Significance
Plants produce structurally diverse specialized metabolites, many of which have been exploited in medicine or as pest control agents, whereas some have been incorporated in our daily lives, such as nicotine. In nature, these metabolites serve complex functions for plants’ ecological adaptations to biotic and abiotic stresses. By analyzing two high-quality wild tobacco genomes, we provide an in-depth genomic study that directly associates genome evolution with the assembly and evolution of the nicotine biosynthetic machinery. These results demonstrate the importance of the interplay of gene duplications and transposable element insertions in the evolution of specialized metabolism biosynthetic pathways and illuminate how complex adaptive traits could evolve.
Keywords: Nicotiana genomes, genome-wide multiplications, transposable elements, nicotine biosynthesis, expression divergence
Abstract
Nicotine, the signature alkaloid of Nicotiana species responsible for the addictive properties of human tobacco smoking, functions as a defensive neurotoxin against attacking herbivores. However, the evolution of the genetic features that contributed to the assembly of the nicotine biosynthetic pathway remains unknown. We sequenced and assembled genomes of two wild tobaccos, Nicotiana attenuata (2.5 Gb) and Nicotiana obtusifolia (1.5 Gb), two ecological models for investigating adaptive traits in nature. We show that after the Solanaceae whole-genome triplication event, a repertoire of rapidly expanding transposable elements (TEs) bloated these Nicotiana genomes, promoted expression divergences among duplicated genes, and contributed to the evolution of herbivory-induced signaling and defenses, including nicotine biosynthesis. The biosynthetic machinery that allows for nicotine synthesis in the roots evolved from the stepwise duplications of two ancient primary metabolic pathways: the polyamine and nicotinamide adenine dinucleotide (NAD) pathways. In contrast to the duplication of the polyamine pathway that is shared among several solanaceous genera producing polyamine-derived tropane alkaloids, we found that lineage-specific duplications within the NAD pathway and the evolution of root-specific expression of the duplicated Solanaceae-specific ethylene response factor that activates the expression of all nicotine biosynthetic genes resulted in the innovative and efficient production of nicotine in the genus Nicotiana. Transcription factor binding motifs derived from TEs may have contributed to the coexpression of nicotine biosynthetic pathway genes and coordinated the metabolic flux. Together, these results provide evidence that TEs and gene duplications facilitated the emergence of a key metabolic innovation relevant to plant fitness.
The pyridine alkaloid nicotine, whose addictive properties are well known to humans, is the signature compound of the genus Nicotiana (Solanaceae). In nature, nicotine is arguably one of the most broadly effective plant defense metabolites, in that it poisons acetylcholine receptors and is thereby toxic to all heterotrophs with neuromuscular junctions. Field studies using genetically modified Nicotiana attenuata (coyote tobacco) plants, an annual wild diploid native to western North America, have revealed that this toxin fulfills multifaceted ecological functions that contribute to plant fitness (1, 2). The strong transcriptional up-regulation of the nicotine biosynthetic machinery in roots in response to herbivore attack of the shoot combined with the active translocation and storage of this toxin provides N. attenuata plants with an inducible protection mechanism against a broad spectrum of herbivores (2). In addition, the transport and nonhomogenous distribution of nicotine in the nectar of flowers within an inflorescence modifies the trap-lining behavior of humming bird pollinators to maximize outcrossing rates (3). These two facets of the ecological utility of nicotine result from the prolific production of this toxin, which can accumulate up to 1% of the leaf dry mass in wild tobacco species (4). This prodigious biosynthetic ability is based on an efficient biochemical machinery composed of multiple genes coexpressed in roots (5, 6). In contrast to the deep knowledge on nicotine’s biosynthesis and ecological functions, the evolution of genomic features that facilitated the assembly of a pathway so critical for the survival of Nicotiana species has remained largely unknown.
Gene duplication and transposable element (TE) insertions continuously shape the evolutionary landscape of genomes and can affect the function of genes with adaptive consequences (7, 8). Whereas whole-genome and local gene duplications provide the raw material for the evolution of novel traits, TE mobility can broadly remodel gene expression by redistributing transcription factor binding sites, shaping epigenetic marks and/or providing target sequences for small regulatory RNAs (7–10). Hence, the combination of gene duplications and TE activity is thought to facilitate the evolution of novel adaptive traits (8). However, the details of this process, in particular its role in the evolution of metabolic complexity through the assembly of novel multigene pathways, remains unclear.
Results and Discussion
Genome Sequencing, Assembly, and Annotation.
We sequenced and assembled the genome of N. attenuata, using 30× Illumina short reads, 4.5× 454 reads, and 10× PacBio single-molecule long reads. We assembled 2.37 Gb of sequences representing 92% of the expected genome size. We further generated a 50× optical map and a high-density linkage map for superscaffolding (SI Appendix, Figs. S1 and S2), which anchored 825.8 Mb to 12 linkage groups and resulted in a final assembly with a N50 contig equal to 90.4 kb and a scaffold size of 524.5 kb (SI Appendix, Fig. S3). Likewise, using ∼50× Illumina short reads, we assembled the Nicotiana obtusifolia genome with a 59.5-kb and 134.1-kb N50 contig and scaffold N50 size, respectively. The combined annotation pipeline integrating both hint-guided AUGUSTUS and MAKER2 gene prediction pipelines predicted 33,449 gene models in the N. attenuata genome. More than 71% of these gene models are fully supported by RNA-sequencing (RNA-seq) reads and 12,617 and 18,176 of these genes are orthologous to Arabidopsis and tomato genes, respectively.
