mice (n = 3) were killed at 0.5 h after i.p. injection of MIND4 at 50 mg/kg dose, and isolated cortical samples were subjected to HPLC analysis.
Frozen mouse cortical tissues were extracted in ice-cold methanol containing 0.4% acetic acid (∼1:6 ratios), probe sonicated for 5 s, and centrifuged at 22,000 × g for 20 min at 4 °C. The supernatant was dried under centrifugation and vacuum and reconstituted with running buffer. Reverse-phase HPLC/electrochemical array separation was accomplished on Shiseido MG240 C18 HPLC column (4.6 × 250 mm, 3-μ particle size), flow rate of 1 mL/min, using 50% methanol in 0.1 M sodium phosphate buffer, pH 3; isocratic elution was performed for 20 min using four-channel electrochemical array. Levels of MIND4 were determined using integrated peak areas and the standard probe.
Analysis of TNFα Levels in Cortex of MIND4-Treated R6/2 Mice.
Female R6/2 mice used in the study were generated by backcrossing R6/2 males from The Jackson Laboratory with C57BL/6 × CBA F1 females and maintained at Massachusetts General Hospital animal facility. Mice were genotyped by PCR using tail-tip DNA and were housed five per cage under standard conditions with ad libitum access to water and food. Mice were administered with MIND4 at the dose of 50 mg/kg by i.p. injections for 2 wk. The drug suspension at 5 mg/mL in 7.5% Cremophor EL (BASF)/2.375% ethanol in PBS, was made fresh daily. At 10 wk of age, R62 and age-matching WT control mice were killed 2 h after last injections, and brains were removed and snap frozen in liquid nitrogen and kept frozen at −80 °C until analyzed. All animal experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (98) and were approved by local animal care committee.
SIRT2 Deacetylase Activity Test.
Deacetylation reaction was performed at 37 °C for 1 h in the presence of human recombinant enzymes: SIRT2 (BioMol-SE-251) 5 units per reaction, compound of interest, standard buffer, 50 μM substrate, and 500 μM NAD+ according to manufacturer’s protocol and as described previously (55).
Compound Activity Test in the Rat Embryonic Striatal Cell Lines ST14A.
ST14A cells (a generous gift of E. Cattaneo, University of Milan, Milan) (92) stably express either a mutant expanded repeat (128Q) or WT (26Q) 546-aa huntingtin (HTT) fragment and were treated with compounds for 24 h as described (93). Protein levels of NQO1, GCLM, and α-tubulin (loading control) were detected in ST14A by immunoblotting with NQO1 (Sigma; 1:1,000), GCLM (Abcam; 1:800), and GAPDH (Millipore; 1:10,000) antibodies.
Nuclear-Cytoplasmic Extract Separation.
WT MEFs (500,000 per dish) were grown for 24 h on 6-cm plates, and then treated with solvent control [0.1% DMSO (vol/vol)] or 0.5 μM MIND4-17. At the end of each treatment time, cells were washed twice in PBS and lysed in buffer A [10 mM KCl, 5 mM MgCl2, 50 mM Tris⋅HCl (pH 7.5), 0.5% Nonidet P-40, 1 mM DTT], containing 1 protease inhibitor and 1 phosphatase inhibitor tablets (Roche) per 10 mL of buffer. Lysates were subjected to centrifugation at 1,000 × g for 5 min at 4 °C, and supernatant (cytoplasmic fraction) was transferred to a fresh microfuge tube. The pellet (nuclear fraction) was washed three times in buffer A before being dissolved in buffer B [2% SDS, 150 mM NaCl, 65 mM Tris⋅HCl (pH 8.0)] and sonicated for 30 s. Finally, the lysates were subjected to Western blot analysis with NRF2 antibody and with GAPDH (rabbit polyclonal; 1:5,000) and lamin A/C (rabbit polyclonal; 1:1,000; Gene Tex) to confirm fraction purity and equal protein loading.
Design and Synthesis of MIND4-17 Analogs.
MIND4-17 analogs 2286, 2291, and 2907 were synthesized as shown in Scheme 1 (Fig. S3). Compound A was prepared following a previously reported method (61). Compound A was converted to 2291, 2907, and MIND4-17 by reacting with 4-nitroflurobenzene, 2-chloro-5-nitropyridine, and 2-chloropyridine, respectively. The nitro group in MIND4-17 was then reduced with stannous chloride to give 2286. 2909 was synthesized as shown in Scheme 2 (Fig. S3). Compound B was synthesized by coupling 2-chloro-5-nitropyridine and tert-butyl methyl malonate (60). Treatment of compound B with hydrazine gave compound C, which upon reaction with 3-phenoxy acetyl chloride gave compound D. 2909 was obtained by reacting compound D with aniline and phosphorous trichloride. 2872 was prepared as shown in Scheme 3 (Fig. S3). Compound E was obtained by reacting phenol with ethyl bromoacetate under basic conditions. The ester group in compound E was converted the hydrazide F by reacting with hydrazine. Compound F was reacted with phenylisocyanate and cyclized under basic condition to yield compound H, which was reacted with 2-chloro-5-nitropyridine to give 2872. Synthesis of MIND4-17 and respective analogs was conducted by a drug discovery company Aurigene (coauthors V.P. and S.S.); NMR analysis shows an acceptable 95% purity. The activities of MIND4-17, synthesized by Aurigene, and commercially purchased from Cambridge (multiple batches) were identical and activities of analogs consistent with established mechanism.
Compound GSH Sensitivity Assay.
Each compound was incubated in cell culture medium in the presence or absence of 1 mM GSH at 37 °C for 30 min before administering to Hepa1c1c7 cells grown on 96-well plates. After a 48-h exposure, the NQO1 enzyme activity was determined in cell lysates. Results are shown as average values of eight replicate wells. The SD in each case was less than 5%.
Fluorescence Lifetime Imaging Analysis of MIND4-17 Effect on NRF2:KEAP1 Complex.
Fluorescence lifetime imaging microscopy (FLIM) was performed as described (94) using an inverted multiphoton laser-scanning microscope (Zeiss LSM780 confocal) with a 63× oil-immersion N.A. 1.4 Plan-Apochromat objective from Zeiss. The software used to run the LSM780 multiphoton microscope was Zen 2010. The microscope was equipped with a black incubation chamber suitable to maintain the live cells and optics at constant temperature 37 °C. Two-photon excitation of EGFP was achieved using a Coherent Scientific Chameleon Ultra II Ti:sapphire laser (tuning range, 680–1080 nm) at 890 nm. The fluorescence lifetime imaging capability was provided by time-correlated single photon-counting electronics (SPC-830; Becker and Hickl). Laser power was adjusted to give a mean photon count rate of the order 104 to 105 photons per s. Fluorescence lifetime measurements were acquired over 90 s. Fluorescence lifetimes were calculated for all pixels in the field of view (256 × 256 pixels), and then a particular region of interest (e.g., cytoplasm) was selected using SPCImage software (Becker and Hickl). The analysis of the FLIM measurements from a particular region of interest was performed by using SPCImage software. The best-fitting model (k2 of the fit close to 1) to extract an accurate fluorescence lifetime value from the decays was applied by adjusting several parameters such as the number of exponential components, the background threshold, the shift index, and the scatter parameter (amount of scattered excitation light detected).
