Abstract
Homeobox a1 (Hoxa1) is one of the most rapidly induced genes in ES cell differentiation and it is the earliest expressed Hox gene in the mouse embryo. In this study, we used genomic approaches to identify Hoxa1-bound regions during early stages of ES cell differentiation into the neuro-ectoderm. Within 2 h of retinoic acid treatment, Hoxa1 is rapidly recruited to target sites that are associated with genes involved in regulation of pluripotency, and these genes display early changes in expression. The pattern of occupancy of Hoxa1 is dynamic and changes over time. At 12 h of differentiation, many sites bound at 2 h are lost and a new cohort of bound regions appears. At both time points the genome-wide mapping reveals that there is significant co-occupancy of Nanog (Nanog homeobox) and Hoxa1 on many common target sites, and these are linked to genes in the pluripotential regulatory network. In addition to shared target genes, Hoxa1 binds to regulatory regions of Nanog, and conversely Nanog binds to a 3′ enhancer of Hoxa1. This finding provides evidence for direct cross-regulatory feedback between Hoxa1 and Nanog through a mechanism of mutual repression. Hoxa1 also binds to regulatory regions of Sox2 (sex-determining region Y box 2), Esrrb (estrogen-related receptor beta), and Myc, which underscores its key input into core components of the pluripotential regulatory network. We propose a model whereby direct inputs of Nanog and Hoxa1 on shared targets and mutual repression between Hoxa1 and the core pluripotency network provides a molecular mechanism that modulates the fine balance between the alternate states of pluripotency and differentiation.
Keywords: gene regulation, Hox genes, pluripotency, Nanog, regulatory networks
Pluripotency and differentiation are two key opposing states integral to the processes of cellular homeostasis. Regulating the proper progression and balance of these two states is not only important in early embryos and embryonic stem (ES) cells, but also critical during organogenesis and morphogenesis for controlling the formation of differentiated cell populations from multipotential progenitors. A variety of studies have defined a core pluripotency gene regulatory network (GRN) that consists of Nanog (Nanog homeobox), Oct4 (octamer-binding transcription factor 4), and Sox (sex-determining region Y box)2, with Klf4 (Kruppel-like factor 4), c-Myc (v-Myc avian myelocytomatosis viral oncogene homolog), Sall4 (Spalt-like transcription factor 4), Esrrb (estrogen-related receptor beta), Utf1 (undifferentiated embryonic cell transcription factor 1), Tet2 (Tet methylcytosine dioxygenase 2), and Glis1 (GLIS family zinc finger 1) representing additional key components of this network (1–4). The core GRN factors actively maintain the pluripotential state by positively modulating the expression of diverse pathways essential for this state, but they also interact with many repressors, like the NuRD (nucleosome remodeling deacetylase) complex, REST (RE1 silencing transcription factor) and co-REST (REST corepressor 1), to inhibit differentiation pathways and maintain the pluripotent state (5–7). Major signaling pathways, such as Wnt, bone morphogenetic protein (BMP)4, and TGF-β, also feed into this GRN to maintain cells in a pluripotency state by modulating expression of core network components (8–11). The expression of the core pluripotency network involves the extensive deployment of auto- and cross-regulatory feedback interactions. For example, Nanog is known to directly activate Oct4 and Sox2, whereas they in turn positively cross-regulate Nanog. Oct4 and Sox2 positively feedback to maintain their own expression via direct autoregulation, whereas Nanog modulates its level of gene expression by negative autoregulation mediated by interactions with Zfp281 (zinc finger protein 281), which recruits the NuRD repressor complex (12–17).
In embryonic stem cells and developing embryos the processes of differentiation and morphogenesis are initiated through the differential response of cells to overlapping and opposing signaling gradients, such as retinoic acid (RA), Fgfs (fibroblast growth factors), and Wnts (18–21). These signaling pathways in turn induce and modulate the expression of master regulators of cellular fate, such as homeobox (Hox) genes, in a spatiotemporal manner. Hox genes play highly conserved roles in modulating regional identity and programs of cellular differentiation in a temporally and spatially restricted manner (21). During RA-induced differentiation of murine ES cells, Hoxa1 is a direct target of RA signaling and is one of the most rapidly induced genes through mechanisms involving control of elongation of paused Pol II (RNA polymerase II) (22–24). This finding is consistent with it being an important early determinant in ES cell differentiation. In murine embryogenesis, Hoxa1 is the earliest the expressed Hox gene (25, 26) and along with Hoxb1, its group 1 paralog, is a direct target of RA signaling in neural and mesodermal tissues through the presence of multiple RA response elements (RAREs) in their flanking cis-regulatory regions (27–30). Loss-of-function and lineage studies have implicated Hoxa1 in development of the inner ear, heart, neural crest specification, and hindbrain patterning (31–33). Growing evidence suggests roles for Hoxa1 in etiology of various cancers through modulation of cell proliferation, invasion, and metastasis (34–37). These diverse functional roles for Hoxa1 appear to be at least partially related to its ability to influence key signaling pathways in differentiating cells (35).
Despite expanding data characterizing the nature of both the pluripotential regulatory network and Hox-dependent differentiation and developmental programs, there is a lack of understanding of how these two distinct yet interrelated programs for controlling cell states interact with each other to maintain an appropriate balance. In this study, we use genome-wide binding analyses of Hoxa1 and Nanog over very early stages of programmed differentiation of murine ES cells. We find evidence that suggests Hoxa1 and Nanog reciprocally regulate a common set of downstream target genes of the pluripotential regulatory network in early stages of differentiation and are involved in direct mutual repression of each other’s expression. This finding suggests a model for Hoxa1–Nanog regulatory interactions that provides insight into how these two independent GRNs may coordinate modulation of the fine balance between the state of pluripotency and differentiation in ES cells.
Results
Dynamic Genome-Wide Occupancy of Hoxa1 and Co-Occupancy of Nanog in Hoxa1-Bound Regions in Early Differentiating ES Cells.