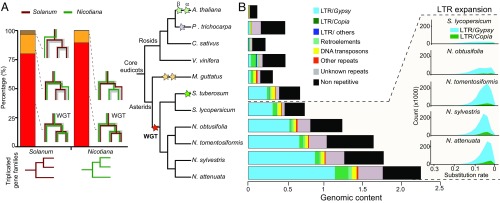
To investigate the evolutionary history of the different Nicotiana genomes, we inferred 23,340 homologous groups using protein sequences from 11 published genomes (SI Appendix, Table S1). A phylogenomic analysis of the identified homologous groups demonstrated that Nicotiana species share a whole-genome triplication (WGT) event with solanaceous species, such as tomato, potato, and Petunia (11), but not with Mimulus (Fig. 1), which is consistent with a large number of duplication events at the Solanaceae branch (SI Appendix, Fig. S4), fourfold degenerate sites (4dTV) between duplicated paralogs (SI Appendix, Fig. S5), and an analysis of the evolutionary origin of threonine deaminase (TD) in Solanaceae (SI Appendix, Fig. S6). At least 3,499 gene pairs originating from this WGT were retained in both Nicotiana and Solanum. Among all retained gene pairs in N. attenuata that resulted from WGT but did not further duplicate in this species, more than 53.7% showed expression divergence (fold change greater than two) in at least one tissue, indicating that these WGT-derived duplicated genes may have evolved divergent functions through neofunctionalization or subfunctionalization.
Fig. 1.
WGT in Nicotiana genomes is shared with other Solanaceae species but the Gypsy retrotransposons expansions are Nicotiana specific. Nicotiana genomes share the WGT with other Solanaceae species. (A, Left) Shared WGT event between Nicotiana and Solanum as revealed by superimposing species tree on the gene tree for the triplicated gene families in Nicotiana and Solanum. Red bar represents the percentage of triplication shared between Nicotiana and Solanum. Yellow bar represents shared duplication events or paralogous loss in one species. Brown bar represents Solanum-specific duplication events. (Right) Phylogenetic tree of 11 plant species and different colored stars indicate previously characterized whole-genome multiplication events. (B) Expansion of Gypsy transposable elements contributes substantially to genome size evolution in Nicotiana. (Left) Genomic content (1C content in gigabases) of repetitive versus nonrepetitive sequences in the 11 plant genomes. Black bar indicates nonrepetitive sequences, whereas other colors indicate repetitive sequences. (Right) Visualization of the expansion history of LTR retrotransposons in four Nicotiana genomes in comparison with tomato. The x axis (number of substitutions per site) refers to the divergence of a LTR from its closest paralog in the genome, with smaller numbers indicating more recent amplification events.
Expansion of Transposable Elements in Nicotiana.
Polyploidization is often associated with a burst of TE activity as a hypothesized consequence of “genomic shock” (12, 13). TEs, especially long terminal repeats (LTRs) are highly abundant in Nicotiana and account for 81.0% and 64.8% of the N. attenuata and N. obtusifolia genomes, representing significantly higher proportions than in other sequenced Solanaceae genomes, such as tomato and potato (Fig. 1). An analysis of the history of TE insertions revealed that all Nicotiana species experienced a recent wave of Gypsy retrotransposon expansion. However, this expansion of Gypsy copies was less pronounced in N. obtusifolia compared with other Nicotiana species analyzed, which accounts for the smaller genome size of N. obtusifolia. A recent study showed that Capsicum species also experienced a large expansion of their Gypsy repertoire (14), albeit earlier than in Nicotiana, indicating that after WGT, the different Solanaceae lineages independently experienced the processes of Gypsy proliferation.
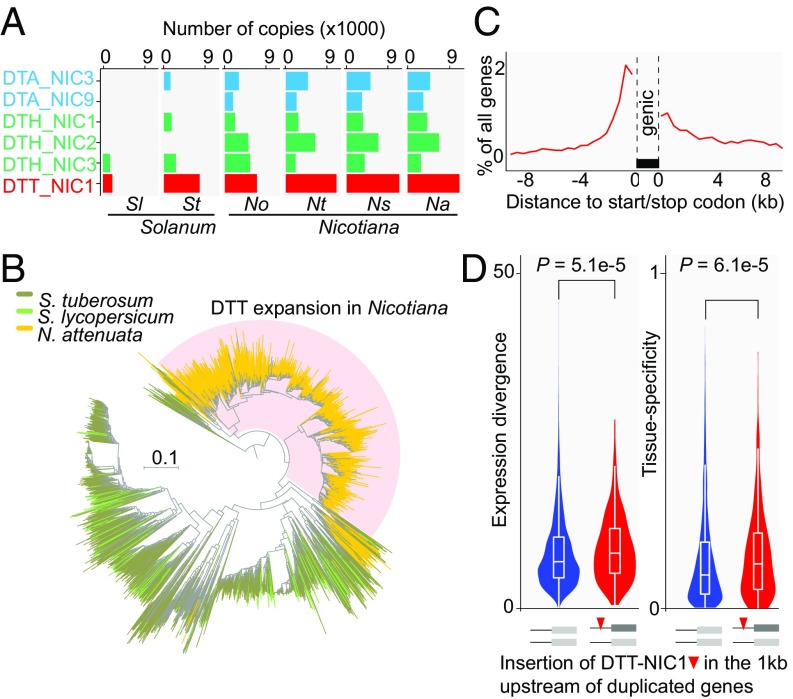
In addition to LTRs, miniature inverted repeat transposable elements (MITEs), which are derived from truncated autonomous DNA transposons, may also play evolutionary roles. Although the size of MITEs is generally small, typically less than 600 bp, MITEs are often located adjacent to genes and are often transcriptionally active. As such, they have been hypothesized to contribute to the evolution of gene regulation (15, 16). In total, we identified 13 MITE families in the genome of N. attenuata, several of them having rapidly and specifically expanded in Nicotiana species (Fig. 2 A and B). Among these expanded MITE families, a Solanaceae-specific subgroup of the Tc1/Mariner defined by DTT-NIC1 is the most abundant. By analyzing insertion positions of this subgroup, we found that DTT-NIC1 copies, similar to other DNA transposons, are significantly enriched within a 1-kb region upstream of the genes in N. attenuata (Fig. 2C). Analyses on the herbivory-induced conserved transcriptomic responses in Nicotiana further showed that insertions of DTT-NIC1 are significantly enriched within the 1-kb upstream region of herbivory-induced early defense signaling genes in N. attenuata, and may have contributed to the recruitment of genes into the induced defense signaling network by introducing WRKY transcription factor binding sites (17).