The FRET efficiency percentage was calculated using SPCImage software for the same region defined to calculate the mean fluorescence lifetime. The image was then analyzed using a two-component exponential decay model where the lifetime t2 (noninteracting proteins lifetime from EGFP-NRF2 plus free mCherry cells) was fixed, whereas the t1 value (lifetime associated to the donor proteins interacting with the acceptor) was left unchecked. Pixel enrichment in the respective FRET populations related to open (0–15%) or closed (15–30%) conformation was quantified from the lifetime distribution histograms. The lifetime histogram for each vehicle and MIND4-17–treated cells was normalized by the total number of pixels Fi = fi/Ni, where fi and Ni are the lifetime histogram and the total number of pixels, respectively.
MS Analysis of KEAP1 BTB Modification by MIND4-17.
The BTB domain of Keap1 (residues 48–190) was cloned into a modified pET19 vector with N-terminal 6×His-tag cleavable with TEV protease and a single point mutation was introduced at position 182 (S182A). The full-length KEAP1 with deletion of the first 54 aa was cloned in similarly in the same expression vector. The overexpression and purification were performed as described in ref. 66.
For analysis of the potential modification of the BTB domain, 10 µM BTB (48–190 S182A) were incubated with 2 mM MIND4-17 (or DMSO, 2% final concentration, for control) for 1 h on ice in 20 mM Tris⋅HCl, pH 8.0, and 150 mM NaCl. Intact protein masses were determined through HPLC-coupled ESI-MS on an AB Sciex TripleTOF 5600+ mass spectrometer (Sciex). Intact protein were first concentrated and washed on a Piccolo Proto 200 C4 5-µm 2.5 × 0.5-mm trap column (Higgins Analytical) and subsequently switched in line with, and separated on, a Jupiter C4 5-µm 300-Å 150 × 1-mm analytical column (Phenomenex) mounted onto a Shimadzu Prominence UFLC (Shimadzu) at a 70 µL⋅min−1 flow rate with the following buffers: A—5% ACN, 5% DMSO, and 0.1% FA; B—90% ACN, 5% DMSO, and 0.1% FA. Proteins were then eluted over with a gradient of 3 min of 1% B to 55% B followed by 1 min of 55% B to 90% B. Mass analysis was performed by ESI-TOF-MS on an AB Sciex TripleTOF 5600+ mass spectrometer (Sciex) with a DuoSpray Ion Source with the following settings: floating voltage of 5,500 V, temperature of 350 °C, declustering potential of 120 with four separate TOF experiments, each respectively with 4, 12, 20, and 40 time bins summed. Data analysis was performed as follows: spectra were integrated over a retention time period, and the summed TOF experiment with the greatest resolution selected. The raw data were then converted and deconvoluted using the MaxEnt I algorithm (Waters) at a resolution of 0.1 Da.
Proteomic Analysis by Quantitative MS.
Preparation of MS samples for reactive cysteine profiling.
HeLa soluble protein lysates (500 μL, 2 mg⋅mL−1) were aliquoted, and MIND4-17 (10 μM), or DMSO was added to the appropriate samples at the designated concentration. Two aliquots were made for each inhibitor concentration or DMSO equaling four tubes for one final sample. The samples were incubated at room temperature (RT) for 1 h. IA-alkyne (100 μM) was added to the samples and incubated at RT for 1 h. Inhibitor-treated and DMSO samples were functionalized with Azo-H tag or Azo-L tag (100 μM, 50× stock in DMSO), respectively (68). All samples were then treated with TCEP (1 mM, 50× fresh stock in water), TBTA ligand (100 μM, 17× stock in DMSO:t-butanol = 1:4), and copper(II) sulfate (1 mM, 50× stock in water) followed by incubation at RT for 1 h. Samples were combined pairwise to combine inhibitor and DMSO-treated samples and centrifuged (6,500 × g, 4 min, 4 °C) to pellet the precipitated proteins. The pellets were resuspended in cold methanol by sonication, and the two samples were combined. Centrifugation was followed by a second methanol wash, after which the pellet was solubilized in Dulbecco’s PBS (DPBS) containing 1.2% SDS via sonication and heating (90 °C, 5 min). The two samples resulting from inhibitor pretreatment, IA-alkyne incubation, and Azo-H or Azo-L labeling are as follows: heavy (DMSO, 100 μM IA-alkyne)/light (10 μM MIND4-17, 100 μM IA-alkyne).
The SDS-solubilized proteome samples were diluted by 5 mL of DPBS for a final SDS concentration of 0.2%. The solution was incubated with 100 μL of streptavidin-agarose beads (Thermo Scientific; washed three times with DPBS to remove storage buffer) overnight at 4 °C. Samples were rotated at RT for 2 h before washed by 5 mL of 0.2% SDS/DPBS, 3 × 5 mL of DPBS, and 3 × 5 mL of water. The beads were pelleted by centrifugation (1,400 × g, 3 min) between washes.
The washed beads were suspended in 500 μL of 6 M urea/DPBS and 10 mM DTT (from 20× stock in water) and placed in a 65 °C heat block for 15 min. Iodoacetamide (20 mM from 50× stock in water) was then added, and the samples were allowed to react at 37 °C for 30 min. Following reduction and alkylation, the beads were pelleted by centrifugation and resuspended in 200 μL of 2 M urea/DPBS, 1 mM CaCl2 (100× stock in water), and trypsin (2 μg). The digestion was allowed to proceed overnight at 37 °C. The beads were pelleted by centrifugation and washed with 3 × 500 μL of DPBS and 3 × 500 μL of water. The azobenzene cleavage was carried out by incubating the beads with 50 μL of 25 mM sodium dithionite at RT for 1 h. The cleavage process was then repeated twice with 75 μL of 25 and 50 mM sodium dithionite, and all of the supernatants were combined. The beads were then washed twice with 75 μL of water, and the wash was combined with the supernatant from the cleavage step to reach 350 μL final. Formic acid (17.5 μL) was added to the sample, which was stored at −20 °C until MS analysis.
Quantitative MS analysis.
LC/LC-MS/MS analysis was performed on an LTQ-Orbitrap Discovery mass spectrometer (Thermo Fisher) coupled to an Agilent 1200 series HPLC. Peptide digests were pressure loaded onto a 250-μm fused silica desalting column packed with 4 cm of Aqua C18 reverse phase resin (Phenomenex). The peptides were eluted onto a biphasic column [100-μm fused silica with a 5-μm tip, packed with 10-cm C 18 and 4-cm Partisphere strong cation exchange resin (SCX, Whatman)] using a gradient 5–100% Buffer B in Buffer A (Buffer A: 95% water, 5% acetonitrile, 0.1% formic acid; Buffer B: 20% water, 80% acetonitrile, 0.1% formic acid). The peptides were then eluted from the SCX onto the C18 resin and into the mass spectrometer using four salt steps previously described (71). The flow rate through the column was set to ∼0.25 μL/min, and the spray voltage was set to 2.75 kV. One full MS scan (FTMS) (400–1,800 Mr) was followed by eight data-dependent scans (ITMS) of the nth most intense ions.