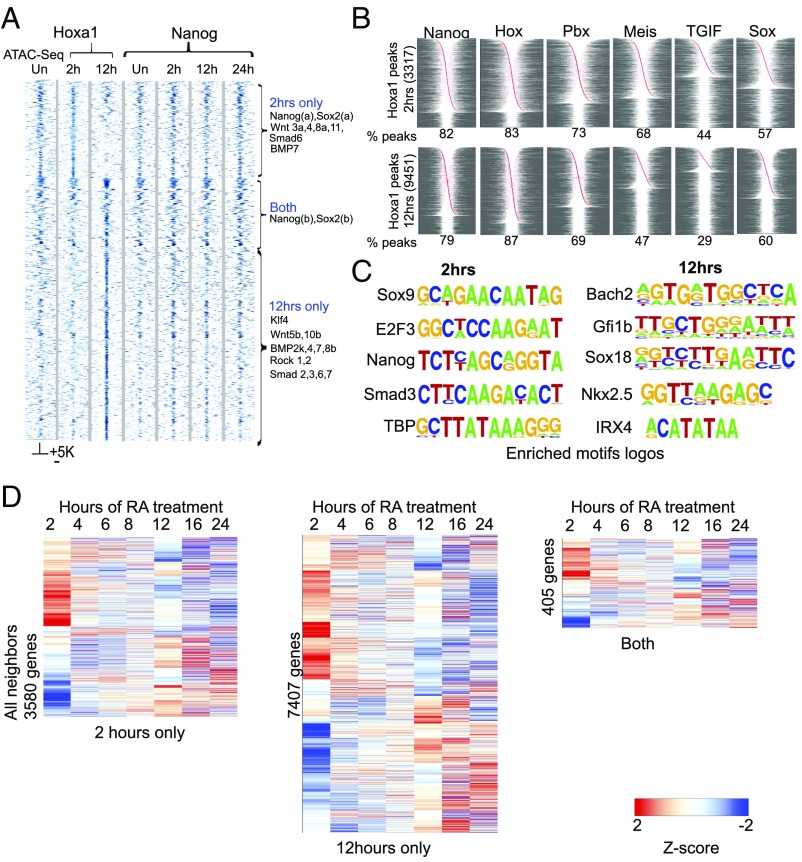
Hox genes are not expressed in ES cells. During RA-induced neuro-ectodermal differentiation of murine ES cells, Hoxa1 is one of the most rapidly induced genes (22, 24). In this RA-induced differentiation paradigm, monitoring events at the level of single cells using methods for single-molecule RNA FISH, we found a robust and relatively uniform RA response by the most rapidly induced genes, including Hoxa1 (22). Following initial activation of Hoxa1, we investigated its early downstream target genes after 2 and 12 h of RA-induced differentiation. We performed chromatin immunoprecipitation and deep sequencing (ChIP-seq) using KH2 ES cells carrying an inducible epitope-tagged version of Hoxa1 (3XFLAG-MYC) (Fig. 1A). At 2 h after RA treatment triggering the initial induction of Hoxa1 expression, there is evidence for genome-wide occupancy of the Hoxa1 protein. In duplicate experiments, we identified 3,317 reproducibly bound regions (Dataset S1). These peaks include occupancy near many genes that play important roles in the regulation of pluripotency [e.g., Nanog, Sox2, Smad6 (SMAD family member 6), BMP7, and Wnts] (Fig. 1A). Surprisingly, by 12 h after RA treatment there is a dramatic shift in the genome-wide binding profile compared with 2 h. There is a loss of occupancy on many of the early target regions bound at 2 h and an expansion of total Hoxa1-binding peaks (9,451), primarily through gain of new sites (Fig. 1A, Fig. S1A, and Dataset S2). Many of these new sites are also adjacent to genes involved in the control of pluripotency (e.g., Klf4, Wnts, and TGF-β pathway members). Only 301 targets are occupied at both the 2- and 12-h time points (Fig. 1A and Fig. S1), but this cohort includes the core pluripotency genes Nanog and Sox2. In light of the relatively homogenous early differentiation of the ES cells (22), these shifts in binding distribution likely reflect differences in the progressive differentiation states between 2 and 12 h, rather than heterogeneity in the population.
Fig. 1.
Dynamic shifts in occupancy of Hoxa1 and co-occupancy of Hoxa1 and Nanog during RA-induced differentiation. (A) Heatmap showing occupancy of Hoxa1 on genomic targets after 2 and 12 h of RA treatment. ATAC-seq for open chromatin states in uninduced ES cells and a time series of Nanog occupancy during differentiation on these same targets is also shown. A ±5-kb region around core binding peaks are shown in the heatmap. Three distinct binding classes designated 2-h only, both, and 12-h only plus examples of genes in each classes are indicated at the right. (B) The panel shows the fraction and relative position of select enriched motifs for Nanog, Hox, Pbx, Meis, TGIF, and Sox in Hoxa1-bound regions at 2 and 12 h of RA treatment. Red dots indicate relative position of the respective associated motif. Peaks are centered on the midpoint of the Hoxa1-bound region and arranged according to distance from center and left to right from center. In cases with the presence of multiple motifs of the same sequence, these are collapsed to a single site. The percentage of Hoxa1 peaks enriched with each motif is shown at the bottom of each graph. (C) Examples of enriched motif logos in regions bound by Hoxa1 after 2 and 12 h of RA treatment. (D) Heatmaps showing differential expression of genes neighboring Hoxa1-bound regions at 2 h, 12 h, or both time points. Values shown are z-scores of log2 expression values from Affymetrix expression profiling over a time course of differentiation compared with levels in uninduced ES cells (22). Hierarchical clustering was done with Euclidean distance, average linkage was thresholded on [−2, 2].
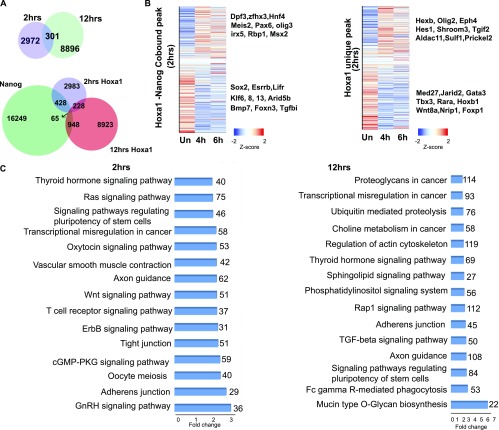
Fig. S1.
Genes and enriched pathways associated with Hoxa1-bound regions in early stages of ES differentiation. (A) Venn diagrams showing common and uniquely bound Hoxa1 regions and Hoxa1 and Nanog co-occupied regions at 2 and 12 h of RA treatment. (B) Heatmaps displaying changes in levels of expression for Hoxa1 and Nanog cobound genes and Hoxa1 unique genes. The heatmap shows z-scores sorted on log2 fold-change. Examples of up- and down-regulated genes are indicated at the right. Hours of RA treatment for the time points are indicated at the bottom. (C) KEGG analysis for the enrichment of signaling pathways among genes adjacent to Hoxa1-bound regions. Enrichment of key signaling pathways related to stem cells and pluripotency is noticeable in this list.
Analysis of sequences within the Hoxa1-bound regions reveals that there are both unique and overlapping cohorts of enriched consensus binding motifs associated with occupancy of Hoxa1 at 2 and 12 h (Fig. 1 B and C and Datasets S3 and S4). Consistent with occupancy of Hoxa1, analysis using TransFac revealed that 83% of the peaks at 2 h and 87% of the peaks at 12 h are enriched for consensus Hox binding motifs positioned near the center of Hoxa1 occupancy (Fig. 1B). In addition to these Hox motifs, we also observed enrichments for a set of other known consensus binding motifs [e.g., Nanog, Pbx (Pre–B-cell leukemia homeobox), Meis (myeloid ecotropic viral integration site 1 homolog), TGIF, and Sox] shared at both of these time points. This finding reveals that there are common motif features in many of the Hoxa1-bound regions and these include consensus sites for Hox, cofactors, and pluripotential transcription factors (TFs) (Fig. 1B). However, in accord with the dynamic differences in occupancy and increased number of peaks bound at 12 h vs. 2 h, de novo motif discovery using Homer analysis revealed differences in both the scope and nature of enriched motifs at these time points (Fig. 1C and Datasets S3 and S4). At 2 h, there is more enrichment for TF consensus binding sites of many core pluripotency factors [e.g., Sox9, Nanog, Smad3, TBP (TATA-binding protein), and E2F3 (E2F transcription factor 3)] (Fig. 1C and Dataset S3). At 12 h we observe a greater degree of enrichment for other motifs. This finding is illustrated by a larger number of peaks with consensus bipartite Hox-Pbx motifs and enrichment for motifs of TFs associated with differentiation [Bach2 (BTB domain and CNC homolog 2), Gfi1b (growth–factor-independent 1B transcriptional repressor), Sox18, Nkx2.5 (NK2 homeobox 5), and Irx4 (Iroquois homeobox 4)] (Fig. 1C and Dataset S4).