Fig. 2.
Expansion of transposable elements of the family DTT-NIC1 increased genome-wide gene expression divergence among duplicated genes in Nicotiana. (A) Copy number of the six most abundant Nicotiana MITE families of transposable elements in Nicotiana and Solanum. Each bar depicts the total number of copies in each species for the six main MITE TEs. MITE families are visualized by different colors: light blue, DTA (hAT); green, DTH (PIF/Harbinger); red, DTT (Tc1/Mariner). DTT-NIC1 from the Tc1/Mariner family is the most abundant of all MITE TEs. Nicotiana species: Na, N. attenuata; No, N. obtusifolia; Ns, N. sylvestris; Nt, N. tomentosiformis. Solanum species: Sl, Solanum lycopersicum; St, Solanum tuberosum. (B) DTT-NIC1 insertions are enriched in the upstream regions of coding sequences. The line indicates the percentage of genes, among all predicted protein coding genes, that contain DTT-NIC1 insertions within a given 500-bp sliding window. (C) Expansion of the DTT-NIC1 family in Nicotiana species. Neighbor joining (NJ) tree of the DTT-NIC1 family in N. attenuata, tomato, and potato. The scale indicates the branch length. The shaded clade highlights the pronounced expansion of DTT-NIC1 in N. attenuata. (D) Insertions of DTT-NIC1 within the 1-kb upstream region of duplicated genes increased tissue-level gene expression divergence (Wilcoxon rank sum test). (Left and Right) Violin plots of the divergences between gene pairs at expression and tissue specificity levels, respectively. Gene expression divergence was calculated based on the Euclidian distance between the expression profiles of gene pairs. Tissue specificity divergence was calculated based on changes of the τ-index between gene pairs. Because tissue specificity (τ-index) and expression divergence were calculated based on log2 transformed transcripts per million (TPM) values, a small shift in the mean represents a relatively large effect. Red bars indicate duplicated pairs, of which one copy has at least one DTT-NIC1 insertion and the other does not. Blue bars indicate duplicated pairs, both of which lack DTT-NIC1 insertions. The width of the probability density represented by the violin plots along the bars corresponds to the number of duplicate gene pairs.
Innovations in metabolic and signaling network architecture are thought to result from the rapid rewiring of tissue-level gene expression patterns following duplication events (18). To examine this inference, we compared the genome-wide expression divergence between gene pairs that resulted from only one round of gene duplication and analyzed the effects of DTT-NIC1 insertions into 1-kb upstream regions of each member of the gene pairs. Insertions of the DTT-NIC1 family were associated with significant divergences in expression and tissue specificity between the gene pairs (Fig. 2D), consistent with the hypothesis that the expansion of this TE family was a critical determinant of genome-wide rewirings of gene regulation occurring postduplication in these Nicotiana species.
Evolution of Nicotine Biosynthesis.
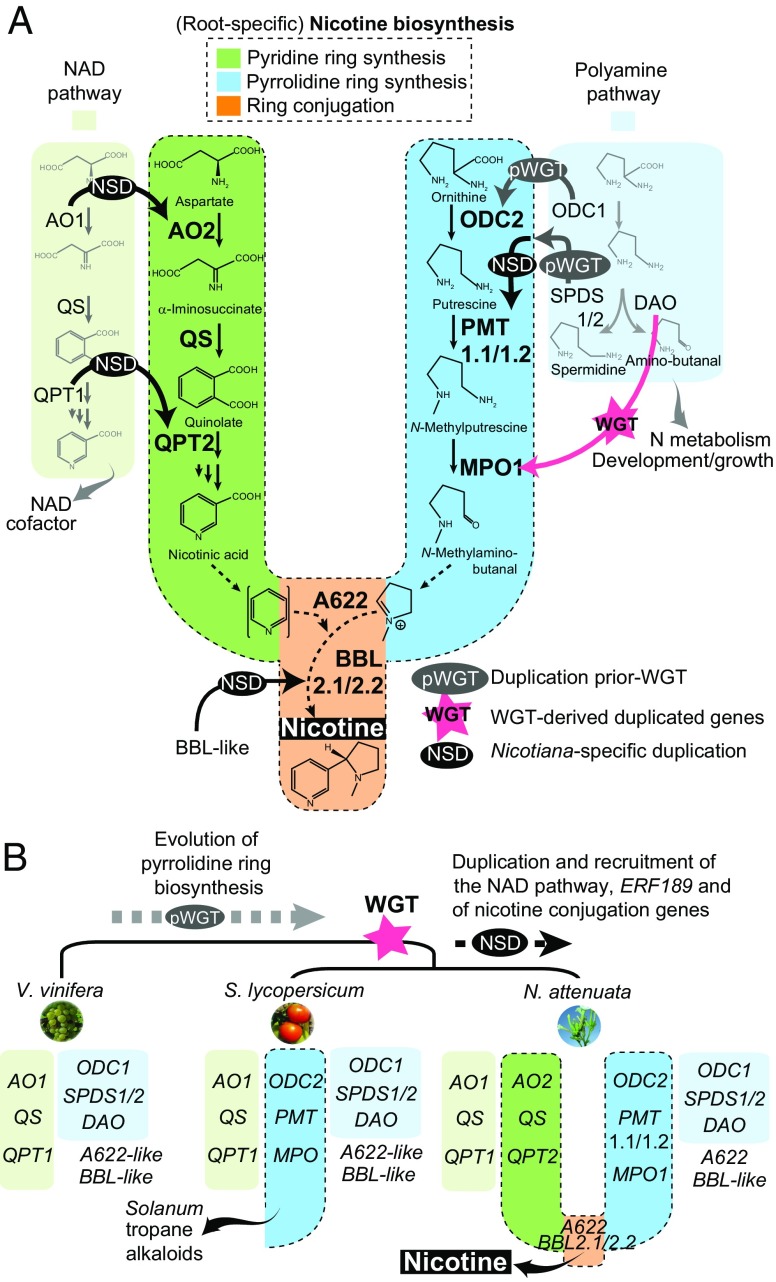
To further understand the role of gene duplication and TE insertions on the evolution of Nicotiana adaptive traits, we reconstructed the evolutionary history of the nicotine biosynthesis pathway, a key defensive innovation of the Nicotiana genus. Nicotine biosynthesis is restricted to the roots and involves the synthesis of a pyridine ring and a pyrrolidine ring, which are coupled most likely via the action of genes coding for an isoflavone reductase-like protein, called A622, and berberine bridge enzyme-like (BBL) enzymes (19, 20) (Fig. 3A). Phylogenomic analyses revealed that genes involved in the biosynthesis of the pyridine and pyrrolidine rings evolved from the duplication of two primary metabolic pathways that are ancient across all major plant lineages: the nicotinamide adenine dinucleotide (NAD) cofactor and polyamine metabolism pathways, respectively (Fig. 3A).