The tandem MS data were searched using the SEQUEST algorithm (69) using a concatenated target/decoy variant of the human IPI databases. A static modification of +57.02146 on cysteine was specified to account for alkylation by iodoacetamide, and differential modifications of +462.2987 (IA-alkyne and cleaved Azo-H) and +456.2849 (IA-alkyne and cleaved Azo-L) were specified on cysteine to account for probe modifications. SEQUEST output files were filtered using DTASelect (70). Quantification of heavy/light ratios (RH/L) was performed using the CIMAGE quantification package as previously described (67).
Quantitative MS data for reactive-cysteine profiling upon MIND4-17 treatment.
Cysteine reactivity in proteomes pretreated with MIND4-17 (heavy-labeled) were compared with DMSO-treated samples (light-labeled). For every cysteine-containing peptide, a heavy/light ratio was calculated that reflects the extent of cysteine reactivity in the inhibitor-treated vs. DMSO samples. A ratio of 1 indicates that there was no change in cysteine reactivity, whereas ratio values >>1 show a loss in cysteine reactivity upon inhibitor treatment. Cysteine-containing peptides with ratio >2 for MIND4-17 are highlighted in green in Dataset S1.
Macrophages for Drug Testing Experiment.
Immortalized mouse bone marrow-derived macrophages were a kind gift of Katherine Fitzgerald (University of Massachusetts Medical School, Worcester, MA). Primary cells were immortalized by infection with J2 retrovirus as described (77). The LPS activation of macrophages, drug treatment, and transcriptional profiling (Fig. S3) were conducted similar to experimental conditions described for microglia BV2 cells.
Compound Transcriptional Expression Profiling Microglia BV2 Cells and Macrophages.
The following qPCR primers were selected using PrimerBank (https://pga.mgh.harvard.edu/primerbank/) database: mIL-1β, for, 5′-GCAACTGTTCCTGAACTCAACT-3′; mIL-1β, rev, 5′-ATCTTTTGGGGTCCGTCAACT-3′; mIL-6, for, 5′-TAGTCCTTCCTACCCCAATTTCC-3′; mIL-6, rev, 5′-TTGGTCCTTAGCCACTCCTTC-3′; mGAPDH, for, 5′-TGTGTCCGTCGTGGATCTGA-3′; mGAPDH, rev, 5′-GGTCCTCAGTGTAGCCCAAG-3′; mNQO1, for, 5′-AGGATGGGAGGTACTCGAATC-3′; mNQO1, rev, 5′-AGGCGTCCTTCCTTATATGCTA-3′; mGCLM, for, 5′-AGGAGCTTCGGGACTGTATCC-3′; mGCLM, rev, 5′-GGGACATGGTGCATTCCAAAA-3′; mTNFα, for, 5′-CCCTCACACTCAGATCATCTTCT-3′; mTNFα, rev, 3′-GCTACGACGTGGGCTACAG-3′; mMCP-1, for, 5′-TTAAAAACCTGGATCGGAACCAA-3′; mMCP-1, rev, 5′-GCATTAGCTTCAGATTTACGGGT-3′.
Isolation and Culturing Primary Astrocytes and Microglia Cells.
Whole brains were obtained from postnatal 1- to 3-d-old WT and YAC128 mouse pups on the FVB/N strain background and placed in Hanks’ balanced salt solution (Invitrogen) on ice. Meninges were removed, and the remaining brain tissue was placed into growth medium (DMEM, 10% FBS, 1% l-glutamine, 1% penicillin/streptomycin) and homogenized. Cells from each brain were pelleted, resuspended in growth medium, transferred into a T150 flask, and cultured at 37 °C. Growth medium was replaced after 24 h and thereafter every 7 d. After 18–21 d in culture, loosely attached microglia were harvested and seeded at 1.4 × 105 cells per mL with preincubated conditioned media into 96-well tissue culture plates. Adherent astrocytes were plated at 1.4 × 105 cells per mL in 10% FBS media for 24 h in 96-well tissue culture plates.
MIND4-17 Activity Test on Cytokine Expression in Primary Human HD and Nondisease Monocytes.
Cells were isolated from whole blood, as previously described (40). In brief, monocytes were sorted via magnetic cell separation columns using anti-CD14 microbeads (Miltenyi Biotec). Monocytes were seeded at 1 × 105 cells per well in 96-well tissue culture plates with R10 media (RPMI culture medium supplemented with 10% FCS, 2 mM l-glutamine, 50 U/mL penicillin, and 50 mg/mL streptomycin). After resting for 16 h, the culture media was replaced with R10 media containing either vehicle (DMSO) or MIND4-17 at different concentrations (0.3, 1, and 3 μM). After 24 h of compound treatment, media was changed again using R10 containing both MIND4-17 at the same concentration and, to stimulate cytokine production, 10 ng/mL IFN-γ (R&D Systems) and 2 mg/mL lipopolysaccharide (LPS) (Sigma-Aldrich). Supernatants were collected at 24 h and stored at −70 °C. Supernatants were analyzed using the human proinflammatory II (four-plex) MSD assay measuring IL-1β, IL-6, IL-8, and TNFα. Cytokine levels were normalized to total protein concentration in each well. Cells were lysed in 50 mM Tris, pH 8, 150 mM NaCl, 0.5% SDS, and 0.5% Triton X-100 and assayed for total protein concentration using a BCA assay (Thermo Fisher). Cytokine production in vehicle-treated controls peaked at a DMSO concentration that was needed to achieve 1 μM compound treatment dose, in an apparent MIND4-17–independent DMSO-mediated effect. Despite this unexplained stimulation by this DMSO concentration, inhibitory effects of MIND4-17 on cytokine production in primary human control and HD monocytes were readily detectable.
To test for toxic effects of MIND4-17 on primary human leukocyte cultures, monocytes were treated with MIND4-17 but not IFN-γ and LPS, mimicking the treatment duration used in the cytokine profiling experiments (Fig. S5). Cell death was measured using a lactate dehydrogenase (LDH) assay (CytoTox-Fluor Cytotoxicity Assay, Promega).