In light of the finding that a large number of Hoxa1-bound peaks are enriched for Nanog motifs (82% at 2 h and 79% at 12 h) (Fig. 1B), we investigated physical occupancy of Nanog on these regions by ChIP-seq in a time course of ES cell differentiation (Fig. 1A). There is clear evidence for co-occupancy of Nanog on many of the early and later Hoxa1-bound regions. Unlike Hoxa1, occupancy of Nanog shows little dynamic variation over a 24-h time course of RA treatment the (Fig. 1A). This finding suggests that functionally relevant Nanog motifs are coassociated with many Hoxa1-bound regions in early differentiating ES cells, implying that they may share many common downstream target genes. Furthermore, to explore whether Hoxa1 is remodeling chromatin as a pioneer factor to facilitate accessibility for binding, we used assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) to monitor the openness of chromatin on future Hoxa1 targets in uninduced ES cells. We found that the majority of Hoxa1-bound regions observed at 2 and 12 h are already accessible or open in uninduced cells (Fig. 1A). Consistent with this idea, Nanog is already bound to most of these same regions in uninduced ES cells, which suggests that Hoxa1 is binding to regions that are already open and occupied by other TFs and not acting primarily as a pioneer factor to mediate opening of chromatin.
To examine how occupancy of Hoxa1 correlates with the expression, we analyzed changes in the expression of nearest-neighbor genes over a time course of differentiation (Fig. 1D and Fig. S1B). Over the 24-h period of differentiation, a large number of these genes show dynamic and differential expression compared with uninduced ES cells. It is striking that many of the genes adjacent to Hoxa1-bound regions at 2 h, 12 h, or both time points display large increases or decreases in levels of expression at 2 h (Fig. 1D). Other changes appear over the first 8 h of differentiation, but there is another major shift in the expression profiles that appears at 12 h and continues to dynamically change in later time points. The near adjacent genes cobound by Hoxa1 and Nanog at 2 h also show significant changes in levels of expression over the initial 4- to 6-h period of RA treatment compared with uninduced ES cells (Fig. S1B). Up-regulated genes in this cobound cohort correlate with differentiation pathways [e.g., Meis2, Pax6 (paired box homeotic gene 6), and Msx2 (Msh homeobox 2)] whereas down-regulated genes [e.g., Sox2, Esrrb, and Lifr (leukemia inhibitory factor receptor alpha)] are enriched for components of the pluripotency GRN (Fig. S1B). This finding is in agreement with the idea of dynamic and reciprocal inputs of Hoxa1 and Nanog into common targets. Genes associated with peaks bound by Hoxa1 independent of Nanog also display differential gene expression. This finding suggests additional, Nanog-independent inputs of Hoxa1 into the pluripotential and differentiation GRNs. The genome-wide data indicate that both differentiation and pluripotential genes appear to be downstream targets of Hoxa1 and Nanog, providing a potential regulatory link between the pluripotency and differentiation pathways.
Stem Cell-Signaling Pathways Are Enriched Downstream Targets of Hoxa1.
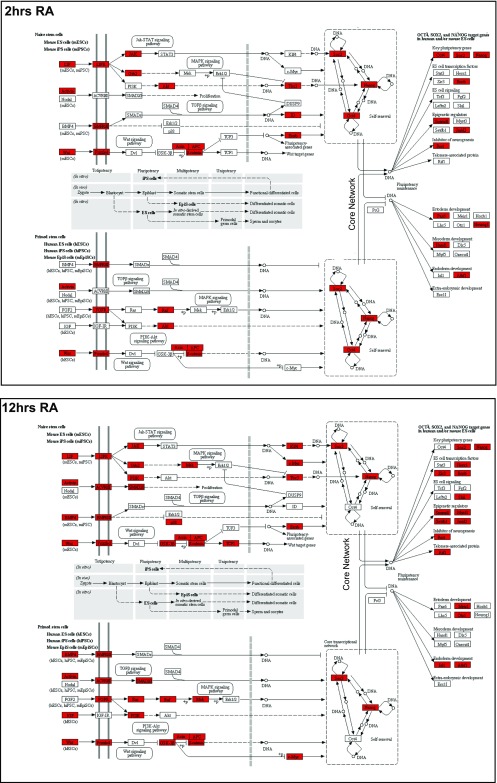
At both the 2- and 12-h time points, Kyoto Encycolpedia of Genes and Genomes (KEGG) pathway analysis of the near adjacent genes bound by Hoxa1 reveals that signaling pathways regulating pluripotency of stems cells and processes of neural differentiation are highly enriched (Figs. S1 and S2 and Dataset S5). Fig. S2 illustrates that many genes associated with both the naïve and primed stem cell-signaling pathways are directly bound by Hoxa1. In accord with the changes in genome-wide binding observed between 2 and 12 h, there are dynamic changes in the occupancy of Hoxa1 near genes in these pathways, as evidenced by the gain and loss of multiple targets (Figs. S1 and S2).
Fig. S2.
Hoxa1 directly binds regions associated with many genes in pluripotency network. The KEGG pathway for pluripotency genes in naïve and primed stem cells is shown and red shaded genes indicate a nearby Hoxa1 binding region. A comparison is show for analysis at 2 h (Upper) and 12 h (Lower). In accord with dynamics of occupancy, a number of new genes acquire Hoxa1 binding in their vicinity at 12 h, whereas some of the genes bound at 2 h are lost.
Even though it is a narrow and early window into the neuronal differentiation program, comparing the putative target genes using KEGG pathway analysis reveals that there are some major differences in the enriched pathways between the two time points (Fig. S1). This finding most likely reflects rapid and progressive changes in the state of differentiation and cellular identity, as illustrated by the dominance of pathways associated more with neuronal differentiation at 12 h (Fig. S1). This analysis emphasizes the wide repertoire and temporal dynamics of Hoxa1 downstream target genes and pathways in one context.
Nanog and Sox2 Are Direct Targets of Hoxa1.