Fig. 3.
Prolific nicotine production in the genus Nicotiana evolved from stepwise duplications of two primary metabolic pathways. (A) Metabolic organization (brightly colored and dashed line outlined branches) and evolution of Nicotiana-specific nicotine biosynthesis via pathway and single gene duplications. Light green and light blue branches on the side indicate the two ancient gene modules with housekeeping functions in plants for the biosynthesis of the NAD cofactor and metabolism of polyamines. Different gene duplication types are indicated by arrows, annotated as follows: NSD, Nicotiana-specific duplications; WGT, whole-genome triplication in Solanaceae; pWGT, gene duplication occurring before WGT. Phylogenetic analyses are presented in SI Appendix, Figs. S7–S14. Nicotiana QS (quinolinate synthase) and A622 did not duplicate but experienced an increase in root expression compared with their tomato homologs (SI Appendix, Figs. S13 and S15). (B) Phylogenomic analysis of grape, tomato, and N. attenuata gene sets highlighting the gradual assembly of the nicotine biosynthetic pathway. Duplication patterns of ODC, PMT, and MPO support the ancient origin of the ornithine-derived N-methyl-Δ1-pyrrolinium, which is notably used as a building block for the biosynthesis of tropane alkaloids in Solanum. AO, aspartate oxidase; BBL, berberine bridge enzyme-like; DAO, diamine oxidase; MPO, N-methylputrescine oxidase; ODC, ornithine decarboxylase; PMT, putrescine methyltransferase; SPDS, spermidine synthase.
However, the timing and mode of duplications of these two pathways differ and reflect the expansion and recruitment of gene sets required for the diversification of alkaloid metabolism in the Solanaceae (Fig. 3B). Duplications that gave rise to the branch extension of the polyamine pathway required for the biosynthesis of the signature alkaloids of Solanaceae and Convolvulaceae (e.g., tropane alkaloids in many genera and nicotine in Nicotiana) are shared among Nicotiana, Solanum, and Petunia with individual gene members recruited from the Solanaceae WGT or earlier duplication events. Genes encoding ornithine decarboxylase (ODC2) and N-putrescine methyltransferase (PMT) duplicated before the shared Solanaceae WGT from their ancestral copies in polyamine metabolism, ODC1 and spermidine synthase (SPDS), respectively (SI Appendix, Figs. S7 and S8). Whereas ODC2 likely retained its ancestral enzymatic function, PMT (derived from SPDS) acquired the capacity to methylate putrescine to form N-methylputrescine through neofunctionalization (21). The N-methylputrescine oxidase (MPO) from the polyamine metabolism pathway evolved from diamine oxidase (DAO) (22) through whole-genome multiplication. Both copies were retained in Nicotiana, Solanum, and Petunia (SI Appendix, Fig. S9), presumably to sustain the flux of N-methyl-Δ1-pyrrolinium required for alkaloid biosynthesis. Duplication patterns of ODC, PMT, and MPO therefore support the ancient origin of the ornithine-derived N-methyl-Δ1-pyrrolinium, which is used as a common building block for the biosynthesis of most alkaloid groups in the Solanaceae and Convolvulaceae (Fig. 3B).
In contrast to the relatively ancient origin of pyrrolidine ring biosynthesis, duplications of the NAD pathway genes, encoding aspartate oxidase (AO) and quinolinic acid phosphoribosyl transferase (QPT), responsible for pyridine ring biosynthesis are Nicotiana specific and likely occurred through local duplication events (SI Appendix, Figs. S10 and S11). BBLs are thought to be involved in the late oxidation step in nicotine biosynthesis that couples the pyridine and pyrrolidine rings, and therefore constitute a key innovation in the Nicotiana-specific synthesis of pyridine alkaloids. BBLs exhibit clear root expression specificity and likely evolved through neofunctionalization after gene duplications (SI Appendix, Fig. S12).