Human iPS Cell Lines and Differentiation of Neuronal Stem Cells.
iPSCs were maintained on MEF feeders prepared using mitomycin C. Normal growth medium was as follows: knockout DMEM/F12 (Life Technologies; 12660-012) with 20% knockout serum replacement (Life Technologies; 10828-028), 1 mM l-glutamine (Life Technologies; 35050-061), 1× MEM nonessential amino acids (Life Technologies; 11140-050), 100 µM 2-mercaptoethanol, and 50 units/mL penicillin/streptomycin (Life Technologies; 15070-063). Genomic DNA was prepared using DNeasy Mini Kit (Invitrogen) from ∼106 cells pelleted after accutase passage. Twenty-five microliters of reaction mix included 0.25 µg of genomic DNA, 1× GoTaq Buffer (Promega), 1× GoTaq DNA Polymerase (Promega), 200 µM DNTPs, 200 nM forward/reverse primers, and 5% DMSO. PCR products were run on a 4% NuSieve 3:1 Agarose gel (Lonza) at 95 V for 3 h. Primers and PCR amplification protocol of CAG triplet repeats have been used as previously described (99). Genotype was confirmed by PCR amplification of genomic DNA (Fig. S6A).
NSCs were established based on a protocol described for mouse ES cells (82). iPSCs were passaged with 1 mg/mL collagenase type I (Life Technologies; 17018-029) in DMEM (Life Technologies; 11995-073) onto 0.1% gelatin-coated plates (Sigma; G1393) with NSC media: advanced DMEM/F12 (Life Technologies; 12634-028) with 2 mM l-glutamine, 50 units/mL penicillin/streptomycin, retinoic acid-free B27 supplement (Life Technologies; 12587-010), N2 supplement; supplemented with 100 ng/mL FGF (Invitrogen; PHG0023), 100 ng/mL EGF (Invitrogen; PMG8041), 5 µg/mL heparin (Sigma; H3149), and 1% knockout serum replacement. Over 5–14 d, bipolar cells were observed to stream out of attached colonies. When cells had formed a 60–80% confluent monolayer of bipolar cells, cells were passaged with Accutase (Sigma; A6964) for 2–5 min until detached, and then centrifuged for 3 min, and the remaining clumps of cells were disrupted with a p1000 pipet tip. Cells were then replated as a single-cell suspension 1:1 onto 0.1% gelatin-coated plastic, in NSC media, without additional knockout serum and with 20 ng/mL FGF, 20 ng/mL EGF, and 5 µg/mL heparin. Cells were maintained in NSC media, without knockout serum and with 20 ng/mL FGF, 20 ng/mL EGF, and 5 µg/mL heparin on 0.1% gelatin-coated plastic and passaged with Accutase as previously described 1:2–1:4 for the first four following passages, then 1:2–1:10 during later passages. NSCs were routinely tested for expression of the radial glial marker nestin by immunofluorescence, and by immunoblotting for presence of neuronal marker β-III-tubulin, absence of astrocytic marker glial fibrillary acid protein (GFAP), and for expression of normal and mutant huntingtin using anti-huntingtin antibody Ab1 (against amino acids 1–17) by Western blot (Fig. S6 B and C).
Discussion
In the present study, we have developed and determined the mechanism of action of a unique and highly selective small-molecule inducer of NRF2 signaling, MIND4-17. The NRF2 signal transduction cascade is initiated by MIND4-17 through covalent modification of the KEAP1 sensor-cysteine C151, mimicking effects of oxidative and electrophilic stress (10, 12, 53, 63). This chemical modification of KEAP1 leads to conformational change and arrest of the KEAP1/NRF2 complex in the closed conformation, disrupting the cycle of degradation of NRF2 (13, 66, 74). Subsequently, this results in accumulation (stabilization) and nuclear translocation of de novo-synthesized NRF2, followed by NRF2-mediated activation of gene expression. Proteomic analysis showed high MIND4-17 selectivity for the KEAP1 target, which makes this compound an attractive probe for investigating the activation of NRF2 signaling (74).
A highly specific KEAP1-modyfing inducer probe such as MIND4-17 has key advantages over genetic manipulations or broadly unspecific oxidants or electrophiles for investigating endogenous biological responses specific to NRF2 signaling in stress. For example, the gene silencing of KEAP1, whose expression is NRF2 dependent, would disrupt an autoregulatory loop (84) and, due to the absence of natural formation of the substrate adaptor and E3 ubiquitin ligase KEAP1/CUL3 complex, would result in overactivation of the NRF2 pathway (47). Moreover, in contrast to small organic inducers like DMF and SFP, which lack selectivity and “SAR-ability,” the unique MIND4-17 scaffold presented here is drug-like and amenable to further chemical modifications to optimize its pharmaceutical properties. In this context, a key pharmacokinetic property requiring further optimization will be brain permeability, which is minimal for MIND4-17 itself and precludes its direct consideration as a clinical candidate.
Although mutant HTT is ubiquitously expressed and impacts multiple cell types across all bodily tissues, we next investigated NRF2 signaling in the nonneuronal cell populations that are the principal mediators of neuroinflammation, namely, microglia, astrocytes, and peripheral monocytes, and in the context of both mouse and human HD model systems. Our results showed that the selective NRF2 inducer MIND4-17 repressed expression of proinflammatory cytokines in primary microglia and astrocytes from HD and WT mice (40, 78). Similar effects were observed in human primary monocytes from HD patients and normal subjects, in which MIND4-17 mediated repression of proinflammatory cytokines in both disease and nondisease cells. Based on these results, we concluded that NRF2 and downstream antiinflammatory pathways are intact and not affected by the HD mutation in these nonneuronal populations.
This finding is especially important as dysfunction in nonneuronal cell types is recognized as a potential contributor to neurodegeneration, in which a critical role for activated microglia in HD has emerged (85). The release of proinflammatory cytokines, associated with the harmful effects of activated microglia in brain, creates a cytotoxic environment for neighboring neurons (34–36). The potential therapeutic benefits of NRF2 activation are increasingly appreciated to originate in glial and CNS-relevant immune cells including microglia and peripheral macrophages (86). Thus, the results from our studies highlight the therapeutic potential of NRF2 signaling to repress proinflammatory processes in the brain parenchyma including astroglia, microglia, and infiltrating peripheral macrophages.
NRF2 activation responses were evaluated in human NSCs derived from patient iPSCs. NSCs can give rise to neurons, astrocytes, and oligodendroglia, and in HD exhibit phenotypes that may be pathologically relevant (83, 87). Moreover, there is evidence that the pool of adult NSCs is depleted in the striatum of HD patient brains (88) and may further contribute to HD pathology over time. Our results showed that, in HD NSCs with a CAG repeat expansion in the range typical for adult-onset disease, NRF2 responses were muted compared with nondisease counterparts. Induction of NRF2 responses were essentially silent in NSCs with an extreme CAG repeat length associated with juvenile onset, but were restored to nondisease levels upon isogenic genetic correction of the CAG expansion. Our data showed that, in the context of human HD NSCs, NRF2 signaling is muted, which was consistent with similar observation in the striatal STHdh mHTTQ111/Q111 cell line (89), although it is yet unclear whether mHTT is directly involved. The results imply a high stress-sensitivity of HD NSCs in adult brain, which could offer an explanation for depletion of these cells in the course of disease (88).