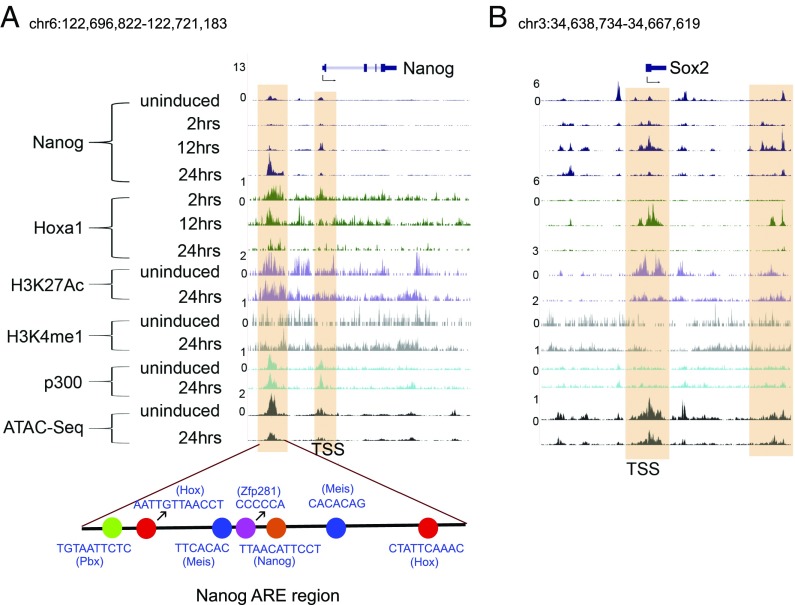
In light of the roles for Nanog and Sox2 as key components of the core pluripotency GRN (6), we looked in more detail at the binding of Hoxa1 to these loci. Fig. 2 shows browser shots of ChIP-seq analyses comparing the binding of Hoxa1, Nanog, p300 (E1A binding protein p300), and epigenetic states as marked by H3K27Ac, H3K4me1, and ATAC-seq. It is interesting that Hoxa1 shows occupancy on both the Nanog transcription start site (TSS) and a 4-kb region upstream (Fig. 2A), which corresponds to a previously identified autoregulatory enhancer (ARE) of Nanog (12). Binding of Nanog and Zfp281 to adjacent sites recruits the NuRD complex and mediates autorepression through this ARE in stem cells to maintain the appropriate levels of Nanog (16, 17). The published work correlates well with the detection of active enhancer marks, p300, and open chromatin over the ARE (Fig. 2A). Multiple Hox and Hox cofactor (Pbx and Meis) sites are located adjacent to the core Nanog motif in the ARE (Fig. 2A). Hoxa1 is recruited to both the TSS and ARE at 2 h and is progressively reduced over a 24-h time course of differentiation. This finding suggests that Hoxa1 directly contributes to the repression and regulation of Nanog during early ES cell differentiation.
Fig. 2.
Regulatory regions of the Nanog and Sox2 core pluripotency genes are bound by Hoxa1. (A) Nanog and (B) Sox2 genomic loci are indicated at the top and below are displayed ChIP-seq and ATAC-seq results for the position of regions bound by a variety of factors, epigenetic marks and open chromatin states in uninduced and differentiated ES cells. The respective time points, factors, and marks are indicated at the left. For Nanog, the distribution of key cis-motifs in a known Nanog ARE enhancer region is shown at the bottom. The shaded regions indicate the multiple domains of Hoxa1 binding in regulatory regions of Nanog and Sox2, such as TSS and distal enhancers. There is evidence for rapid recruitment of Hoxa1 to these regions.
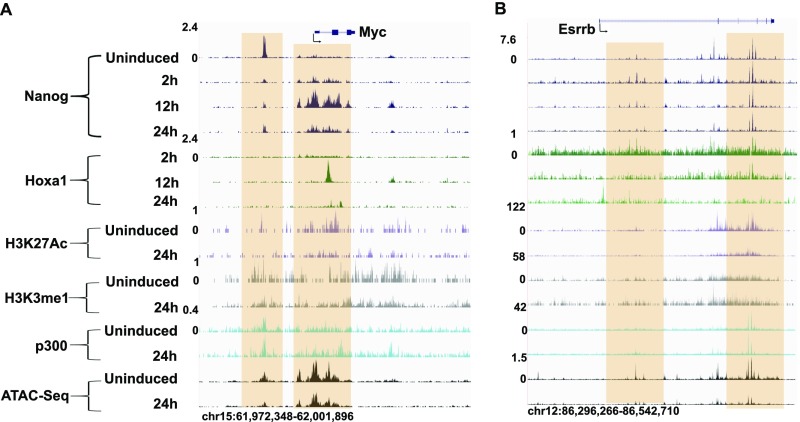
In an analogous manner, Hoxa1 displays transient and early occupancy at both the TSS and a downstream region of Sox2 (Fig. 2B). These sites of Hoxa1 binding are coassociated with Nanog binding and correlate with known regulatory inputs of Nanog into Sox2 regulation (13). A similar pattern of co-occupancy between Nanog and Hoxa1 is also observed on the Myc and Essrb genes (Fig. S3). Together, these data are consistent with the idea that Hoxa1 is rapidly recruited to cis-regulatory regions of major core regulators of the pluripotential GRN and plays a role in both modulating their expression and on common downstream target genes as a part of its function in promoting differentiation.
Fig. S3.
Regulatory regions of the Myc and Esrrb pluripotency genes are bound by Hoxa1. (A) Myc and (B) Esrrb genomic loci are indicated at the top and below are displayed ChIP-seq and ATAC-seq results for the position of regions bound by a variety of factors, epigenetic marks, and open chromatin states in uninduced and differentiated ES cells. The respective time points, factors and marks are indicated at the left. The shaded regions indicate the multiple domains of Hoxa1 binding in putative regulatory regions.
Cross-Regulatory Interactions Between Nanog and Hoxa1.
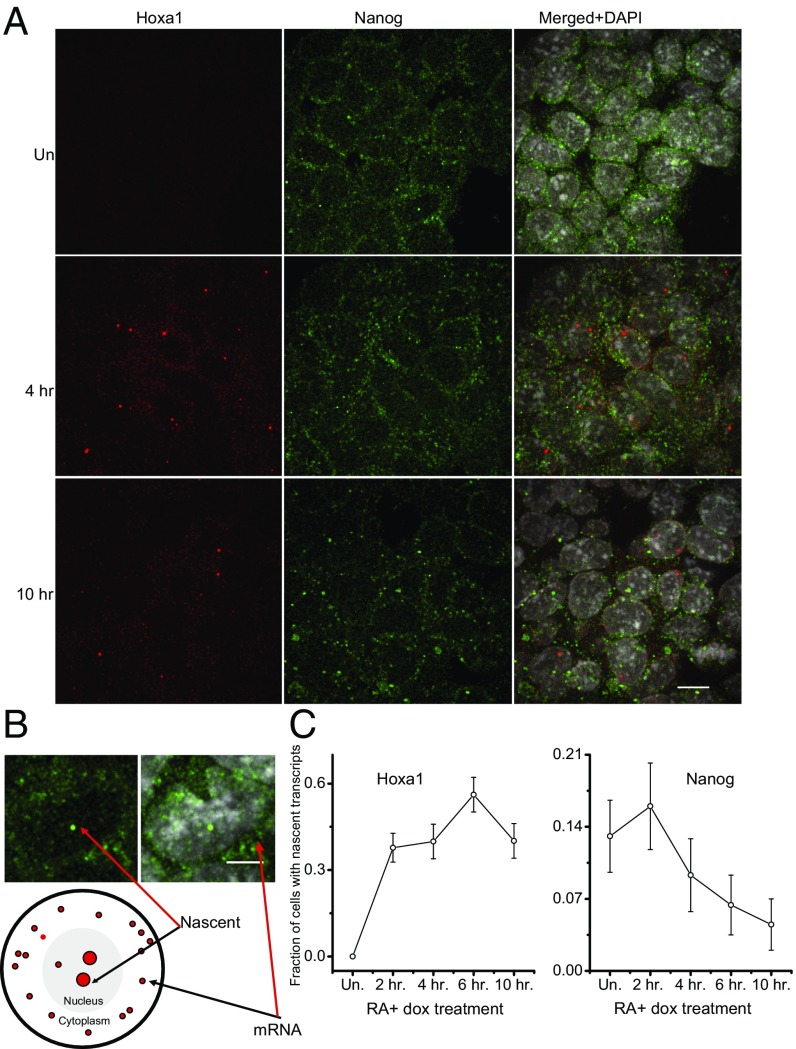
To examine whether the binding of Hoxa1 to the Nanog locus affects its transcriptional activity, we used single-molecule RNA FISH to quantify expression levels of Nanog and Hoxa1 in individual cells. The advantage of this approach is that it permits the quantification of both total steady-state levels of RNA and nascent transcriptional activity per cell. We find that the total steady-state levels of Nanog remain relatively stable over the early time course of RA-induced differentiation as measured by RNA-seq, quantitative PCR, and FISH (22). In contrast, there is a progressive decline in the levels of nascent transcripts from 2 h onward, with fewer transcripts per cell and fewer cells with nascent transcripts (Fig. 3). These data suggest there is an early and rapid decrease in transcriptional activity of Nanog. The timing of this decrease reciprocally correlates with the rapid induction of Hoxa1 for both steady state and nascent transcripts (Fig. 3) (22). This finding suggests a model whereby Hoxa1 is rapidly induced during differentiation and, through binding to the TSS and ARE of Nanog, contributes to the down-regulation of its transcriptional activity.