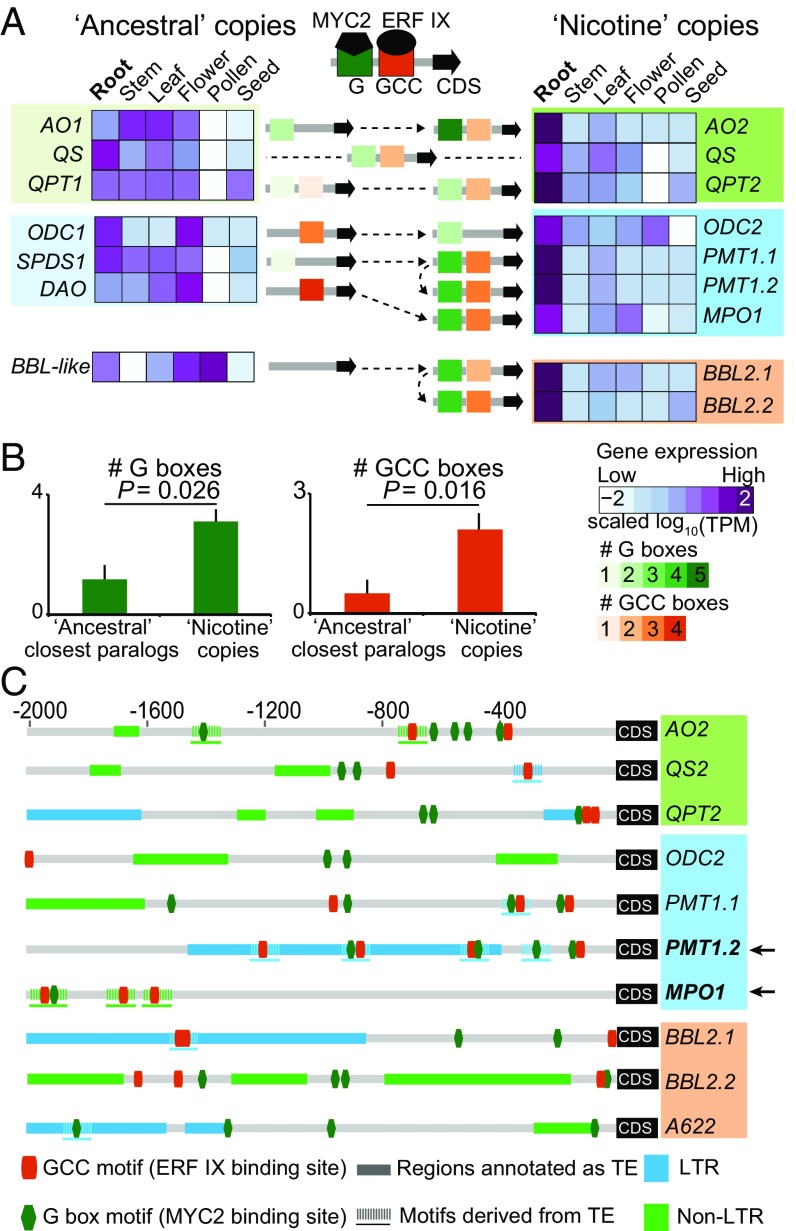
Tissue-level RNA-seq transcriptome analyses in N. attenuata confirmed that, whereas ancestral copies exhibit diverse expression patterns among different tissues, all of the duplicated gene copies recruited for nicotine biosynthesis are specifically expressed in roots (Fig. 4A) as well as transcriptionally up-regulated in response to herbivory via the jasmonate signaling pathway (23). Experimental work has shown that the transcription factors of the ethylene response factor (ERF189) subfamily IX and MYC2, play central roles in the up-regulation of nicotine genes (5). Analyzing the evolutionary history of MYC2 revealed that this gene duplicated at the base of the Solanaceae via genome-wide multiplication or segmental duplications, and two duplicated copies were retained in the genomes of Nicotiana and several Solanaceae species (SI Appendix, Fig. S16). Interestingly, ERF189 is located within the ERF IX cluster (6, 24) as a result of ancestral tandem duplications shared among Nicotiana, Solanum, Capsicum, and Petunia after WGT (SI Appendix, Fig. S17). After the split between Nicotiana and Solanum, ERF189 underwent independent tandem duplications in each linage, but root-specific expression for these duplicated genes is only found in diploid Nicotiana and not in tomato (SI Appendix, Fig. S17). Due to its essential role in regulating the expression of all nicotine genes (6), the acquisition of a root-specific expression by ERF189, likely in the ancestor of Nicotiana species, might have played a critical role for the coordinated root expression of nicotine biosynthesis genes.
Fig. 4.
Coordinated transcriptional regulation of nicotine biosynthetic genes in roots was likely facilitated by transposon-derived transcription factor binding site insertions. (A) Acquisition of transcription factor binding motifs and root-specific expression evolution of nicotine biosynthesis genes (5, 23). Heatmaps depict the scaled expression of nicotine biosynthetic genes and their ancestral copies or closest paralogs across six distinct tissues. Light to dark violet coloration denotes low to high tissue-level expression (TPM, transcripts per million). A622, which likely neofunctionalized without being duplicated in solanaceous species, was not included in this analysis. Gene color categorizations are as used in Fig. 3: light colors correspond to NAD and polyamine primary metabolic pathways and brighter colors to subbranches of the nicotine pathway. The root-specific expression of nicotine biosynthetic genes and dramatic transcriptional up-regulation during insect herbivory is coordinated by the action of MYC2 and ERF IX transcription factors, which respectively target G- and GCC-type boxes in the promoters. Numbers of GCC and G-box motifs detected within 2-kb upstream region of nicotine biosynthetic genes and their ancestral copies are represented using specific color gradients (from light to dark green for increasing number of G boxes and light to dark orange for GCC boxes). (B) Average numbers of G and GCC boxes for the “ancestral” closest paralogs and “nicotine” copies of gene sets. P-values were calculated based on Wilcoxon rank sum test. (C) Many GCC and G-box motifs from nicotine biosynthesis genes are likely derived from TE insertions. Each row depicts the motif and TE annotation of the 2-kb upstream region of an individual nicotine biosynthesis gene. The predicted GCC and G-box motifs are shown in dark orange and dark green small boxes, respectively. The regions that were annotated as TEs from RepeatMasker are shown as rectangles with two different colors. Light blue, LTR; green, non-LTR. Motif sequences and their 150-bp flanking region showed significant homology (E-value less than 1e-5) to annotated TE sequences in N. attenuata are shown in dashed lines. In the case of PMT1.2 and MPO1 (highlighted by black arrows), almost all G and GCC boxes are apparently derived from TE insertions.