Taken together, our results highlight a differential and complex NRF2-dependent stress response in human brain and emphasize potential therapeutic benefits of NRF2 activation for HD treatment.
Materials and Methods
Compound Source and Storage.
Compounds were procured from ChemBridge Corporation (purity quality control ensured by provided NMR spectroscopy data), dissolved in molecular-biology–grade DMSO to 10 mM stock concentrations, aliquoted, and stored at −80 °C. MIND4-17 was resynthesized (purity, >95%) and showed essentially identical potency of NRF2 activation to MIND4-17 in multiple batches purchased from ChemBridge. Design and schemes for synthesis of MIND4-17 analogs 2872, 2286, 2291, 2907, and 2909 can be found in SI Materials and Methods. DMF was purchased from Sigma and SFP from LKT Laboratories.
Drug Test in MPTP Mouse Model.
MIND4 was solubilized at 5 mg/mL in 7.5% Cremophor EL (BASF)/2.375% ethanol in PBS. See SI Materials and Methods for details on tolerability and brain-permeability studies. Male C57BL/6 mice (∼25 g) from Charles River Laboratories were housed in temperature- and humidity-controlled rooms with a 12-h dark/light cycle and had free access to food and water. Subacute MPTP paradigm (20 mg/kg i.p. injection once daily for 4 d) was used to test dose–response of MIND4 (90) at 30 or 60 mg/kg (i.p. 10 min before and 50 min after each MPTP injection). The effective dose of 60 mg/kg was used in the subsequent experiments. Control animals received saline (control for MPTP) and 10% propylene glycol and 90% dextrose as vehicle for MIND4 by i.p. Mice were killed 7 d after the last MPTP administration and striatal DA and metabolite HVA were determined by high-performance liquid chromatography (HPLC) coupled with electrochemical detection (ECD) (90). Immunostaining for tyrosine hydroxylase (TH), a marker for dopaminergic neurons, was performed using mouse anti-TH antibody (Sigma). Total numbers of TH-positive neurons in the substantia nigra (SN) were counted under blinded conditions using the Bioquant Image Analysis System (R&M Biometrics) (91).
NQO1 Bioassay.
Inducer potency was quantified by use of the NQO1 bioassay in Hepa1c1c7 cells as described (50, 51). In brief, cells grown in 96-well plates were exposed to serial dilutions of each compound for 48 h, and the NQO1 enzyme activity was determined in cell lysates. Results are shown as average values of eight replicate wells. The SD in each case was less than 5%.
Compound Activity Test in the Rat Embryonic Striatal Cell Lines ST14A.
ST14A cells (a generous gift from E. Cattaneo, University of Milan, Milan) (92) stably express either a mutant expanded repeat (128Q) or WT (26Q) 546-aa HTT fragment and were treated with compounds for 24 h as described (93). See SI Materials and Methods for information on antibodies.
Compound Activity Test in Mouse Embryonic Fibroblasts.
MEFs from WT, NRF2-knockout (NRF2-KO), or KEAP1-knockout (KEAP1-KO) mice (59) were cultured in plastic dishes (Invitrogen) coated for 30 min with 0.1% (wt/vol) gelatin. For experiments, cells (250,000 per well) were grown for 24 h on six-well plates, and then treated with solvent control [0.1% DMSO (vol/vol)] or compounds for 24 h. For Western blot analysis, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing 1 protease inhibitor mixture tablet (Roche) per 10 mL of buffer. Proteins were resolved by SDS/PAGE and immunoblotted with specific antibodies against NQO1 (1:1,000) or NRF2 (1:1,000), both gifts from John D. Hayes (University of Dundee, Dundee, UK), and β-actin (Sigma; mouse monoclonal; 1:10,000). Conventional method was used to separate nuclear and cytoplasmic fractions to test NRF2 induction and stabilization (SI Materials and Methods for details).
Cell-Based KEAP1-Target Validation Assay.
COS1 cells were plated 16 h before transfection, followed by cotransfection with plasmids encoding WT, mutant C151S, or C226S/C613S double-mutant KEAP1 and NRF2-V5 (generous gifts from M. MacMahon and John D. Hayes, University of Dundee, Dundee, UK) at 1:1 ratios. A plasmid encoding β-galactosidase was cotransfected to monitor transfection efficiency. Twenty-four hours posttransfection, cells were exposed to MIND4-17 for 3 h, and extracts were prepared and loaded on SDS/PAGE normalized by β-galactosidase activity. Samples were resolved on SDS/PAGE and immunoblotted with antibodies against KEAP1 (1:2,000; rabbit polyclonal; a kind gift from John D. Hayes, University of Dundee, Dundee, UK) and V-5 (1:5,000; mouse monoclonal; Invitrogen).
Fluorescence Lifetime Imaging Analysis of NRF2:KEAP1 Complex.
HEK293 cells (200,000 per dish) grown in standard media in 6-cm glass dishes were cotransfected with constructs encoding EGFP-NRF2 and KEAP1-mCherry using Lipofectamine 2000 (Invitrogen) as described (13). Cells were imaged 24 h posttransfection before and after 1-h exposure to 1 μM MIND4-17. All images were acquired by confocal microscopy using a laser-scanning confocal microscope (LSM780; Carl Zeiss). The microscope was equipped with a thermostatic chamber suitable to maintain the live cells and optics at constant 37 °C. Imaging was performed using a 63× oil-immersion, N.A. 1.4, Plan-Apochromat objective from Zeiss. FLIM was performed as described (94). Specific details of analysis are provided in SI Materials and Methods.
MS Analysis of KEAP1 Modification(s).
Expression and purification of recombinant BTB and full-length KEAP1 proteins are described in detail in SI Materials and Methods. To analyze covalent modification(s) of the BTB domain, 10 µM BTB (48-190 S182A) were incubated with 2 mM MIND4-17 (or 2% DMSO final concentration for control) for 1 h on ice in 20 mM Tris⋅HCl, pH 8.0, and 150 mM NaCl. Intact protein masses were determined through HPLC-coupled electrospray ionization (ESI)–MS on an AB Sciex TripleTOF 5600+ mass spectrometer (Sciex) as described in detail in SI Materials and Methods.
NRF2-Dependent Transcriptional Profiling of MIND4-17 in BV2 Microglia Cells.
The BV2 mouse microglial cell line was a generous gift from Michael Whalen (Massachusetts General Hospital, Boston). Cells were maintained in DMEM media (Invitrogen) supplemented with 10% FBS (Sigma) and antibiotic–antimycotic mix (Invitrogen). Cells were seeded into 12-well plates at the density of 3 × 105 cells per well. Cells were treated with the indicated concentrations of MIND4-17 for 24 h. Cells were then stimulated with 10 ng/mL LPS (Escherichia coli; Sigma) for 2 h. Total RNA was isolated using ZR Miniprep kit (Zymo Research). One milligram of total RNA was used to prepare cDNA (ProtoScript kit; New England Biolabs). Gene expression levels for NQO1, GCLM, IL1, IL6, TNFα, and MCP1 were analyzed by the VeriQuest SYBR green assay (Affymetrix) using a Roche 480 thermocycler. The sequences of gene-specific primers for quantitative PCR (qPCR) can be found in SI Materials and Methods.