Fig. 3.
Analysis of transcriptional dynamics Hoxa1 and Nanog using single-molecule RNA FISH. (A) Expression of Hoxa1 and Nanog in differentiating ES cells. Hoxa1 is shown in red channel andNanog is seen in green channel. Nuclei are stained with DAPI. (B) The one-to-two bright foci per cell represent nascent nuclear transcripts, whereas transcripts outside nuclei are steady-state stable mRNAs. (C) Quantification of nascent transcripts of Nanog and Hoxa1. Hoxa1 shows rapid up-regulation, whereas Nanog undergoes a progressive decrease after 2 h of RA treatment. (Scale bars: A, 10 μm; B, 5 μm.)
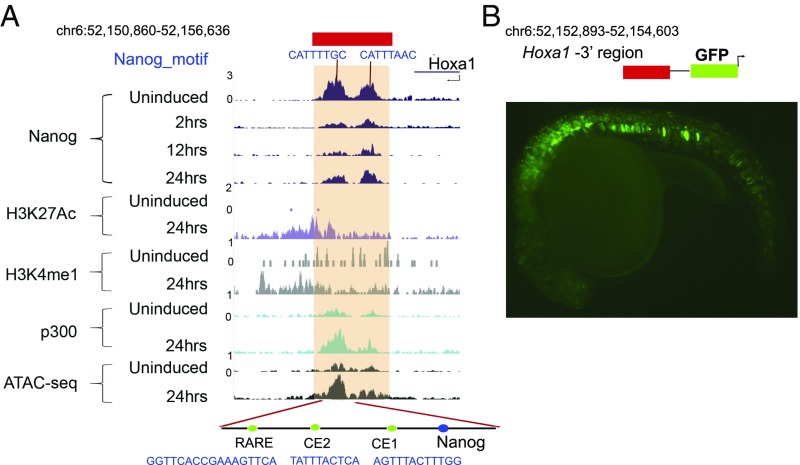
In the Nanog ChIP-seq experiments during the time course of ES cell differentiation (Fig. 1A), we noted that there are two binding peaks in a region located 2 kb 3′ of Hoxa1 (Fig. 4A). One of these peaks contains a Nanog binding site located near two conserved elements (CE1 and CE2) and a RARE (Fig. 4A). CE2 and the RARE have been previously shown to have regulatory activity in assays where large genomic fragments from the Hoxa1 locus were tested in transgenic mouse reporter assays (29, 38). In addition, the RARE has been mutated in the endogenous Hoxa1 locus and shown to be required for early expression (27). To test the activity of this smaller region with the two Nanog binding peaks, we linked a 1.8-kb fragment to a GFP reporter vector and scored for regulatory activity in F0 transgenic zebrafish embryos (Fig. 4B). Reporter expression is detected in mesodermal and neuro-ectodermal cells in a manner similar to mouse Hoxa1 expression, suggesting that this is a functional core of the enhancer.
Fig. 4.
Cross-regulation of Hoxa1 by Nanog. (A). Occupancy of Nanog on a 3′ enhancer of Hoxa1. The Hoxa1 genomic locus is indicated at the top and below are displayed ChIP-seq and ATAC-seq results for the position of regions bound by a variety of factors, epigenetic marks and open chromatin states in uninduced and differentiated ES cells. The respective time points, factors, and marks are indicated at the left. The shaded regions indicate the two adjacent peaks of Nanog binding, which correlate with the presence of Nanog consensus sites, near Hoxa1. These Nanog sites fall within a region which harbors two functionally important elements, CE2 and a RARE, as indicated at the bottom. (B) Regulatory analysis of the Nanog-bound region downstream of Hoxa1 using a transient transgenic assay in zebrafish. The diagram at the top indicates that a 1.8-kb region containing two Nanog sites were linked to a GFP reporter vector. Below is a lateral view of a 20 h postfertilization (hpf) embryo displaying mesodermal and neuro-ectodermal reporter expression. Image dimensions are 1.3 mm × 0.9 mm.
Nanog binding is highest in uninduced ES cells and is rapidly lost by 2 h. In uninduced ES cells the region is relatively closed based on ATAC-seq and low occupancy of p300 (Fig. 4A). By 24 h, the region is open, occupied by p300, and contains active enhancer marks. This result suggests that the Hoxa1 enhancer is not active in uninduced ES cells and that Nanog may be involved in repressing its activity and hence the expression of Hoxa1. The loss of Nanog occupancy on these sites by 2 h correlates with the rapid induction of Hoxa1. Together, these experiments suggest that Hoxa1 and Nanog are involved in direct cross-regulatory interactions and mutually repress their expression to modulate the balance between pluripotency and differentiation.
Discussion
Temporal Dynamics of Hoxa1 Binding.
In this study we have uncovered a dynamic nature in occupancy of Hoxa1 on genome-wide targets during the RA programmed differentiation of mouse ES cells. Hoxa1 is rapidly induced and binds a large number of targets by 2 h of differentiation (Fig. 1A). Many of the genes adjacent to these sites are associated with stem cell-signaling pathways and show rapid up- or down-regulation compared with uninduced ES cells (Fig. 1 and Fig. S1C). By 12 h of differentiation, the genome-wide profile is very different, with a loss of most of the targets observed at 2 h and the appearance of new binding sites (Fig. 1A). Despite this shift in occupancy, the sites bound at 12 h also correlate with genes associated with stem cell and other signaling pathways, but there is a change to include pathways associated with differentiation processes (Fig. S1C). The striking shift in binding-site distribution of Hoxa1 correlates with temporal changes in expression of markers for cell lineage and differentiation processes that reveal different cellular states exist at 2 and 12 h (22). Interestingly, a recent study on single-cell transcriptome analysis of mouse ES cells differentiated with RA over a 96-h time course also showed similar progressive changes in differentiation and that 12 h was an important transition point for exit from pluripotency (39). Transient dynamics of Hoxa1 binding on downstream targets implies that its regulatory influence varies with the temporal progression of the differentiation state of the cell.