Regulation of nicotine genes by MYC2 and ERF189 relies in part on the presence of two transcription factor binding sites, the GCC and G-box elements in their promotor regions (5, 6, 25). Nicotine biosynthetic genes harbor significantly more GCC and G-box elements in their 2-kb upstream regions than do their ancestral copies (Fig. 4B), consistent with the hypothesis that the accumulation of GCC and G-box elements in promoter regions contributed to the evolution of the coordinated transcriptional regulation required for high-flux nicotine biosynthesis. Investigating the origin of GCC and G-box motifs in upstream regions of 10 nicotine biosynthesis genes showed that at least 43% and 29% of GCC and G-box motifs, respectively (Fig. 4C), are likely derived from TE insertions. Whereas it is unclear whether all of these predicted TE-derived GCC and G-box motifs are involved in regulating the expression of nicotine genes, some likely are. For example, in the case of PMT1, a previous experimental study revealed that the 650-bp upstream region, which specifically contained additional TE-derived GCC motifs, had a much larger capacity to drive the expression of a reporter gene in Nicotiana roots than did the 111-bp upstream region that lacked these motifs (26). Furthermore, all GCC and G-box motifs within the 2-kb 5′ region of MPO1 that is likely under control of ERF189 (22) are derived from TEs (Fig. 4C).
The mechanisms of genome organizational evolution, such as genome-wide multiplications and TE expansions, facilitated the evolution of several aspects of the antiherbivore defense arsenal, including a key metabolic innovation in Nicotiana species. These results are consistent with the hypothesis that TEs, which have often been considered as “junk” DNA, can be important orchestrators of the gene expression remodeling that is required for the evolution of adaptive traits. Because native Nicotiana species do not survive in nature without the ability to produce large quantities of nicotine to ward off attackers (27), it might be that this junk has facilitated innovations essential for their survival.
Conclusion
We sequenced, de novo assembled, and annotated the genomes of two Nicotiana species. Several species of this genus have been developed as model systems for studying plant–environment interactions (N. attenuata and N. obtusifolia sequenced in the present study), whereas others are widely used molecular biology experimental systems (Nicotiana benthamiana and Nicotiana tabacum). The fully annotated gene models, transposable elements, smRNAs, and transcriptomic atlas (SI Appendix, Tables S1–S5 and Datasets S1–S9) of N. attenuata will enable additional comparative analyses to illuminate the evolution of specialized metabolites and adaptive traits, not only for this genus, but also more generally within the Solanaceae.
Materials and Methods
Genome Sequencing and Assembly.
For N. attenuata, the Illumina HiSeq2000 system was used to generate a high-coverage whole-genome shotgun sequencing (WGS) of the genome based on short reads (2 × 100 bp or 2 × 120 bp). Different paired-end libraries were constructed using the Illumina TruSeq DNA sample preparation kit v2. The fragment size distribution maxima were observed at 180, 250, 600, and 950 bp. Additionally, two mate-pair libraries were constructed using Illumina mate-pair library preparation kit v2, which had their maxima at 5,500 and 20,000 bp of the fragment size frequency distribution, respectively. A lower genome-wide coverage of long reads (median read length 780 bp) was generated by the Roche/454 GS FLX (+) pyro-sequencing technology using Roche rapid library prep kit v2. For N. obtusifolia, two paired-end libraries and a single mate-pair library were constructed using the same material. The two paired-end libraries had fragment size distribution maxima at 480 and 1,050 bp, respectively. The mate-pair library had a maximum at 3,500 bp. The 20-kb mate-pair libraries were constructed at Eurofins/MWG using the Cre-recombinase circularization approach from Roche (Roche Diagnostics). These were both sequenced with 454 technology and Illumina HiSeq2000 (by removing Roche adaptor sequences and replacing them by Illumina adaptor sequences). The PacBio reads were sequenced at the Cold Spring Harbor Laboratory. The overall workflow and detailed methods are shown in SI Appendix.
Analyses of 248 conserved core eukaryotic genes using the CEGMA (28) pipeline indicated that both N. attenuata and N. obtusifolia assemblies were only slightly less complete in full-length gene contents than that of the tomato genome (86.7%) and similar to the assembly of the potato genome (83.9%) (SI Appendix, Table S2).
Annotation of Transposable Elements.
De novo annotation of repeated elements was performed with RepeatModeler version open-4-0-5 with the parameters “-engine ncbi.” We identified 667 consensus repeat sequences (1.3 Mb total size) in the N. attenuata genome. To classify these consensus repeat sequences, additional annotation using TEclass (29) was applied for repeats that were not classified by RepeatModeler. Among all identified repeats, LTRs, DNA transposons, and long interspersed nuclear elements contributed most, representing 47.5, 28.3, and 9.3%, respectively. The annotated repeats were used for masking repeat sequences using RepeatMasker (open-4.0.5) using parameter “-e ncbi –norna.” We further reannotated transposable elements using the N. attenuata repeat library for two Nicotiana additional genomes: Nicotiana sylvestris and Nicotiana tomentosiformis (30). To make the results comparable, we used the same approach to de novo identify the TE library of Solanum lycopersicum (24) and Solanum tuberosum (31) genomes.
MITEs in Nicotiana were annotated in two steps. First, MITE-Hunter (32) was used to find MITE families in the N. attenuata genome using default parameters, except “-P 0.2.” Following the manual of MITE-Hunter, the identified MITE candidate families were first subjected to coverage evaluation using TARGeT. The output results were manually inspected and only the MITE families that showed even distribution of coverage were selected. Then these selected candidate MITE families were manually checked for their terminal inverted repeats (TIRs) and target site duplications (TSDs). In total, 15 MITE families were initially identified. We then assigned these 15 MITE families to different superfamilies and classes based on sequence homology to a P-MITE database. Two MITE families that showed no homology to any known MITE sequences were excluded from the downstream analysis. Second, using these 13 MITE consensus sequences as a library, we identified the copy number of each MITE family using RepeatMasker with parameters “-nolow -no_is -s -cutoff 250.” A complete MITE sequence was defined as being no more than 3 bp shorter than the representative sequence. The multiple sequence alignment and neighbor joining tree construction of the DTT-NIC1 family were performed using ClustalW.