MIND4-17 Activity Test on Cytokine IL-6 Release in Primary Mouse Astrocytes and Microglia.
Whole brains were obtained from postnatal 1- to 3-d-old WT and YAC128 mouse pups on the FVB/N strain background and placed in Hanks’ balanced salt solution (Invitrogen) on ice. Meninges were removed, and the remaining brain tissue was placed into growth medium (DMEM, 10% FBS, 1% l-glutamine, 1% penicillin/streptomycin), and homogenized. See details on cell isolation and culturing in SI Materials and Methods. After the initial isolation, microglia and astrocytes were seeded at a density of 1.4 × 105 cells per mL into 96-well tissue culture plates. Twenty-four hours later, the culture medium was replaced with medium containing MIND4-17 at different concentrations in growth medium containing 1% FBS. Twenty-four hours later, medium containing IFN-γ (final concentration, 10 ng/mL; R&D Systems) with CSE (Associates of Cape Cod) at a final concentration of 100 ng/mL was added. Supernatants were collected at 9 h and stored at −20 °C. Cells were lysed, and total protein levels were determined using the micro-BCA kit (Thermo Scientific). Supernatants were analyzed using mouse IL-6 ELISA (e-Bioscience) and IL-6 levels normalized to total protein levels.
MIND4-17 Activity Test on Cytokine Expression in Primary Human HD and Nondisease Monocytes.
All human experiments were performed in accordance with the Declaration of Helsinki and approved by University College London (UCL)/UCL Hospitals Joint Research Ethics Committee. All subjects provided informed written consent. Blood samples were obtained from control subjects and genetically diagnosed, symptomatic HD patients. Patients were classed as having early or moderate-stage disease using the total functional capacity scale (13–7, early; 6–3, moderate) (95). Subjects with inflammatory or infective conditions were excluded.
Information on Cohorts of HD and Healthy Subjects Who Participated in the Study.
The cohorts of HD and healthy subjects who participated in the study were as follows: HD (n = 13): age, 56.84 ± 9.23; CAG repeats, 42.46 ± 1.80; female/male ratio, 53/47; healthy subjects (n = 10): age, 49.98 ± 15.03; female/male ratio, 53/47.
Cells were isolated from whole blood, as previously described (40) (SI Materials and Methods for details). After resting for 16 h, the culture medium was replaced with R10 medium containing either vehicle (DMSO) or MIND4-17 at different concentrations (0.3, 1, and 3 μM). After 24-h treatment, medium was changed again using R10 containing both MIND4-17 at the same concentration and, to stimulate cytokine production, 10 ng/mL IFN-γ (R&D Systems) and 2 mg/mL LPS (Sigma-Aldrich). Supernatants were collected at 24 h and analyzed using the human proinflammatory II (four-plex) MSD assay measuring IL-1β, IL-6, IL-8, and TNFα. Normalized cytokine levels were analyzed on a logarithmic scale due to their skewed distribution. Statistical analysis was performed using a linear mixed model, to allow for correlation between measurements from the same subject. A constant correlation was assumed between all six measurements from each subject, with robust SEs to allow for deviation from this assumption. An advantage of this approach is that linear mixed models provided unbiased estimates of each comparison even if there were missing data, provided the data were assumed to be missing at random. Contrasts of interest were then calculated using linear combinations of parameter estimates.
MIND4-17 Activity Test in Human NSCs.
iPSCs were obtained from National Institute of Neurological Disorders and Stroke (NINDS) Repository at the Coriell Institute for Medical Research: catalog no. ND38554 (named here WT54), catalog no. ND3855 (WT55), catalog no. ND38551 (HD51), and catalog no. ND38548 (HD48). The HD4 cell line harbors a severe juvenile repeat range 72 CAG mutation; in the isogenic-companion cell line HD116c, the pathological mutant CAG expansion was corrected to a nonpathological 21 CAG repeat by homologous recombination (83). Genotype was confirmed by PCR amplification of genomic DNA (Fig. S6A). Please see SI Materials and Methods for additional details on maintenance iPSCs. NSCs were established based on a protocol described for mouse ES cells (82), and detailed protocol is provided in SI Materials and Methods. For compound treatments, cells were plated at 100,000 cells per well on poly-l-lysine/laminin–coated 24-well plates. Compound was diluted to two times the final concentration in complete medium, and then an equal volume of solution was added to each well. The final concentrations of MIND4-17 were 0–3 µM. An equal volume of carrier (DMSO) was used as a control. Cells were treated for 24 h, and then lysed in buffer and analyzed by SDS/PAGE and Western blot.
Supplementary Material
Acknowledgments
We thank Masayuki Yamamoto (Tohoku University) for providing the WT and Nrf2-knockout mice originally used for isolation of WT and NRF2 KO MEFs and for Keap1-knockout KEAP1 KO MEFs; John D. Hayes (University of Dundee) for antibodies against NQO1, NRF2, and KEAP1; and Michael McMahon (University of Dundee) for expression plasmids encoding KEAP1 WT and mutant proteins. This work was supported by NIH Grants U01-NS066912 and R01NS04528, National Institute of General Medical Sciences (NIGMS) Grant GM080356, Biotechnology and Biological Sciences Research Council Grants BB/J007498/1 and BB/L01923X/1, and Cancer Research UK Grant C20953/A18644. We also acknowledge support from RJG Foundation (L.Q., X.C., and A.G.K.) and CHDI Foundation (M.D. and K.K.-G.), from Alzheimer Forschung Initiative Grant 14834 (to C.S.), and from NINDS Grant NS100529 (to L.M.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614943114/-/DCSupplemental.