At the mechanistic level, this temporal shift in binding-site distribution may be related to differences in the nature of enriched motifs found in bound regions (Datasets S3 and S4) and the presence of Hox cofactors (e.g., Pbx and Meis). There are both unique and overlapping cohorts of enriched binding motifs associated with occupancy of Hoxa1 at 2 and 12 h (Fig. 1 B and C). This finding is illustrated by enrichment at 12 h compared with 2 h for bipartite Hox-Pbx motifs, which serve as elements that integrate combinatorial binding of Hox, Pbx, and Meis (40). It is worth noting that Pbx1 is expressed at low levels in uninduced ES cells but Pbx, Meis, and other TALE (three amino acid loop extension proteins) transcription activator-like effector cofactors, known to partner with Hox proteins, are not generally available early in the differentiation process. Hence, associated with the differences in the state of differentiation at 2 h versus 12 h, there is greater availability of Hoxa1 and the TALE cofactors at the later time point. This finding suggests that there are likely to be temporal differences in the utilization of cofactors that impact site selection and binding specificity/affinity. Therefore, the shifts in binding profiles and sequence preferences may be mediated through temporal differences in the levels of expression or activity of Hoxa1 itself, and through variation of Hox cofactors, such as TALE proteins, chromatin modifiers, or other TFs that modulate Hoxa1 binding (40).
Pluripotency Network as a Target of Hoxa1.
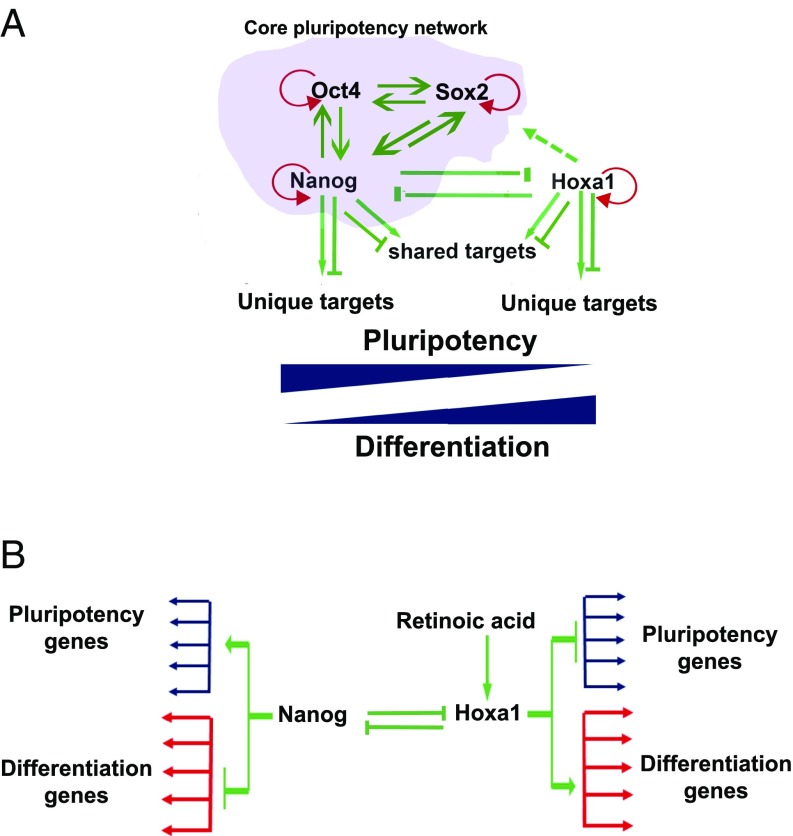
The core pluripotential GRN controls many genes and pathways that actively maintain the pluripotential state and at the same time represses developmental and differentiation pathways, such as those dependent upon Hox genes (6, 7, 41, 42). This result can be achieved through direct activation of downstream genes, such as Esrrb, which in turn play important positive roles in controlling the pluripotent state (1, 6). Repression is mediated through direct recruitment of repressor complexes, such as NuRD (16, 17), or through maintenance of repressed chromatin states, such as those mediated by Polycomb complexes (7). A large number of the putative Hoxa1 downstream target genes in our analyses are part of the pluripotential GRN, including Nanog, Sox2, and Myc, and we found that they are cobound by Nanog (Figs. 1 and 2 and Fig. S1). Furthermore, these common targets show differential expression upon the early induction of Hoxa1 (Fig. S1). This raises the possibility that in a reciprocal manner compared with Nanog and the core pluripotential network, Hoxa1 may repress genes that maintain pluripotency and promote expression of those that facilitate differentiation. The mechanistic basis through which Hoxa1 works as an activator or repressor of transcription on different targets is poorly understood. However, Hox cofactors may dictate functional outcomes in a context-dependent manner and have been suggested to be involved in repression, as well as activation of target genes in Drosophila (43–45). Our data suggest a model whereby direct input of Nanog and Hoxa1 on shared targets regulates the fine balance between the pluripotent and differentiation states (Fig. 5A).
Fig. 5.
Model showing regulatory interactions between the pluripotency and Hoxa1-dependent differentiation GRNs. (A) Summary of extensive regulatory interactions between the core pluripotency GRN and Hoxa1. Shared targets of Nanog and Hoaxa1 illustrate novel regulatory cross-talk between the pluripotency and differentiation pathways. (B) Mutual direct repression of Hoxa1 and Nanog provide regulatory subcircuits for modulating two alternate states. This cis-element subcircuitry represents double-negative gates used to define alternate states of pluripotency and differentiation.
Cross-Regulatory Interactions Between Hoxa1 and Nanog.
In addition to shared target genes, there is another layer of regulatory input between Hoxa1 and Nanog through mutual repression. Hoxa1 binds to an autoregulatory repressor region and the TSS of Nanog (Fig. 2A), whereas Nanog binds to a 3′ enhancer of Hoxa1 (Fig. 4A). We have also found that Hoxa1 binds to regulatory regions of Sox2 and Myc (Fig. 2B and Fig. S3), which shows extensive input of Hoxa1 into core components of the pluripotential regulatory networks. Integrating this in the model for cross-talk between Nanog and Hoxa1, in undifferentiated ES cells, the Hoxa1-mediated repression of pluripotential gene expression is released by direct repression of Hoxa1 by Nanog (Fig. 5B). In terms of cis-regulatory architecture, this type of subcircuit is termed a “double-negative gate,” which is a commonly used efficient mechanism using active repression as a means to spatially or temporally restrict programs of gene expression (46).
This model for extensive cross-talk between Hoxa1 and the pluripotency network may have relevance beyond ES cells. Recent studies in Xenopus have shown that many features of the regulatory program that controls pluripotency in the blastula are also present in neural crest cells (47). Because Hox genes, including Hoxa1, are known to play important roles in formation and patterning of neural crest cells (48, 49), they may have a similar cross-regulatory interaction with the pluripotential network in this in vivo context. The findings in this study underscore the dynamic and diverse inputs of Hoxa1 in modulating gene regulatory networks associated with signaling pathways and pluripotency.
Methods
ES Cell Culture.
KH2 ES cells (50) and KH2 ES cells with epitope-tagged Hoxa1 (3XFLAG-MYC) were grown in feeder-free conditions using N2B27+2i media supplemented with 2,000 U/mL of ESGRO (Millipore). N2B27+2i media consist of neurobasal media (21103-049, Invitrogen), DMEM/F12 media (10565-018, Invitrogen), 0.5× N2 (17502-048, Invitrogen), 1× B27 (17504044, Invitrogen), 1× β-mercaptoethanol (ES-007-E, Millipore), 1× Glutamax (10378-016), 1× NEAA (07600, SCT), 3 µM CHIR99021 (4423, Tocris), PD0325901 (72184, SCT), 0.033% BSA (15260037) Thermo Fisher Scientific) (Qi-Long, 2008). Cells were seeded on a gelatinized plate without a feeder layer. After 48 h the media was changed to differentiation media [DMEM + 10% (vol/vol) Serum + NEAA + 0.03 µM RA] for a requisite length of time. Uninduced ES cells were grown in N2B27+2i up to 80–90% confluency.