Annotation of Protein-Coding Genes.
The N. attenuata genome was annotated using the Nicotiana Genome Annotation (NGA) pipeline, which employs both hint-guided (hg) Augustus (v. 2.7) (33) (hg-Augustus) and MAKER2 (v.2.28) (34), gene prediction pipelines based on genome release v1.0. The detailed descriptions of the pipeline are shown in SI Appendix.
Comparative Genomic Analysis.
We assigned genes to homologous groups (HGs) using a similarity-based method. For this method, we used all genes that were predicted from the 11 genomes, listed in SI Appendix, Table S1. All-vs.-all BLAST analysis was used to compare the sequence similarity of all protein coding genes, and the results were filtered based on the following criteria: e-value less than 1e-20; match length greater than 60 amino acids; sequence coverage greater than 60% and identity greater than 50%. All remaining blast results were then clustered into HGs using a Markov clustering algorithm (MLC).
We constructed a phylogenetic tree for all identified HGs using an in-house developed pipeline (SI Appendix).
Evolution of Nicotine Biosynthesis and GCC and G-Box Transcription Factor Binding Sites.
Genes previously characterized as being involved in nicotine biosynthesis were retrieved from the literature and sequences were downloaded from the National Center for Biotechnology Information. Phylogenetic trees for each nicotine biosynthesis gene were constructed as described above. Sequence alignment and tree structures were then manually inspected. Duplication events of nicotine biosynthetic genes were inferred from the phylogenetic tree structures as well as, when possible, from manual examination of syntenic information from the tomato and potato genomes. The genomic organization of the ERF189 gene cluster was obtained from the assembly of N. obtusifolia, in which all homologs of the ERF189 gene cluster were assembled within one scaffold.
We extracted the GCC and G-box motif matrix from the literature (5, 25), and used FIMO (35) to detect the occurrence of these two motifs within the 2-kb upstream regions of both nicotine biosynthesis genes and of their ancestral/nonroot-specific copies. Only the motifs with e-values less than 1e-3 were considered. The e-value cutoff was selected based on comparing the predicted motifs and the motifs that were experimentally validated in the 2-kb upstream region of QPT2 in N. tabaccum (25). It is indeed conceivable that some of the predicted motifs identified by the above approach might be false positives; however, two independent observations lend credence to the accuracy of the overall estimations of motif numbers: (i) Significantly greater numbers of motifs are found in the upstream regions of nicotine genes in comparison with their ancestor copies. If the false positives are high, the number of motifs would be similar between the groups. (ii) The number of G-box/GCC motifs that we identified are overall consistent with the results that were obtained from in vitro assays (5, 6, 25, 36). For example, our predictions revealed the absence of GCC motifs but the presence of G box (Fig. 4C) within the 2-kb upstream region of A622, exactly as previously reported in an experimental study (36).
Manually inspecting the positions of the annotated motif regions revealed that several motifs overlapped with annotations of TEs, such as in the upstream regions of MPO and PMT, indicating that some of these motifs may be derived from TE sequences. To test this hypothesis, we first searched GCC and G-box motif sequences within the consensus TE sequences. Overall, GCC and G-box motifs could be found in more than 54% of these TE consensus sequences. The number of GCC box and G-box motifs per kilobase of sequences in the TE consensus sequences were as high as 19 and 28, respectively. Permutation tests by randomly shuffling the positions of GCC and G boxes 1,000 times in the N. attenuata genome and then comparing with actual TE locations further revealed that these two motifs were significantly enriched in TE regions (P < 0.001). These data reveal that many TEs contain the GCC and G-box motif sequences. Next, we performed additional analyses to calculate the number of the GCC and G-box motifs that were derived from TE insertions that were located within the upstream regions of nicotine biosynthesis genes. For this analysis, we extracted 150-bp sequences that included left and right flanking sequences and the motif sequence in the middle and compared these with the RepeatMasker annotated TE sequences in the N. attenuata genome using YASS (37), a tool designed to search for diverged sequences. To reduce false positives, only the matches that contained the expected motif sequences and had an e-value lower than 1e-5 were considered. Note that the number of GCC and G-box motifs in the nicotine biosynthesis genes that derive from TEs estimated by this approach is likely highly conservative, because this method fails to identify the corresponding homologous sequences in cases where the motif sequences and their flanking regions have diverged significantly from their ancestral TE sequences.
Supplementary Material
Acknowledgments
We thank members of the Department of Molecular Ecology for assistance with the manual curation of gene models and for scientific discussions; Dr. Sang-Gyu Kim and Dr. Matthias Erb for help with RNA sample collections; Dr. Alex Hastie (BioNano Genomics) for assembling of the BioNano optical map; and Prof. Jonathan Gershenzon, Dr. Ewald Grosse-Wilde, and Nicolas Arning for their constructive comments and suggestions on an earlier version of the manuscript. We acknowledge the following sources for funding: Swiss National Science Foundation (PEBZP3-142886 to S.X.); the Marie Curie Intra-European Fellowship (328935 to S.X.); European Research Council Advanced Grant ClockworkGreen (293926 to I.T.B.); DFG Exzellenzinitiative II to the University of Heidelberg (E.G. and A.N.-Q.); and the Max Planck Society, which provided all of the funds for the sequencing. The CoGe platform (www.genomevolution.org) is supported by the National Science Foundation (IOS 1339156 and IOS 1444490).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Whole Genome Shotgun projects of N. attenuata and N. obtusifolia have been deposited at DDBJ/ENA/GenBank under accession nos. MJEQ00000000, MCOF00000000, and MJEQ00000000, respectively. All short reads and PacBio reads used in this study were deposited in NCBI under BioProject PRJNA317743 (RNA-seq reads of N. attenuata transcriptomes), PRJNA316810 (short genomic sequencing reads of N. attenuata), PRJNA317654 (WGS PacBio long genomic reads of N. attenuata), PRJNA316803 (RNA-seq reads and assembly of N. obtusifolia transcriptomes), and PRJNA316794 (short genomic sequencing reads of N. obtusifolia).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700073114/-/DCSupplemental.