References
- 1.Itoh K, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 3.Baird L, Dinkova-Kostova AT. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol. 2011;85:241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 4.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol Chem. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 6.Tong KI, et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 9.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci USA. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baird L, Llères D, Swift S, Dinkova-Kostova AT. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc Natl Acad Sci USA. 2013;110:15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baird L, Dinkova-Kostova AT. Diffusion dynamics of the Keap1-Cullin3 interaction in single live cells. Biochem Biophys Res Commun. 2013;433:58–65. doi: 10.1016/j.bbrc.2013.02.065. [DOI] [PubMed] [Google Scholar]
- 15.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 16.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi EH, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Dinkova-Kostova AT, Talalay P. Coordinate regulation of enzyme markers for inflammation and for protection against oxidants and electrophiles. Proc Natl Acad Sci USA. 2008;105:15926–15931. doi: 10.1073/pnas.0808346105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DH, Gold R, Linker RA. Mechanisms of oxidative damage in multiple sclerosis and neurodegenerative diseases: Therapeutic modulation via fumaric acid esters. Int J Mol Sci. 2012;13:11783–11803. doi: 10.3390/ijms130911783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knatko EV, et al. Nrf2 activation protects against solar-simulated ultraviolet radiation in mice and humans. Cancer Prev Res. 2015;8:475–486. doi: 10.1158/1940-6207.CAPR-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browne SE, Beal MF. Oxidative damage in Huntington’s disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 22.Möller T. Neuroinflammation in Huntington’s disease. J Neural Transm (Vienna) 2010;117:1001–1008. doi: 10.1007/s00702-010-0430-7. [DOI] [PubMed] [Google Scholar]
- 23.The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 24.Sorolla MA, et al. Protein oxidation in Huntington disease. Biofactors. 2012;38:173–185. doi: 10.1002/biof.1013. [DOI] [PubMed] [Google Scholar]
- 25.Stack EC, Matson WR, Ferrante RJ. Evidence of oxidant damage in Huntington’s disease: Translational strategies using antioxidants. Ann N Y Acad Sci. 2008;1147:79–92. doi: 10.1196/annals.1427.008. [DOI] [PubMed] [Google Scholar]
- 26.Li X, et al. Aberrant Rab11-dependent trafficking of the neuronal glutamate transporter EAAC1 causes oxidative stress and cell death in Huntington’s disease. J Neurosci. 2010;30:4552–4561. doi: 10.1523/JNEUROSCI.5865-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johri A, Beal MF. Antioxidants in Huntington’s disease. Biochim Biophys Acta. 2012;1822:664–674. doi: 10.1016/j.bbadis.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsunemi T, et al. PGC-1α rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorolla MA, et al. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45:667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Ellrichmann G, et al. Efficacy of fumaric acid esters in the R6/2 and YAC128 models of Huntington’s disease. PLoS One. 2011;6:e16172. doi: 10.1371/journal.pone.0016172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stack C, et al. Triterpenoids CDDO-ethyl amide and CDDO-trifluoroethyl amide improve the behavioral phenotype and brain pathology in a transgenic mouse model of Huntington’s disease. Free Radic Biol Med. 2010;49:147–158. doi: 10.1016/j.freeradbiomed.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonifati DM, Kishore U. Role of complement in neurodegeneration and neuroinflammation. Mol Immunol. 2007;44:999–1010. doi: 10.1016/j.molimm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Pizza V, Agresta A, D’Acunto CW, Festa M, Capasso A. Neuroinflamm-aging and neurodegenerative diseases: An overview. CNS Neurol Disord Drug Targets. 2011;10:621–634. doi: 10.2174/187152711796235014. [DOI] [PubMed] [Google Scholar]
- 34.Nagatsu T, Sawada M. Inflammatory process in Parkinson’s disease: Role for cytokines. Curr Pharm Des. 2005;11:999–1016. doi: 10.2174/1381612053381620. [DOI] [PubMed] [Google Scholar]
- 35.Phani S, Loike JD, Przedborski S. Neurodegeneration and inflammation in Parkinson’s disease. Parkinsonism Relat Disord. 2012;18:S207–S209. doi: 10.1016/S1353-8020(11)70064-5. [DOI] [PubMed] [Google Scholar]
- 36.Smith JA, Das A, Ray SK, Banik NL. Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res Bull. 2012;87:10–20. doi: 10.1016/j.brainresbull.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalrymple A, et al. Proteomic profiling of plasma in Huntington’s disease reveals neuroinflammatory activation and biomarker candidates. J Proteome Res. 2007;6:2833–2840. doi: 10.1021/pr0700753. [DOI] [PubMed] [Google Scholar]
- 38.Silvestroni A, Faull RL, Strand AD, Möller T. Distinct neuroinflammatory profile in post-mortem human Huntington’s disease. Neuroreport. 2009;20:1098–1103. doi: 10.1097/WNR.0b013e32832e34ee. [DOI] [PubMed] [Google Scholar]
- 39.Sapp E, et al. Early and progressive accumulation of reactive microglia in the Huntington disease brain. J Neuropathol Exp Neurol. 2001;60:161–172. doi: 10.1093/jnen/60.2.161. [DOI] [PubMed] [Google Scholar]
- 40.Björkqvist M, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wild E, et al. Abnormal peripheral chemokine profile in Huntington’s disease. PLoS Curr. 2011;3:RRN1231. doi: 10.1371/currents.RRN1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavese N, et al. Microglial activation correlates with severity in Huntington disease: A clinical and PET study. Neurology. 2006;66:1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- 43.Tai YF, et al. Imaging microglial activation in Huntington’s disease. Brain Res Bull. 2007;72:148–151. doi: 10.1016/j.brainresbull.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 44.Frank-Cannon TC, Alto LT, McAlpine FE, Tansey MG. Does neuroinflammation fan the flame in neurodegenerative diseases? Mol Neurodegener. 2009;4:47. doi: 10.1186/1750-1326-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Politis M, et al. Microglial activation in regions related to cognitive function predicts disease onset in Huntington’s disease: A multimodal imaging study. Hum Brain Mapp. 2011;32:258–270. doi: 10.1002/hbm.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinti L, et al. SIRT2- and NRF2-targeting thiazole-containing compound with therapeutic activity in Huntington’s disease models. Cell Chem Biol. 2016;23:849–861. doi: 10.1016/j.chembiol.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Williamson TP, Johnson DA, Johnson JA. Activation of the Nrf2-ARE pathway by siRNA knockdown of Keap1 reduces oxidative stress and provides partial protection from MPTP-mediated neurotoxicity. Neurotoxicology. 2012;33:272–279. doi: 10.1016/j.neuro.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaidery NA, et al. Targeting Nrf2-mediated gene transcription by extremely potent synthetic triterpenoids attenuate dopaminergic neurotoxicity in the MPTP mouse model of Parkinson’s disease. Antioxid Redox Signal. 2013;18:139–157. doi: 10.1089/ars.2011.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bezard E, Przedborski S. A tale on animal models of Parkinson’s disease. Mov Disord. 2011;26:993–1002. doi: 10.1002/mds.23696. [DOI] [PubMed] [Google Scholar]