ATAC, ChIP-seq.
ATAC-seq protocol was done as described by Buenrostro et al. (51). ChIP-seq was done according to the Upstate protocol with certain modifications (22, 52). Cells were fixed by adding formaldehyde to media at a final concentration of 1% followed by incubation at 37 °C for 11 min. Immunoprecipitation (IP) experiments were done using monoclonal α-FLAG M2, (F1804, Sigma-Aldrich). Libraries were prepared using the KAPA HTP Library Prep Kit for Illumina and Bio Scientific NEXTflex DNA barcodes. The resulting libraries were purified using the Agencourt AMPure XP system (Beckman Coulter), then quantified using a Bioanalyzer (Agilent Technologies) and a Qubit fluorometer (Life Technologies). Postamplification size selection was performed on all libraries using a Pippin Prep (Sage Science). Libraries were requantified, normalized, pooled, and sequenced on an Illumina HiSEq. 2500 instrument as 50-bp single read. Following sequencing, Illumina Real Time Analysis v1.18.64 and CASAVA v1.8.2 were run to demultiplex reads and generate FASTQ files.
Data Analysis.
Raw reads were aligned to the University of California, Santa Cruz (UCSC) mm10 mouse genome with bowtie2 2.2.0 (53). Primary reads from each bam were converted to bigWig tracks, normalized to reads-per-million, and visualized at the UCSC genome browser (https://genome.ucsc.edu/) (54). Peaks were called with MACS2 2.1.0.20140616 [3], parameters “-p 0.25 –m 5 50.” The top 100,000 peaks by P value, for each replicate, were compared with inconsistency discovery rate (IDR) 1.7.0 (https://sites.google.com/site/anshulkundaje/projects/idr) and valid pairs with IDR P ≤ 0.01 were taken as the sample peak list. Nanog samples produced such an abundance of peaks that the IDR P-value threshold was lowered to 0.0001. IP/input RPM log2 fold-change signal across peak coordinates was visualized with the CoverageView package in R (https://www.bioconductor.org/packages/release/bioc/html/CoverageView.html) using windows ± 5 kb from peak midpoints. Nanog binding was essentially the same across time, so peaks from all four time points were collapsed into a single Nanog peak list of 17,651 peaks. ATAC-seq data has no input, so instead of using IP/input log2 fold-change, we transformed the coverage matrix to percent-of-maximum, so that signal would be within the same range as log2 fold-change. Motif content of peaks was analyzed using the vertebrate motif set from TransFac v2016.2 (55), searched with the FIMO program (56), and also a list of kmers of interest, searched with grep. Each peak set was given a background of 10 random coordinate sets, each with the same length and chromosomal distribution as the original peaks. Motif enrichment in peaks versus background was assessed with Fisher’s Exact Test, using BH P-value correction. For each peak set, all Ensembl 80 protein-coding nearest-neighbor genes upstream and downstream were identified and analyzed for functional enrichments. Briefly, KEGG (57) pathway terms (www.genome.jp/kegg/) and Gene Ontology (GO) (58) terms (geneontology.org/) downloaded May 2016 were compared between gene lists versus the rest of the genome. Terms overenriched in neighbor genes by Fisher Exact Test, BH-adjusted P ≤ 0.05, with at least three genes, were accepted. Raw data for RNA-seq in uninduced, 4- and 6-h RA time points were taken from National Center for Biotechnology Information (NCBI) Gene Expresson Omnibus accession no. GSE61590 (22).
Zebrafish Transgenic Reporter Assay.
For generating constructs, two oligos 5′-ATAACAGGGTAATGAGGGCCGTAGCCCAAGAGTTTCTTC-3′ and 5′-CCTCGAGGATATCGAGCTCGGGAGCTGAGTCTTCATCTTC-3′ were used as forward and reverse primers to amplify a 1,711-bp region 3′ of Hoxa1 from mouse genomic DNA. PCR-purified putative enhancer elements were cloned into the HLC vector (59) using the Gibson Assembly Master Mix kit (New England Biolabs). Correct inserts were confirmed by sequencing. Constructs for injection were diluted to 125-ng/μL concentration. Tol2-mediated transgenesis was performed by microinjection into fertilized Slusarski AB (wild-type) zebrafish eggs, as described previously (60). At least 100 embryos were injected for each reporter construct. Embryos were screened for GFP fluorescence using a Leica M205FA microscope and imaged with a Leica DFC360FX camera and LAS AF imaging software. Images were cropped and adjusted for brightness and contrast using Adobe Photoshop CS5.1. All experiments involving zebrafish were approved by the Institutional Animal Care and Use Committee of the Stowers Institute for Medical Research (Krumlauf Protocol No. 2015-0149).
RNA FISH.
RNA FISH samples were prepared, and data were obtained essentially as previously described, using the Stellaris, multiprobe approach (22). The Hoxa1 probe-set sequences are as published previously (22) and the Nanog probe-set sequence is the DesignReady version offered by Stellaris. For probe labeling, unlabeled probe sets carrying a C-term TEG-Amino tag, were purchased from Biosearch Technologies. Probes (4 nmol) were fluorescently labeled overnight in 0.1 M sodium tetraborate pH 9 at 4 °C. Hoxa1 was labeled with AF647 and Nanog with AF555. Two units of amine reactive succinimidyl ester Decapacks (ThermoFisher) were used for each reaction and following quenching labeled probes were purified using Reverse-Phase HPLC. Probes were separated using an Ettan LC (GE Healthcare) using a 4.6 × 250-mm, 5-µm, C18-EMS end-capped Kinetex column (Phenomenex). Mobile phase A was 0.1 M ammonium acetate (EMD) pH7 and mobile phase B was acetonitrile (Millipore). A linear gradient of 5% B to 100% (vol/vol) B was run over the course of 20 min at 1 mL/min. Peaks were monitored at 280 nm for probe and either 555 or 647 nm, depending on the dye. Dual positive peaks were collected and concentrated by spin vac.
For quantification of the fraction of cells with nascent transcripts, nuclei were manually segmented using the Dapi channel. After blurring with a Gaussian blur of radius 1 pixel, the pixel of maximum intensity over the manually segmented area in 3D was recorded, over a total z dimension of 11.4 μm. For Hoxa1, the histogram of maximum pixel intensity per cell over all data was clearly biphasic. A cut-off was set based on the distribution, and any nuclei with a pixel intensity over the cut-off was considered to have a nascent transcript. For the Nanog data, although nascent transcripts were apparent, the distribution of maximum intensity per nuclei was not sufficient to set a cut-off. Cells with nascent transcripts were counted by manual inspection of the signal in 3D. For determination of total signal per cell, the region of the nuclear segmentation was dilated 10 pixels in each region, and after thresholding of the background signal, the total integrated intensity per cell was recorded over the dilated region in 3D, over a total z dimension of 11.4 μm.
Data Submission.