References
- 1.Kessler A, Halitschke R, Baldwin IT. Silencing the jasmonate cascade: Induced plant defenses and insect populations. Science. 2004;305:665–668. doi: 10.1126/science.1096931. [DOI] [PubMed] [Google Scholar]
- 2.Steppuhn A, Gase K, Krock B, Halitschke R, Baldwin IT. Nicotine’s defensive function in nature. PLoS Biol. 2004;2:E217. doi: 10.1371/journal.pbio.0020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kessler D, et al. Unpredictability of nectar nicotine promotes outcrossing by hummingbirds in Nicotiana attenuata. Plant J. 2012;71:529–538. doi: 10.1111/j.1365-313X.2012.05008.x. [DOI] [PubMed] [Google Scholar]
- 4.Adler LS, Seifert MG, Wink M, Morse GE. Reliance on pollinators predicts defensive chemistry across tobacco species. Ecol Lett. 2012;15:1140–1148. doi: 10.1111/j.1461-0248.2012.01838.x. [DOI] [PubMed] [Google Scholar]
- 5.Shoji T, Hashimoto T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 2011;52:1117–1130. doi: 10.1093/pcp/pcr063. [DOI] [PubMed] [Google Scholar]
- 6.Shoji T, Kajikawa M, Hashimoto T. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell. 2010;22:3390–3409. doi: 10.1105/tpc.110.078543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley M, Oakey RJ. Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet. 2013;9:e1003234. doi: 10.1371/journal.pgen.1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lisch D. How important are transposons for plant evolution? Nat Rev Genet. 2013;14:49–61. doi: 10.1038/nrg3374. [DOI] [PubMed] [Google Scholar]
- 9.Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: Where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 11.Bombarely A, et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat Plants. 2016;2:16074. doi: 10.1038/nplants.2016.74. [DOI] [PubMed] [Google Scholar]
- 12.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 13.Grandbastien MA, et al. Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet Genome Res. 2005;110:229–241. doi: 10.1159/000084957. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 2014;46:270–278. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- 15.Kuang H, et al. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: New functional implications for MITEs. Genome Res. 2009;19:42–56. doi: 10.1101/gr.078196.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, et al. Evolution of herbivore-induced early defense signaling was shaped by genome-wide duplications in Nicotiana. eLife. 2016;5:e19531. doi: 10.7554/eLife.19531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 19.Kajikawa M, Shoji T, Kato A, Hashimoto T. Vacuole-localized berberine bridge enzyme-like proteins are required for a late step of nicotine biosynthesis in tobacco. Plant Physiol. 2011;155:2010–2022. doi: 10.1104/pp.110.170878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajikawa M, Hirai N, Hashimoto T. A PIP-family protein is required for biosynthesis of tobacco alkaloids. Plant Mol Biol. 2009;69:287–298. doi: 10.1007/s11103-008-9424-3. [DOI] [PubMed] [Google Scholar]
- 21.Minguet EG, Vera-Sirera F, Marina A, Carbonell J, Blázquez MA. Evolutionary diversification in polyamine biosynthesis. Mol Biol Evol. 2008;25:2119–2128. doi: 10.1093/molbev/msn161. [DOI] [PubMed] [Google Scholar]
- 22.Naconsie M, Kato K, Shoji T, Hashimoto T. Molecular evolution of N-methylputrescine oxidase in tobacco. Plant Cell Physiol. 2014;55:436–444. doi: 10.1093/pcp/pct179. [DOI] [PubMed] [Google Scholar]
- 23.Shoji T, Ogawa T, Hashimoto T. Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell Physiol. 2008;49:1003–1012. doi: 10.1093/pcp/pcn077. [DOI] [PubMed] [Google Scholar]
- 24.Sato S, et al. Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoji T, Hashimoto T. Recruitment of a duplicated primary metabolism gene into the nicotine biosynthesis regulon in tobacco. Plant J. 2011;67:949–959. doi: 10.1111/j.1365-313X.2011.04647.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu B, Timko M. Methyl jasmonate induced expression of the tobacco putrescine N -methyltransferase genes requires both G-box and GCC-motif elements. Plant Mol Biol. 2004;55:743–761. doi: 10.1007/s11103-004-1962-8. [DOI] [PubMed] [Google Scholar]
- 27.Machado RA, McClure M, Hervé MR, Baldwin IT, Erb M. Benefits of jasmonate-dependent defenses against vertebrate herbivores in nature. eLife. 2016;5:e13720. doi: 10.7554/eLife.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 29.Abrusán G, Grundmann N, DeMester L, Makalowski W. TEclass: A tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics. 2009;25:1329–1330. doi: 10.1093/bioinformatics/btp084. [DOI] [PubMed] [Google Scholar]
- 30.Sierro N, et al. Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol. 2013;14:R60. doi: 10.1186/gb-2013-14-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X, et al. Potato Genome Sequencing Consortium Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 32.Han Y, Wessler SR. MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38:e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309-12. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoji T, Hashimoto T. DNA-binding and transcriptional activation properties of tobacco NIC2-locus ERF189 and related transcription factors. Plant Biotechnol. 2012;29:35–42. [Google Scholar]
- 37.Noé L, Kucherov G. YASS: Enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 2005;33:W540-3. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.