- 50.Prochaska HJ, Santamaria AB. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: A screening assay for anticarcinogenic enzyme inducers. Anal Biochem. 1988;169:328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- 51.Fahey JW, Dinkova-Kostova AT, Stephenson KK, Talalay P. The “Prochaska” microtiter plate bioassay for inducers of NQO1. Methods Enzymol. 2004;382:243–258. doi: 10.1016/S0076-6879(04)82014-7. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: Isolation and elucidation of structure. Proc Natl Acad Sci USA. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takaya K, et al. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic Biol Med. 2012;53:817–827. doi: 10.1016/j.freeradbiomed.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fox RJ, et al. BG-12 (dimethyl fumarate): A review of mechanism of action, efficacy, and safety. Curr Med Res Opin. 2014;30:251–262. doi: 10.1185/03007995.2013.849236. [DOI] [PubMed] [Google Scholar]
- 55.Outeiro TF, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 56.Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 57.Wakabayashi N, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benedict AL, Knatko EV, Dinkova-Kostova AT. The indirect antioxidant sulforaphane protects against thiopurine-mediated photooxidative stress. Carcinogenesis. 2012;33:2457–2466. doi: 10.1093/carcin/bgs293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ludtmann MH, Angelova PR, Zhang Y, Abramov AY, Dinkova-Kostova AT. Nrf2 affects the efficiency of mitochondrial fatty acid oxidation. Biochem J. 2014;457:415–424. doi: 10.1042/BJ20130863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ujjainwalla F, Walsh TF. Total syntheses of 6-and 7-azaindole derived GnRH antagonists. Tetrahedron Lett. 2001;42:6441–6445. [Google Scholar]
- 61.Polucci P, et al. Alkylsulfanyl-1,2,4-triazoles, a new class of allosteric valosine containing protein inhibitors. Synthesis and structure-activity relationships. J Med Chem. 2013;56:437–450. doi: 10.1021/jm3013213. [DOI] [PubMed] [Google Scholar]
- 62.Hur W, et al. A small-molecule inducer of the antioxidant response element. Chem Biol. 2010;17:537–547. doi: 10.1016/j.chembiol.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 63.Eggler AL, Luo Y, van Breemen RB, Mesecar AD. Identification of the highly reactive cysteine 151 in the chemopreventive agent-sensor Keap1 protein is method-dependent. Chem Res Toxicol. 2007;20:1878–1884. doi: 10.1021/tx700217c. [DOI] [PubMed] [Google Scholar]
- 64.Fourquet S, Guerois R, Biard D, Toledano MB. Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation. J Biol Chem. 2010;285:8463–8471. doi: 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Linker RA, et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134:678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- 66.Cleasby A, et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9:e98896. doi: 10.1371/journal.pone.0098896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qian Y, et al. An isotopically tagged azobenzene-based cleavable linker for quantitative proteomics. ChemBioChem. 2013;14:1410–1414. doi: 10.1002/cbic.201300396. [DOI] [PubMed] [Google Scholar]
- 69.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 70.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)—a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 72.Baird L, Swift S, Llères D, Dinkova-Kostova AT. Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnol Adv. 2014;32:1133–1144. doi: 10.1016/j.biotechadv.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bertrand HC, et al. Design, synthesis, and evaluation of triazole derivatives that induce Nrf2 dependent gene products and inhibit the Keap1-Nrf2 protein-protein interaction. J Med Chem. 2015;58:7186–7194. doi: 10.1021/acs.jmedchem.5b00602. [DOI] [PubMed] [Google Scholar]
- 74.Jung KA, Kwak MK. The Nrf2 system as a potential target for the development of indirect antioxidants. Molecules. 2010;15:7266–7291. doi: 10.3390/molecules15107266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Innamorato NG, et al. The transcription factor Nrf2 is a therapeutic target against brain inflammation. J Immunol. 2008;181:680–689. doi: 10.4049/jimmunol.181.1.680. [DOI] [PubMed] [Google Scholar]
- 76.Koh K, et al. Transcription factor Nrf2 suppresses LPS-induced hyperactivation of BV-2 microglial cells. J Neuroimmunol. 2011;233:160–167. doi: 10.1016/j.jneuroim.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 77.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Träger U, et al. JAK/STAT signalling in Huntington's disease immune cells. PloS Curr. 2013 doi: 10.1371/currents.hd.5791c897b5c3bebeed93b1d1da0c0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slow EJ, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 80.Kwan W, et al. Bone marrow transplantation confers modest benefits in mouse models of Huntington’s disease. J Neurosci. 2012;32:133–142. doi: 10.1523/JNEUROSCI.4846-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 82.Ritch JJ, et al. Multiple phenotypes in Huntington disease mouse neural stem cells. Mol Cell Neurosci. 2012;50:70–81. doi: 10.1016/j.mcn.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.An MC, et al. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee OH, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J Biol Chem. 2007;282:36412–36420. doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- 85.Crotti A, et al. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci. 2014;17:513–521. doi: 10.1038/nn.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lastres-Becker I, et al. Fractalkine activates NRF2/NFE2L2 and heme oxygenase 1 to restrain tauopathy-induced microgliosis. Brain. 2014;137:78–91. doi: 10.1093/brain/awt323. [DOI] [PubMed] [Google Scholar]
- 87.HD iPSC Consortium Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ernst A, et al. Neurogenesis in the striatum of the adult human brain. Cell. 2014;156:1072–1083. doi: 10.1016/j.cell.2014.01.044. [DOI] [PubMed] [Google Scholar]
- 89.Jin YN, et al. Impaired mitochondrial dynamics and Nrf2 signaling contribute to compromised responses to oxidative stress in striatal cells expressing full-length mutant huntingtin. PLoS One. 2013;8:e57932. doi: 10.1371/journal.pone.0057932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu K, et al. Estrogen prevents neuroprotection by caffeine in the mouse 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of Parkinson’s disease. J Neurosci. 2006;26:535–541. doi: 10.1523/JNEUROSCI.3008-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen X, et al. Disrupted and transgenic urate oxidase alter urate and dopaminergic neurodegeneration. Proc Natl Acad Sci USA. 2013;110:300–305. doi: 10.1073/pnas.1217296110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ehrlich ME, et al. ST14A cells have properties of a medium-size spiny neuron. Exp Neurol. 2001;167:215–226. doi: 10.1006/exnr.2000.7551. [DOI] [PubMed] [Google Scholar]
- 93.Quinti L, et al. Evaluation of histone deacetylases as drug targets in Huntington's disease models. Study of HDACs in brain tissues from R6/2 and CAG140 knock-in HD mouse models and human patients and in a neuronal HD cell model. PloS Curr. 2010 doi: 10.1371/currents.RRN1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Llères D, James J, Swift S, Norman DG, Lamond AI. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J Cell Biol. 2009;187:481–496. doi: 10.1083/jcb.200907029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marder K, et al. Huntington Study Group Rate of functional decline in Huntington’s disease. Neurology. 2000;54:452–458. doi: 10.1212/wnl.54.2.452. [DOI] [PubMed] [Google Scholar]
- 96.Chopra V, et al. The sirtuin 2 inhibitor AK-7 is neuroprotective in Huntington’s disease mouse models. Cell Rep. 2012;2:1492–1497. doi: 10.1016/j.celrep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen X, et al. The sirtuin-2 inhibitor AK7 is neuroprotective in models of Parkinson’s disease but not amyotrophic lateral sclerosis and cerebral ischemia. PLoS One. 2015;10:e0116919. doi: 10.1371/journal.pone.0116919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 99.Zhang BR, et al. CCG polymorphisms in the huntingtin gene have no effect on the pathogenesis of patients with Huntington’s disease in mainland Chinese families. J Neurol Sci. 2012;312:92–96. doi: 10.1016/j.jns.2011.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.