All raw sequencing data are submitted to the NCBI as Sequence Read Archive accession no. SRP079975. The raw and processed data from a previous study were submitted to the NCBI Gene Expression Omnibus under accession no. GSE61590 were used for transcriptional profiling. All other original source data not available through a public repository have been deposited in the Stowers Institute Original Data Repository and are available online at www.stowers.org/research/publications/odr.
Supplementary Material
Acknowledgments
We thank Tari Parmely and members of the Stowers Institute Tissue Culture Facility for assistance with mouse ES cells and other cell culture needs; the Stowers Institute Molecular Biology Facility for genomics sequencing approaches; and members of the R.K. laboratory for valuable discussions and feedback. This research was supported by funds from the Stowers Institute.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Regulatory Networks and Network Models in Development and Evolution,” held April 12–14, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Gene_Regulatory_Networks.
This article is a PNAS Direct Submission. E.V.R. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA) database, www.ncbi.nlm.nih.gov/sra (accession no. SRP079975).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610612114/-/DCSupplemental.
References
- 1.Festuccia N, et al. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11(4):477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orkin SH, et al. The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb Symp Quant Biol. 2008;73:195–202. doi: 10.1101/sqb.2008.72.001. [DOI] [PubMed] [Google Scholar]
- 4.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 5.Liang J, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 6.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 8.Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465(7299):713–720. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 9.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132(6):1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 10.Fei T, et al. Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell Res. 2010;20(12):1306–1318. doi: 10.1038/cr.2010.158. [DOI] [PubMed] [Google Scholar]
- 11.Liu F. PCTA: A new player in TGF-beta signaling. Sci Signal. 2008;1(46):pe49. doi: 10.1126/scisignal.146pe49. [DOI] [PubMed] [Google Scholar]
- 12.Navarro P, et al. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. EMBO J. 2012;31(24):4547–4562. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132(4):567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26(4):903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 15.Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fidalgo M, et al. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc Natl Acad Sci USA. 2012;109(40):16202–16207. doi: 10.1073/pnas.1208533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fidalgo M, et al. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells. 2011;29(11):1705–1716. doi: 10.1002/stem.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young T, et al. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev Cell. 2009;17(4):516–526. doi: 10.1016/j.devcel.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Diez del Corral R, Breitkreuz DN, Storey KG. Onset of neuronal differentiation is regulated by paraxial mesoderm and requires attenuation of FGF signalling. Development. 2002;129(7):1681–1691. doi: 10.1242/dev.129.7.1681. [DOI] [PubMed] [Google Scholar]
- 20.Diez del Corral R, et al. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40(1):65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- 21.Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344(1):7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Kumar B, et al. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res. 2015;25(8):1229–1243. doi: 10.1101/gr.184978.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bami M, Episkopou V, Gavalas A, Gouti M. Directed neural differentiation of mouse embryonic stem cells is a sensitive system for the identification of novel Hox gene effectors. PLoS One. 2011;6(5):e20197. doi: 10.1371/journal.pone.0020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C, et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25(14):1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy P, Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991;111(1):61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- 26.Hunt P, et al. A distinct Hox code for the branchial region of the vertebrate head. Nature. 1991;353(6347):861–864. doi: 10.1038/353861a0. [DOI] [PubMed] [Google Scholar]
- 27.Dupé V, et al. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE) Development. 1997;124(2):399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- 28.Marshall H, et al. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370(6490):567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 29.Langston AW, Thompson JR, Gudas LJ. Retinoic acid-responsive enhancers located 3′ of the Hox A and Hox B homeobox gene clusters. Functional analysis. J Biol Chem. 1997;272(4):2167–2175. doi: 10.1074/jbc.272.4.2167. [DOI] [PubMed] [Google Scholar]
- 30.Studer M, Pöpperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265(5179):1728–1732. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- 31.Makki N, Capecchi MR. Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev Biol. 2011;357(2):295–304. doi: 10.1016/j.ydbio.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makki N, Capecchi MR. Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev Biol. 2010;341(2):499–509. doi: 10.1016/j.ydbio.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makki N, Capecchi MR. Cardiovascular defects in a mouse model of HOXA1 syndrome. Hum Mol Genet. 2012;21(1):26–31. doi: 10.1093/hmg/ddr434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wardwell-Ozgo J, et al. HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene. 2014;33(8):1017–1026. doi: 10.1038/onc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taminiau A, et al. HOXA1 binds RBCK1/HOIL-1 and TRAF2 and modulates the TNF/NF-κB pathway in a transcription-independent manner. Nucleic Acids Res. 2016;44(15):7331–7349. doi: 10.1093/nar/gkw606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zha TZ, et al. Overexpression of HOXA1 correlates with poor prognosis in patients with hepatocellular carcinoma. Tumour Biol. 2012;33(6):2125–2134. doi: 10.1007/s13277-012-0472-6. [DOI] [PubMed] [Google Scholar]
- 37.Bitu CC, et al. HOXA1 is overexpressed in oral squamous cell carcinomas and its expression is correlated with poor prognosis. BMC Cancer. 2012;12:146. doi: 10.1186/1471-2407-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JR, Chen SW, Ho L, Langston AW, Gudas LJ. An evolutionary conserved element is essential for somite and adjacent mesenchymal expression of the Hoxa1 gene. Dev Dyn. 1998;211(1):97–108. doi: 10.1002/(SICI)1097-0177(199801)211:1<97::AID-AJA9>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Semrau S, et al. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. bioRxiv. August 7, 2016 doi: 10.1101/068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merabet S, Mann RS. To be specific or not: The critical relationship between Hox and TALE proteins. Trends Genet. 2016;32(6):334–347. doi: 10.1016/j.tig.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 42.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450(7173):1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 43.Rauskolb C, Wieschaus E. Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. EMBO J. 1994;13(15):3561–3569. doi: 10.1002/j.1460-2075.1994.tb06663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merabet S, et al. The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev Cell. 2003;4(5):761–768. doi: 10.1016/s1534-5807(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 45.Galant R, Walsh CM, Carroll SB. Hox repression of a target gene: Extradenticle-independent, additive action through multiple monomer binding sites. Development. 2002;129(13):3115–3126. doi: 10.1242/dev.129.13.3115. [DOI] [PubMed] [Google Scholar]
- 46.Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc Natl Acad Sci USA. 2008;105(51):20063–20066. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buitrago-Delgado E, Nordin K, Rao A, Geary L, LaBonne C. NEURODEVELOPMENT. Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science. 2015;348(6241):1332–1335. doi: 10.1126/science.aaa3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gavalas A, Trainor P, Ariza-McNaughton L, Krumlauf R. Synergy between Hoxa1 and Hoxb1: The relationship between arch patterning and the generation of cranial neural crest. Development. 2001;128(15):3017–3027. doi: 10.1242/dev.128.15.3017. [DOI] [PubMed] [Google Scholar]
- 49.Trainor PA, Krumlauf R. Patterning the cranial neural crest: Hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci. 2000;1(2):116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- 50.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44(1):23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 51.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1-9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith KT, Martin-Brown SA, Florens L, Washburn MP, Workman JL. Deacetylase inhibitors dissociate the histone-targeting ING2 subunit from the Sin3 complex. Chem Biol. 2010;17(1):65–74. doi: 10.1016/j.chembiol.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matys V, et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gene Ontology Consortium Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker HJ, Bronner ME, Krumlauf R. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature. 2014;514(7523):490–493. doi: 10.1038/nature13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fisher S, et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1(3):1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.